Abstract
The new release of SchistoDB (http://SchistoDB.net) provides a rich resource of genomic data for key blood flukes (genus Schistosoma) which cause disease in hundreds of millions of people worldwide. SchistoDB integrates whole-genome sequence and annotation of three species of the genus and provides enhanced bioinformatics analyses and data-mining tools. A simple, yet comprehensive web interface provided through the Strategies Web Development Kit is available for the mining and visualization of the data. Genomic scale data can be queried based on BLAST searches, annotation keywords and gene ID searches, gene ontology terms, sequence motifs, protein characteristics and phylogenetic relationships. Search strategies can be saved within a user’s profile for future retrieval and may also be shared with other researchers using a unique web address.
INTRODUCTION
In the last few years, major advances have been made in the sequencing of genomes of the blood flukes of the genus Schistosoma which causes chronic diseases in ∼200 million people globally (1). Thus far, the focus has been on Schistosoma haematobium, Schistosoma japonicum and Schistosoma mansoni. Recently, S. haematobium had its genome sequenced (2) and published as well as a new and improved assembly of the first-sequenced schistosome genome, S. mansoni, was made available (3). The initial version of SchistoDB (http://SchistoDB.net) (4) was implemented to provide easy access and visualization of the S. mansoni genome and features (5), integrated to other data types such as expressed sequence tags (ESTs), proteins and metabolic pathways. In order to interrogate datasets from multiple genomes in a comprehensive manner, a new version of SchistoDB was developed as a resource for genomic data across the genus Schistosoma.
This was done in partnership with the NIAID-funded Eukaryotic Pathogen Bioinformatics Resource Center (http://EuPathDB.org) (6). This enhanced database uses the same database structural framework and uses the graphical Strategies Web Development Kit (WDK) search interface (7). It also provides a data-mining interface for the comparative and functional genomic data of three species of schistsomes and an integrated query system as part of the WDK and Genomics Unified Schema database structure. SchistoDB differs from other resources such as the Wellcome Trust Sanger Institute GeneDB (http://GeneDB.org) which has complementary data query or visualization tools but do not have the data-mining capabilities and broad cross-species comparisons which are possible using the WDK search interface. Data are currently obtained directly from providers at sequencing centers, GenBank and associated functional data repositories.
CONTENTS OF THE CURRENT RELEASE
The current release of SchistoDB contains the latest release of the genome sequence and annotation from S. haematobium (2), S. japonicum (8) and S. mansoni (3) provided by the Faculty of Veterinary Science at the University of Melbourne, the Chinese National Human Genome Center and the Wellcome Trust Sanger Institute, respectively. More information about these genomes is available in Table 1.
Table 1.
Species in the current release of SchistoDB
Genomes and annotation in SchistoDB are processed through the same analysis pipeline, which provides additional data, including InterPro domains (9), gene ontology term association (10), signal peptide predictions (11), transmembrane domain predictions, open reading frame predictions, BLAST against the non-redundant genome database at the National Center for Bioinformatics, orthology prediction based on OrthoMCL (12) and synteny prediction. Pipeline details are available at http://schistodb.net/schisto/showXmlDataContent.do?name=XmlQuestions.Methods.
HOW TO USE SCHISTODB
The home page
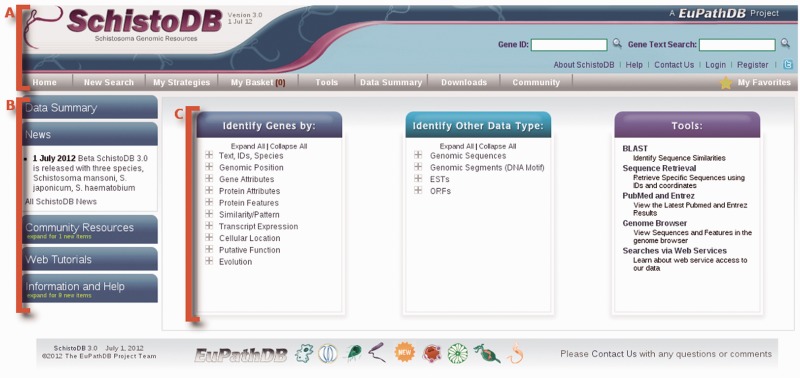
The SchistoDB home page is virtually the same as all EuPathDB pages, with differences only in color, logo and data content. A visitor to these sites will first notice the home page layout, which has been designed to provide the user with convenient and immediate access to data and tools. The home page is divided into three main sections (Figure 1). (i) A top banner section, providing quick access back to the home page, gene ID and text searches, (ii) ‘contact’ and login/registration links and (iii) mouse over menus (Figure 1A). The information and help menus on the left (Figure 1B). The central section provides links to all searches (Figure 1C), and links to tools, such as BLAST (13), the sequence retrieval tool and the Generic Model Organism Database genome browser (14). Creating an account and logging in allows search strategies to be saved and shared, and gene associated annotation comments to be created which are linked to the author.
Figure 1.
Screen shot of the SchistoDB homepage. (A) The banner section present on all SchistoDB webpages provides links to registration, login, and contact us forms, ID and text searches, information and help, and all available searches and tools. (B) The side panel on the homepage provides expandable tabs that reveal links to a data summary table, community news items, links to community resources, web tutorials and additional information and help links. (C) The central portion of the home page includes all available searches and tools—gene-specific searches, searches against other data types such as genomic sequence, open reading frames and ESTs and tools, such as BLAST, sequence retrieval and the genome browser.
Building a search strategy
The search strategy system allows users to filter the results list based on a combined set of criteria and also add or sort columns. After running the first search (Figure 2), a user might elect to add other filtering steps. This can be achieved by sequentially adding new searches to grow the strategy horizontally. Steps in a strategy may be viewed, revised, renamed and developed further by nesting or deleted. Entire search strategies may be renamed, copied, saved and shared with a unique strategy URL or deleted. An example of a complex multi-step search strategy can be seen in Figure 2. Using this strategy, for example, all secretory genes expressed in eggs are identified. This is achieved by finding all genes with predicted secretory signal peptides and/or transmembrane domains (Steps 1 and 2, Figure 2), and that have egg EST libraries mapping evidence (Step 3, Figure 2). As a final step, a transformation is applied on the results to identify all Schistosoma orthologs of the results in Step 3 (Step 4, Figure 2) since there are only egg EST libraries for S. mansoni. Several options can be applied to a whole strategy including renaming, copying, saving, deleting and sharing. The latter allows users to email colleagues a unique URL of a strategy of interest, which enables the receiver to open and modify the strategy in their own workspace (for example, the strategy in Figure 2 can be accessed here: http://schistodb.net/schisto/im.do?s=ea7002f6e5b2996d).
Figure 2.
Screen shot of a search strategy with the first step being the results of running the search shown in Figure 1C (inset). The ‘Add Step’ button reveals a popup with all available searches in SchistoDB. The results of any search are displayed in a dynamic table that allows removing, adding and moving columns, downloading results and adding results to the basket.
Additional features
There is a range of features that allows users to bookmark their favorite genes for quick future access; add genes to a basket in order to combine such gene set in later search; add arbitrary weights to steps to obtain a ranked list or write comments to genes to improve its annotation. Data in SchistoDB are conveniently available for bulk download from the ‘Data Files’ section accessible from the ‘Downloads’ menu item in the gray tool bar (Figure 1A). Data files are in folders organized by database release version number and species. The sequence retrieval tool, accessible from the tools section (Figure 1C), allows users to specify exact coordinates or lists of genes or proteins to be downloaded.
FUNDING
National Institutes of Health (NIH)—Fogarty International Center [TW007012-03]; The Burroughs Wellcome Fund (BWF); CNPq [573839/2008-5]; FAPEMIG [CBB-1181/08485/2009, REDE-56/11] and EC FP7 [241865]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the genome sequencing centers: the Wellcome Trust Sanger Institute, the Faculty of Veterinary Science at the University of Melbourne and the Chinese National Human Genome Center for providing the genome assembly and annotation. Without their generous pre-publication contribution developing this integrated database resource would not have been possible. Special thanks to the EupathDB team, which provided skills, expertise and the technology to accomplish this work.
REFERENCES
- 1.Rollinson DA. Wake up call for urinary schistosomiasis: reconciling research effort with public health importance. Parasitology. 2009;136:1593–1610. doi: 10.1017/S0031182009990552. [DOI] [PubMed] [Google Scholar]
- 2.Young ND, Jex AR, Li B, Liu S, Yang L, Xiong Z, Li Y, Cantacessi C, Hall RS, Xu X, et al. Whole-genome sequence of Schistosoma haematobium . Nat. Genet. 2012;44:221–225. doi: 10.1038/ng.1065. [DOI] [PubMed] [Google Scholar]
- 3.Protasio AV, Tsai IJ, Babbage A, Nichol S, Hunt M, Aslett MA, De Silva N, Velarde GS, Anderson TJC, Clark RC, et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni . PLoS Negl. Trop. Dis. 2012;6:e1455. doi: 10.1371/journal.pntd.0001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zerlotini A, Heiges M, Wang H, Moraes RLV, Dominitini AJ, Ruiz JC, Kissinger JC, Oliveira G. SchistoDB: a Schistosoma mansoni genome resource. Nucleic Acids Res. 2009;37:D579–D582. doi: 10.1093/nar/gkn681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, et al. The genome of the blood fluke Schistosoma mansoni . Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38:D415–D419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer S, Aurrecoechea C, Brunk BP, Gao X, Harb OS, Kraemer ET, Pennington C, Treatman C, Kissinger JC, Roos DS, et al. The strategies WDK: a graphical search interface and web development kit for functional genomics databases. Database. 2011 doi: 10.1093/database/bar027. 2011, bar027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F, Zhou Y, Wang Z-Q, Lu G, Zheng H, Brindley PJ, McManus DP, Blair D, Zhang Q-hua, Zhong Y, et al. The Schistosoma japonicum genome reveals features of host–parasite interplay. Nature. 2009;460:345–351. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gene Ontology Consortium. The Gene Ontology: enhancements for 2011. Nucleic Acids Res. 2012;40:D559–D564. doi: 10.1093/nar/gkr1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 12.Chen F, Mackey AJ, Stoeckert CJ, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]