Abstract
The European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena/) collects, maintains and presents comprehensive nucleic acid sequence and related information as part of the permanent public scientific record. Here, we provide brief updates on ENA content developments and major service enhancements in 2012 and describe in more detail two important areas of development and policy that are driven by ongoing growth in sequencing technologies. First, we describe the ENA data warehouse, a resource for which we provide a programmatic entry point to integrated content across the breadth of ENA. Second, we detail our plans for the deployment of CRAM data compression technology in ENA.
INTRODUCTION
The European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) has for the last 30 years captured, organized and presented the world’s public domain output of sequence information. This fundamental data resource for the life sciences provides both the sequence component of the permanent scientific record and a forum for the global sharing and dissemination of early research data. Comprehensive coverage is assured through the efforts of our thousands of data submitters and our deep and long-established data exchange collaborations under the International Nucleotide Sequence Database Collaboration (http://www.insdc.org) (1).
Continuing rapid technological advance in sequencing has driven its adoption as a near-ubiquitous discovery and assay platform across and beyond the life sciences. Given the commitment of major global public sequencing-based projects, such as those of the International Genome Sequencing Consortium (ICGC; http://www.icgc.org/) and the Earth Microbiome Project (http://www.earthmicrobiome.org/), our expectation is that the sequencing technology industry will continue to drive the technologies forward aggressively over the next several years. As such, we see as inevitable the further broadening of the range of applications to which sequencing can be put and of the diversity of uses of ENA.
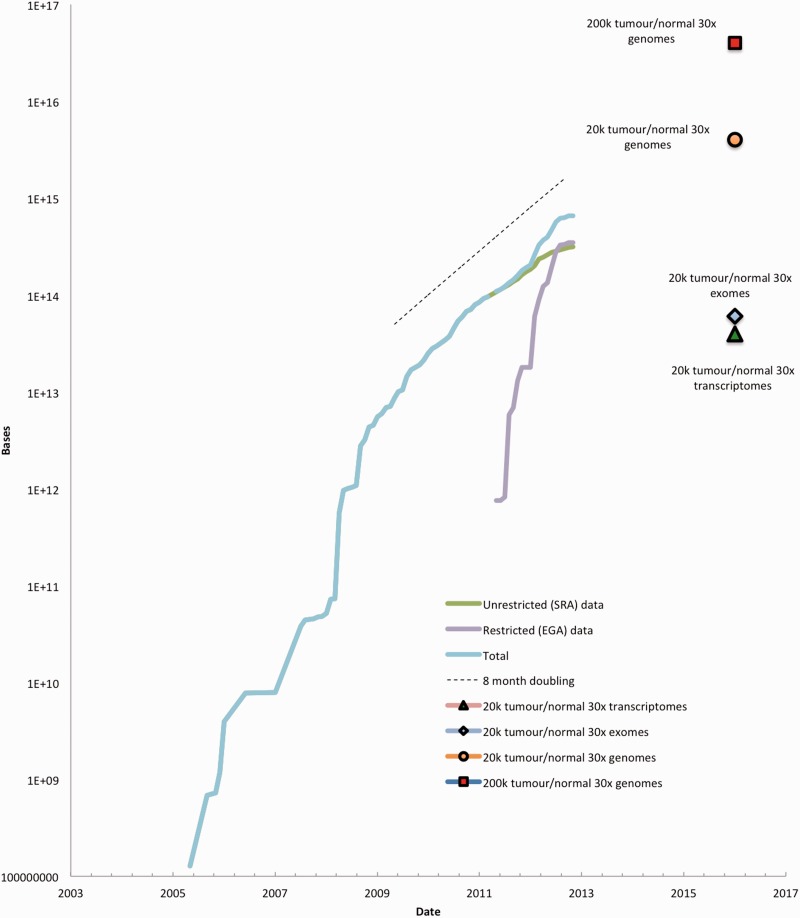
Hand in hand with the spread of sequencing is the ever rising volume of public domain sequence data. We have continued to witness an accumulation of raw next generation data into ENA at an exponential rate with doubling times commonly ∼10 months and, given even modest projections from ICGC projects alone, we expect to continue to see comparable rates of growth over a 5–10 year time window (Figure 1).
Figure 1.
Accumulation of raw data into ENA. The figure shows the accumulation of raw next generation sequence data into public data repositories. In addition to unrestricted data available in ENA, the figure also shows the accumulation of sequence data into the European Bioinformatics Institute's, European Genome-phenome Archive, a repository for human molecular medicine research-related data requiring authorized access for ethical reasons (http://www.ebi.ac.uk/ega/). Data points for expected output of global cancer genome sequencing projects are shown.
During 2011, a continued focus on the usability of ENA has given rise to a number of new services and significant updates to existing services, covering submissions, validation technology, data warehousing, discovery tools and data presentation improvements.
In this article, we review briefly developments in ENA content and highlight a selection of important service developments. We then focus on two of our responses to growth in sequencing: first, our substantial new Advanced search service interface for the discovery of integrated ENA content and, second, our deployment plans for CRAM sequence data compression.
ENA CONTENT
ENA content has continued to grow throughout 2012 in all major areas. In terms of data volume, the data are dominated by raw data from next generation platforms (310 trillion bases from 14 000 sequencing studies; Figure 1) while in terms of submitting scientists, assembled and annotated sequences dominate with ∼1400 submitters using EBI submission services in 2012. Amongst the many genome submissions are a number of notable metazoan cases, including Bonobo (http://www.ebi.ac.uk/ena/data/view/PRJNA49285) (2). An extensive wheat genomics and transcriptomics study has been made available in ENA for which we have various components of the data, including raw data, assembled contigs and high-level assembly information (http://www.ebi.ac.uk/ena/data/view/ERP000319,ERP001415). An extensive study into the epidemiology of Vibrio species has been carried out for which raw data from the 1145 samples have been made available in ENA (http://www.ebi.ac.uk/ena/data/view/ERP000270) (3). An interesting environmental sequencing example has been published, in which shotgun metagenomics has been applied in a study of mutualism between gut microbiota in formula- versus breast-fed infants (http://www.ebi.ac.uk/ena/data/view/ERP001038) (4). In the context of biodiversity, over 2012, we have received submissions of sequence data from 104 species reported by the International Union for the Conservation of Nature to be in ‘at risk’ or ‘endangered’ categories for which no previous sequence data had been available (http://www.iucn.org/).
MAJOR SERVICE DEVELOPMENTS
The European Bioinformatics Institute (EBI) approaches the delivery of the ENA data resource under a programme of continual developments both to its core technologies and its many services. Users are invited to follow our news items on our home page and to subscribe to our announcement mailing list (http://listserver.ebi.ac.uk/mailman/listinfo/ena-announce) to keep abreast of developments. We encourage users to contact us, through datasubs@ebi.ac.uk, for specific assistance and to provide feedback on our services; our services are shaped by user requirements and we value these inputs. Table 1 lists a selection of major service developments in 2012.
Table 1.
Major service developments in 2012
| Development | Utility |
|---|---|
| 11 face-to-face training events and creation of new training materials | http://www.ebi.ac.uk/ena/about/training |
| Application-specific checklists | Framework for community developers to develop specifications for external applications and pipelines for ENA validation and presentation of data (e.g. http://www.ebi.ac.uk/ena/about/epigenomics_submissions) |
| Checklists for annotated sequence submissions | Checklists that define Webin submission templates for annotated sequences (https://www.ebi.ac.uk/embl/genomes/submission/app/login/login.jsf) |
| Creation of genome assembly storage layer | Improved and more granular support for assembly information including compliance with AGP2 (http://www.ebi.ac.uk/ena/about/genome_assembly_database) |
| Creation of search warehouse and launch of ENA query builder and search | See below and http://www.ebi.ac.uk/ena/about/browser |
| Extension of ENA sequence search to include reference sequence data directly from Ensembl and Ensembl Genomes | Provision of rapid interactive and programmatic sequence search service to all sequences in ENA and the Ensembl and Ensembl Genomes resources (http://www.ebi.ac.uk/ena/search/#Search) |
| Inclusion of capillary sequencing instruments into the raw sequence storage layer (SRA) | Raw capillary sequence submissions are now supported in the Sequence Read Archive (SRA) storage layer in ENA (http://www.ebi.ac.uk/ena/about/sra_preparing_metadata) |
| Inclusion of sample checklist support for raw data submissions in Webin | Provision of tabular sample checklist functionality in the Webin system that greatly simplifies the submission of sample annotation for different sample types (https://www.ebi.ac.uk/ena/submit/sra/#home) |
| Launch of raw sequence data file uploader/downloader utility | Support for simple and managed upload and download of raw sequence data files—available as interactive and command line clients (e.g., http://www.ebi.ac.uk/ena/data/sradownload/SRA-FileDownloader.jnlp) |
| New services for genome assembly submissions | Tools and workflow for the submission of assembly information (http://www.ebi.ac.uk/ena/about/genome_assembly_submissions) |
| Release of CRAM sequence data compression technology | See below and http://www.ebi.ac.uk/ena/about/cram_toolkit |
| Simplification of transcript shotgun assembly submissions | Simplification of submission process for the growing body of transcriptomes assembled from shotgun data (http://www.ebi.ac.uk/ena/about/TSA-submission-instructions) |
ENA ADVANCED SEARCH SERVICE INTERFACE AND QUERY BUILDER
For a general purpose data resource such as ENA, the ever broadening adoption of sequencing as a discovery and assay platform brings the challenge of a user base that is both growing and diversifying. To rise to the challenge that this creates, we operate a highly collaborative model for the provision of services to specialist users. Under this model, we scale the utility of ENA through the coordinated development of core services from EBI (such as technologies for submissions, data management, compression, storage, search and presentation) with more specialist activities (such as standards development, visualization, analysis and specialist presentation) operated by partners where the specialist expertise already exists.
Central to this approach is a robust and versatile programmatic service layer at ENA upon which secondary services and applications can be built. Adding to the existing portfolio of webservices for search and retrieval of ENA content we have developed and deployed in late 2012 a powerful data warehouse, accompanying webservice and interactive query builder interface that integrates the many classes of ENA content and supports highly granular but rapid discovery and retrieval.
ENA content is a challenge for data warehousing for three reasons. First, ENA holds a large number of distinct records (on the scale of billions), which challenge many systems traditionally used for the management of biological information. Second, ENA data are highly dynamic; the flow of new data and updated records into ENA is continuous, with production databases perhaps changing every few seconds, and our aim is to provide daily distributions of data from most parts of ENA. Third, the warehouse must support interactive use through the web; for the most common uses, this requires rapid (within a few seconds) responses to searches and requests for data.
The design of the warehouse that we have deployed responds to these challenges and provides a balance between search granularity (depth of indexing), dynamic updates and performance. Our implementation combines a custom document-based indexing and retrieval system and an analytical database. The warehouse is made available under a RESTful service to which user queries are despatched under an expressive query language. A ‘query builder’ interactive web interface is also provided that assists the user in assembling a required search. Our focus for further enhancements to this service in 2013 will be the development of interactive web interfaces that are intuitive, but exploit much of the power that queries against the warehouse can support. We look forward to working with our users to develop these important services.
Search query language
A search against ENA content requires the definition of a ‘domain’ (a pre-determined partition of ENA content) and one or more filters (conditions that need to be met) that can be combined using Boolean operators. A ‘domain’ comprises a number of ‘results’ which are deeper partitions of ENA content. For queries based on these more granular ‘result’ partitions, display/download format and pagination options are available. Although a ‘domain’ is a partition of content based on the conceptual nature of content (for example raw sequence data versus assembly versus annotation), a ‘result’ is a partition that also takes into account the structure of the underlying content. Because diverse structures are used in ENA for optimal management of different data that lie even within a single ‘domain’, it is only at the level of ‘results’ that some format options can be made available; to access these options for ‘domain’-wide queries, the user is required to express a number of ‘result’-level queries.
At the time of going to press, 8 domains have been made available that comprise 14 results and cover, e.g. sample information, information on raw data content, assembly information and annotation (Table 2). Filters available cover controlled vocabularies, dates, numbers, text fields, taxonomic classifications and georeference information (Table 2). For usage examples and a more complete description of the service, please refer to http://www.ebi.ac.uk/ena/about/browser.
Table 2.
Search ‘domains’ and ‘results’
| Domain | Domain description | Result | Result data structure | Result description |
|---|---|---|---|---|
| Assembly | Genome and transcriptome assembly information | Assembly | EMBL-Bank and assembly database | Genome assemblies from contig upwards |
| Sequence | Assembled and (optionally) annotated sequence | Sequence_release | EMBL-Bank | Sequence from the latest EMBL-Bank release |
| Sequence_update | EMBL-Bank | Sequences from the EMBL-Bank update product, covering and modified and new entries since last release | ||
| Study | Information relating to a sequencing-based scientific investigation; a unit of scientific output | Study | SRA study | Large-scale sequencing project that comprises assembled/annotated sequences |
| Coding | Sequences believed to encode proteins | Sequence_coding | ENA-CDS | Submitter-annotated protein-coding sequences |
| Analysis | Primary interpretations of raw read data, such as alignments, OTU tables and genome tracks | Analysis | SRA analysis | Read analysis objects |
| Analysis_sample | SRA analysis | Read analysis objects grouped by sequenced sample | ||
| Analysis_study | SRA analysis | Read analysis objects grouped by study | ||
| Read | Raw sequencing data from next generation platforms | Read_run | SRA run | Read data |
| Read_experiment | SRA run | Read data grouped by experiment objects | ||
| Read_sample | SRA run | Read data grouped by sequenced samples | ||
| Read_study | SRA run | Read data grouped by study objects | ||
| Taxon | Information relating to taxa | Taxon | ENA-taxonomy | Data in the ENA Taxonomy |
| Trace | Raw sequencing data from capillary platforms | Read_trace | Trace archive | Raw capillary read data |
Query builder interface
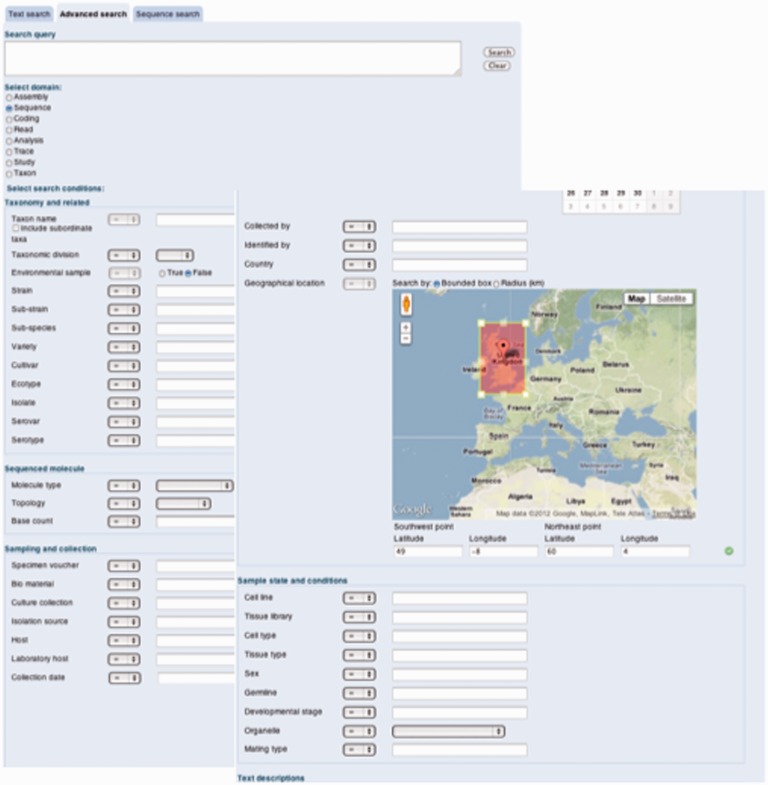
The query builder interface, available from the ‘Advanced search’ tab on the ENA website (http://www.ebi.ac.uk/ena/data/view), supports the full range of warehouse queries (Figure 2). It is provided to simplify the process of refining query syntax, but its use requires some knowledge of the nature of content partitioning and filter options (discussed earlier). A more intuitive interface will be the focus of these developments in early 2013 and we will continue to offer the ‘Text search’ tab that is built upon the EBI Search service.
Figure 2.
Query builder interface screenshot. The figure shows the query builder graphical user interface for the ENA warehouse search service. This interface is available from the ‘Advanced search’ tab visible across the ENA browser.
CRAM SEQUENCE DATA COMPRESSION
At the time of writing, ENA raw sequencing data show exponential growth with doubling time 10 months. Although most of the technologies required to handle the informatics around sequencing (such as network, disk and compute) also grow exponentially, they do so at markedly higher doubling times. Growth in the capacity to produce sequence is therefore out-stripping growth in the technologies that must be provisioned to respond to the data. In 2012, we have developed and deployed a robust and versatile format and software toolkit, CRAM, for the reference-based compression of raw sequence read data. Although as a sequence data repository, ENA faces the data growth phenomenon primarily as a challenge of rising cost of disk, our focus in the design and implementation of the CRAM software is as a general technology for the handling of raw sequence data that addresses issues around the movement of data across networks and memory provision as well as storage.
Technical progress
The early concept and proof of principle for CRAM has been published in 2011 (5). Although Python code from this study has been made available, we have released a vastly expanded version as a Java toolkit together with a full description of the CRAM format. Our design principle for CRAM has been full integration with existing software stacks and workflows, such that CRAM can be deployed globally with minimal effort. CRAM has been built from the start to be compatible with the Binary Alignment Format (BAM) (6), currently the most widely used community format for raw sequence storage, to allow effortless interoperability between these two formats. Furthermore, because we have integrated CRAM technology into SAM-JDK (upon which the popular GATK and Picard tools are built), any users of these tools can read and write to CRAM formats in place of BAM.
CRAM supports the storage of sequence read calls and per-call qualities. In lossless mode, where both calls and qualities can be precisely reconstructed, significant compression can be achieved over currently used formats. In lossy mode, where calls can be reconstructed precisely, but a variety of models can be chosen for data reduction in qualities, far greater compression can be achieved.
CRAM Toolkit is available from http://www.ebi.ac.uk/ena/about/cram_toolkit and further details about the format can be found at http://www.ebi.ac.uk/ena/about/cram-format-specification. We are currently drafting a full description to be published in due course.
Deployment in ENA production services
ENA production will have increasing dependencies on CRAM technology over 2013. CRAM will become the preferred submission format alongside BAM during 2013. For submissions, our current recommendation at the time of going to press is for data to be prepared under the lossless mode of CRAM, for which the format can provide a direct alternative to such formats as BAM. Data submitted in this way are presented in CRAM format and we expect to continue to roll out CRAM presentation across appropriate content.
Importantly, we wish to highlight our policy in relation to the use of CRAM lossy compression. Although we expect that lossy modes of CRAM will be required for certain data sets in the future to contain their volume, it is our aim with CRAM to provide a framework in which data producers, the broad scientific community that consumes ENA data, and the funding agencies are empowered to make decisions about the level of compression that can appropriately be applied to different data sets (7). ENA will not apply lossy compression on submitted data without prior announcement and prior consultation with principal stakeholders. In addition, for legacy data already submitted and loaded into ENA, we will not seek to apply lossy compression without discussion with data owners. Please refer to http://www.ebi.ac.uk/ena/about/ena_compression_policy for further details of this policy.
FUNDING
Funding for open access charge: European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1.Karsch-Mizrachi I, Nakamura Y, Cochrane G. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2012;40:D33–D37. doi: 10.1093/nar/gkr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prufer K, Munch K, Hellmann I, Akagi K, Miller JR, Walenz B, Koren S, Sutton G, Kodira C, Winer R, et al. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz S, Friedberg I, Ivanov IV, Davidson LA, Goldsby JS, Dahl DB, Herman D, Wang M, Donovan SM, Chapkin RS. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol. 2012;13:r32. doi: 10.1186/gb-2012-13-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsi-Yang Fritz M, Leinonen R, Cochrane G, Birney E. Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res. 2011;21:734–740. doi: 10.1101/gr.114819.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochrane G, Cook CE, Birney E. The future of DNA sequence archiving. GigaScience. 2012;1 doi: 10.1186/2047-217X-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]