Abstract
The naturally occurring mannopeptimycins (formerly AC98-1 through AC98-5) are a novel class of glycopeptide antibiotics that are active against a wide variety of gram-positive bacteria. The structures of the mannopeptimycins suggested that they might act by targeting cell wall biosynthesis, similar to other known glycopeptide antibiotics; but the fact that the mannopeptimycins retain activity against vancomycin-resistant organisms suggested that they might have a unique mode of action. By using a radioactive mannopeptimycin derivative bearing a photoactivation ligand, it was shown that mannopeptimycins interact with the membrane-bound cell wall precursor lipid II [C55-MurNAc-(peptide)-GlcNAc] and that this interaction is different from the binding of other lipid II-binding antibiotics such as vancomycin and mersacidin. The antimicrobial activities of several mannopeptimycin derivatives correlated with their affinities toward lipid II, suggesting that the inhibition of cell wall biosynthesis was primarily through lipid II binding. In addition, it was shown that mannopeptimycins bind to lipoteichoic acid in a rather nonspecific interaction, which might facilitate the accumulation of antibiotic on the bacterial cell surface.
Multiply drug-resistant pathogenic bacteria continue to emerge, and this situation poses a great threat to effective antimicrobial chemotherapy and infection control. Particularly threatening is the emergence of clinical strains of Staphylococcus aureus and Enterococcus spp. that are gradually building resistance to vancomycin, often considered to be the drug of last resort. These resistance issues emphasize the importance of the development of new antibiotics that would be effective for the treatment of bacterial infections by blocking essential pathogen-specific molecular processes. These include both newly discovered processes and those already targeted by antimicrobial chemotherapy. The biosynthesis of the bacterial cell wall is unique to bacteria and thus remains an important target for the screening and development of new drugs.
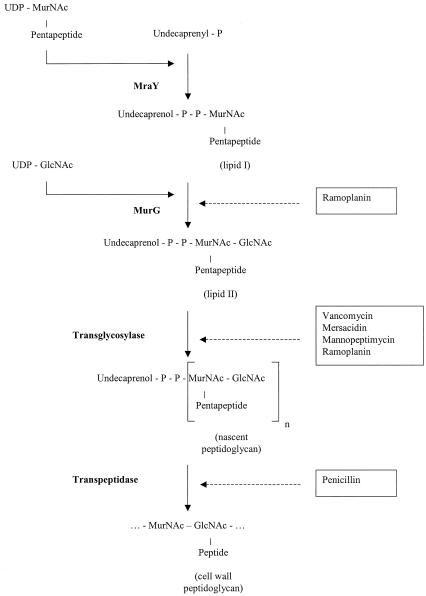
The biosynthesis of peptidoglycan (PG), the major component of the bacterial cell wall, consists of three major stages. This process originates in the cytoplasm by the synthesis of the UDP-linked precursors UDP-N-acetylglucosamine (UDP-GlcNAc) and a species-specific UDP-N-acetylmuramic acid (UDP-MurNAc)-peptide catalyzed by the MurA-MurF enzymes (21, 22, 32, 33, 49). In the second stage, which is catalyzed by the membrane-bound enzymes MraY and MurG, nonnucleotide portions of the two precursors are sequentially transferred to the undecaprenylphosphate carrier, resulting in the formation of two lipid intermediates, lipid I and lipid II (19, 45, 49). The final stage involves the incorporation of the MurNAc-peptide-GlcNAc subunit into the nascent PG chain (transglycosylation) and the subsequent attachment of the nascent PG to the preexisting cell wall by the formation of bonds between peptide chains (transpeptidation) (46, 49, 51, 53-55). In Escherichia coli, both of these reactions are catalyzed by distinct domains of two bifunctional penicillin-binding proteins (PBPs), PBP1A and PBP1B (11, 20, 31). However, in S. aureus the role of individual PBPs in transglycosylation and transpeptidation remains controversial. Although the nucleotide sequence of PBP2 suggests that it has bifunctionality (30), it has been reported that the transglycosylase activity is not associated with the transpeptidase activity in S. aureus (35).
Known inhibitors of bacterial cell wall biosynthesis target various stages of this process. Cycloserine and fosfomycin inhibit the formation of cytoplasmic precursors (23, 25, 44). Bacitracin inhibits the recycling of the undecaprenol carrier and consequently blocks the formation of lipid I (42, 43). Ramoplanin blocks the conversion of lipid I to lipid II (40) and, as proposed recently (27, 28), might sequester lipid II and thereby inhibit transglycosylation as well. Most of the known inhibitors of the transglycosylation reaction, including glycopeptides (vancomycin, teicoplanin) and lantibiotics (nisin, mersacidin), act by binding to lipid II, the substrate of transglycosylase (10, 12, 36). However, moenomycin interacts directly with transglycosylase (50). Penicillin and other β-lactam antibiotics are inhibitors of transpeptidation (7). The later stages of cell wall biosynthesis and the steps inhibited by various antimicrobial agents are shown in Fig. 1.
FIG. 1.
Late stages of membrane-associated PG biosynthesis and the effects of various antibiotics. The target reactions of the antibiotics used in this study are depicted. The catalyzing enzymes are shown in boldface. The antibiotics are boxed. Ramoplanin is shown twice, reflecting recent findings on inhibition of the transglycosylation reaction by this antibiotic (27, 28).
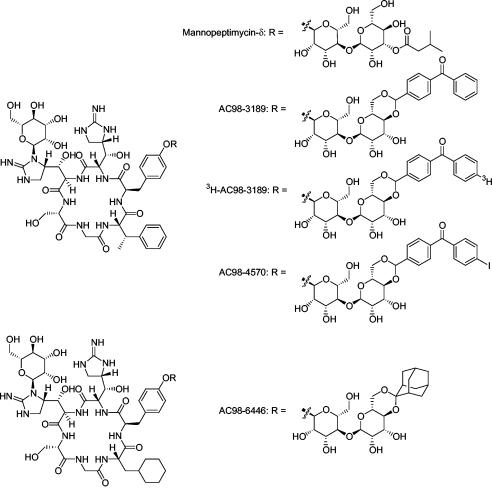
Because of the emergence of resistance to many of the existing antibiotics, a search for new cell wall-specific inhibitors continues. The mannopeptimycins are novel glycopeptide antibiotics that were originally isolated as a complex of five natural products (mannopeptimycins α through ɛ, formerly known as AC98-1 through AC98-5, respectively) from a strain of Streptomyces hygroscopicus, LL-AC98 (18). Subsequently, various semisynthetic modifications that enhanced the antibacterial properties were made; among the most promising derivatives is AC98-6446, an accessible ketal derivative of a modified mannopeptimycin core (Fig. 2) (R. G. Dushin, T. Z. Wang, G. Fortier, S. Iera, M. Paramichelakis, L. Richard, J. Sellstedt, and S. Shah, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. F352, 2002; P. J. Petersen, H. Hartman, T. Wang, R. G. Dushin, and P. A. Bradford, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. F354, 2002; P. J. Petersen, P. Labthavikul, T. Wang, R. G. Dushin, and P. A. Bradford, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. F353, 2002). The mannopeptimycins are active against a wide variety of gram-positive bacteria, including well-characterized antibiotic-resistant pathogens such as vancomycin-resistant enterococci, penicillin-resistant Streptococcus pneumoniae, and glycopeptide-intermediate and methicillin-resistant S. aureus (Petersen et al., 42nd ICAAC, abstr. F354; Petersen et al., 42nd ICAAC, abstr. F353; P. J. Petersen, W. J. Weiss, E. B. Lenoy, H. He, R. T. Testa, and P. A. Bradford, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1148, 2001). This broad spectrum of activity as well as the complete absence of spontaneous resistance upon in vitro selection makes the mannopeptimycins promising candidates as future therapeutic agents. Preliminary mechanism-of-action studies suggested that the mannopeptimycins are similar to other glycopeptide antibiotics in that they target cell wall biosynthesis, more specifically, the transglycosylation reaction (13, 39). However, the activity of the mannopeptimycins against vancomycin-resistant organisms indicated that they might have a unique mode of action. The aim of the present study was to investigate the mechanism of antimicrobial activity of the mannopeptimycins.
FIG. 2.
Structures of mannopeptimycins.
MATERIALS AND METHODS
Bacterial strains and growth conditions
S. aureus ATCC 29213, S. aureus Smith, and E. coli ATCC 25922 were used in this study and are well-characterized reference strains. Staphylococcus epidermidis PT 5036 is a clinical isolate. S. aureus 12873-10 was described previously and has increased resistance to both vancomycin and teicoplanin (37). Bacterial cells were grown in either brain heart infusion broth (BHI) or Trypticase soy broth (TSB) at 37°C with aeration.
Chemicals
The following commercially available compounds were purchased from the indicated manufacturers: UDP-[14C]GlcNAc (250 mCi/mmol) from American Radiolabeled Chemicals, Inc.; vancomycin, pentapeptide (Ala-d-γ-Glu-Lys-d-Ala-d-Ala), UDP-GlcNAc, and lipoteichoic acid (LTA) of S. aureus from Sigma; and BHI and TSB from Difco Laboratories. Mersacidin was kindly provided by Hans-Georg Sahl. Ramoplanin was kindly provided by Franco Parenti. Mannopeptimycin-δ, AC98-3189, [3H]AC98-3189, AC98-0708, AC98-4800, AC98-5370, and AC98-6446 were used in this study. They were synthesized at Wyeth Research and have been described previously (Dushin et al., 42nd ICAAC). Mannopeptimycin-δ and AC98-4800 (mannopeptimycin aglycone) have been described previously (18).
Selection and synthesis of a mannopeptimycin photoaffinity probe
To facilitate the study of the mechanism of action of the mannopeptimycins, a radiolabeled benzophenone (BP)-based photoaffinity probe was prepared. BP derivatives are excited upon exposure to long-wave (350- to 360-nm) UV light and preferentially form covalent bonds with unreactive CH groups in the biological target. A radiolabeled BP derivative was chosen with the assumption that the BP moiety would form cross-links between the probe molecule and its biological target in a relatively lipophilic environment (1, 3, 14, 47). BP affinity probes have been used to label targets within membranes (16, 17), while BP moieties linked through acetal functionality have previously been reported in the context of steroids (24). Hence, a BP moiety was attached to mannopeptimycin-α via an acetal linkage. The resulting derivative, AC98-3189 (Fig. 2), had good antibacterial activity (MIC for S. aureus, 0.5 to 2 μg/ml). A complete description of the preparation of this material will be reported elsewhere (R. G. Dushin, A. G. Sutherland, A. Minnick, and M. May, unpublished data).
PBP-binding competition assay
The cell membranes from S. aureus ATCC 29213 and E. coli ATCC 25922 used in the PBP assays were prepared as described previously (41, 56), with the exception that a glass-bead cell disrupter instead of sonication was used to break the cells. An overnight bacterial culture of S. aureus ATCC 29213 in BHI was diluted 1:100 in 8 liters of BHI, grown with shaking to an A600 of 0.6, and centrifuged. The cell pellet was washed twice in 200 ml of sonication buffer (50 mM Tris-HCl [pH 8.0], 1 mM MgCl2) and was resuspended in 100 ml of lysis buffer (sonication buffer supplemented with 2 mM β-mercaptoethanol). The cells were broken with a glass-bead cell disrupter by using 0.1-mm-diameter glass beads (Biospec Products, Bartlesville, Okla.); and the unbroken cells, cell debris, and beads were removed by centrifugation at 3,600 × g for 10 min, followed by two additional centrifugations at 9,300 × g for 20 min each. The supernatant was then subjected to ultracentrifugation (30,000 rpm [66,200 × g], 1 h, 10°C; Beckman 70 Ti rotor). The pellet (membrane fraction) was then resuspended in 4 ml of lysis buffer and passed several times through an 18-gauge needle to obtain a homogeneous suspension.
In the competition assay, the solubilized membranes were incubated with different concentrations of mannopeptimycin-δ and control antibiotics for 10 min at 30°C. Aliquots (10 μl) of [3H]benzylpenicillin were then added, and the mixture was incubated for an additional 10 min. The reaction was terminated by the addition of 500 μl of cold acetone. The precipitated protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE), followed by fluorography, as described previously (56).
Competition of soluble pentapeptide for mannopeptimycin-δ
A competitive binding bioassay was performed by previously described methods (5), except that S. aureus 12873-10 was used as the indicator organism (38). The pentapeptide Ala-d-γ-Glu-Lys-d-Ala-d-Ala, vancomycin, and mannopeptimycin-δ were diluted in water and mixed in fixed pentapeptide-to-glycopeptide molar ratios of 0:1, 5:1, 2.5:1, 1:1, 0.5:1, and 0.25:1. Following inoculation of the organism onto a square bioassay plate (245 by 245 mm) containing Mueller-Hinton agar (BBL), 20 μl of each mixture was spotted onto sterile filter paper disks. Each mixture was tested in quadruplicate. The plate was incubated overnight at 37°C, and the zone diameter (in millimeters) was measured and recorded.
Isolation of PG nucleotide precursors from S. aureus
The pool of cytoplasmic UDP-linked PG precursors was extracted by a modification of a previously described method (34). S. aureus Smith was grown to mid-logarithmic phase in either TSB or TSB supplemented with AC98-6446 (final concentration, 2 μg/ml), chilled, harvested by centrifugation, washed in 0.9% NaCl, and extracted with cold trichloroacetic acid (final concentration, 5%) for 30 min at 4°C. The extract was desalted on a Sephadex G-25 column and concentrated by rotary evaporation. High-pressure liquid chromatography (HPLC) separation of the precursors was performed essentially by the method of Flouret et al. (15), with some modifications. The precursors were separated by reverse-phase HPLC on a μBondapak C18 column (3.9 by 300 mm; Waters) in 50 mM ammonium formate (pH 3.9) at a flow rate of 0.5 ml/min. Elution of the precursors was monitored at 254 nm. The peak corresponding to UDP-MurNAc-pentapeptide was harvested, and its identity was confirmed by electrospray ionization mass spectrometry with a Micromass Q-ToF mass spectrometer equipped with a nanoelectrospray source.
Isolation of PG nucleotide precursors from S. epidermidis
Cytoplasmic PG precursors were extracted and purified from S. epidermidis PT 5036 by a previously described method (6, 8). S. epidermidis PT 5036 was grown to mid-logarithmic phase either in BHI or in BHI supplied with mannopeptimycin-δ (final concentration, 8 μg/ml) and was then harvested by centrifugation at 8,000 × g for 10 min. Cytoplasmic PG precursors were extracted with 1.1 M formic acid on ice for 30 min and were then centrifuged at 15,000 × g for 20 min at 4°C. The supernatant was collected and neutralized to pH 8.0 with NH4OH. The precursors were then purified by three cycles of lyophilization followed by chromatography on a Sephadex G-25 column. Fractions were eluted with water at 4°C, monitored at 262 nm, and then pooled and lyophilized. The sample was then analyzed by HPLC with an isocratic mobile phase consisting of 70% buffer A (10 mM CH3COOHNH4 [pH 5.0]) and 30% buffer B (2% acetonitrile). The identity of UDP-MurNAc-pentapeptide was confirmed by electrospray ionization mass spectrometry analysis with a Micromass Q-ToF mass spectrometer equipped with a nanoelectrospray source.
PG biosynthesis in vitro
The biosynthesis of PG in vitro, which yields both lipid II and PG, was performed as described previously (2, 40), with some modifications, including the addition of 0.1% Triton X-100 to the reaction mixture and preincubation of the membranes both prior to the addition of UDP-MurNAc-pentapeptide (30 min on ice) and prior to the addition of UDP-[14C]GlcNAc (10 min, room temperature). The procedure used for the preparation of cell membranes for thin-layer chromatography (TLC) was identical to the procedure described above for the PBP-binding assay. The reaction mixture contained 0.05 M Tris-HCl (pH 7.8), 10 mM MgCl2, and 0.1% Triton X-100 in a final volume of 14 μl; cell membranes (20 μg of protein); 33 μM UDP-MurNAc-pentapeptide; and 5.7 μM either UDP-GlcNAc or UDP-[14C]GlcNAc. When required, the samples were preincubated with either various concentrations of mannopeptimycin-δ or 50 μg of ramoplanin per ml for 10 min at room temperature prior to the addition of UDP-[14C]GlcNAc. Upon the addition of all reagents, the reaction mixture was incubated for 30 min at room temperature and was then placed into a boiling-water bath for 2 min to inactivate the enzymes, thus preventing degradation of lipid II. Aliquots (2 μl) of the samples were separated by TLC on silica gel plates (K6; Whatman) for 2 h in isobutyric acid-1 M NH4OH (5:3; vol/vol). After separation, the plates were dried and exposed to X-ray film (BioMax MS; Kodak).
Preparative TLC and extraction of lipid II
Preparative quantities (200 μl) of both unlabeled and 14C-labeled samples from the in vitro PG biosynthesis reactions were applied to a preparative TLC plate (PK6F; Whatman) and separated as described above. The 14C-labeled sample was applied on both sides of the plate to serve as a marker for lipid II isolation. After separation, the plates were dried and exposed to X-ray film (BioMax MS; Kodak). The area of silica gel containing lipid II was identified by matching of the gel with an autoradiographed standard and was scraped off the plate. Lipid II was extracted from the silica gel with 30 ml of a mixture of 90% (vol/vol) methanol and 10% (vol/vol) acetic acid, followed by incubation for 5 min at room temperature on a rotating platform (100 rpm). The silica gel was removed by centrifugation at 3,000 × g for 5 min and was extracted twice more under identical conditions. The supernatants were combined in a 500-ml round-bottom flask and concentrated to the minimal possible volume under vacuum on a rotary evaporator (Bü chi; Flawil, Switzerland). The volume was brought up to 30 ml with methanol, and the mixture was evaporated again. This procedure was repeated seven times to minimize traces of acetic acid. The resulting pellet was redissolved in 30 ml of methanol, transferred to a 50-ml conical flask, and evaporated to dryness. The new pellet was dissolved in 600 μl of methanol, centrifuged at 13,000 × g for 1 min to remove fine silica gel particles, and used for subsequent experiments. As judged by extraction of [14C]lipid II under the conditions described above and subsequent analytical TLC, a rate of recovery of intact lipid II of up to 75% was achieved.
TLC-based binding assay
Aliquots (2 μl) from nonradioactive heat-inactivated PG biosynthesis reaction mixtures (prepared either in the absence or in the presence of ramoplanin [see above]) or various amounts of lipid II extract (from 0 to 20 μl, evaporated to dryness by gentle airflow) were placed in glass tubes and mixed with 50 ng of [3H]AC98-3189 in 40 mM Tris-HCl (pH 8.0)-0.8 mM MgCl2-0.02% Triton X-100 buffer. To cross-link the photoaffinity moiety to lipid II, reaction mixtures (total volume, 10 μl) were transferred to individual wells of a 96-well polypropylene plate, and the plates were incubated for 10 min at 30°C and exposed to long-wave (wavelength, 350 to 360 nm) UV light for 30 min on ice. The samples were separated by TLC on silica gel plates (K6; Whatman) as described above. After separation, the plates were dried and exposed to X-ray film (BioMax MS; Kodak) for 10 to 14 days. Spot densitometry of the autoradiographs was performed by using a GS-710 densitometer (Bio-Rad).
Gel-based binding assay
A gel-based mannopeptimycin-lipid II binding assay was developed by modification of a previously described assay (12). Various amounts (from 0 to 10 μl) of the lipid II TLC extract mentioned above were placed in glass tubes (12 by 35 mm) and evaporated to dryness by gentle airflow. The dried samples were preincubated with different amounts of various competing drugs in 50 mM Tris-HCl (pH 7.5)-1 mM MgCl2 buffer for 10 min at 30°C. [3H]AC98-3189 was added to each tube, and the tubes were incubated again for 10 min at 30°C. Glycerol and bromophenol blue were added prior to electrophoresis. In addition to lipid II (with no lipid II included in the control sample), a typical sample contained 15 μl of buffer, 5 μl of competing drug solution (various concentrations), 125 ng of [3H]AC98-3189 (5 μl of a 25-μg/ml solution), and 6 μl of a mixture of 50% glycerol and 0.125% bromophenol blue. Samples were subjected to nondenaturing PAGE on an 18% polyacrylamide gel (Novex). Following electrophoresis, the gel was treated with an autoradiography enhancer (Entensify; New England Nuclear Corp., Boston, Mass.) according to the instructions of the manufacturer, dried, and exposed to X-ray film (X-OMAT AR; Kodak) for 2 days at −70°C.
RESULTS
Effect of mannopeptimycins on PG biosynthesis
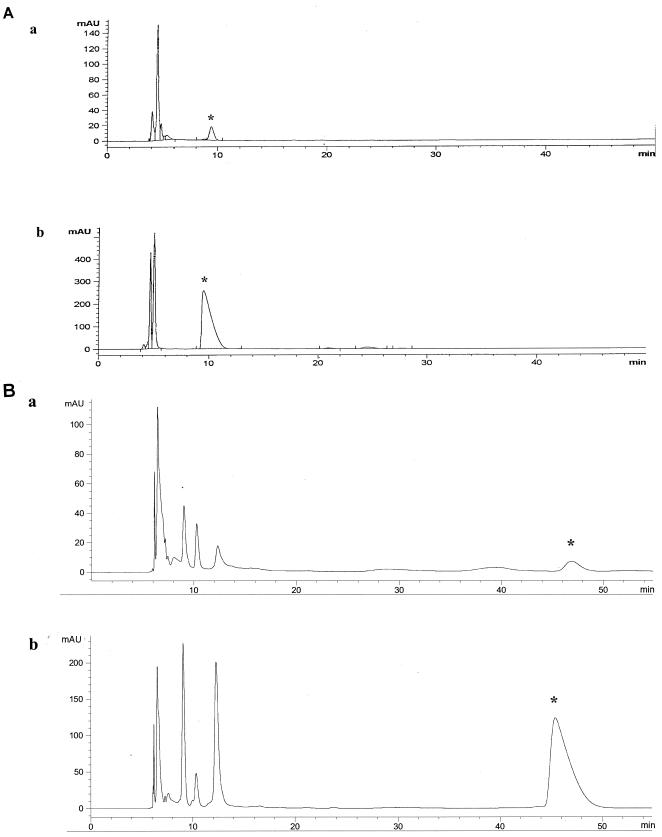
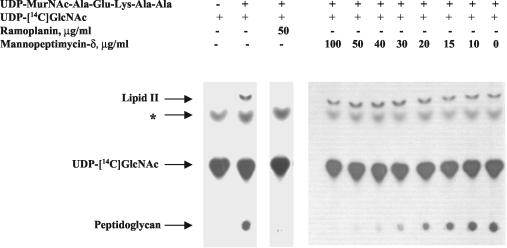
Treatment with mannopeptimycin-δ caused the accumulation of UDP-MurNAc-Ala-Glu-Lys-Ala-Ala (referred to as UDP-MurNAc-pentapeptide) in S. epidermidis strain PT 5036 (Fig. 3A). On the basis of the peak area, there was at least a 30-fold increase in the amount of UDP-MurNAc-pentapeptide in mannopeptimycin-δ-treated cells compared with the amount in untreated cells. Similarly, the more active mannopeptimycin, AC98-6446 (Fig. 2), caused the accumulation of UDP-MurNAc-pentapeptide in S. aureus Smith (Fig. 3B). These results indicate that the cytoplasmic enzymes MurA through MurF are not affected and that the mannopeptimycins most likely impair cell wall biosynthesis at the time of or after the formation of the undecaprenol-bound intermediates, lipid I and lipid II. The data presented in Fig. 4 confirm the results of the experiments performed by DeCenzo et al. (13) and show that, like ramoplanin, the mannopeptimycins inhibit PG formation. However, in contrast to ramoplanin, the mannopeptimycins have no effect on the formation of lipid II. This suggests that the mannopeptimycins act on one of the final steps of PG synthesis, such as transglycosylation or transpeptidation, or both. A competition PBP-binding assay with labeled benzylpenicillin showed that mannopeptimycin-δ does not bind to any of the staphylococcal or E. coli PBPs (data not shown). In contrast, ampicillin, imipenem, and aztreonam, the β-lactam antibiotics with known affinities for specific PBPs, showed reduced levels of binding of the labeled benzylpenicillin. This suggests that mannopeptimycin-δ does not bind to the transpeptidation domain of classical PBPs and favors the hypothesis that the mannopeptimycins, similar to vancomycin, most likely target transglycosylation rather than the transpeptidation reaction.
FIG. 3.
Effect of mannopeptimycins on the accumulation of cytoplasmic PG precursors in S. epidermidis and S. aureus. (A) HPLC profile of cytoplasmic UDP-linked PG precursors isolated from S. epidermidis PT 5036 that was grown either in the absence (a) or in the presence (b) of mannopeptimycin-δ (final concentration, 8 μg/ml). The peak that corresponds to UDP-MurNAc-pentapeptide is marked by an asterisk. (B) HPLC profile of cytoplasmic UDP-linked PG precursors isolated from S. aureus Smith that was grown either in the absence (a) or in the presence (b) of AC98-6446 (final concentration, 2 μg/ml). The peak that corresponds to UDP-MurNAc-pentapeptide is marked by an asterisk.
FIG. 4.
Effects of mannopeptimycin-δ and ramoplanin on lipid II and PG formation. Whole-cell membranes of S. aureus ATCC 29213 were prepared and supplied with UDP-MurNAc-pentapeptide and UDP-[14C]GlcNAc as described in the Materials and Methods. Mannopeptimycin-δ and ramoplanin were added as indicated prior to the addition of UDP-[14C]GlcNAc. Samples were separated by TLC and autoradiographed. The band of an unknown nature, which is marked by an asterisk and that is detected regardless of the presence or absence of UDP-MurNAc-pentapeptide, might result from the previously described translocation of [14C]N-acetylglucosamine-1-phosphate from UDP-[14C]GlcNAc to the C55-lipid carrier (9, 29).
Competition of soluble pentapeptide for mannopeptimycin-δ
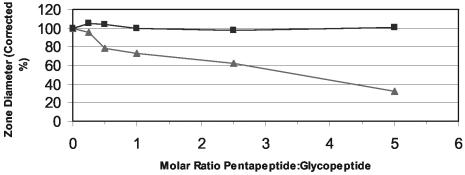
Vancomycin interferes with late stages of cell wall biosynthesis by binding to the d-Ala-d-Ala terminus of the lipid II pentapeptide moiety (36). The ability of mannopeptimycin-δ to bind to the same site was assessed by a competition experiment with d-Ala-d-Ala-containing pentapeptide. As shown in Fig. 5, as the molar ratio of pentapeptide to vancomycin increased, there was a concomitant decrease in the diameter of the zone of inhibition for S. aureus 12873-10, demonstrating that soluble pentapeptide competes with the cell-based target of vancomycin. In contrast, there was no decrease in the zone diameter when the molar ratio of pentapeptide to mannopeptimycin-δ was increased. This suggests that, unlike the mechanism of action of vancomycin, the terminal d-Ala-d-Ala of the pentapeptide is not the target for mannopeptimycin-δ.
FIG. 5.
Competition of pentapeptide with the glycopeptide antibiotics mannopeptimycin-δ and vancomycin against S. aureus 12873-10. ▴, vancomycin; ▪, mannopeptimycin-δ. Data are plotted as the inhibition zone diameter around a 7-mm-diameter filter paper disk corrected to 100% for the sample with no pentapeptide.
Interaction of [3H]AC98-3189 with isolated lipid II
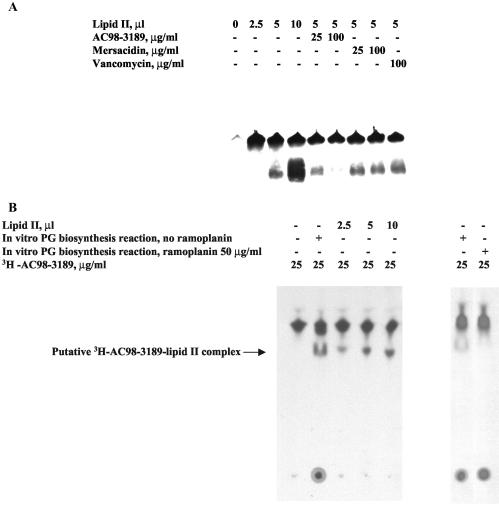
In order to determine whether the mannopeptimycins bind to lipid II at a site(s) different from that to which vancomycin binds, lipid II was isolated by a preparative TLC procedure, mixed with [3H]AC98-3189, and separated by gel electrophoresis under nondenaturing conditions. As shown in Fig. 6A, part of the labeled mannopeptimycin migrated into the gel in the presence of lipid II (lower band), while another part precipitated into the well (upper band). Neither of these signals was detected in the absence of lipid II, indicating that the lipid II-mannopeptimycin interaction is the reason that these additional two bands were observed. The intensity of the lower band was dependent on the concentrations of both lipid II and [3H]AC98-3189, was decreased in the presence of AC98-3189, and was unaffected by the addition of either vancomycin or mersacidin in a competition-type assay (Fig. 6A). The intensity of the lower band was not antagonized by the addition of various compounds that represent or mimic parts of lipid II, such as UDP-MurNAc-pentapeptide, MurNAc, and GlcNAc; and these compounds did not cause entry of [3H]AC98-3189 into the gel (data not shown). The presence of two different signals (upper and lower bands) might be explained either by the formation of several types of mannopeptimycin-lipid II complexes with different molar ratios of components or by the oligomerization of the complex.
FIG. 6.
Mannopeptimycin-lipid II binding assay. (A) Native PAGE. Samples containing a mixture of [3H]AC98-3189, TLC-extracted lipid II, and a competing antibiotic were prepared as described in Materials and Methods. Samples were separated by nondenaturing PAGE on an 18% polyacrylamide gel (Novex), and the gel was dried and autoradiographed. (B) Analytical TLC. Either 2-μl aliquots of nonradioactive heat-inactivated PG biosynthesis reaction mixtures (prepared either in the absence or in the presence of ramoplanin) or various amounts of TLC-purified lipid II were supplied with [3H]AC98-3189, as indicated. Following incubation and UV-cross-linking, the samples were separated by TLC, and the plates were dried and autoradiographed.
Interaction of [3H]AC98-3189 with lipid II in whole-cell membranes
In order to investigate the specificity of the mannopeptimycin-lipid II binding observed, [3H]AC98-3189 was incubated with the components of an in vitro PG biosynthesis reaction. To form a covalent bond between [3H]AC98-3189 and its ligand(s) in the cell membrane, the [3H]AC98-3189-cell membrane mixture was exposed to long-wave UV light to cross-link the BP group. The mixture was then separated by TLC. As shown in Fig. 6B, in the absence of the membrane preparation, the labeled mannopeptimycin was detected as one band, whereas in the presence of the membrane preparation, two additional signals were observed. One band was seen on the TLC baseline, while the other band showed decreased mobility relative to that of free [3H]AC98-3189. The retardation of mannopeptimycin mobility was most likely caused by the interaction of the drug with some membrane-bound ligand(s). On the basis of the results of the gel-based binding assay (see above), the species represented by the upper retarded band most likely represents a mannopeptimycin-lipid II complex. To test this hypothesis, purified lipid II was substituted for the membrane preparation in the TLC binding assay. The band represented by the shift in the mobility of [3H]AC98-3189 in the presence of purified lipid II was identical to the upper band observed with whole membranes (Fig. 6B). In addition, the upper retarded species was not observed with the membranes that were depleted of lipid II by treatment with ramoplanin (Fig. 6B). In contrast, treatment with ramoplanin did not affect the retention of [3H]AC98-3189 at the TLC baseline (Fig. 6B).
The retention of a portion of radiolabeled mannopeptimycin on the baseline observed with membranes might possibly suggest an interaction with some high-molecular-weight membrane-bound compound(s). Among possible candidates for mannopeptimycin binding are LTA and nascent teichoic acid (TA), both of which contain multiple negatively charged phosphate groups that might interact electrostatically with the positively charged amino groups of the mannopeptimycins. Treatment of the membranes with both proteinase K and muramidase had no effect on mannopeptimycin binding (data not shown). Therefore, it is unlikely that other membrane-bound substances such as proteins or nascent PG contribute significantly to the interaction with the mannopeptimycins.
Competition assay with mannopeptimycins and binding of mannopeptimycin to LTA
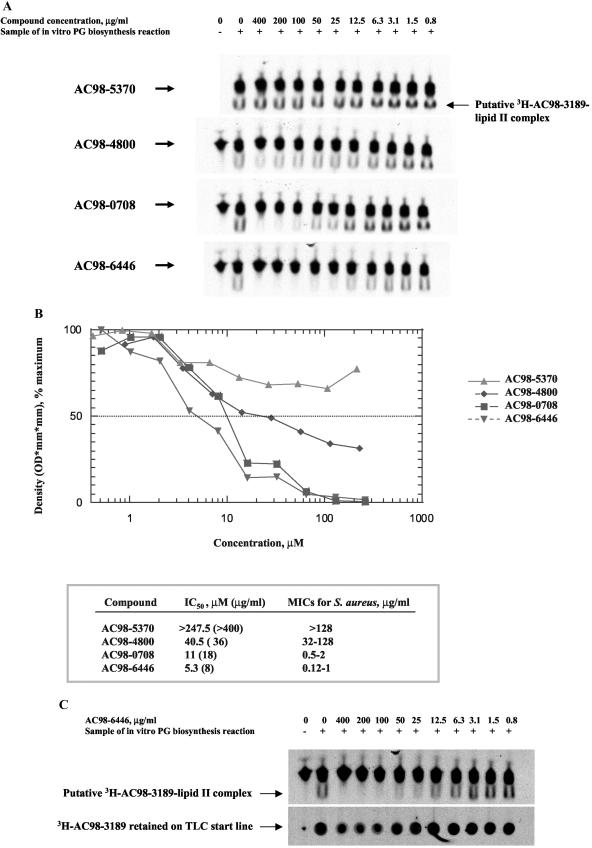
In order to determine whether the binding of mannopeptimycins to lipid II is related to their antimicrobial activity, several mannopeptimycins with different MICs were tested for their ability to compete with [3H]AC98-3189 for the lipid II interaction in the TLC binding assay. As shown in Fig. 7A, the intensity of the [3H]AC98-3189-lipid II band decreased to various extents with increasing concentrations of different competing compounds, indicating the differences in the affinities of the mannopeptimycins for lipid II. As shown in Fig. 7B, the antimicrobial activities of the mannopeptimycins (MICs) correlated with their affinities for lipid II (50% inhibitory concentrations), which further supports the hypothesis that lipid II serves as a primary target for the mannopeptimycins. In contrast, the amount of [3H]AC98-3189 retained on the baseline TLC was not significantly affected by the most active mannopeptimycin, AC98-6446 (Fig. 7C), suggesting that the binding of the mannopeptimycins to membrane components other than lipid II is rather nonspecific.
FIG. 7.
Competition assay with mannopeptimycins. (A) TLC assay. The assay was done essentially as described in the legend to Fig. 6B. Prior to the addition of [3H]AC98-3189, the samples were supplied and preincubated with various mannopeptimycins, as indicated. (B) Quantitative analysis of TLC assay. Shown are a graph derived from densitometry analysis of spots that correspond to putative [3H]AC98-3189-lipid II complex (A) and a comparison of the 50% inhibitory concentrations (IC50s) versus the MICs of various mannopeptimycins. A log scale is used for the compound concentration. (C) Effect of AC98-6446 on binding of [3H]AC98-3189 to membrane components. Shown is a comparison of the competition effects of AC98-6446 on the amount of both putative [3H]AC98-3189-lipid II complex and [3H]AC98-3189 that was retained on the TLC baseline.
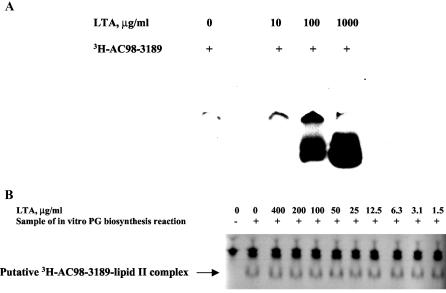
To test whether the mannopeptimycins interact with LTA, LTA was used in the aforementioned gel-based binding and TLC-based competition assays. As shown in Fig. 8A, the presence of LTA resulted in the entry of [3H]AC98-3189 into the gel. No entry of [3H]AC98-3189 into the gel was observed in the absence of LTA, indicating that such an entry is a result of an interaction of LTA with mannopeptimycin. The relatively high concentrations of LTA that are required to observe the entry of mannopeptimycin into the gel indicate the low affinity of binding of mannopeptimycins to LTA. The addition of LTA did not affect mannopeptimycin-lipid II binding in the TLC assay (Fig. 8B), supporting the notion that the interaction between the mannopeptimycins and lipid II has a higher affinity than that between the mannopeptimycins and LTA.
FIG. 8.
Mannopeptimycin-LTA binding assay. (A) Native PAGE. Samples containing a mixture of [3H]AC98-3189 and LTA were separated by nondenaturing PAGE on an 18% polyacrylamide gel (Novex), and the gel was dried and autoradiographed. (B) Effect of LTA on mannopeptimycin-lipid II binding. The assay was done essentially as described in the legend to Fig. 6B. Prior to the addition of [3H]AC98-3189, the samples were supplied and preincubated with various amounts of LTA as indicated.
DISCUSSION
The ability of the mannopeptimycins to inhibit the growth of a wide range of gram-positive pathogens, including those resistant to methicillin and vancomycin, provided the incentive for the mechanism-of-action studies described here. The results obtained in this study indicate that the mannopeptimycins inhibit PG synthesis by blocking the transglycosylation reaction. It was demonstrated that the mannopeptimycin [3H]AC98-3189 bound to both the isolated and the membrane-bound PG precursor, lipid II, and that the affinity of this interaction correlated with the antimicrobial activities of other mannopeptimycins. This suggests that, similar to other glycopeptide antibiotics, such as vancomycin and teicoplanin, the lantibiotic mersacidin, and likely, ramoplanin, the mannopeptimycins inhibit transglycosylase by binding to its substrate rather than by binding to the enzyme itself.
It should be noted that the extraction of lipid II from TLC plates described in this study did not yield 100% pure lipid II, as has been the case for other lipid II purification protocols based on radioactivity tracing (48, 52). It has been reported that traces of other lipids, particularly lipid I, are present in the final preparations of lipid II (48, 52). Therefore, the results of the experiments, which were performed with partially purified lipid II, did not exclude the possibility that the mannopeptimycins might interact with lipid I and/or other membrane lipids. However, the addition of ramoplanin, a known inhibitor of lipid II biosynthesis that does not affect the formation of lipid I (40), eliminated the upper retarded species in the TLC binding assay (Fig. 6B), suggesting that the mannopeptimycins primarily interact with lipid II.
It was also demonstrated that, in addition to lipid II, [3H]AC98-3189 interacts with LTA. It is likely that the mannopeptimycin-LTA interaction is of an electrostatic nature due to the presence of multiple negatively charged phosphate groups in TAs. This finding might point to some additional inhibitory effects of the mannopeptimycins. The biosyntheses of both TA and LTA are linked, as LTA serves as an alanyl donor for TA and is reported to act as an LTA carrier involved in the transfer of ribitol phosphate (26). It is possible that by binding to LTA, the mannopeptimycins interfere with the proper transfer of functional groups from the LTA carrier to TA and as a result block TA biosynthesis. Another possibility is that the mannopeptimycins might act similarly to the lantibiotics nisin and Pep5, which disrupt the electrostatic interactions between autolytic enzymes and LTA, resulting in cell lysis (4). Alternatively, either LTA or TA, or both, might serve as the initial binding site(s) for the mannopeptimycins, which could allow the accumulation of antibiotic on the bacterial cell surface and promote a specific mannopeptimycin-lipid II interaction.
It was shown that the addition of soluble pentapeptide does not affect the antimicrobial activity of mannopeptimycins and that vancomycin does not compete with mannopeptimycin for lipid II binding (Fig. 5 and 6A). This ruled out the involvement of the pentapeptide portion of lipid II in the mannopeptimycin-lipid II interaction and might explain the activity of the mannopeptimycins against vancomycin-resistant organisms. Mannopeptimycins might interact with other components of lipid II, such as the disaccharide unit of MurNAc-GlcNAc and the pyrophosphate moiety. Both of these groups were proposed to be binding sites for another lipid II-binding antibiotic, mersacidin (12). However, mersacidin does not compete with the mannopeptimycins for lipid II binding, indicating that these two antibiotics might recognize lipid II differently as well. This hypothesis is perhaps supported by the fact that the mannopeptimycins are active against the mersacidin-producing organism, a Bacillus sp. (data not shown). Alternatively, the lack of an effect of mersacidin on mannopeptimycin-lipid II binding might result from a higher affinity of the mannopeptimycins toward lipid II, which would not preclude at least a partial overlap in the binding sites of these two antibiotics. Additional experiments are required to determine the exact sites involved in the mannopeptimycin-lipid II interaction.
Acknowledgments
We thank Hans-Georg Sahl for providing mersacidin and Franco Parenti for providing ramoplanin.
REFERENCES
- 1.Abe, I., Y. F. Zheng, and G. D. Prestwich. 1998. Photoaffinity labeling of oxidosqualene cyclase and squalene cyclase by a benzophenone-containing inhibitor. Biochemistry 37:5779-5784. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. S., P. M. Meadow, M. A. Haskin, and J. L. Strominger. 1966. Biosynthesis of the peptidoglycan of bacterial cell walls. I. Utilization of uridine diphosphate acetylmuramyl pentapeptide and uridine diphosphate acetylglucosamine for peptidoglycan synthesis by particulate enzymes from Staphylococcus aureus and Micrococcus lysodeikticus. Arch. Biochem. Biophys. 116:487-515. [DOI] [PubMed] [Google Scholar]
- 3.Andrus, M. B., T. M. Turner, Z. E. Sauna, and S. V. Ambudkar. 2000. Synthesis and preliminary analysis of a P-glycoprotein-specific [3H]-benzophenone photoaffinity label based on (−)-stipiamide. Bioorg. Med. Chem. Lett. 10:2275-2278. [DOI] [PubMed] [Google Scholar]
- 4.Bierbaum, G., and H. G. Sahl. 1985. Induction of autolysis of staphylococci by the basic peptide antibiotics Pep 5 and nisin and their influence on the activity of autolytic enzymes. Arch. Microbiol. 141:249-254. [DOI] [PubMed] [Google Scholar]
- 5.Billot-Klein, D., L. Gutmann, D. Bryant, D. Bell, J. Van Heijenoort, J. Grewal, and D. M. Shlaes. 1996. Peptidoglycan synthesis and structure in Staphylococcus haemolyticus expressing increasing levels of resistance to glycopeptide antibiotics. J. Bacteriol. 178:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billot-Klein, D., L. Gutmann, E. Collatz, and J. van Heijenoort. 1992. Analysis of peptidoglycan precursors in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 36:1487-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumberg, P. M., and J. L. Strominger. 1974. Interaction of penicillin with the bacterial cell: penicillin-binding proteins and penicillin-sensitive enzymes. Bacteriol. Rev. 38:291-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochner, B. R., and B. N. Ames. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257:9759-9769. [PubMed] [Google Scholar]
- 9.Bracha, R., and L. Glaser. 1976. An intermediate in teichoic acid biosynthesis. Biochem. Biophys. Res. Commun. 72:1091-1098. [DOI] [PubMed] [Google Scholar]
- 10.Breukink, E., I. Wiedemann, C. van Kraaij, O. P. Kuipers, H. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 11.Broome-Smith, J. K., A. Edelman, S. Yousif, and B. G. Spratt. 1985. The nucleotide sequences of the ponA and ponB genes encoding penicillin-binding protein 1A and 1B of Escherichia coli K12. Eur. J. Biochem. 147:437-446. [DOI] [PubMed] [Google Scholar]
- 12.Brotz, H., G. Bierbaum, K. Leopold, P. E. Reynolds, and H. G. Sahl. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeCenzo, M., M. Kuranda, S. Cohen, J. Babiak, Z. D. Jiang, D. Su, M. Hickey, P. Sancheti, P. A. Bradford, P. Youngman, S. Projan, and D. M. Rothstein. 2002. Identification of compounds that inhibit late steps of peptidoglycan synthesis in bacteria. J. Antibiot. (Tokyo) 55:288-295. [DOI] [PubMed] [Google Scholar]
- 14.Dorman, G., and G. D. Prestwich. 1994. Benzophenone photophores in biochemistry. Biochemistry 33:5661-5673. [DOI] [PubMed] [Google Scholar]
- 15.Flouret, B., D. Mengin-Lecreulx, and J. van Heijenoort. 1981. Reverse-phase high-pressure liquid chromatography of uridine diphosphate N-acetylmuramyl peptide precursors of bacterial cell wall peptidoglycan. Anal. Biochem. 114:59-63. [DOI] [PubMed] [Google Scholar]
- 16.Haas, M., and B. Forbush III. 1988. Photoaffinity labelling of a 150 kDa (Na + K + Cl)-cotransport protein from duck red cells with an analog of bumetanide. Biochim. Biophys. Acta 939:131-144. [DOI] [PubMed] [Google Scholar]
- 17.Haas, M., and B. Forbush III. 1987. Photolabeling of a 150-kDa (Na + K + Cl) cotransport protein from dog kidney with a bumetanide analogue. Am. J. Physiol. 253:C243-C252. [DOI] [PubMed] [Google Scholar]
- 18.He, H., R. T. Williamson, B. Shen, E. I. Graziani, H. Y. Yang, S. M. Sakya, P. J. Petersen, and G. T. Carter. 2002. Mannopeptimycins, novel antibacterial glycopeptides from Streptomyces hygroscopicus, LL-AC98. J. Am. Chem. Soc. 124:9729-9736. [DOI] [PubMed] [Google Scholar]
- 19.Higashi, Y., J. L. Strominger, and C. C. Sweeley. 1967. Structure of a lipid intermediate in cell wall peptidoglycan synthesis: a derivative of a C55 isoprenoid alcohol. Proc. Natl. Acad. Sci. USA 57:1878-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishino, F., K. Mitsui, S. Tamaki, and M. Matsuhashi. 1980. Dual enzyme activities of cell wall peptidoglycan synthesis, peptidoglycan transglycosylase and penicillin-sensitive transpeptidase, in purified preparations of Escherichia coli penicillin-binding protein 1A. Biochem. Biophys. Res. Commun. 97:287-293. [DOI] [PubMed] [Google Scholar]
- 21.Ito, E., and J. L. Strominger. 1962. Enzymatic synthesis of the peptide in bacterial uridine nucleotides. I. Enzymatic addition of l-alanine, d-glutamic acid, and l-lysine. J. Biol. Chem. 237:2689-2695. [Google Scholar]
- 22.Ito, E., and J. L. Strominger. 1962. Enzymatic synthesis of the peptide in bacterial uridine nucleotides. II. Enzymatic synthesis and addition of d-alanyl-d-alanine. J. Biol. Chem. 237:2696-2703. [Google Scholar]
- 23.Kahan, F. M., J. S. Kahan, P. J. Cassidy, and H. Kropp. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann. N. Y. Acad. Sci. 235:364-386. [DOI] [PubMed] [Google Scholar]
- 24.Kym, P. R., K. E. Carlson, and J. A. Katzenellenbogen. 1993. Progestin 16-alpha,17-alpha-dioxolane ketals as molecular probes for the progesterone receptor: synthesis, binding affinity, and photochemical evaluation. J. Med. Chem. 36:1111-1119. [DOI] [PubMed] [Google Scholar]
- 25.Lambert, M. P., and F. C. Neuhaus. 1972. Mechanism of d-cycloserine action: alanine racemase from Escherichia coli W. J. Bacteriol. 110:978-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert, P. A., I. C. Hancock, and J. Baddiley. 1977. Occurrence and function of membrane teichoic acids. Biochim. Biophys. Acta 472:1-12. [DOI] [PubMed] [Google Scholar]
- 27.Lo, M. C., J. S. Helm, G. Sarngadharan, I. Pelczer, and S. Walker. 2001. A new structure for the substrate-binding antibiotic ramoplanin. J. Am. Chem. Soc. 123:8640-8641. [DOI] [PubMed] [Google Scholar]
- 28.Lo, M. C., M. Hongbin, A. Branstrom, J. Helm, N. Yao, R. Goldman, and S. Walker. 2000. A new mechanism of action proposed for ramoplanin. J. Am. Chem. Soc. 122:3540-3541. [Google Scholar]
- 29.McArthur, H. A., F. M. Roberts, I. C. Hancock, and J. Baddiley. 1978. Lipid intermediates in the biosynthesis of the linkage unit between teichoic acids and peptidoglycan. FEBS Lett. 86:193-200. [DOI] [PubMed] [Google Scholar]
- 30.Murakami, K., T. Fujimura, and M. Doi. 1994. Nucleotide sequence of the structural gene for the penicillin-binding protein 2 of Staphylococcus aureus and the presence of a homologous gene in other staphylococci. FEMS Microbiol. Lett. 117:131-136. [DOI] [PubMed] [Google Scholar]
- 31.Nakagawa, J., S. Tamaki, S. Tomioka, and M. Matsuhashi. 1984. Functional biosynthesis of cell wall peptidoglycan by polymorphic bifunctional polypeptides. Penicillin-binding protein 1Bs of Escherichia coli with activities of transglycosylase and transpeptidase. J. Biol. Chem. 259:13937-13946. [PubMed] [Google Scholar]
- 32.Neuhaus, F. C. 1962. The enzymatic synthesis of d-alanyl-d-alanine. I. Purification and properties of d-alanyl-d-alanine synthetase. J. Biol. Chem. 237:778-786. [PubMed] [Google Scholar]
- 33.Neuhaus, F. C. 1962. The enzymatic synthesis of d-alanyl-d-alanine. II. Kinetic studies on d-alanyl-d-alanine synthetase. J. Biol. Chem. 237:3128-3135. [PubMed] [Google Scholar]
- 34.Park, J. T. 1966. Membrane associated reaction involved in bacterial cell wall mucopeptide synthesis. Methods Enzymol. 8:466-472. [Google Scholar]
- 35.Park, W., and M. Matsuhashi. 1984. Staphylococcus aureus and Micrococcus luteus peptidoglycan transglycosylases that are not penicillin-binding proteins. J. Bacteriol. 157:538-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds, P. E. 1989. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8:943-950. [DOI] [PubMed] [Google Scholar]
- 37.Shlaes, D. M., and J. H. Shlaes. 1995. Teicoplanin selects for Staphylococcus aureus that is resistant to vancomycin. Clin. Infect. Dis. 20:1071-1073. [DOI] [PubMed] [Google Scholar]
- 38.Shlaes, D. M., J. H. Shlaes, S. Vincent, L. Etter, P. D. Fey, and R. V. Goering. 1993. Teicoplanin-resistant Staphylococcus aureus expresses a novel membrane protein and increases expression of penicillin-binding protein 2 complex. Antimicrob. Agents Chemother. 37:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh, M. P., P. J. Petersen, W. J. Weiss, J. E. Janso, S. W. Luckman, E. B. Lenoy, P. A. Bradford, R. T. Testa, and M. Greenstein. 2002. Mannopeptimycins, new cyclic glycopeptide antibiotics: antibacterial and mechanistic activities. Antimicrob. Agents Chemother. 47:62-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somner, E. A., and P. E. Reynolds. 1990. Inhibition of peptidoglycan biosynthesis by ramoplanin. Antimicrob. Agents Chemother. 34:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spratt, B. G. 1977. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur. J. Biochem. 72:341-352. [DOI] [PubMed] [Google Scholar]
- 42.Stone, K. J., and J. L. Strominger. 1971. Mechanism of action of bacitracin: complexation with metal ion and C55 -isoprenyl pyrophosphate. Proc. Natl. Acad. Sci. USA 68:3223-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storm, D. R., and J. L. Strominger. 1973. Complex formation between bacitracin peptides and isoprenyl pyrophosphates. The specificity of lipid-peptide interactions. J. Biol. Chem. 248:3940-3945. [PubMed] [Google Scholar]
- 44.Strominger, J. L., E. Ito, and R. H. Threnn. 1960. Competitive inhibition of enzymatic reactions by oxamycin. J. Am. Chem. Soc. 82:998-999. [Google Scholar]
- 45.Strominger, J. L., K. Izaki, M. Matsuhashi, and D. J. Tipper. 1967. Peptidoglycan transpeptidase and d-alanine carboxypeptidase: penicillin-sensitive enzymatic reactions. Fed. Proc. 26:9-22. [PubMed] [Google Scholar]
- 46.Tipper, D. J., and J. L. Strominger. 1965. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc. Natl. Acad. Sci. USA 54:1133-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turek, T. C., I. Gaon, D. Gamache, and M. D. Distefano. 1997. Synthesis and evaluation of benzophenone-based photoaffinity labeling analogs of phenyl pyrophosphates containing stable amide linkages. Bioorg. Med. Chem. Lett. 7:2125-2130. [Google Scholar]
- 48.Umbreit, J. N., and J. L. Strominger. 1972. Isolation of the lipid intermediate in peptidoglycan biosynthesis from Escherichia coli. J. Bacteriol. 112:1306-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Heijenoort, J. 1996. Murein synthesis, p. 1025-1034. In F. C. Neidhardt, I. Curtis, R., J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology Press, Washington, D.C.
- 50.Van Heijenoort, J., Y. van Heijenoort, and P. Welzel. 1988. Moenomycin: inhibitor of peptidoglycan polymerization in Escherichia coli, p. 549-557. In P. Actor, L. Daneo-Moore, M. L. Higgins, M. R. J. Salton, and G. D. Shockman (ed.), Antibiotic inhibition of bacterial cell surface assembly and function. American Society for Microbiology, Washington, D.C.
- 51.Van Heijenoort, Y., M. Derrien, and J. van Heijenoort. 1978. Polymerization by transglycosylation in the biosynthesis of the peptidoglycan of Escherichia coli K 12 and its inhibition by antibiotics. FEBS Lett. 89:141-144. [DOI] [PubMed] [Google Scholar]
- 52.van Heijenoort, Y., M. Gomez, M. Derrien, J. Ayala, and J. van Heijenoort. 1992. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J. Bacteriol. 174:3549-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward, J. B., and H. R. Perkins. 1973. The direction of glycan synthesis in a bacterial peptidoglycan. Biochem. J. 135:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward, J. B., and H. R. Perkins. 1974. Peptidoglycan biosynthesis by preparations from Bacillus licheniformis: cross-linking of newly synthesized chains to preformed cell wall. Biochem. J. 139:781-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waxman, D. J., R. R. Yocum, and J. L. Strominger. 1980. Penicillins and cephalosporins are active site-directed acylating agents: evidence in support of the substrate analogue hypothesis. Phil. Trans. R. Soc. 289:257-271. [DOI] [PubMed] [Google Scholar]
- 56.Yang, Y., N. Bhachech, and K. Bush. 1995. Biochemical comparison of imipenem, meropenem and biapenem: permeability, binding to penicillin-binding proteins, and stability to hydrolysis by β-lactamases. J. Antimicrob. Chemother. 35:75-84. [DOI] [PubMed] [Google Scholar]