Abstract
In 2007, Comparative Fungal Genomics Platform (CFGP; http://cfgp.snu.ac.kr/) was publicly open with 65 genomes corresponding to 58 fungal and Oomycete species. The CFGP provided six bioinformatics tools, including a novel tool entitled BLASTMatrix that enables search homologous genes to queries in multiple species simultaneously. CFGP also introduced Favorite, a personalized virtual space for data storage and analysis with these six tools. Since 2007, CFGP has grown to archive 283 genomes corresponding to 152 fungal and Oomycete species as well as 201 genomes that correspond to seven bacteria, 39 plants and 105 animals. In addition, the number of tools in Favorite increased to 27. The Taxonomy Browser of CFGP 2.0 allows users to interactively navigate through a large number of genomes according to their taxonomic positions. The user interface of BLASTMatrix was also improved to facilitate subsequent analyses of retrieved data. A newly developed genome browser, Seoul National University Genome Browser (SNUGB), was integrated into CFGP 2.0 to support graphical presentation of diverse genomic contexts. Based on the standardized genome warehouse of CFGP 2.0, several systematic platforms designed to support studies on selected gene families have been developed. Most of them are connected through Favorite to allow of sharing data across the platforms.
INTRODUCTION
Fungal genome sequencing has rapidly increased since the release of the genome sequences of Saccharomyces cerevisiae in 1996 (1). With the current and anticipated advances in sequencing technology (2,3), the rate of fungal genome sequencing will continue to accelerate. Currently, there exist more than 300 fully sequenced fungal genomes in the public domain (4,5), with many species and several isolates of previously sequenced species being sequenced (6,7). In addition, the 1000 Fungal Genome project (F1000; http://1000.fungalgenomes.org/) will greatly help us to uncover genomic underpinnings of fungal evolution and life styles via large-scale comparative genomics studies. In combination with genomes from plants and animals as well as fungi, in-depth comparative genomics across multiple eukaryotic kingdoms will be facilitated (8–10). To efficiently support such large-scale, genome-based inquiries, it is critical to archive the available genome sequences and annotation information in a standardized format so that they can be easily and efficiently retrieved and analyzed.
To address this need, in 2007, the first version of Comparative Fungal Genomics Platform (CFGP) was released with 65 fungal and Oomycete genomes (11). The CFGP was founded on a new user interface (UI) called Data-driven User Interface (DUI), which made use of its bioinformatics tools and the management of task histories easy and efficient. Since then, the number of genomes archived and bioinformatics tools have grown substantially. (Supplementary Table S1 and Table 1) Furthermore, the standardized genome data warehouse of CFGP has supported the development of multiple platforms that are specialized for supporting the archiving and analysis of specific gene families and functional groups (Table 2). Some of these platforms share the Favorite of CFGP to provide an efficient mechanism for sharing data with CFGP and to enable the use of its bioinformatics tools for a variety of analyses. In this article, we outline the improvements made in CFGP 2.0 and how its standardized genome warehouse has been exploited in development of other comparative genomics platforms.
Table 1.
List of bioinformatics tools available in CFGP 2.0
| Category | Name | Input data | References |
|---|---|---|---|
| Sequence search | BLAST | Sequences | (12) |
| BLAST2 | Sequences | (12) | |
| BLASTMatrix | Sequences | (11) | |
| ID of functional domains | InterPro Scan | Sequences | (13) |
| Phylogenetic analysis | ClustalW | Sequences | (14) |
| DNAML | Alignment | (15) | |
| PROML | Alignment | (15) | |
| DNAPARS | Alignment | (15) | |
| PROTPARS | Alignment | (15) | |
| PHYML | Alignment | (16) | |
| ID of secreted proteins | SignalP 3.0 | Sequences | (17) |
| SigCleave | Sequences | (18) | |
| SigPred | Sequences | (19) | |
| RPSP | Sequences | (20) | |
| SecretomeP | Sequences | (21) | |
| Subcellular localization | PSortII | Sequences | (22) |
| predictNLS | Sequences | (23) | |
| ChloroP | Sequences | (24) | |
| TargetP | Sequences | (25) | |
| Prediction of trans-membrane helices | TMHMM2 | Sequences | (26) |
| Prediction of RNA secondary structure | tRNAScan-SE | Sequences | (27) |
| mFold 3.2 | Sequences | (28) | |
| Post-translational modification | NetCGlyc | Sequences | (29) |
| NetNGlyc | Sequences | – | |
| NetOGlyc | Sequences | (30) | |
| NetPhos | Sequences | (31) | |
| Conserved motif search | MEME | Sequences | (32) |
A total of 27 tools in nine categories are available via the Favorite browser of CFGP 2.0.
Table 2.
List of online platforms and tools supporting studies on specific gene families or functional groupsa
| Name | URL | Reference |
|---|---|---|
| Cyber-infrastructure for Fusarium | http://www.fusariumdb.org/ | (33) |
| Fungal Transcription Factor Database | http://ftfd.snu.ac.kr/ | (34) |
| Fungal Cytochrome P450 Database | http://p450.riceblast.snu.ac.kr/ | (35) |
| Seoul National University Genome Browser | http://genomebrowser.snu.ac.kr/ | (36) |
| The Systematic Platform for Identifying Mutated Proteins | http://pimp.starflr.info/ | (37) |
| Insect Mitochondrial Genome Database | http://www.imgd.org/ | (38) |
| Fungal Secretome Database | http://fsd.snu.ac.kr/ | (39) |
| Eukaryotic DNAJ/K Database | http://edd.snu.ac.kr/ | – |
| Cell Wall-degrading Enzyme Database | http://www.cwde.org/ | – |
aThese platforms were developed using the standardized genome warehouse and the web template engine of CFGP 2.0.
METHODS
System design
The structure and core databases of CFGP 2.0 are basically identical to those used for the first version. The system consists of databases including a standardized genome warehouse, wrapper programs written by the Perl and C languages and DUI. To balance the server load so as to ensure a more efficient operation of the system, its core databases were distributed in multiple servers, and more web servers were added. The MySQL relational database management system was used to manage and curate the data. Its web interfaces were written in PHP with javascript, and analysis functions in Favorite Browser were relayed by Perl scripts and automatically coordinated by the system monitoring servers.
Mining orthologs
The source code of InParanoid 4.1 was used for the identification of orthologs in the archived proteomes. First, the genomes of 35 species that are frequently utilized were subjected to ortholog identification (Supplementary Table S1). All pairwise comparisons of data from these 35 species were carried out. The latest version of InParanoid 7 provides orthologs from 100 eukaryotic genomes. However, some genomes that we have used were not included in the latest version; data for those species were downloaded from CFGP 2.0 and subsequently subjected to ortholog identification.
EXPANDED GENOME DATA WAREHOUSE
The standardized genome warehouse of CFGP has been substantially expanded in both the number of species and taxonomic coverage. In addition to 283 genomes corresponding to 152 fungal and Oomycete species, 39 plant and 105 animal genomes have also been archived (Figure 1 and Supplementary Table S2). The animal and plant genomes were incorporated to enable comparative evolutionary genomics studies across multiple eukaryotic kingdoms.
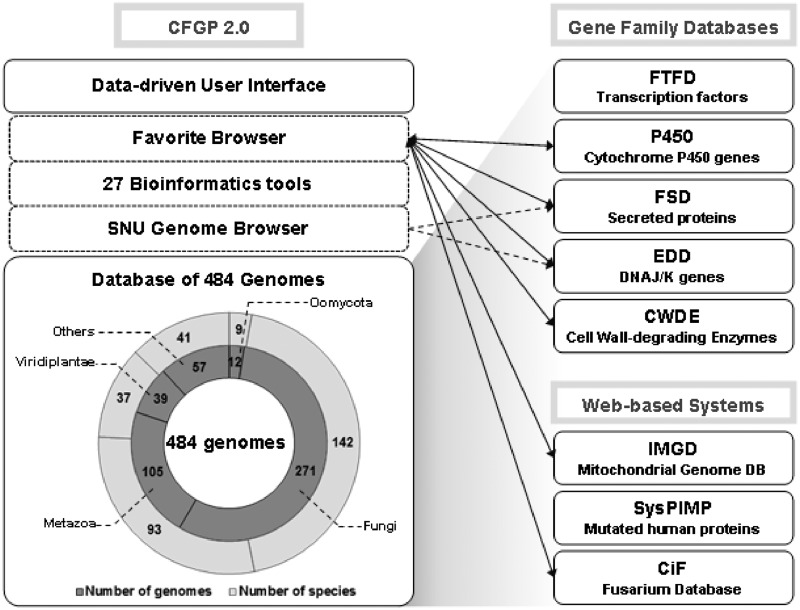
Figure 1.
A diagram illustrating the system architecture and the content of the genomes archived in CFGP 2.0. Key features of CFGP 2.0 were depicted on the left. The web-based platforms that have been developed based on the standardized genome warehouse of CFGP 2.0 are listed on the right. Bidirectional arrows indicate that they support the Favorite Borwser, which synchronizes with CFGP 2.0. Dashed arrows denote that SNUGB was integrated in two platforms, FSD and EDD. In the pie graph, the inner and outer circles represent the number of genomes and species for each taxon, respectively.
ENHANCED UTILITY AND NEW FEATURES
Improved UI
The UI of CFGP 2.0 was greatly improved to provide better user experience. All modifications followed the HTML5 and CSS3 standards, on which most widely used web browsers are based. Three main application/utility frames were rearranged to make the switch from one frame to another more intuitive. The Favorite and presentation frames can be toggled for more flexible browsing (Figure 2). All web pages have been thoroughly tested with multiple web browsers, including Chrome, Firefox, Internet Explorer and Safari.
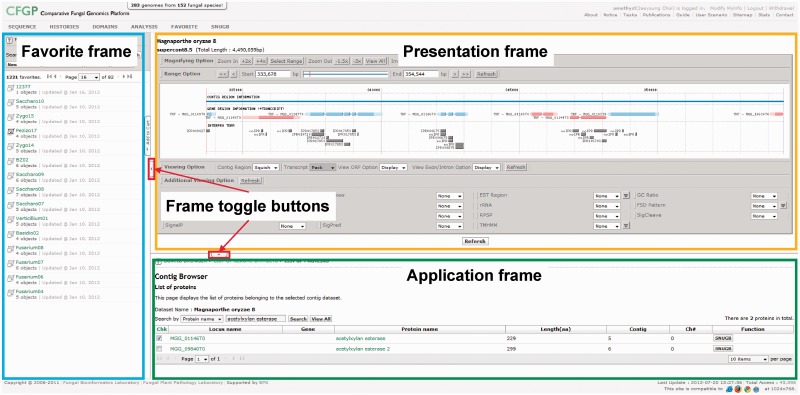
Figure 2.
Outline of the improved DUI of CFGP 2.0. The frames outlined in blue, orange and purple correspond to the Favorite, presentation and application frame, respectively. The buttons boxed in red are to hide or show Favorite and presentation frames, respectively.
Taxonomy browser
As the number of species representing diverse taxa continues to increase, a browsing tool based on simple text search (e.g. species name) was not sufficient to help users to efficiently explore available genomes. The Taxonomy Browser implemented in CFGP 2.0 provides predictive text feature as well as a hierarchical tree-based taxon structure to show the taxonomic position of a chosen species. Once a specific species is selected, the number of genomes and available sequence types are listed with direct links to corresponding sequences.
Seoul National University Genome Browser
The first version of CFGP did not offer a graphical interface to present the genomic context and notable features such as GC content, functional domains and the signal peptide. This new genome browser implemented in CFGP 2.0 enabled users to view such information in the chosen region. The target region of viewing can be selected by assigning the start and end positions with a mouse or by typing in its genome coordinate (Figure 3).
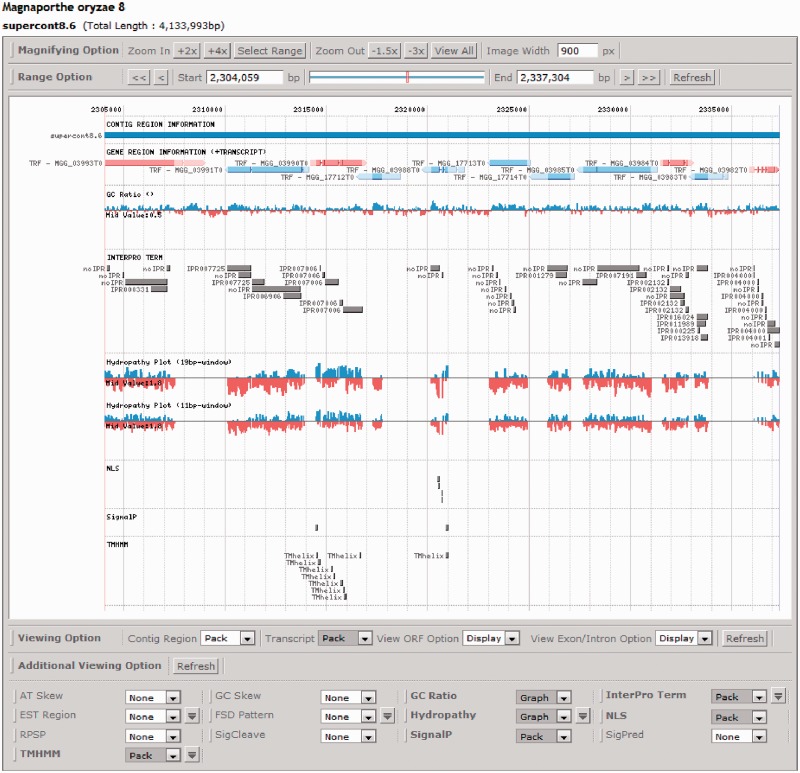
Figure 3.
A screenshot of SNUGB implemented in CFGP 2.0. SNUGB allows users to view 13 biological features and the gene structures in the selected genome region. Those features include GC contents, functional domains, nuclear localization signals, signal peptides and trans-membrane helixes.
New bioinformatics tools added to the Favorite Browser
Compared with the first version that only provided six tools, CFGP 2.0 is equipped with 27 tools covering nine categories of data analysis or viewing (Table 1). This addition enables users to perform more analyses without leaving CFGP.
Ortholog browsing function
Finding orthologs for a specific gene in multiple species often requires numerous BLAST searches and validation processes. In the first version of CFGP, we tried to eliminate copy-and-paste of sequences by incorporating the DUI (11). In CFGP 2.0 we simplified the identification and collection of orthologs by offering pre-computed data. There are several ortholog identification programs such as InParanoid (40), Ortholog, MSOAR (41) and THOR (42). We adopted InParanoid to identify orthologs via pairwise comparisons among 35 frequently accessed genomes. For every protein sequence encoded by each of these 35 genomes, orthologous genes in the other 34 genomes are provided to allow a quick overview of its distribution among these species and also to support their further analyses by saving them into a Favorite on the fly.
FUNCTIONAL/EVOLUTIONARY GENOMICS PLATFORMS DEVELOPED BASED ON THE STANDARDIZED GENOME WAREHOUSE OF CFGP 2.0
Via the use of the standardized genome warehouse of CFGP 2.0, a number of platforms that aim to support comparative analyses of specific gene families and/or functional groups have been developed: (i) Cyber-infrastructure for Fusarium (CiF; http://www.fusariumdb.org/) (33), (ii) Fungal Transcription Factor Database (FTFD; http://ftfd.snu.ac.kr/) (34), (iii) Fungal Cytochrome P450 Database (FCPD; http://p450.riceblast.snu.ac.kr/) (35), (iv) Fungal Secretome Database (FSD; http://fsd.snu.ac.kr/) (39), (v) Eukaryotic DNAJ and DNAK Database (EDD; http://edd.snu.ac.kr/) (Cheong et al., manuscript in preparation) and (vi) Cell Wall-degrading Enzymes Database (CWDE; http://www.cwde.org/) (Choi et al., manuscript in preparation). The Seoul National University Genome Browser (SNUGB) (http://genomebrowser.snu.ac.kr/) (36) was also implemented in FSD and EDD. The Insect Mitochondrial Genome Database (IMGD; http://www.imgd.org/) (38) employs the Species-driven UI, which enables intuitive and fast taxonomical browsing with multiple add-on analysis functions. Finally, the Systematic Platform for Identifying Mutated Proteins (SysPIMP; http://pimp.starflr.info/) (37) was developed to support the identification of mutations related to human diseases. The Favorite Browser of CFGP 2.0 is connected with many of those platforms to efficiently support data exchange and sharing across multiple platforms. All the data saved in the Favorite Browser are synchronized in real-time so that users can fully exploit data and functions provided by these platforms.
FUTURE DIRECTIONS
To keep up with the rapidly released and updated eukaryotic genomes, CFGP 2.0 will be updated on a regular basis. We will integrate more useful modules, software or interface scheme to continuously improve the environment for users conducting comparative and evolutionary genomics studies. In order to support efforts to uncover possible functions of many hypothetical genes, the ortholog information database will be expanded by adding the corresponding information from more species.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2.
FUNDING
National Research Foundation of Korea grant funded by the Korea government [2012-0001149 and 2012-0000141]; TDPAF [309015-04-SB020]; Next-Generation BioGreen 21 Program of Rural Development Administration in Korea [PJ00821201]; a graduate fellowship through the Brain Korea 21 Program (to J.C., K.C. and J.J.). Funding for open access charge: Seoul National University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:546, 563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 2.Metzker ML. Sequencing technologies — the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 3.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat. Rev. Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40:D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keyhani NO. Fungal genomes and beyond. Fungal. Genom. Biol. 2011;1:e101. [Google Scholar]
- 6.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornell MJ, Alam I, Soanes DM, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Talbot NJ, Oliver SG. Comparative genome analysis across a kingdom of eukaryotic organisms: specialization and diversification in the fungi. Genome Res. 2007;17:1809–1822. doi: 10.1101/gr.6531807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards TA, Soanes DM, Foster PG, Leonard G, Thornton CR, Talbot NJ. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell. 2009;21:1897–1911. doi: 10.1105/tpc.109.065805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Dam TJ, Rehmann H, Bos JL, Snel B. Phylogeny of the CDC25 homology domain reveals rapid differentiation of Ras pathways between early animals and fungi. Cell. Signal. 2009;21:1579–1585. doi: 10.1016/j.cellsig.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Park J, Park B, Jung K, Jang S, Yu K, Choi J, Kong S, Kim S, Kim H, Kim JF, et al. CFGP: a web-based, comparative fungal genomics platform. Nucleic Acids Res. 2008;36:D562–D571. doi: 10.1093/nar/gkm758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Seattle: Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- 16.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 17.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 18.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 19.Bradford JR. Thesis (MRes) Leeds, UK: University of Leeds (School of Biochemistry and Molecular Biology); 2001. Protein design for biopharmaceutical development at glaxosmithkline: in silico methods for prediction of signal peptides and their cleavage sites, and linear epitopes. [Google Scholar]
- 20.Plewczynski D, Slabinski L, Ginalski K, Rychlewski L. Prediction of signal peptides in protein sequences by neural networks. Acta Biochim. Pol. 2008;55:261–267. [PubMed] [Google Scholar]
- 21.Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 22.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emanuelsson O, Nielsen H, Von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 26.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 27.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julenius K. NetCGlyc 1.0: prediction of mammalian C-mannosylation sites. Glycobiology. 2007;17:868–876. doi: 10.1093/glycob/cwm050. [DOI] [PubMed] [Google Scholar]
- 30.Hansen JE, Lund O, Tolstrup N, Gooley AA, Williams KL, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj. J. 1998;15:115–130. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- 31.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 32.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park B, Park J, Cheong KC, Choi J, Jung K, Kim D, Lee YH, Ward TJ, O'Donnell K, Geiser DM, et al. Cyber infrastructure for Fusarium: three integrated platforms supporting strain identification, phylogenetics, comparative genomics and knowledge sharing. Nucleic Acids Res. 2011;39:D640–D646. doi: 10.1093/nar/gkq1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park J, Jang S, Kim S, Kong S, Choi J, Ahn K, Kim J, Lee S, Park B, Jung K, et al. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics. 2008;24:1024–1025. doi: 10.1093/bioinformatics/btn058. [DOI] [PubMed] [Google Scholar]
- 35.Park J, Lee S, Choi J, Ahn K, Park B, Kang S, Lee YH. Fungal cytochrome P450 database. BMC Genomics. 2008;9:402. doi: 10.1186/1471-2164-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung K, Park J, Choi J, Park B, Kim S, Ahn K, Choi D, Kang S, Lee YH. SNUGB: a versatile genome browser supporting comparative and functional fungal genomics. BMC Genomics. 2008;9:586. doi: 10.1186/1471-2164-9-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xi H, Park J, Ding G, Lee YH, Li Y. SysPIMP: the web-based systematical platform for identifying human disease-related mutated sequences from mass spectrometry. Nucleic Acids Res. 2009;37:D913–D920. doi: 10.1093/nar/gkn848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee W, Park J, Choi J, Jung K, Park B, Kim D, Lee J, Ahn K, Song W, Kang S, et al. IMGD: an integrated platform supporting comparative genomics and phylogenetics of insect mitochondrial genomes. BMC Genomics. 2009;10:148. doi: 10.1186/1471-2164-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi J, Park J, Kim D, Jung K, Kang S, Lee YH. Fungal secretome database: integrated platform for annotation of fungal secretomes. BMC Genomics. 2010;11:105. doi: 10.1186/1471-2164-11-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Z, Chen X, Vacic V, Nan P, Zhong Y, Jiang T. MSOAR: a high-throughput ortholog assignment system based on genome rearrangement. J. Comput. Biol. 2007;14:1160–1175. doi: 10.1089/cmb.2007.0048. [DOI] [PubMed] [Google Scholar]
- 42.Bainbridge MN, Warren RL, He A, Bilenky M, Robertson AG, Jones SJ. THOR: targeted high-throughput ortholog reconstructor. Bioinformatics. 2007;23:2622–2624. doi: 10.1093/bioinformatics/btl513. [DOI] [PubMed] [Google Scholar]