Abstract
The Human Metabolome Database (HMDB) (www.hmdb.ca) is a resource dedicated to providing scientists with the most current and comprehensive coverage of the human metabolome. Since its first release in 2007, the HMDB has been used to facilitate research for nearly 1000 published studies in metabolomics, clinical biochemistry and systems biology. The most recent release of HMDB (version 3.0) has been significantly expanded and enhanced over the 2009 release (version 2.0). In particular, the number of annotated metabolite entries has grown from 6500 to more than 40 000 (a 600% increase). This enormous expansion is a result of the inclusion of both ‘detected’ metabolites (those with measured concentrations or experimental confirmation of their existence) and ‘expected’ metabolites (those for which biochemical pathways are known or human intake/exposure is frequent but the compound has yet to be detected in the body). The latest release also has greatly increased the number of metabolites with biofluid or tissue concentration data, the number of compounds with reference spectra and the number of data fields per entry. In addition to this expansion in data quantity, new database visualization tools and new data content have been added or enhanced. These include better spectral viewing tools, more powerful chemical substructure searches, an improved chemical taxonomy and better, more interactive pathway maps. This article describes these enhancements to the HMDB, which was previously featured in the 2009 NAR Database Issue. (Note to referees, HMDB 3.0 will go live on 18 September 2012.).
INTRODUCTION
The human metabolome can be defined as the complete collection of small molecule metabolites found in the human body (1). These small molecules include peptides, lipids, amino acids, nucleic acids, carbohydrates, organic acids, vitamins, minerals, food additives, drugs, toxins, pollutants and just about any other chemical (with a molecular weight <2000 Da) that humans ingest, metabolize, catabolize or come into contact with. Together with the genome and the proteome, the human metabolome essentially defines who and what we are. However, in contrast to the genome and proteome, the metabolome itself is not easily defined. This is because the human metabolome is not solely dictated by our genes. Our environment (what we eat, breathe, drink) and our microflora (the bacteria that live in our intestinal tract) contribute to the metabolome. In other words, the human metabolome consists of a mix of both endogenous and exogenous compounds. Endogenous metabolites are small molecules synthesized by the enzymes encoded by our genome or our microfloral genomes, and exogenous metabolites are ‘foreign’ or xenobiotic chemicals consumed as foods, drinks, drugs or other ‘consumables’. The fact that so many different chemicals from so many different sources can potentially appear in the human metabolome has made its characterization difficult. Nevertheless, several concerted efforts have been made to decipher the human metabolome—or, more appropriately, the human metabolomes. Beginning in 2005 and continuing to the present, the Human Metabolome Project (HMP) (2) has been using a variety of high-throughput metabolomic studies in combination with comprehensive literature surveys to compile as much information about the human metabolome(s) as possible. This information is released publicly through the Human Metabolome Database or HMDB (3,4).
First introduced in 2007, the HMDB is currently the world’s largest and most comprehensive, organism-specific metabolomics database. It contains spectroscopic, quantitative, analytic and physiological information about human metabolites, their associated enzymes or transporters, their abundance and their disease-related properties. Since its initial release, the HMDB website has been accessed more than 5 million times and its associated papers cited nearly 1000 times. Feedback from users has led to many excellent suggestions on how to expand and enhance HMDB’s offerings. Likewise, continuing advances in the field of metabolomics along with ongoing data collection and curation by the HMP team has led to a substantial expansion of the HMDB’s content. Here, we wish to report on these developments as well as the many additions and improvements appearing in the latest release of the HMDB (version 3.0).
DATABASE ENHANCEMENTS
Details regarding the HMDB’s overall layout, navigation structure, data sources, curation protocols, data management system and quality assurance criteria have been described earlier (3,4). These have largely remained the same for this release. Here, we shall focus primarily on describing the changes and improvements made to the HMDB since 2009. More specifically, we will describe the: (i) enhancements to the HMDB’s content, completeness and coverage, (ii) improvements to the HMDB’s interface and MetaboCard layout, (iii) improvements to its spectral databases and spectral viewing tools and (iv) improvements to the HMDB’s data downloads, data formats and data structures.
Expanded database content, completeness and coverage
A detailed content comparison between the HMDB (release 1.0 and 2.0) versus the HMDB (release 3.0) is provided in Table 1. As seen in this Table 1, the latest release of the HMDB now has detailed information on >40 000 metabolites, representing an expansion of nearly 600% over what was previously contained in the database. This increase is primarily a result of the significant expansion of both ‘detected’ metabolites [separated into two categories: (i) detected and quantified and (ii) detected not quantified] and ‘expected’ metabolites (those for which biochemical pathways are known or human intake/exposure is frequent but the compound has yet to be detected in the body). In previous releases of HMDB, no clear distinction was made between these categories. In this release, the distinction is made quite explicitly using a ‘status’ data field that is now part of HMDB’s new chemical ontology. This new chemical ontology also distinguishes the source (or probable source) of these compounds using the ‘origin’ data field. Using this data field (for both detected and expected metabolites), compounds are further classified as being endogenous, microbial, drug derived, a toxin/pollutant, food derived or different combinations of the above.
Table 1.
Content comparison of HMDB 1.0 with HMDB 2.0 and HMDB 3.0
| Database feature or content status | HMDB | HMDB | HMDB |
|---|---|---|---|
| (version 1.0) | (version 2.0) | (version 3.0) | |
| Number of metabolites | 2180 | 6408 | 40 153 |
| Number of unique metabolite synonyms | 27 700 | 43 882 | 199 668 |
| Number of compounds with disease links | 862 | 1002 | 3105 |
| Number of compounds with biofluid or tissue concentration data | 883 | 4413 | 5027 |
| Number of compounds with chemical synthesis references | 220 | 1647 | 1943 |
| Number of compounds with experimental reference 1H and or 13C NMR spectra | 385 | 792 | 1054 |
| Number of compounds with reference MS/MS spectra | 390 | 799 | 1249 |
| Number of compounds with reference GC-MS reference data | 0 | 279 | 1220 |
| Number of human-specific pathway maps | 26 | 58 | 442 |
| Number of compounds in Human Metabolome Library | 607 | 920 | 1031 |
| Number of HMDB data fields | 91 | 102 | 114 |
| Number of predicted molecular properties | 2 | 2 | 10 |
| Metabolite search/browse | Yes | Yes | Yes |
| Pathway search/browse | No | Yes | Yes |
| Disease search/browse | No | Yes | Yes |
| Chemical class search/browse | No | Yes | Yes |
| Biofluid browse | No | Yes | Yes |
| Metabolite library browse | No | Yes | Yes |
| Protein/transporter browse | No | No | Yes |
Among the ‘detected’ metabolites, the number has grown from 4413 (in version 2.0) to 20 900 (in version 3.0), or roughly by 450%. In many cases, these additions do not represent the discovery of new compounds, but simply reflect improvements in the HMDB curation team’s ability to experimentally measure or to identify (with the assistance of text mining tools) metabolites previously reported in the literature. Among the ‘expected’ metabolites, their numbers have grown much more significantly, from 1995 (in version 2.0) to more than 19 000 (in version 3.0). This figure includes more than 450 dipeptides, >1500 drugs and drug metabolites, >13 000 food-derived compounds and more than 2000 other compounds. Historically, there have been two reasons for including ‘expected’ molecules in the HMDB. First, based on what is known about human biochemistry along with what is known about human food and drug consumption patterns, we have every reason to believe that these compounds exist in the ‘collective’ human metabolome (but not necessarily in every human metabolome) and that they could be or will eventually be detected in human biofluids and/or tissues. Second, we believe the HMDB needs to provide resources, particularly, for the mass spectrometry (MS)-based metabolomics and lipidomics communities, to assist with the putative identification of novel or ‘yet-to-be-seen’ compounds through mass matching.
The HMDB is not the first or only metabolome database to include ‘expected’ compounds. Indeed, METLIN (5) has been including all possible di- and tripeptide combinations (∼8400 compounds) for many years, whereas LipidMaps (6) has included many 1000s of lipid species that have not yet been formally identified or measured. We chose not to include tripeptides in this release of HMDB as the experimental evidence for stable, long-lived tripeptides is not yet as strong as it is for stable, long-lived dipeptides. As a general rule, the HMDB’s expected metabolites do not contain the level of detail found in the experimentally detected HMDB entries. On average, expected metabolites in the HMDB contain 30–50 completed data fields, whereas detected metabolites typically have 80–120 completed data fields. Furthermore, users can readily select which sets of compounds (detected and quantified, detected, not quantified and expected) they wish to browse or search against. Nevertheless, with this substantial growth of metabolites in the HMDB, the database now has many more compounds than most other commonly used metabolite/metabolome databases [KEGG (7) = 16 843, HumanCyc (8) = 1321, BiGG (9) = 1509 or LipidMaps (6) = 37 127]. Furthermore, more than 25 000 compounds in the HMDB are (not yet) in other chemical databases, including PubChem (10) and ChEBI (11).
In addition to substantially increasing the number of metabolite entries, we have also increased the completeness of the HMDB’s annotations for hundreds of metabolites by adding many more detailed compound descriptions, including more compound synonyms (∼450% increase), more compounds with biofluid concentration or tissue location data (∼20% increase) and a greater number of compounds with synthesis records (∼20% increase). Furthermore, HMDB now includes data on metabolite transporters, channels, symporters in addition to the usual enzyme-specific data. This is especially true for the ‘Detected’ metabolites.
Over the past 2 years, a significant effort has also been undertaken to upgrade the number and quality of reference nuclear magnetic resonance (NMR), tandem mass spectrometry (MS/MS) and gas chromatography-mass spectrometry (GC-MS) spectra in the HMDB. In particular, hundreds of additional reference MS and NMR spectra were collected, assigned and/or annotated by the curation team, whereas hundreds of additional annotated/assigned MS and NMR reference spectra were obtained from the BioMagResBank (12), METLIN (5) and MassBank (13). This effort has led to a 30–50% increase in the number of compound reference spectra in the HMDB.
In the 2009 release of HMDB, we described the addition of ∼30 new pathway diagrams to the database which supplemented a number of KEGG pathway diagrams. Since then, these early pathway diagrams have been substantially upgraded in terms of quality, quantity and viewability, particularly through the inclusion of our own SMPDB pathways (14). With 442 hand-drawn, zoomable, fully hyperlinked human metabolic pathway maps now in the HMDB, the total number of pathway diagrams (including disease and metabolic signaling pathways) in the HMDB has increased by ∼800%. Unlike most online metabolic maps, these HMDB/SMPDB pathway maps are quite specific to human metabolism and explicitly show the compound structures, protein quaternary structures, enzyme cofactors and subcellular compartments where specific reactions are known to take place. All chemical structures in these pathway maps are hyperlinked to HMDB MetaboCards and all enzymes are hyperlinked to UniProt data cards. By making use of the infrastructure originally built for the SMPDB, metabolite, protein and/or gene transcript concentrations may now be interactively mapped onto each pathway diagram. Furthermore, each pathway diagram in the HMDB is fully searchable (via PathSearch) and each pathway now has a short synoptic description.
For a number of years the HMDB has been manually classifying all compounds in the HMDB into chemical ‘kingdoms’, ‘classes’ and ‘families’. This chemical taxonomy has proven to be useful for many researchers, but it has also been quite challenging to maintain and we have found a number of inconsistencies. Furthermore, there are now a growing number of alternate chemical taxonomies being used by other databases, such as BioCyc/MetaCyc (15), ChEBI (11) and LipidMaps (6). In an effort to both standardize our own chemical classification scheme and to integrate other chemical taxonomies/ontologies, we have completely redone the chemical taxonomy in HMDB. For the new version, a similar hierarchical classification system is still being used (kingdom, super class, class, subclass, other descriptors, substituents, direct parent), but the classification process is now fully automated (using thousands of structure rules), fully defined (with thousands of definitions) and fully self-consistent. The new taxonomy also uses a ‘consensus’ terminology or naming convention that makes compound classification more consistent and more easily mapped to other databases’ classification schemes. Furthermore, under the field ‘other descriptors’ most of the known classification or taxonomic terms from other chemical databases, if available for that compound, are now included. A more detailed description of the chemical taxonomy software is being prepared and will be submitted for publication shortly. In addition to this chemical taxonomy, the HMDB also has a chemical ontology. This ontology includes status (detected or expected), origin (endogenous, microbial, drug, etc.), biofunction (energy, signalling, buffer, etc.), application (nutrient, industrial chemical, pharmaceutical, etc.) and cellular location (membrane, cytoplasm, extracellular). We are hopeful that this new and improved chemical taxonomy and ontology will help to provide a common language for large-scale mammalian metabolome comparisons.
Thanks to the feedback provided by HMDB’s user community, a number of new data fields have been added to each MetaboCard. For instance, HMDB 3.0 now provides an additional set of 10 computed/predicted property descriptors including (i) water solubility, (ii) LogP (two different algorithms), (iii) LogS, (iv) pKa, (v) hydrogen acceptor count, (vi) hydrogen donor count, (vii) polar surface area, (viii) rotatable bond count, (ix) refractivity and (x) polarizability. While more computed structure descriptors are certainly available, these represent the most frequently used descriptors and should allow HMDB users to perform much more detailed computed structure queries, analyses and comparisons.
We believe that one of the most important and unique features of the HMDB is the information it contains on metabolite concentrations (normal and abnormal), disease associations and tissue locations. Relative to the previous release, the HMDB now has 20% more of these clinically or physiologically relevant details. Over the past 3 years, several comprehensive studies conducted by the HMP’s metabolomic analysis team have led to the experimental validation and accurate concentration measurements of thousands of compounds in cerebrospinal fluid (16), serum (17) and urine (18). These data have been supplemented with recently published metabolome-specific studies on saliva (19), prostate (20) and other tissues/biofluids. Disease association and abnormal concentrations have also been added via the HMP’s literature curation team. These additions should make version 3.0 of the HMDB substantially more useful for clinical chemists and physicians. They should also aid many metabolomics researchers in clarifying what should (and what shouldn’t) be routinely found in certain human biofluids or tissues.
In addition to significantly expanding the data content in HMDB, a major effort has been directed at improving the quality of HMDB’s existing data. Over the past 3 years, thousands of compound names, synonyms and descriptions were expanded or corrected. Likewise, hundreds of new compound synthesis and concentration references were collected, checked and validated. Similarly, extensive checks were performed on all of HMDB’s small molecule structures to confirm that they exhibit the correct structure, chirality and stereochemistry. In particular, we developed a custom structure-checking program that used direct structure comparison (via a Mol file) of each of HMDB’s structures against the corresponding structures in other databases (PubChem, ChEBI, ChemSpider, etc.). Any HMDB structure that did not match with the corresponding structure in one or more of these external databases was flagged. Each of these was assessed and/or corrected manually by a team of trained chemists. In many cases, the HMDB structure was correct and the external database structure was found to be in error; in other cases, the HMDB structure was determined to be in error and was subsequently corrected. In addition to these changes, a substantial effort has also been put into identifying and correcting minor structural, image format, naming, annotation and spectral assignment errors in the HMDB. While a number of internal checking and editing procedures are used by the HMDB curation team [see (4) for details], we are particularly grateful to external users who identified particularly subtle errors or offered suggestions to improve the data quality. Overall, we are quite confident that the quality of the data in HMDB (version 3.0) is much better than previous releases.
User interface improvements
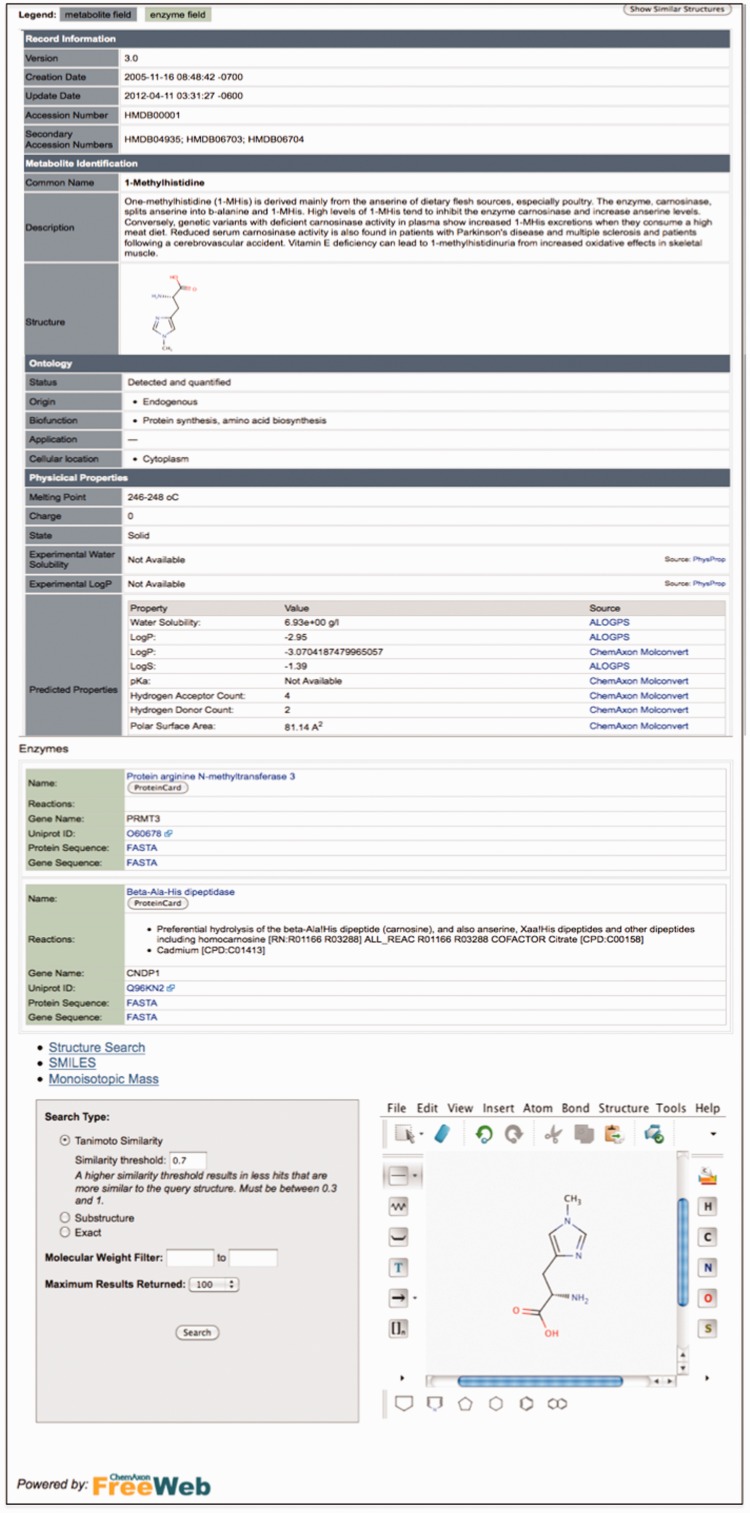
Relative to previous releases, the user interface improvements for this year’s release of the HMDB are relatively modest. The most obvious change is the restructuring of the MetaboCard into 14 distinctive categories or superfields with clearly demarcated titles (Figure 1). These superfields include: (i) record information, (ii) metabolite identification, (iii) chemical taxonomy, (iv) chemical ontology, (v) physical properties, (vi) spectra, (vii) biological properties, (viii) normal concentrations, (ix) abnormal concentrations, (x) associated disorders, (xi) external links, (xii) references, (xiii) enzymes and (xiv) transporters. We believe this should make the data easier to view and browse. In addition, most of the detailed enzyme and transporter data that used to be displayed on the MetaboCard, is now in a separate ProteinCard which can be accessed by clicking the button under each enzyme or transporter name. To further improve the browsing in HMDB, we have also implemented sortable tables in the Metabolite Browse menu. Clicking on the up/down arrows in the table identifiers allows one to resort metabolites by HMDB ID (numerically), Name (alphabetically), International Union of Pure and Applied Chemists (IUPAC) Name (alphabetically), Formula (Alphanumerically) and Mass (Numerically), going from largest to smallest, from A–Z or vice versa. In addition to the usual browse options (metabolite, disease, pathway, biofluid, chemical class and metabolite library), we have also added a Protein Browse option to version 3.0. Protein Browse allows users to browse through, view or sort the list of human proteins, enzymes and transporters that are associated with each metabolite. Each protein is linked to a ‘ProteinCard’, which contains additional molecular, physiological or ontological data (∼30 data fields) about that protein as derived from a variety of databases or calculated from its sequence. Small improvements have also been made to HMDB’s Chem Query tool. Chem Query is the HMDB’s chemical structure search utility. It can be used to sketch (through ChemAxon’s freely available chemical sketching applet) or paste a SMILES string of a query compound into the Chem Query window. Submitting the query launches a structure similarity search that looks for common substructures from the query compound that matches the HMDB’s database of known or expected human compounds. Users can also select the type of search (exact or Tanimoto score) to be performed. High-scoring hits are presented in a tabular format with hyperlinks to the corresponding MetaboCards. The Chem Query tool allows users to quickly determine whether their compound of interest is a known human metabolite or chemically related to a known human metabolite. The same ChemAxon structure querying applet is also used with the ‘Find Similar Structures’ button located at the top of every MetaboCard.
Figure 1.
A screenshot montage of the different HMDB views showing the new layout of the MetaboCards and the new structure search tool.
Enhancements to spectral databases and spectral searching
In metabolomics, most compounds are identified via spectral comparisons against libraries of known compound spectra. Consequently, there is a critical need by many metabolomics researchers for comprehensive, publicly accessible libraries of reference compound spectra. As mentioned earlier, the number and type of reference spectra in release 3.0 is now much greater than in earlier releases. But so too is the utility of these spectra. Earlier versions of the HMDB’s NMR and MS spectral assignments, peak lists or annotation files were kept in an unconventional format (for NMR) or not available for download (for MS). With a growing consensus on data exchange formats for MS and NMR spectra and improved methods becoming available for viewing spectra we have undertaken a substantive update to the HMDB’s spectral resources. All NMR spectral assignments are now available for download in an NMR-STAR (12) format. This format captures all relevant spectral features, spectral collection conditions, assignments and chemical structure information. As before, all NMR spectra continue to be available as raw (FID) files and as simple images (PNG format). In addition to these changes for HMDB’s NMR spectra, nearly all MS/MS spectra in the HMDB are now available in mzML format (21). The mzML format is rapidly becoming the preferred format for MS data exchange as it robustly captures all relevant spectral features, MS spectral collection conditions and associated annotations. As before, all MS spectra in the HMDB are also available as simple images (PNG format) and as simple mass list files. Significant improvements have also been made for HMDB’s spectral viewing and searching tools. In particular, users can now select between expected (∼33 000), detected (∼7000) or combined (40 000) metabolites for their MS search routines. A simple adduct calculator generates more than 25 possible adducts (containing Na+, K+, NH4+ and dimer adducts) for three different modes (positive ion, negative ion, neutral) for each compound, creating an effective database of more than 1 million masses for high resolution parent ion matching.
Improvements in HMDB’s data formats and data structures
While many visible ‘front-end’ enhancements have been implemented, HMDB’s back-end has also been significantly enhanced. First, the HMDB information management system (MetaboLIMS) has been substantially rewritten, allowing the HMP curation team to perform a far more facile uploading and monitoring of newly entered data. Second, the HMDB user interface has been completely rewritten using Ruby-on-Rails (changed from the original Perl). Third, all the structure information in the HMDB has been consolidated into a central chemical structure database (MolDB) that now allows more rapid querying, retrieval, monitoring and updating of structural data. Fourth, all the spectral in formation in the HMDB has been consolidated into a central spectral structure database (SpecDB) that now allows much more rapid querying, retrieval, monitoring and updating of spectral data. Fifth, as mentioned earlier, all the downloadable spectral data have been converted into files that conform to standard exchange formats (NMR-STAR for NMR data and mzML for MS/MS data). Of course, all chemical structural data in the HMDB is already in standard SDF, PDB and MOL formats, while all sequence data is in a standard FASTA format. The adoption of data exchange standards for all spectral, structural and sequence data should make data dumps and data downloads much easier. Finally, most of the HMDB data has been converted to an easily parsed XML format. This should make data downloads of the annotated database and the development of data extraction routines much simpler and far faster for programmers and database developers.
CONCLUSION
The HMDB is a comprehensive, web-accessible metabolomics database that brings together quantitative chemical, physical, clinical and biological data about all experimentally ‘detected’ and biologically ‘expected’ human metabolites. Over the past 3 years, a significant expansion to the content and a significant enhancement to the database’s capabilities have taken place. Many of these content additions and content corrections are the result of continued experimental and literature mining efforts by the HMDB curatorial and analytical staff. Likewise, many of the graphical interface and spectral viewing/downloading improvements, which arose primarily from external user suggestions, are the result of significant programming efforts by the HMDB software development team. Overall, we believe these improvements should make the HMDB much more useful to a much wider collection of metabolomics and clinical researchers. This report is certainly not the final word on the human metabolome, nor is it the final version of the HMDB. What this particular release of the HMDB provides is a ‘snapshot’ of the human metabolome as of 1 January 2013. No doubt the size of the human metabolome will continue to grow as will the need for additional resources or additional data fields to handle the ever-expanding needs of the community that the HMDB serves.
FUNDING
Genome Alberta (a division of Genome Canada); Canadian Institutes of Health Research (CIHR); Alberta Ingenuity Centre for Machine Learning (AICML); Alberta Innovates BioSolutions (AIBS). Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pearson H. Meet the human metabolome. Nature. 2007;446:8. doi: 10.1038/446008a. [DOI] [PubMed] [Google Scholar]
- 2. The Human Metabolome Project website http://www.metabolomics.ca/ (29 August 2012, date last accessed)
- 3.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 6.Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M, Karp PD. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BØ. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl Acad. Sci. USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, Cheng T, Wang Y, Bryant SH. PubChem as a public resource for drug discovery. Drug Discov. Today. 2010;15:1052–1057. doi: 10.1016/j.drudis.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 14.Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, Guo AC, et al. SMPDB: the small molecule pathway database. Nucleic Acids Res. 2010;38:D480–D487. doi: 10.1093/nar/gkp1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandal R, Guo AC, Chaudhary KK, Liu P, Yallou FS, Dong E, Aziat F, Wishart DS. Multi-platform characterization of the human cerebrospinal fluid metabolome: a comprehensive and quantitative update. Genome Med. 2012;4:38. doi: 10.1186/gm337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, et al. The human serum metabolome. PLoS One. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stretch C, Eastman T, Mandal R, Eisner R, Wishart DS, Mourtzakis M, Prado CM, Damaraju S, Ball RO, Greiner R, et al. Prediction of skeletal muscle and fat mass in patients with advanced cancer using a metabolomic approach. J. Nutr. 2012;142:14–21. doi: 10.3945/jn.111.147751. [DOI] [PubMed] [Google Scholar]
- 19.Takeda I, Stretch C, Barnaby P, Bhatnager K, Rankin K, Fu H, Weljie A, Jha N, Slupsky C. Understanding the human salivary metabolome. NMR Biomed. 2009;22:577–584. doi: 10.1002/nbm.1369. [DOI] [PubMed] [Google Scholar]
- 20.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 21.Deutsch E. mzML: a single, unifying data format for mass spectrometer output. Proteomics. 2008;8:2776–2777. doi: 10.1002/pmic.200890049. [DOI] [PubMed] [Google Scholar]