Abstract
TIGRFAMs, available online at http://www.jcvi.org/tigrfams is a database of protein family definitions. Each entry features a seed alignment of trusted representative sequences, a hidden Markov model (HMM) built from that alignment, cutoff scores that let automated annotation pipelines decide which proteins are members, and annotations for transfer onto member proteins. Most TIGRFAMs models are designated equivalog, meaning they assign a specific name to proteins conserved in function from a common ancestral sequence. Models describing more functionally heterogeneous families are designated subfamily or domain, and assign less specific but more widely applicable annotations. The Genome Properties database, available at http://www.jcvi.org/genome-properties, specifies how computed evidence, including TIGRFAMs HMM results, should be used to judge whether an enzymatic pathway, a protein complex or another type of molecular subsystem is encoded in a genome. TIGRFAMs and Genome Properties content are developed in concert because subsystems reconstruction for large numbers of genomes guides selection of seed alignment sequences and cutoff values during protein family construction. Both databases specialize heavily in bacterial and archaeal subsystems. At present, 4284 models appear in TIGRFAMs, while 628 systems are described by Genome Properties. Content derives both from subsystem discovery work and from biocuration of the scientific literature.
INTRODUCTION
In multiple sequence alignments, information emerges as to which residues positions are important to the nature of a protein family, versus which vary freely but could be assigned undue importance during the scoring of pairwise alignments. From these multiple alignments, profile hidden Markov Models (HMMs) are built. These probabilistic models allow exquisitely sensitive searches for proteins related by homology to the aligned sequences. The TIGRFAMs database is a collection of these HMMs constructed with the purpose of letting automated annotation pipelines attach specific functional annotations to proteins encoded by newly sequenced microbial genomes. The HMM search produces evidence, and the logic of the annotation software exploits the evidence. But the HMM evidence itself is persistent, and based on fixed cutoff scores for consistency from use to use, and may be put to additional purposes. Genome Properties is a collection of rules for interpreting evidence, most in the form of HMM hits, to make judgments about the likely presence or absence of complex biological traits in an organism according to whether or not its genome encodes the components necessary for that trait. The process of systems reconstruction in Genome Properties provides guidance for protein family construction in TIGRFAMs, so the two databases develop in concert.
TIGRFAMs
TIGRFAMs as an annotation-driving database
Publications that report specific protein characterization data typically describe one or two proven examples of the protein in question but stop short of providing any rule for recognizing all examples from all organisms. Subsequent biocuration at either specialized or comprehensive value-added databases ties articles to sequences, attaches meaningful functional names, and adds feature tables (signal peptides, active sites, modified sites, etc.) and other searchable content such as EC numbers and GO terms. But for only limited numbers of protein families has this curation matured into tools sufficient to identify and annotate all functionally equivalent sequences. Most proteins belong to large superfamilies whose memberships are heterogeneous in function, complicating such efforts. The right granularity for dividing a superfamily into subgroups by function seems to differ from case to case, and no one set of heuristics always works. New tools are needed. TIGRFAMs is built to address this need by serving as an annotation-driving database, a library of protein family definitions that becomes a tool set used by automated genome analysis pipelines.
Each TIGRFAM as a proxy for a curatorial expert in that protein family
The TIGRFAMs database provides manually reviewed definitions of protein families with the following characteristics to make them useful for automated genome annotation, pathway reconstruction, creation of phylogenetic profiles and any number of other computationally driven studies. First, all annotation is traced to its original source without ever relying on transitive annotations from public sequence databases. Second, every protein is considered in the context of any related protein families that differ in function, if they exist, with examination of molecular phylogenetic trees. Third, only sequences that can be assigned with high confidence are selected as exemplars for the seed alignment. Multiple sequence alignments are examined for misalignments, inconsistent domain architecture, altered active or binding sites, faulty gene models, long branches in phylogenetic trees that may suggest neofunctionalization, etc. Biocuration during model construction includes review of local synteny, metabolic context and phenotypic data. Fourth, each model is searched against several protein databases, including the CharProtDB collection of explicitly characterized proteins (1), sequences with known structures in PDB and NCBI’s non-redundant protein sequence collection, which pulls annotation from multiple sources, including value-added databases such as UniProt and archives of sequences as originally submitted. Each completed HMM is intended to serve as a proxy for an expert curator, emulating expertise developed at the time of model construction but operating at BLAST-like speeds (2).
TIGRFAMs models describe the level of their specificity
Each entry in TIGRFAMs carries a designation that describes how the set of proteins in the family vary in function. If all members of a protein family perform the same function, the family is designated equivalog. We previously introduced the term ‘equivalog’ to describe a relationship of conserved function among homologs (3), noting that the term ‘ortholog’ is doubly wrong since orthology does not imply conserved function and laterally transferred genes (xenologs) may be equivalogs. The isology type informs automated annotation pipelines which of two different HMMs that matches the same region of the same protein gets priority. Each TIGRFAMs equivalog model provides a high-precedence instruction to automated annotation pipelines to transfer the protein name, enzyme commission (EC) number if present, gene symbol and Gene Ontology (GO) terms to any protein whose match to the HMM meets the score cutoff. An equivalog model assigns more specific annotations than a subfamily model, which in turn outranks a domain model. The general principle is that models built with narrower scope tend to outrank models with broader scope where they hit the same proteins. In many cases, a subfamily model is created in TIGRFAMs, with deliberately generic annotation attached, to prevent automated annotation pipelines from propagating an overly specific annotation from a more distant homolog according to evidence of lower rank.
TIGRFAMs content complements the Pfam collection by emphasizing protein function over domain architecture
The TIGRFAMs database is designed to be used in conjunction with other sources of annotation, especially Pfam (4). Both databases contain HMMs built with and searchable by the same freely available software package HMMER3, developed by Eddy (2). Pfam models frequently describe domains of homologous sequence that occur as finite regions within larger proteins and are shared across sets of proteins that range widely in function (4). A single protein often has multiple Pfam domains. A TIGRFAMs equivalog model, by contrast, typically identifies fewer proteins, covers a larger fraction of total sequence length, and provides more specific annotation that should be given higher precedence by automated annotation pipelines. For example, Pfam model PF04055 describes the radical SAM domain, found in over 40 000 recognizable member sequences in public databases. Many of these have additional domains, for a wide variety of domain architectures and even wider array of functions, most of which remain unknown. TIGRFAMs describes over 100 functionally distinct protein families that share the radical SAM domain, including methyltransferases that modify structural RNAs, cofactor biosynthesis enzymes performing complex rearrangements, lipid metabolism enzymes, peptide maturases for natural products biosynthesis and enzyme activases. These models resolve many subgroups of the radical SAM superfamily by function in a way that the domain decomposition provided by Pfam cannot. New TIGRFAMs work avoids construction of models that duplicate the scope and extent of existing Pfam models, but it will include construction of domain or repeat models for homology regions that have never before been described.
TIGRFAMs has grown by over 40% since the previous database update paper (5), from 3000 to 4284 models. Most new models describe less common proteins, so coverage by TIGRFAMs for the average microbial genome has increased only about 20%. Overall coverage of microbial genomes is variable, averaging about 33%. Table 1 shows illustrative levels of coverage by TIGRFAMs for five bacterial and two archaeal genomes. The table distinguishes hits in aggregate by all TIGRFAMs models from hits just to equivalog-type HMM hits, where the HMM matches spans nearly the full protein length and members of that family should share just one function. As expected, TIGRFAMs identifies more proteins in absolute terms in larger genomes, but matches a greater percentage of proteins for species with smaller genomes or from more intensively studied lineages. The current release, 13.0 (July 2012), is the fourth release to use HMMER3 instead of HMMER2. A web server page kindly provided by Janelia Farm, http://hmmer.janelia.org/search/hmmscan, provides high-speed searching of HMM libraries (6), with links provided to TIGRFAMs pages for models meet the criterion of scoring better than the trusted cutoff. The TIGRFAMs home pages do not provide searching, but instead provide a link to the Janelia Farm hmmscan page.
Table 1.
TIGRFAMs HMM hit coverage for five bacterial and two archaeal genomes
| Species | No. of proteins | No. of matched, (%) | No. of equivalog-level, (%) |
|---|---|---|---|
| Mycoplasma genitalium G-37 | 473 | 260 (55) | 220 (47) |
| Streptococcus sp. SK140 | 1856 | 672 (36) | 489 (26) |
| Haemophilus parainfluenzae HK262 | 2001 | 935 (47) | 730 (36) |
| Burkholderia mallei ATCC 23344 | 5031 | 1379 (27) | 1009 (20) |
| Fusobacterium necrophorum ATCC 51357 | 2060 | 680 (33) | 506 (25) |
| Methanocaldococcus jannaschii DSM 2661 | 1783 | 584 (33) | 462 (26) |
| Haloferax volcanii DS2 | 4074 | 699 (17) | 415 (10) |
A project goal is to make negative results from searches informative
The goal during construction of each TIGRFAMs entry is to create a HMM with cutoff scores calibrated to collect comprehensively the set of all proteins that merit the annotations provided with the model, with vanishingly few false-positives but also very few false-negatives (other than proteins left incomplete through sequencing or assembly errors). The advantage of few false negatives becomes clear during the analysis of complete genomes. If six key protein families represent six essential steps in some biochemical pathway, and the six corresponding TIGRFAMs entries all report no match during the analysis of a complete microbial genome, then the repeated absence of evidence for that cohort of genes becomes fairly strong evidence that the cohort is in fact absent.
TIGRFAMs may be used through representations in other resources
TIGRFAMs is incorporated into InterPro (7), a hierarchical collection composed of entries from multiple databases of protein family signatures. InterPro distributes an integrated software package that performs searches for all member databases at once, including TIGRFAMs. Similarly, the Conserved Domain Database (CDD) (8) incorporates TIGRFAMs models, so domain searches through NCBI may show corresponding matches. Note, however, that CDD first converts TIGRFAMs models from HMMER3 to RPS-BLAST models, so the calibration of cutoff scores usually is lost in favor of simpler heuristics. A CDD entry built from a TIGRFAMs model, e.g. CDD:188483 derived from TIGR03968 (‘mycofactocin system transcriptional regulator’), may attach a region feature to many proteins that we were careful to exclude when our model was constructed. A region feature showing TIGRFAMs information through a database cross-reference (db_xref) to a CDD entry is showing sequence homology that may be useful know, but true membership in the TIGRFAMs family should be verified based on actual HMM search scores.
TIGRFAMs web pages
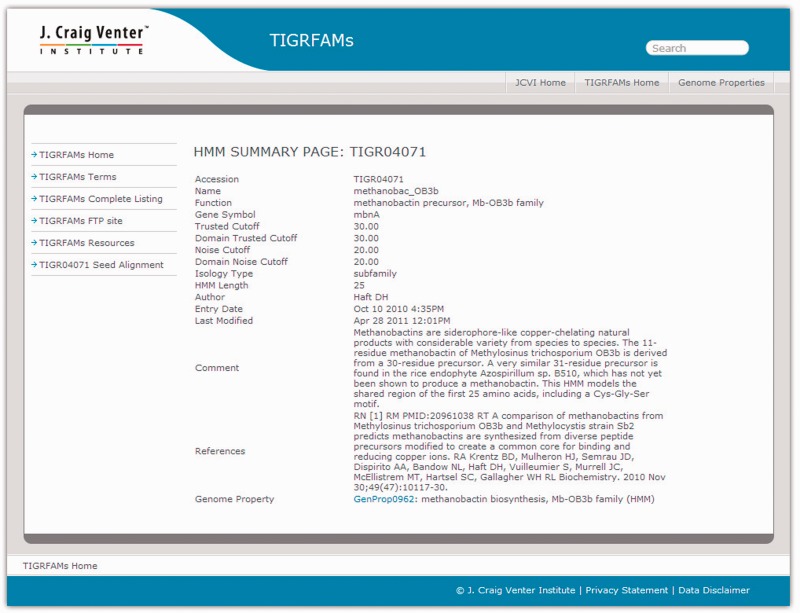
Web pages for TIGRFAMs was provided originally through the Comprehensive Microbial Resource (9,10), or CMR. TIGRFAMs switched to an independent home page at www.jcvi.org/tigrfams during the conversion from using HMMER2 to using HMMER3. Legacy data for TIGRFAMs releases 9.0 and earlier, and Genome Properties results relying on HMMER2-generated data for over 550 genomes, remain accessible through the CMR. Figure 1 provides an illustration of a summary page for a single TIGRFAMs entry, TIGR04071, representing a family of precursor peptides that are post-translationally modified to become methanobactin Mb-OB3b (a copper chelator analogous to iron-chelating siderophores) or another member of the Mb-OB3b class (11).
Figure 1.
Example of a TIGRFAMs web page. The TIGRFAMs homepage and all its web pages have five links located on the left navigational sidebar. These go to the TIGRFAMs Home page itself, a glossary of key terms used in TIGRFAMs, a complete listing of all models (4284 in release 13.0), the TIGRFAMs FTP Site, and a page with links to additional resources, including rapid searching of a protein sequence against HMM libraries. The page shown in the figure is summary page for a single TIGRFAMs entry, the Mb-OB3b-class methanobactin precursor family TIGR04071. Each summary page provides details about a single TIGRFAM, including cutoffs, HMM length, references and any assigned GO terms. The summary page also displays links to Genome Properties, if any, associated with that HMM. Users can view and download the HMM seed alignment through the HMM Seed Alignment link found on the left-hand side of the HMM Summary page. The TIGRFAMs complete listing page (not shown) displays accessions and functional names for all models, as well as length, isology type and EC number.
GENOME PROPERTIES
Providing high-level views of attributes inferred from genome sequences
Genome Properties is a collection of definitions for the higher-level attributes that may be ascribed to a species when a sufficient set of molecular markers are detected in its genome, or else reported jointly absent (12). If all enzymes listed as essential markers of biotin biosynthesis are detected (by HMMs in the TIGRFAMs database), then the Genome Property ‘biotin biosynthesis’ is set by rule to ‘YES’. Assertions made by Genome Properties are useful to summarize high-level traits of species biology from genome analysis, to understand metabolic context while trying to understand the roles of other proteins from the same species, and for comparative genomics based on the whole biological processes rather than single genes.
Genome Properties entries describe subsystems
The linkage between TIGRFAMs and Genome Properties is paralleled in a kindred effort, the SEED from the Fellowship for the Interpretation of Genomes (FIG) (13). FIG selected the term ‘subsystem’ to describe the biological role in aggregate that a set of marker proteins working in concert enable. We support usage of that term, using ‘subsystem’ here to refer in general to any emergent property in species biology that is understood more clearly by viewing the collection of component genes together rather than separately. We use the term ‘Genome Property’ for any subsystem described as an entry in the Genome Properties database.
Metabolic reconstruction is based on evidence rather than annotation
Many sequences in public protein sequence databases are annotated incorrectly as biotin synthase (EC 2.8.1.6), to give one example, through errors of transitive annotation, in part because biotin synthase was one of the first radical SAM superfamily (14) members sequenced, characterized and named (15). In fact, misannotations that attach overly specific functions are a particularly abundant class of error (16). Any metabolic reconstruction system that takes annotations at face value runs a risk of going badly astray. Genome Properties follows a principle that whether or not some genome encodes a subsystem should be determined by evidence sufficient to drive annotation, not by pre-existing annotations of uncertain provenance. Following this principle means that creating a new Genome Property often necessitates building new entries for TIGRFAMs database of HMMs, or else confirming that existing models in either TIGRFAMs or Pfam behave suitably.
Genome Properties emphasizes non-pathway subsystems
Genome Properties includes simple biochemical pathways, of course, but describes many additional types of subsystem. When Genome Properties describes a subsystem whose core is an enzymatic pathway, it often represents non-enzymatic proteins such as transcriptional regulators and molecular chaperones as additional components. Computation of a Genome Property may depend on a protein, a feature within a protein (such as a sorting signal), or a genetic element that does not even code for a protein, such as an array of CRISPR repeats or the selenocysteine tRNA. It describes physical complexes such as transporters and flagella. It describes systems in which one protein acts upon another, including secretion systems, protein-sorting systems, and post-translational modification systems.
Methanobactins are generating considerable interest because neither their diversity nor their biosynthesis is well understood yet (17). The very small size of the only known class of precursor peptide has meant missed gene calls (at least initially) in every species so far with an example of the system, originally just Azospirillum sp. B510 and Methylosinus trichosporium OB3b (11), but now including Gluconacetobacter oboediens 174Bp2, Gluconacetobacter sp. SXCC-1, Tistrella mobilis KA081020-065, Pseudomonas extremaustralis and Methylocystis sp. SC2. Model TIGR04071 for the methanobactin precursor family belongs to a Genome Property, GenProp0962, which explains that the gene should occur in the context of two other, larger molecular markers, members of families TIGR04159 and TIGR04160. These two companion proteins are much easier to find when prospecting in newly sequenced genomes for new examples of methanobactin-like natural products.
The architecture of Genome Properties
Each Genome Property is described completely by records in a series of tables stored in a relational database. The tables are now made available for downloading by ftp through the TIGRFAMs/Genome Properties web pages, at ftp.jcvi.org/pub/data/TIGRFAMs/GEN_PROP. The table structure and meanings of all fields are described in the release notes. The logic of the table structure is discussed below.
Each Genome Property has a basic definition in the prop_def table that includes a name (e.g. ‘urease’), a unique accession (e.g. ‘GenProp0051’), a paragraph-long description, and some ancillary information. Because the first subsystems described in Genome Properties were simple enzymatic pathways, components are referred to in relational database tables as ‘steps.’ Thus, the components that belong to a given Genome Property are enumerated in the prop_step table. Note that one enzymatic activity, a single entity in the typical pathway definition, will be represented by multiple components if the enzyme is comprised of multiple subunits.
A Genome Property is judged complete, and may be assigned the state ‘YES’, if every component listed as required is found. Nearly every Genome Property has two or more components, largely because the comparative genomics of subsystem reconstruction is often essential for creating and establishing trust in the protein family definitions that a Genome Property requires.
Property definitions include genes that are not necessarily essential
Some protein families occur exclusively as part of some subsystem, yet comparative genomics shows they are not always present, and not required for all instances of the subsystem to operate (although they may be required in some cases). Such a protein may be listed as a part of the system by entry into the prop_step table, but is marked as non-essential, meaning not core to the definition of the Genome Property and not used to compute its completeness. A protein that is absolutely required, but for which no reliable detection tool is available, similarly must be marked as non-essential to prevent the scoring system from calling the subsystem incomplete. Models may be unavailable because an activity has not yet been matched to any sequence, or a single known sequence example is not easily extended into a whole protein family definition, or the role tends to be filled by members of different proteins in different species (as is common for phosphatases and aminotransferases), or the function may be hard to assign based on full-length homology rather than select specificity-determining residues (as seems to occur with transporters).
Genome Properties allows multiple lines of evidence for each step
Genome Properties defines types of evidence that will be treated as sufficient to show that a required component is encoded within a genome. In most cases, the evidence is a TIGRFAMs or Pfam HMM that scores above the model’s cutoffs to some protein. Trusted cutoffs are used rather than gathering thresholds (these differ only for Pfam). For a given canonical protein function (e.g. glutamate–cysteine ligase, EC 6.3.2.2), several different known families may be known, and assignment to any one of these may constitute evidence. Thus, a separate linking table, step_ev_link, defines what evidence is sufficient to satisfy a step.
In some cases, Genome Properties requires that there be at least one member encoded in a genome of some family of proteins, without implying that all members found from that family necessarily participate in the Genome Property in question. However, recording sets of such proteins during evaluations of Genome Properties across large numbers of genomes, and distinguishing those found in genomes encoding the other candidate markers from the rest supports data mining approaches that may lead to the construction of new protein families. Genome Properties allows an evidence type designated HMM-CLUST, meaning that a protein must be a member of the designated protein family but also within 3000 base pairs of another marker of the same system. The HMM-CLUST method may identify, for example, members of the radical SAM domain family (PF04055 in Pfam) found in close proximity to a precursor gene for post-translationally modified peptide. This co-clustering may mark a subfamily or equivalog group within radical SAM, which can then be ascribed a role in peptide modification. Such approaches let Genome Properties computation support both protein family development and discovery of new types of subsystems.
Thresholds
In the ideal case, a Genome Property has complete evidence, or no evidence, in nearly every genome. If most components are found, but not all, the YES-leaning state ‘some evidence’ may be assigned. But some proteins can play a role in any of several different properties. Finding such a marker, but no other, for a given property in question does not suggest the property is actually present. The enzyme selenide, water dikinase (SelD, TIGRFAMs entry TIGR00476), for example, is essential to at least three traits (18): selenocysteine incorporation (GenProp0016), the selenouridine tRNA modification (GenProp0692) and post-translational activation of selenium-dependent molybdenum hydroxylases (GenProp0726). For any one of these systems, finding SelD only is very weak evidence. Similarly, a Genome Property may rely in part on an HMM that was available (perhaps from Pfam) but that hits a number of homologs beyond the set that actually carry the function of interest. Absence of any member of that family from a genome would be informative, so it is useful to require a hit as an additional constraint for recognizing a subsystem to be present. But finding a member of that family as the only evidence would be very weak evidence the entire subsystem is encoded. For these reasons, each Genome Property has a threshold value. If the number of components found does not exceed the threshold, evidence that the Genome Property on the whole is encoded by a genome should be considered weak, and the state ‘not supported’ will be assigned instead of ‘some evidence.’
Genome Properties is hierarchical
A component required for a Genome Property to be complete may itself be a Genome Property. Urea utilization (GenProp0814), for example, consists of a urea uptake system and a urea degradation pathway. For urea degradation, either of two pathways is sufficient, urease (GenProp0051) or the urea carboxylase/allophanate hydrolase pathway (GenProp0481). Whichever of the two is the more complete is used to score the urea utilization property.
Genome Properties and TIGRFAMs usually are constructed in concert
Where a set of proteins cooperate to form an enzymatic pathway, or some other type of subsystem with a fixed set of required components, each successfully completed protein family definition gives contextual clues that help identify trusted exemplars for the remaining protein families. Some protein families are straightforward to construct because essentially every detectable homolog appears to perform the identical function. The very clear boundaries to such families provide information that can guide construction of additional protein families. It appears, for example, that every detectable homolog of the first described PqqA, a peptide whose role is to serve as the precursor of pyrroloquinoline quinone (PQQ), likewise serves as a PQQ precursor peptide. In contrast to PqqA, the PQQ biosynthesis enzyme PqqE belongs to radical SAM, a family so abundant that 1 genome may encode over 30 members, each different in function. Correctly separating all true PqqE from their functionally distinct homologs would be difficult except that the PqqA model (TIGR02107) assured that the PqqE model (TIGR02109) would be constructed with no false-positives among its seed members. The PqqE model, in turn, identifies all PQQ biosynthesis systems where the small (∼23 amino acid) PqqA peptide was missed because of faulty gene calling.
Genome Properties made available for the current release, designated 3.0, number 628, a marked expansion over the ∼200 Genome Properties released at the time of the last published database description. The current total includes new subsystem definitions plus previously unreleased ones likely to benefit from additional development.
GENOME PROPERTIES WEB PAGES
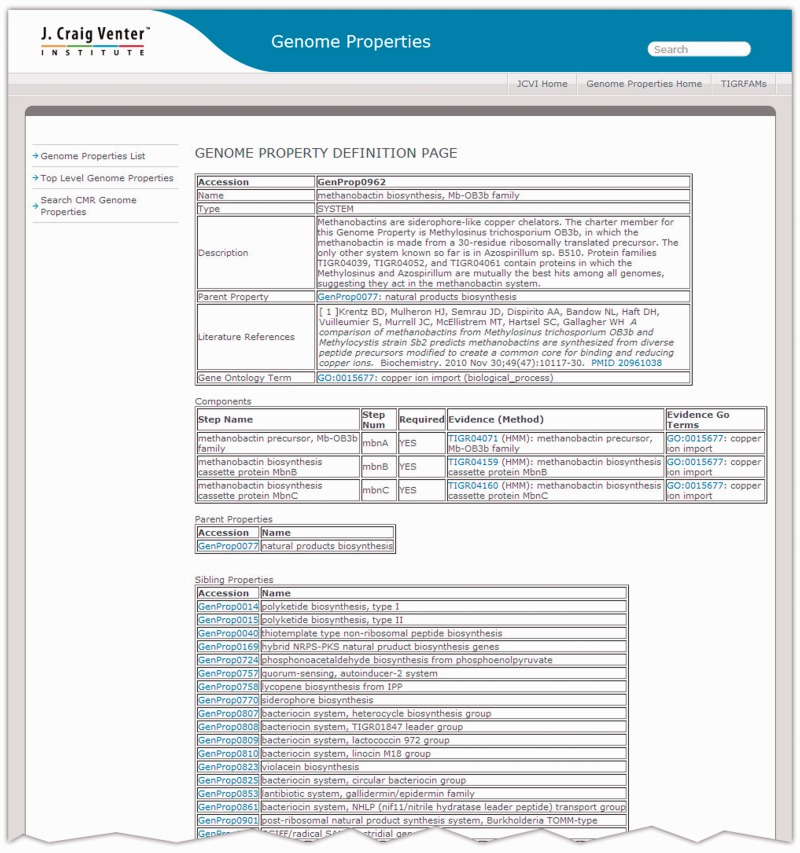
Like TIGRFAMs, Genome Properties was originally hosted through the CMR (9,10). At last update, the CMR contained over 500 microbial genomes. Genomes in the CMR show results from evaluating Genome Properties as implemented based on HMMER2 models, using TIGRFAMs models up through release 9.0. Genome Properties is now hosted independently of the CMR. A sample page, describing Genome Property GenProp0962 (‘methanobactin biosynthesis, Mb-OB3b family’), is shown in Figure 2. The new Genome Properties pages provide a link to the legacy version of Genome Properties in the CMR, giving access to computed Genome Properties results in a comparative genomics browser environment.
Figure 2.
Example of a Genome Properties Definition Page. The Genome Properties home page provides three links on the left navigation sidebar: Genome Properties List, Top Level Genome Properties and Search CMR Genome Properties. The Genome Properties List shows all current Genome Properties along with their property type and property name. Clicking on a Genome Property will display the Genome Property Definition page. The Definition Page for GenProp0962 (‘methanobactin biosynthesis, Mb-OB3b family’) is illustrated. The Genome Property Definition page displays the property’s name, description, parent/child/sibling properties, literature references and associated GO terms. The components of the property are listed with the step names, and whether or not the step is required (used to evaluate completeness). The evidence required to detect that a component has been found is described, usually the accession of a TIGRFAMs HMM with a working link to HMM Summary Page. Users can browse through the Genome Properties through the Top Level Genome Properties page linked on the home page. Users traverse through the properties by first selecting a top level property (e.g. Metabolism, Taxonomy). Associated parent, child and sibling properties are displayed for selection. The Search CMR Properties link directs users to the CMR genome property search.
SPECIAL FOCUS AREAS FOR TIGRFAMS AND GENOME PROPERTIES
TIGRFAMs focuses heavily, and Genome Properties almost exclusively, on subsystems encoded in bacterial and archaeal genomes.
Successful construction of a TIGRFAMs equivalog-level model defines a molecular marker, and makes it possible to evaluate the presence/absence of that marker across the set of all complete microbial genomes. The result, a truth table of 1’s and 0’s for a large number of genomes, comprises a phylogenetic profile (19). The profile serves as input to the Partial Phylogenetic Profiling algorithm (20), which performs data mining to find (and guide construction of new HMMs for) additional protein families that belong to the same subsystem, even if the families in question have never before been defined and fall within large superfamilies. New models contribute to improved Genome Property definitions, more accurate comparative genomics, and iteratively improved descriptions of subsystems biology.
Prokaryotic protein-sorting systems is an area of special focus for TIGRFAMs and Genome Properties, containing 37 such properties. Seventeen of these, including one involving a rhomboid family intramembrane serine protease (21), one featuring a sorting signal unique to the deltaProteobacteria (22), and all subclasses of exosortase-based and archaeosortase-based sorting/processing systems (23), represent discoveries made via TIGRFAMs/Genome Properties development.
TIGRFAMs and Genome Properties created the original nomenclature for CRISPR subtypes, defining eight variants (Ecoli, Dvulg, Ypest, Hmari, Tneap, Nmeni, Mtube and the RAMP module) (24). More recently, a reanalysis using sensitive detection methods demonstrated remote homologies that grouped CRISPR systems into type I (where Ecoli is type I-E, Ypest is type I-F, etc), type II and type III (25). Most existing TIGRFAMs models for CRISPR-associated protein families now show multilevel naming that reflects both schemes. Since our original report of the first eight subtypes, we have detected conserved architectures for additional CRISPR subtypes, and described them through TIGRFAMs and Genome Properties. These include the Aferr subtype (GenProp0670, the unique CRISPR system in Acidithiobacillus ferrooxidans ATCC 23270), the Pging subtype (GenProp0768, named after Porphyromonas gingivalis W83), the Myxan subtype (GenProp0922, named after Myxococcus xanthus), and the PreFran subtype (GenProp1061, named for genera Prevotella and Francisella).
Lastly, development of these databases has introduced many Genome Properties in which a subfamily of radical SAM represents a key marker, and many examples of natural product biosynthesis systems in which a ribosomally translated precursor from a novel protein family is post-translationally modified. These classes overlap heavily for radical SAM proteins with a C-terminal SPASM domain (TIGR04085), where the acronym SPASM signifies roles in the maturation of Subtilosin, PQQ, Anaerobic Sulfatases and Mycofactocin (26).
CONCLUSION
TIGRFAMs and Genome Properties continue to be developed, in parallel, as annotation-driving databases for microbial genome analysis. Context provided by Genome Properties gives feedback for the process of making the fixed cutoffs of each HMM in TIGRFAMs as accurate as possible. The intended result is that classification of proteins into families with identified functions provides a trustworthy basis for many types of subsequent analysis. The most immediate of these is whether or not some genome in question encodes the necessary sets of components for each of over 600 different subsystems.
AVAILABILITY
TIGRFAMs resources and Genome Properties resources are available through home pages at http://www.jcvi.org/cgi-bin/tigrfams/index.cgi and http://www.jcvi.org/cgi-bin/genome-properties/index.cgi, respectively.
FUNDING
This project has been funded in whole or part with federal funds from the National Human Genome Research Institute [R01 HG004881] and from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services [N01-AI30071, HHSN272200900007C]. Funding for open access charge: National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract numbers N01-AI30071 and/or HHSN272200900007C.
Conflict of interest statement. None declared.
REFERENCES
- 1.Madupu R, Richter A, Dodson RJ, Brinkac L, Harkins D, Durkin S, Shrivastava S, Sutton G, Haft D. CharProtDB: a database of experimentally characterized protein annotations. Nucleic Acids Res. 2012;40:D237–D241. doi: 10.1093/nar/gkr1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 3.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. The Comprehensive Microbial Resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidsen T, Beck E, Ganapathy A, Montgomery R, Zafar N, Yang Q, Madupu R, Goetz P, Galinsky K, White O, et al. The comprehensive microbial resource. Nucleic Acids Res. 2010;38:D340–D345. doi: 10.1093/nar/gkp912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krentz BD, Mulheron HJ, Semrau JD, Dispirito AA, Bandow NL, Haft DH, Vuilleumier S, Murrell JC, McEllistrem MT, Hartsel SC, et al. A comparison of methanobactins from Methylosinus trichosporium OB3b and Methylocystis strain Sb2 predicts methanobactins are synthesized from diverse peptide precursors modified to create a common core for binding and reducing copper ions. Biochemistry. 2010;49:10117–10130. doi: 10.1021/bi1014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haft DH, Selengut JD, Brinkac LM, Zafar N, White O. Genome Properties: a system for the investigation of prokaryotic genetic content for microbiology, genome annotation and comparative genomics. Bioinformatics. 2005;21:293–306. doi: 10.1093/bioinformatics/bti015. [DOI] [PubMed] [Google Scholar]
- 13.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohsawa I, Speck D, Kisou T, Hayakawa K, Zinsius M, Gloeckler R, Lemoine Y, Kamogawa K. Cloning of the biotin synthetase gene from Bacillus sphaericus and expression in Escherichia coli and Bacilli. Gene. 1989;80:39–48. doi: 10.1016/0378-1119(89)90248-5. [DOI] [PubMed] [Google Scholar]
- 16.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenney GE, Rosenzweig AC. Chemistry and biology of the copper chelator methanobactin. ACS Chem. Biol. 2012;7:260–268. doi: 10.1021/cb2003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haft DH, Self WT. Orphan SelD proteins and selenium-dependent molybdenum hydroxylases. Biol. Direct. 2008;3:4. doi: 10.1186/1745-6150-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu MK, Selengut JD, Haft DH. ProPhylo: partial phylogenetic profiling to guide protein family construction and assignment of biological process. BMC Bioinform. 2011;12:434. doi: 10.1186/1471-2105-12-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haft DH, Varghese N. GlyGly-CTERM and rhombosortase: a C-terminal protein processing signal in a many-to-one pairing with a rhomboid family intramembrane serine protease. PLoS One. 2011;6:e28886. doi: 10.1371/journal.pone.0028886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLoS Genet. 2012;8:e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haft DH, Payne SH, Selengut JD. Archaeosortases and exosortases are widely distributed systems linking membrane transit with posttranslational modification. J. Bacteriol. 2012;194:36–48. doi: 10.1128/JB.06026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haft DH, Basu MK. Biological systems discovery in silico: radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J. Bacteriol. 2011;193:2745–2755. doi: 10.1128/JB.00040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]