Abstract
This article summarizes our progress with RegulonDB (http://regulondb.ccg.unam.mx/) during the past 2 years. We have kept up-to-date the knowledge from the published literature regarding transcriptional regulation in Escherichia coli K-12. We have maintained and expanded our curation efforts to improve the breadth and quality of the encoded experimental knowledge, and we have implemented criteria for the quality of our computational predictions. Regulatory phrases now provide high-level descriptions of regulatory regions. We expanded the assignment of quality to various sources of evidence, particularly for knowledge generated through high-throughput (HT) technology. Based on our analysis of most relevant methods, we defined rules for determining the quality of evidence when multiple independent sources support an entry. With this latest release of RegulonDB, we present a new highly reliable larger collection of transcription start sites, a result of our experimental HT genome-wide efforts. These improvements, together with several novel enhancements (the tracks display, uploading format and curational guidelines), address the challenges of incorporating HT-generated knowledge into RegulonDB. Information on the evolutionary conservation of regulatory elements is also available now. Altogether, RegulonDB version 8.0 is a much better home for integrating knowledge on gene regulation from the sources of information currently available.
INTRODUCTION
Escherichia coli K-12 is one of the best-characterized microorganisms. RegulonDB is a relational database that serves the scientific community involved in the study of bacteria, offering in an organized and computable form, knowledge on transcriptional regulation that has been manually curated from original scientific publications. This includes curated information on known mechanisms of regulation of transcription initiation through the activation and repression of transcription factors (TFs), which bind to individual sites around promoters; the organization of operons and their various transcription units (TUs) and the integration of regulons as gensor units (GUs). The RegulonDB team also continues to perform high-throughput (HT) experimental identification of promoters in the E. coli genome. Our mission has been to be the compilers and editors of the knowledge generated by the international scientific community regarding the regulatory elements of transcriptional regulation of gene expression in E. coli K-12. Our work maintains up-to-date information in both the RegulonDB and EcoCyc databases [(1,2) and an update by Keseler et al. in this issue].
We should emphasize that any piece of knowledge is curated with its associated reference(s) and the corresponding evidence code on which unified criteria have been defined, enabling distinctions between strong versus weakly supported objects. As detailed later, this classification has been enriched, initiating the process to integrate multiple sources of evidence to define gold standards.
High-quality expanded encoded mechanistic knowledge from different sources
In the main menu ‘About RegulonDB’, we show the historical increase of all objects through the years. During the past 2 years, the number of publications supporting the corpus of knowledge encoded in RegulonDB has increased to 4667. We have increased the number of known functional and non-functional conformations of TFs from 232 to 298, corresponding to a total of 103 TFs (see historical increase in RegulonDB web site). By ‘functional’ we mean the conformations that bind to DNA and exert their regulatory effect. The analysis of the repertoire of regulatory mechanisms focusing on the architecture of signal recognition, specifically, the functional conformation (holo or apo) of a TF, its function or mode of regulation (activator, repressor or dual) and the anabolic or catabolic nature of its regulated genes, enables searches at a genomic level for design principles under the framework of the demand theory of gene regulation, which we discuss elsewhere (Balderas-Martínez et al., submitted for publication). All conformations are supported by experimental methods that have been classified into strong or weak evidence types (see the new Evidence page in RegulonDB).
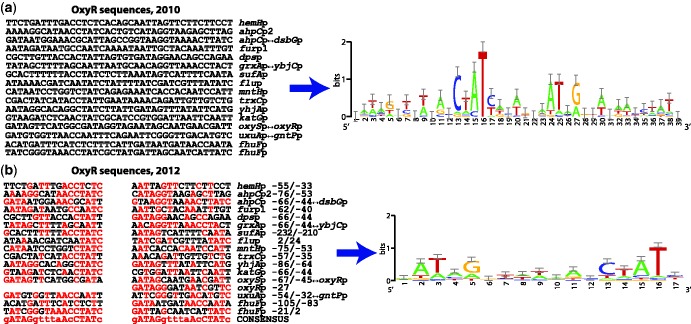
A constant effort focused on detailed correction of TF-binding site (TFBS) properties, such as the length, symmetry, precise position, strand and orientation, is now reflected in new improved alignments for ∼130 TFs. This has been a demanding and time-consuming effort of continuous curation that has strongly enhanced the quality of the evidence for the DNA-binding sites of the TF collection, a core element of the mechanistic and genomic imprint of transcriptional regulation. See the OxyR example in Figure 1. This effort started in 2009, and it is already providing fruits in terms of improved computational TF-DNA models.
Figure 1.
Analysis of TFBSs to improve the quality of PWMs in the RegulonDB database. OxyR binds in tandem, covering regions of ∼40 bp (a). We identified within these regions, two inverted-repeat motifs of 17 bp, separated by 5 bp (b). Therefore, we now propose a new consensus sequence, GATAGGTTnAACCTATC, for the binding sites of OxyR. This new annotation has improved the quality of the matrices (b) and, therefore, also the predictions of binding sites for OxyR.
The number of TFs that possess at least four binding sites has increased from 71 to 86 in the past 2 years, enabling the construction of position weight matrix (PWM) bioinformatics models. Since 2011, we have proposed the use of four independent criteria to assess the quality of matrices: (i) information content conservation of at least 1.5 bits in at least six positions in the matrix; (ii) a low false-positive rate (<1e−4) for recovering 70% of the annotated sites; (iii) an observed distribution of scores in the upstream regions on E. coli K-12 that shows overrepresentation of high scores compared with the theoretical distribution and (iv) not overfitting the matrix to the sequences that were used to build it (3). For details of these four criteria, see the documentation on PWMs in RegulonDB. Based on these criteria, the current collection of 86 TFs contains 50% high-quality models. The low-quality models are mostly those for TFs with a reduced number of sites. For instance, when counting only matrices with eight or more sites, 58% are of high quality. In 2008, only 33% of the 60 TFs with a PWM had a high-quality matrix, whereas currently 56% of these 60 TFs have a high-quality matrix, reflecting the importance of our curation and correction efforts.
The increased quality of the PWM collection is reflected in the number of false-positives that might be generated from a whole-genome computational prediction of binding sites. Overall, the known versus predicted fraction of sites when assessing all our computational predictions in the genome has diminished from ∼1 to 40 in 2008, to 1 to 5 in 2010, and to 1 to 3 in the current version.
The improved PWMs were used to initiate curation of regulatory interactions that had no binding site identified, despite the availability of experimental evidence that supported them. Our current manual curation of the predicted sites has identified TFBSs for 35 interactions. In seeking consistency of evaluation of knowledge irrespective of its source, we used similar criteria to assess the quality of binding sites identified by chromatin immunoprecipitation (ChIP)-Seq experiments (see ‘Enriched classifications based on classic and HT evidence’ and Supplementary Data).
We have expanded our curation to include factors that bind allosterically to RNA polymerase directly. The two currently known mechanisms for E. coli regarding allosteric binding involve ppGpp and DksA. We curated regulatory interactions in which the nucleotide guanosine 5′-diphosphate, ppGpp (referred to as both tetraphosphate and as its precursor, pppGpp) (4,5) and the small protein DksA (6,7) bind to the RNA polymerase alone or form a complex with each other, affecting transcription in either a positive or negative manner, or act antagonistically on the same promoter (8,9) (see Supplementary Figure S1 in the Supplementary Data). Currently, 70 promoter interactions regulated by ppGpp, as well as some that include regulation by DksA, have been curated. The growth conditions under which the promoters are regulated are also included in each reaction of regulation (see Supplementary Figure S1 in the Supplementary Data).
HIGH-LEVEL CURATION
We believe that the integration of knowledge to facilitate an understanding at different levels of abstraction and detail is a major challenge for genomic databases. In the following section, we describe two directions of our efforts towards obtaining higher integration levels: (i) GUs and (ii) the organization of multiple TFBSs into regulatory phrases.
Fur, a complex GU
In 2011, we described the new concept of genetic sensory-response units, or ‘gensor units’, which are composed of four components: (i) the signal, (ii) the signal-to-effector reactions that end with activation or inactivation of the TF, (iii) the regulatory switch (resulting in activation or repression of transcription of target genes) and (iv) the consequence, or effects and roles of the regulated genes (1). RegulonDB contains 25 completed GUs, which are organized into two categories: carbon source utilization and metabolism of amino acids. These are all GUs for local TFs and small regulons. We decided to curate a much larger GU as a first step towards eventually compiling information on GUs of global regulators.
Certainly, the size and complexity of the Fur (ferric uptake regulator) GU poses new challenges in its representation. Fur regulates transcription initiation of 66 TUs, including nine TFs, a regulatory small RNA (sRNA) and two sigma factors (σ19 and σ38). It includes >200 reactions and close to 300 nodes. To facilitate interpretation of this GU, we included a high-level illustration that provides an overview of all classes of genes and functions subject to Fur regulation (see Figure 2). Search ‘gensor unit’ in the main menu in RegulonDB and select Fur overview.
Figure 2.

Overview of the GU of the Fur TF. In the presence of Fe+, Fur represses genes involved in transport and release of Fe+ from siderophores and genes for biosynthesis and assembly of FeS clusters; in addition, it activates genes involved in Fe+ storage and activates/represses genes that encode proteins that contain Fe+ or a group heme as a cofactor. In the presence of the signal, Fur also regulates transcription of nine TFs, the σ19 and σ38 factors and a regulatory sRNA, RhyB, submaps of which are depicted as dark gray squares that can be expanded to see their corresponding GU. In addition, Fur regulates genes that encode metal-binding proteins (other than Fe+) and other proteins that apparently have no direct relationship with Fe+ or other metals.
Regulatory phrases
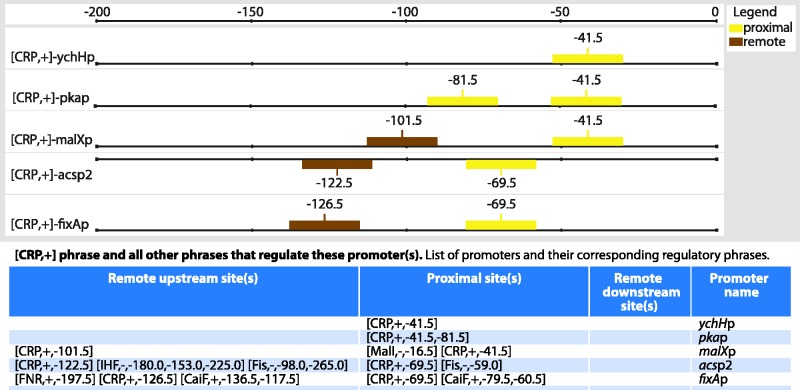
Another area that will clearly benefit from a more integrated description of the genome is the encoding of the organization and functioning of regulatory regions governing transcription. Previously, we displayed the collection of sites in upstream regions affecting each promoter, leaving it to the user to decipher how these multiple sites, which bind the same or different TFs, work in a coordinated fashion, or not, to regulate transcription. For instance, regulation of the acsp2 promoter is affected by two activator sites for CRP, three repressor sites for Fis and three for IHF. The functions and positions of these eight sites are listed one by one in RegulonDB, when in fact it is known, first, that both in case of Fis and IHF, the multiple sites work together, and, second, that each group of sites represses the acsp2 promoter independently: FIS in log phase and IHF in stationary phase. Both proteins work as anti-activators of CRP during the transition from log-phase to stationary-phase growth (10,11). Briefly, the aim is to then group sites that work together in a ‘regulatory phrase’, or module. This integration of many sites into a reduced number of phrases will contribute to the understanding of complex regulation. Thus, phrases working independently that affect the σ70 family of promoters should have at least one proximal site, where the position of a proximal site guarantees direct interaction with the RNA polymerase (12–14).
It has been known for years that the possible arrangements of sites and their functioning can vary for each TF, or each TF family. In addition to showing this higher organization within individual promoters, we also generated a new page within RegulonDB that groups all possible arrangements described in the genome for each TF, and even for complex phrases with sites of different TFs, that support coordinated regulation of multiple TFs working together to affect transcription initiation (See Figure 3). For instance, the [CRP +] phrase offers the list of all precise positions found in E. coli, with either one or several sites used by CRP to activate transcription (15,16). It will then be easier to see that the CRP pair of sites activating acsp2 occurs also at similar positions in fixAp, which is subject to CaiF and FNR activation, or that the proximal −69.5 CRP activating position also occurs at the csiDp, gntKp and prpRp promoters in the context of regulation by other TFs. This first version of regulatory phrases was based on the identification of proximal sites first and then on detailed curation of cases of multiple TFs known to work jointly [e.g. CytR with CRP; or MelR with CRP (17)], as well as on an exhaustive identification of regulatory phrases with no proximal site, mostly from TFs known to bend the DNA and function as architectural elements [e.g. IHF, Fis and other proteins (18,19)].
Figure 3.
The [CRP,+] regulatory phrase. The graph shows sites of the [CRP,+] phrase for five promoters, and the table includes all additional sites that regulate these promoters. Each promoter name is a link to the page in RegulonDB presenting all phrases for that promoter. Proximal sites are those within the interval from −93 to +20, from which the TF can directly interact with RNA polymerase. All other sites are considered remote, either upstream or downstream.
THE CHALLENGE OF ENCODING KNOWLEDGE GENERATED BY NOVEL ‘OMIC’ TECHNOLOGIES
As HT methodologies have more frequently become a source of information regarding gene regulation, we have had to address several conceptual and practical issues for their easier inclusion in RegulonDB. We have expanded our classification scheme for the various degrees of confidence in these different methodologies. In addition, we have analysed how independent the different methods are (i.e. their different potential sources of false-positives); from this information, we are able to then propose which methods upgrade the quality of evidence to ‘strong’ for objects with two types of weak evidence, and to ‘confirmed’ evidence for objects with two independent strong types of evidence.
We implemented tracks that facilitate the display of HT data, and we have also implemented formats for investigators to submit their HT data sets. Furthermore, we report the results of our RNA sequencing (RNA-Seq)-based identification of transcription start sites (TSSs), which have increased considerably the collection of TSSs for the E. coli genome.
Enriched classifications based on classic and HT evidence
Since the release of version 6.0 of RegulonDB, we have classified evidence associated with the objects annotated in RegulonDB as strong or weak, depending on the confidence level of the associated experimental or computational methodologies. This two-tier rating system quickly distinguishes reliable from less reliable knowledge, contributing to better comparisons, interpretations and selection of gold standards.
However, this classification was not defined for other sources of knowledge beyond classic methodologies; in addition, the different types of evidence do not add up. We had not previously addressed the analyses from different sources of knowledge that, if independent, should increase the degree of confidence for a given piece of knowledge, object or interaction.
To facilitate adding evidence from HT methodologies without losing track of the highly reliable manually curated knowledge supporting RegulonDB, we had to expand our classification to the rapidly growing number of HT methodologies used for the identification of TFBSs, TSSs and TUs (20). These new technologies have generated a flood of new data, as they have allowed analysis of putative targets in parallel, but they are also associated with a high risk of false-positives due to new sources of stochastic effects, ‘batch’ errors and experimental artifacts (21–23). Therefore, the majority of HT methods, for instance, RNA-Seq and ChIP-Seq, generate evidence classified as weak within RegulonDB. Strong evidence requires efficient measures to exclude false-positives as well as the reliability of the evidence based on biologically congruent replicates. The results of the detailed analyses of the different HT methodologies are reflected in the expanded evidence classifications shown in Table 1 of the new Evidence page in RegulonDB web site.
The global character of HT approaches makes it natural to compare their results with equally global computational predictions. However, the analysis of HT data sets involves bioinformatics and biostatistics processing, which, given the diversity of strategies, may limit their comparison until more standardized procedures have been established. A final outcome when these issues are addressed will be the combination not only of the different experiments and HT data sets, but also of all sources of knowledge, computational and evolutionary predictions, classic methodologies and HT strategies, to keep track of each contribution and to assign an appropriate level of confidence to each object and interaction.
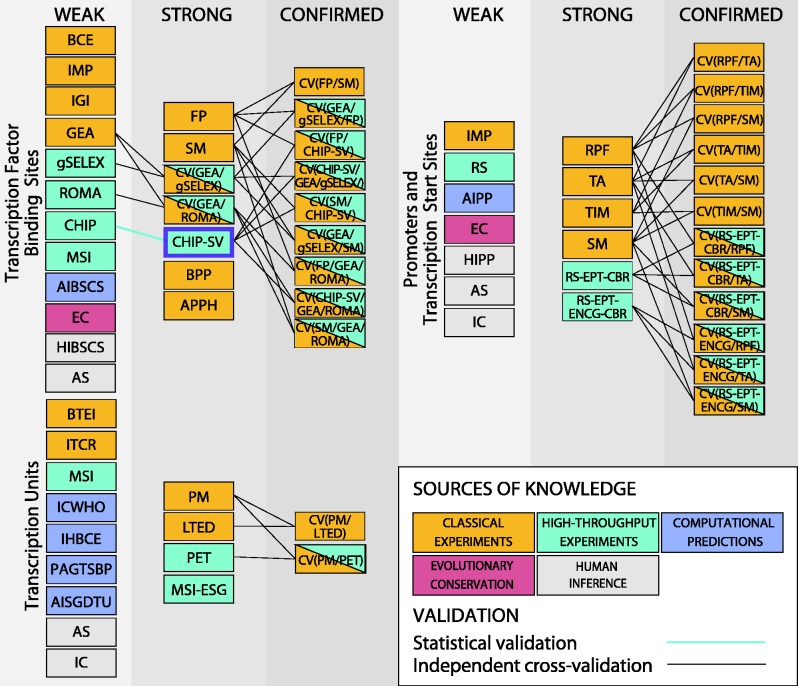
In an initial step in this direction, independent cross-validation has been applied for promoters and regulatory interactions. This new concept integrates multiple types of evidence with the intention of mutually excluding false-positive results. The classification of ‘strong evidence’ is assigned to data that are supported by at least two independent weak types of evidence, provided that the two sources of knowledge do not share major sources of false-positives and do not use common raw materials or common experimental steps. For instance, TSSs that have been identified by transcription initiation mapping can be cross-validated with in vitro transcription assays. Similarly, TFBSs that have been identified by genomic SELEX can be cross-validated by in vivo gene expression data. Moreover, by applying this new concept to data that are supported by strong evidence, we can extend our two-tier rating system to three tiers. To this end, we have introduced a third confidence score, ‘confirmed’. Data supported by confirmed evidence, that is, by at least two types of independent strong evidence, have a high reliability and can be considered gold standard data in RegulonDB. For instance, TFBSs that have been identified by footprinting analysis and, in addition, have been validated by mutational analysis of the binding site, are now classified as data with confirmed evidence. The detailed analysis of this improvement will appear in a publication elsewhere (20). The results of this cross-validation are summarized in Table 2 of the Evidence page in RegulonDB web site (See Figure 4).
Figure 4.
Schematic drawing of the classification of evidence in RegulonDB. Evidence codes for classical experiments: BCE, binding of cellular extracts; IMP, inferred by mutant phenotype; IGI, inferred by genetic interaction; GEA, gene expression analysis; FP, footprinting; SM, site mutation; BPP, binding of purified protein; APPH, assay with protein purified to homogeneity; RPF, RNA-polymerase footprinting; TA, in vitro transcription assays; TIM, transcription initiation mapping; PM, effect of polar mutation on neighboring genes; LTED, length of transcript experimentally determined; BTEI, identification of the boundaries of a transcript, ITCR, inferred through co-regulation; Evidence codes for HT experiments; gSELEX, genomic SELEX; ROMA, run-off transcription microarray analysis; ChIP, chromatin immunoprecipitation; GEA, microarray or RNA-seq gene expression analysis; IMP, inferred from mutant phenotype; RS, RNA-seq; RS-EPT-CBR, RNA-seq with at least two different enrichment strategies for primary transcripts and consistent biological replicates; RS-EPT-ENCG-CBR, RNA-seq with at least two different enrichment strategies for primary transcripts and evidence for a ncRNA, consistent biological replicates; MSI, mapping of signal intensities by microarray analysis or RNA-seq; PET, paired-end di-tagging; MSI-ESG, mapping of signal intensities and evidence for a single gene; Computational approaches: AIBSCS, automated inference based on similarity to consensus sequences; AIPP, automated inference of promoter position; ICWHO, inferred computationally without human oversight; IHBCE, inferred by a human based on computational evidence; PAGTSBP, products of adjacent genes in the same biological process; AISGDTU, automated inference that a single-gene direction is a TU; EC, evolutionary conservation; Human inference: HIBSCS, human inference based on similarity to consensus sequences; AS, author statements; HIPP, human inference on promoter position; IC, inferred by curator. CHIP-SV describes statistical validation of ChIP data sets. CV(A/B) describes independent cross-validation of evidence A and B.
We evaluated the confidence levels of HT and classic methodologies through a more detailed curation process, which included independent cross-validation and/or statistical validation. Statistical validation was used to evaluate the confidence for TFBSs discovered by ChIP technology, by using a strategy that was consistent with the evaluation of PWMs from manually curated binding sites, as described previously. To this end, we are implementing a pipeline to assess the quality of the ChIP-Seq/chip experimental data. We initiated analysing PurR-binding sites, which were identified by ChIP-chip (24) (see the Supplementary Data). The strategy was divided into three main evaluation steps: (i) assessing the enrichment of TFBSs with high scores for the aimed TF in the set of ChIP-identified regions based on matrix quality (3) (see Supplementary Figure S2 in the Supplementary Data). (ii) Discovery of overrepresented motifs in the set of ChIP-identified regions, as well as detection of secondary motifs that could be related to cofactors that bind the targeted TF from the ChIP experiment. We have used peak motifs (25) to rediscover the PWMs for TFs by comparing the discovered motifs with those annotated in RegulonDB (see Supplementary Figure S3 in the Supplementary Data). (iii) If any result from these two steps reveals an uncommon behavior, the set of ChIP-identified regions is not annotated in the RegulonDB core, and rather only as an independent track in the genome browser. If the set of ChIP-identified regions satisfies both evaluations, the exact binding sites are identified with the annotated matrix in RegulonDB by using the program matrix scan (26). The sites are then analysed by a curator to classify quality as high or low, depending on stringent threshold parameters and the context where the sites appear. Statistical validation of the PurR-binding sites confirmed 13 binding sites that had been previously known and annotated as having strong evidence within RegulonDB; one site was upgraded from weak to strong evidence, and three new sites identified in the ChIP analysis were validated as having strong evidence.
We offer the results of this example, step by step, using a bioinformatics pipeline with tools publicly available for those experimentalists interested in using it. See Supplementary Tables S2, S3 and S4 in section III of the Supplementary Data. Currently, we are applying this approach to other recently published ChIP data to further improve the evaluation process using this pipeline. Our intention is to provide a standardized analysis platform to enable consistent comparisons across multiple experiments from different laboratories. Alternatively, provided that the raw sequences are available, we could perform such an analysis ourselves.
TSSs and promoter mapping by using RNA-Seq
Due to the high sensitivity of next-generation sequencing technologies and the highly dynamic nature of the bacterial transcriptome, several thousands of 5′ RNA ends will be detected in any given RNA-Seq experiment. The great majority of them, however, correspond to processed or degraded products. Therefore, in an effort to enrich for primary unprocessed transcripts, different methods have been attempted. Mainly, the TEX exoribonuclease enzyme, which has a preference for 5′-monophosphate (5′-MP) ends (27), and ligation of synthetic RNA adapters to the whole RNA pool for 5′-MP elimination (1) have been used. However, these methods still leave a great number of processed products, as indicated by the presence of a large fraction of rRNA and tRNA sequences in the supposedly 5′-triphosphate-enriched libraries [(28) and unpublished results]. Therefore, to achieve more reliable TSS mapping, a combination of these two methods was chosen, and only the 5′ RNA ends consistently detected in several independent experiments are reported here as highly likely TSSs, consistent with the evidence classification for HT data sets discussed in the previous section.
We prepared six Illumina sequencing libraries, each one from a culture grown to mid-log phase in minimal medium with glucose. After standard rRNA removal, the remaining RNA from each culture was either directly used for library preparation (MT libraries) and/or treated to generate at least one of the following library types: 5′-MP only (M), triphosphate adapter (TA) or triphosphate exonuclease (TE), as reported before (1). Three of our library sets contained all of these library types, and other three do not have TE. Additionally, we generated two MT libraries from cultures grown in LB medium and MM with acetate as carbon source. The resulting libraries allowed us to test the consistency for TSS detection despite experimental noise and the imposed technical and growth condition variations.
A total of 77 628 858 non-rRNA Illumina sequences from the sum of all libraries were mapped to 821 789 positions in the E. coli genome. Among this position set, we found 67% and 86% of the 1418 TSSs reported in RegulonDB (with classic methodologies) with an exact position coincidence and within three nucleotides, respectively. It is important to remember that the RegulonDB set has promoters that have been identified under a large variety of conditions. As anticipated, positions conserved in an increasing number of libraries tend to be located at the upstream regions of genes (which represent about 20% of the genome sequence), as 15% of the positions present in a single library map to upstream regions, but this number increases to 71% for the positions present in 22 libraries.
A total of 5197 positions were consistently observed in at least half of the MT, TA and TE libraries. As these libraries were enriched for 5′ triphosphorylated RNA, the selected positions are considered highly reliable TSSs. Some of these positions were also detected in M libraries, probably representing dephosphorylated 5′ mRNA ends (mRNA degradation intermediates). Of the 5197 positions, 53% mapped in upstream regions and 551 mapped within ±3 nucleotides of one of the TSSs reported in RegulonDB. That is, 99.37% reduction of the original positions maintained 45% of the TSSs reported in RegulonDB detected in the complete data set. As expected for bona fide TSSs, only a few, 12, positions were present in convergent gene regions, and some of them were regions large enough to contain sRNAs. It is remarkable that transcripts in the antisense orientation, which have been reported to be highly abundant in bacteria (1,29–31), dramatically decreased as the number of experiments increased. Of the 5197 highly conserved positions, only 80 (1.5%) were located in the antisense orientation. These results strongly suggest that a large fraction of the antisense transcripts detected in RNA-Seq experiments are artifacts of the methodologies or are not consistently expressed in the cells, as recently suggested by Ochman and coworkers (32).
In conclusion, the highly conserved positions in our combined libraries detected 5197 putative TSSs, 53% of them located up to 150 bp upstream of genes, 0.2% in convergent regions, 1.5% in the antisense orientation and 44% within the coding region. Of the latter, it is unknown how many of them could be TSSs for genes located further downstream than our arbitrary 150-bp threshold. All these positions are included in RegulonDB with their predicted promoters annotated, and they are also available as a data set for track display. A detailed data analysis will be published elsewhere.
Tracks display of HT data sets and submission forms for HT data sets
Initially motivated by the need to display data sets from HT experiments, we implemented a new tool in the main menu for use of a browser with the option of several tracks, based on GBrowser v.248 (33,34). In an initial step in this direction, independent cross-validation has been applied for promoters and regulatory interactions. We have also included a mechanism that enables the display of the variety of ‘Data Sets’ in GBrowser. On the GBrowser page, a user can proceed to ‘Select tracks’ to see the full set of options currently available, classified by type of object, including operons, regulators (TFs, and sRNAs), TFBSs,(ChIP-Seq and RegulonDB data sets), HT-mapped TSSs and RegulonDB promoter data sets, manually curated as well as computational predictions, among others. An additional category called ‘Genome regions’, for genes as well untranslated regions of 5′ and 3′ends of TUs, is also included.
Every single data set can be documented as requested when authors submit their experimental data, with specific formats for each type of source (i.e. TSS, ChIP-Seq). The display of some icons has been adapted to those we use in RegulonDB. A web form is available for those interested in submitting their data sets directly online. After careful analysis and curation (see the PurR example in Supplementary Data), those individual objects with strong evidence will be added individually to RegulonDB. Additionally, the full data set will be available as such. Data sets with weak evidence will be available for display through tracks but will not be incorporated as individual objects into RegulonDB.
Evolutionary conservation of promoters and regulatory interactions
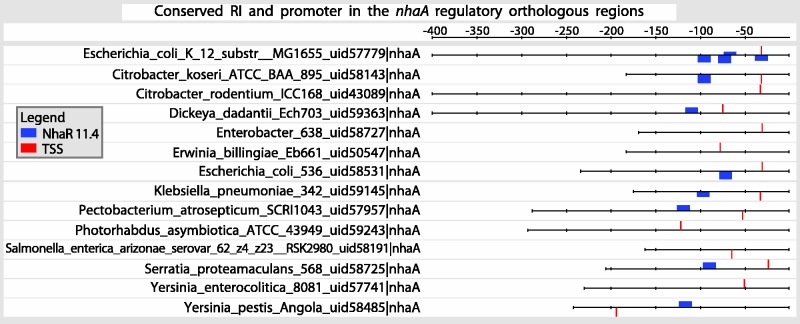
Given the availability of completed genomes, it makes sense to estimate and add the evolutionary conservation of regulatory elements as an additional relevant source of knowledge for the regulatory network. For the first time, we have added the evolutionary evidence for promoters and TFBSs in RegulonDB, and we will add information on conservation of operon organization. We have assessed the evolutionary evidence to conservation within gammaproteobacteria because enterobacteria being evolutionary closer show a higher fraction of redundant upstream regions. Our results are available from the gene and regulon pages, with graphics showing a summary of the number of genomes where conservation is found and the alignment and conserved sequences available as multiple alignments. See a subset of nhaA orthologous upstream regions and conservation of promoters and NhaR sites in Figure 5).
Figure 5.
Evolutionary conservation of regulatory interactions and promoters. The figure shows the conservation of both promoters and regulatory interactions in a subset of orthologous regulatory regions corresponding to nhaA. The complete set can be directly searched in RegulonDB. Orthologs were selected by using a bidirectional best hit method (35), and sequences are masked to eliminate sequence redundancy to assess an unbiased conservation score. Nonetheless, the maps like the one shown here show all the sites found in all orthologous regions including masked sequences. Blue boxes correspond to the detected binding sites for the NhaR TF; the width of the boxes correspond to the scores of the sites, with the highest obtained score indicated in the symbol key (11.4). Red vertical lines indicate the predicted position for the TSSs based on identification of the promoter −10 and −35 boxes. The zero coordinate is that of the beginning of the gene.
Currently, there are 375 sequenced gammaproteobacterial genomes, from which 160 are enterobacteria and 30 are part of the Escherichia subclassification. Due to the close evolutionary distance of these genomes, we decided to mask redundant sequences longer than 30 bp with two mismatches, to avoid overestimating conservation in sequences of orthologous promoters for one gene. On average, 32% of a set of orthologous upstream regions per gene contributed to the assessment of conservation.
We added and updated the conservation for all σ70 promoters in RegulonDB, based on the strategy reported in reference (36), in which we analysed the conservation of clusters of overlapping σ70 putative promoters across enterobacterial genomes. We have shown that 74% of the functional promoters are embedded in clusters of ∼80 pb containing 4.82 signals on average (36,37).
RegulonDB version 8.0 has 811 σ70 promoters that were identified from manual curation and have been classified by evidence type: 630 with strong evidence and 181 with weak evidence. Of these, 678 promoters (523 with strong evidence and 155 with weak evidence) were found to be conserved in at least one orthologous gene, with an average of conservation observed in 18% of orthologs. Thus, 83% of σ70 promoters showed evolutionary conservation of the promoter sequence (P < 0.0001) and/or of its position relative to the start of the orthologous gene.
We found no correlation between the percentage of conservation and the type of evidence; instead, we found a strong correlation between the score of the sequences recognized by the σ70 factor and the degree of evolutionary conservation. Promoters with sequences more similar to the consensus sequence of σ70 are more conserved: 67% versus 7.5% conservation is observed for high-similarity versus low-similarity promoters.
For the sake of determining conservation of TFBSs, we only considered the regulatory regions of orthologous target genes (if there was an ortholog for the TF gene in the same organism). We determined the conservation of the TF–target gene regulatory interaction if there was a higher number of TFBSs than expected by chance in the set of orthologous promoters for the target gene of the TF, based on the regulog assumption of an interaction: that is, that the TF and target orthologs are present and there is a TFBS upstream of the regulated gene (38). Overall, we observed that 41% of the regulatory interactions showed conservation within gammaproteobacteria and 38% in enterobacterial genomes. Interestingly, we observed higher conservation of regulatory interactions supported by strong evidence (27%), compared with 10% for interactions with weak evidence.
A new regulon page: addressing user needs and suggestions
Based on comments and suggestions offered by RegulonDB users, we decided to modify the page displaying information about regulons and simplified the search for all TFBSs of a single TF.
In close collaboration with interested users, we redesigned the page that displays the information on regulons, through the participation of our team and a web design expert, and we generated a new interface that is more user-friendly and better integrated.
The new page for regulons includes an icon linking a regulon to the GU when its GU has been curated, the detailed summary text prepared by curators for the TF, followed by a section displaying the functional and non-functional conformation(s), a classification of the signal based on its source as internal, external or dual (39); a category for the TF based on its connectivity, the target regulated genes and the operon where the TF gene belongs. Subsequent sections describe functional properties of the regulon, the set of TFBSs and their organization patterns and phrases, logos, PWMs and additional properties.
Users’ requests sent to regulondb@ccg.unam.mx are answered immediately. We implemented a ‘Contact Us’ form under ‘About RegulonDB’ and at the bottom of every page in the RegulonDB portal, which provides a more user-friendly means of submission of questions or comments.
CONCLUSIONS AND PERSPECTIVES
We are aware that RegulonDB is not the sole source for information on regulation in E. coli, as we share our manual curation of transcriptional regulation with EcoCyc, in addition to several other existing resources for E. coli with which users can search for knowledge beyond transcriptional regulation [e.g. PortEco (http://porteco.org/); M3D (http://m3d.bu.edu/cgi-bin/web/array/index.pl?section=home); COLOMBOS (http://bioi.biw.kuleuven.be/colombos/), among several others].
There is a large number of bioinformatics resources with information on gene regulation, gene expression and related knowledge. A compendium of ∼100 selected resources of >240 was made available since 2009 (40). They are classified in nine major categories (e.g. gene expression; TFs/gene regulation; RNA, etc) with their link and short description. See the ‘Additional resources’ link in the main page of RegulonDB for more details.
Some of the unique guidelines of the encoding of knowledge in RegulonDB regarding gene regulation are our focus on high-level curation, currently illustrated by GUs and by the organization of binding sites into regulatory phrases, the search for clearly defining gold standards based on enabling the combination of independent sources of evidence into higher levels of confidence and the addition of evolutionary conservation as another source of knowledge on gene regulation.
Furthermore, significant progress in these past 2 years is summarized as follows. We have significantly increased the alternative functional and non-functional conformations, documented now for 103 TFs, a data set that has provoked a discussion of the demand theory of gene regulation within a genomic perspective (to be published elsewhere). The sustained effort of detailed curation of relevant properties of TFBSs for 130 TFs has significantly enhanced the precision of our encoding of the anchoring of mechanisms in the genome, improving the PWMs and their predictions. Our next step, already initiated, is the grouping of binding sites into phrases.
Addressing the challenge of omics technologies and the assessment of the confidence levels for their results have been crucial in this field. We have proposed criteria for the classification of the degree of confidence that may be useful for any bacterial study. We have illustrated the use of a bioinformatics pipeline with tools publicly available that can provide a standardized analysis platform to enable consistent comparisons across multiple experiments from different laboratories. We are making available the results of the highly reproducible HT whole-genome mapping of ∼5000 TSSs from the group of Enrique Morett. In addition to this data set, their results show that inclusion of sufficient independent experiments, together with the use of more than one enrichment method for primary transcripts, is essential to increase the confidence in reliably detecting TSSs, as indicated by the enrichment of upstream positions and the high proportion of previously reported TSSs also detected and included in RegulonDB.
All these efforts, together with the distinctive availability and display of HT data sets, our combined bioinformatics and manual sustained curation, the inclusion of evolutionary conservation and the structuring of computable and high-level encoding, makes RegulonDB a well-designed home for integrating up-to-date knowledge on gene regulation from all, or most, of the relevant sources of knowledge currently available.
Escherichia coli K-12 will certainly keep our group and many other research groups busy for a while!
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–3 and Supplementary References [41–46].
FUNDING
National Institute of General Medical Sciences of the National Institutes of Health [GM071962 and GM077678]; Consejo Nacional de Ciencia y Tecnología (CONACyT) [103686 and 179997]; Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT-UNAM) [IN210810 and IN209312]. Funding for open access charge: National Institute of General Medical Sciences of the National Institutes of Health [GM071962].
Conflict of interest statement. None declared.
ACKNOWLEDGMENTS
The authors acknowledge Jacques van Helden for his participation in the design of the new regulon page; Ingrid Keseler for periodically sending us selected literature references for curation; Ruth Martínez-Adame for her help in functionality testing; Ricardo Grande for Illumina’s libraries preparation and sequencing; Romualdo Zayas for technical support and Altamira Studio for their contributions to web design issues.
REFERENCES
- 1.Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muniz-Rascado L, Solano-Lira H, Jimenez-Jacinto V, Weiss V, Garcia-Sotelo JS, Lopez-Fuentes A, et al. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39:D98–D105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muniz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medina-Rivera A, Abreu-Goodger C, Thomas-Chollier M, Salgado H, Collado-Vides J, van Helden J. Theoretical and empirical quality assessment of transcription factor-binding motifs. Nucleic Acids Res. 2011;39:808–824. doi: 10.1093/nar/gkq710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 5.Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- 6.Vassylyeva MN, Perederina AA, Svetlov V, Yokoyama S, Artsimovitch I, Vassylyev DG. Cloning, expression, purification, crystallization and initial crystallographic analysis of transcription factor DksA from Escherichia coli. Acta. Crystallogr. D Biol. Crystallogr. 2004;60:1611–1613. doi: 10.1107/S0907444904015033. [DOI] [PubMed] [Google Scholar]
- 7.Mallik P, Paul BJ, Rutherford ST, Gourse RL, Osuna R. DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli. J. Bacteriol. 2006;188:5775–5782. doi: 10.1128/JB.00276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyzen R, Kochanowska M, Wegrzyn G, Szalewska-Palasz A. Transcription from bacteriophage lambda pR promoter is regulated independently and antagonistically by DksA and ppGpp. Nucleic Acids Res. 2009;37:6655–6664. doi: 10.1093/nar/gkp676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potrykus K, Vinella D, Murphy H, Szalewska-Palasz A, D'Ari R, Cashel M. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J. Biol. Chem. 2006;281:15238–15248. doi: 10.1074/jbc.M601531200. [DOI] [PubMed] [Google Scholar]
- 10.Browning DF, Beatty CM, Sanstad EA, Gunn KE, Busby SJ, Wolfe AJ. Modulation of CRP-dependent transcription at the Escherichia coli acsP2 promoter by nucleoprotein complexes: anti-activation by the nucleoid proteins FIS and IHF. Mol. Microbiol. 2004;51:241–254. doi: 10.1046/j.1365-2958.2003.03824.x. [DOI] [PubMed] [Google Scholar]
- 11.Beatty CM, Browning DF, Busby SJ, Wolfe AJ. Cyclic AMP receptor protein-dependent activation of the Escherichia coli acsP2 promoter by a synergistic class III mechanism. J. Bacteriol. 2003;185:5148–5157. doi: 10.1128/JB.185.17.5148-5157.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collado-Vides J, Magasanik B, Gralla JD. Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collado-Vides J. Towards a unified grammatical model of sigma 70 and sigma 54 bacterial promoters. Biochimie. 1996;78:351–363. doi: 10.1016/0300-9084(96)84767-5. [DOI] [PubMed] [Google Scholar]
- 14.Ushida C, Aiba H. Helical phase dependent action of CRP: effect of the distance between the CRP site and the -35 region on promoter activity. Nucleic Acids Res. 1990;18:6325–6330. doi: 10.1093/nar/18.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belyaeva TA, Rhodius VA, Webster CL, Busby SJ. Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: organisation of the RNA polymerase alpha subunits. J. Mol. Biol. 1998;277:789–804. doi: 10.1006/jmbi.1998.1666. [DOI] [PubMed] [Google Scholar]
- 16.Murakami K, Owens JT, Belyaeva TA, Meares CF, Busby SJ, Ishihama A. Positioning of two alpha subunit carboxy-terminal domains of RNA polymerase at promoters by two transcription factors. Proc. Natl Acad. Sci. USA. 1997;94:11274–11278. doi: 10.1073/pnas.94.21.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belyaeva TA, Wade JT, Webster CL, Howard VJ, Thomas MS, Hyde EI, Busby SJ. Transcription activation at the Escherichia coli melAB promoter: the role of MelR and the cyclic AMP receptor protein. Mol. Microbiol. 2000;36:211–222. doi: 10.1046/j.1365-2958.2000.01849.x. [DOI] [PubMed] [Google Scholar]
- 18.Browning DF, Grainger DC, Busby SJ. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 2010;13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Rimsky S, Travers A. Pervasive regulation of nucleoid structure and function by nucleoid-associated proteins. Curr. Opin. Microbiol. 2011;14:136–141. doi: 10.1016/j.mib.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Weiss V, Medina-Rivera A, Huerta AM, Santos-Zavaleta A, Salgado H, Morett E, Collado-Vides J. Evidence Classification of High-Throughput Protocols and Confidence Integration in RegulonDB. Database. 2013 doi: 10.1093/database/bas059. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho BK, Federowicz SA, Embree M, Park YS, Kim D, Palsson BO. The PurR regulon in Escherichia coli K-12 MG1655. Nucleic Acids Res. 2011;39:6456–6464. doi: 10.1093/nar/gkr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas-Chollier M, Herrmann C, Defrance M, Sand O, Thieffry D, van Helden J. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res. 2012;40:e31. doi: 10.1093/nar/gkr1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, van Helden J. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39:W86–W91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 28.Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl Acad. Sci. USA. 2012;109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, Taboada B, Jimenez-Jacinto V, Salgado H, Juarez K, Contreras-Moreira B, et al. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in. E. coli. PLoS One. 2009;4:e7526. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu. Rev. Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghavan R, Sloan DB, Ochman H. Antisense transcription is pervasive but rarely conserved in enteric bacteria. MBio. 2012;3:e00156–12. doi: 10.1128/mBio.00156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donlin MJ. Using the generic genome browser (GBrowse) Curr. Protoc. Bioinformatics. 2009 doi: 10.1002/0471250953.bi0909s28. Chapter 9, Unit 9.9. [DOI] [PubMed] [Google Scholar]
- 35.Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- 36.Huerta AM, Collado-Vides J, Francino MP. Proceedings of the SMBE Tri-National Young Investigators' Workshop 2005. Positional conservation of clusters of overlapping promoter-like sequences in enterobacterial genomes. Mol. Biol. Evol. 2006;23:997–1010. doi: 10.1093/molbev/msk004. [DOI] [PubMed] [Google Scholar]
- 37.Huerta AM, Collado-Vides J. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 2003;333: 261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 38.Alkema WB, Lenhard B, Wasserman WW. Regulog analysis: detection of conserved regulatory networks across bacteria: application to Staphylococcus aureus. Genome Res. 2004;14:1362–1373. doi: 10.1101/gr.2242604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Antonio A, Janga SC, Salgado H, Collado-Vides J. Internal-sensing machinery directs the activity of the regulatory network in Escherichia coli. Trends Microbiol. 2006;14:22–27. doi: 10.1016/j.tim.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Collado-Vides J, Salgado H, Morett E, Gama-Castro S, Jimenez-Jacinto V, Martinez-Flores I, Medina-Rivera A, Muniz-Rascado L, Peralta-Gil M, Santos-Zavaleta A. Bioinformatics resources for the study of gene regulation in bacteria. J. Bacteriol. 2009;191:23–31. doi: 10.1128/JB.01017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turatsinze JV, Thomas-Chollier M, Defrance M, van Helden J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat. Protoc. 2008;3:1578–1588. doi: 10.1038/nprot.2008.97. [DOI] [PubMed] [Google Scholar]
- 42.Nygaard P, Smith JM. Evidence for a novel glycinamide ribonucleotide transformylase in Escherichia coli. J. Bacteriol. 1993;175:3591–3597. doi: 10.1128/jb.175.11.3591-3597.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danielsen S, Kilstrup M, Barilla K, Jochimsen B, Neuhard J. Characterization of the Escherichia coli codBA operon encoding cytosine permease and cytosine deaminase. Mol. Microbiol. 1992;6:1335–1344. doi: 10.1111/j.1365-2958.1992.tb00854.x. [DOI] [PubMed] [Google Scholar]
- 44.Karatza P, Frillingos S. Cloning and functional characterization of two bacterial members of the NAT/NCS2 family in Escherichia coli. Mol. Membr. Biol. 2005;22:251–261. doi: 10.1080/09687860500092927. [DOI] [PubMed] [Google Scholar]
- 45.Maier C, Bremer E, Schmid A, Benz R. Pore-forming activity of the Tsx protein from the outer membrane of Escherichia coli. Demonstration of a nucleoside-specific binding site. J. Biol. Chem. 1988;263:2493–2499. [PubMed] [Google Scholar]
- 46.Qi F, Turnbough CL., Jr Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J. Mol. Biol. 1995;254:552–565. doi: 10.1006/jmbi.1995.0638. [DOI] [PubMed] [Google Scholar]