Abstract
ZiFDB (Zinc Finger Database, http://zifdb.msi.umn.edu) is a web-accessible database that houses information on individual C2H2 zinc fingers (ZFs) and engineered zinc finger arrays (ZFAs). ZiFDB serves as a resource for biologists interested in engineering ZFAs for use as sequence-specific DNA-binding reagents. Here, we describe four new features of ZiFDB: (i) the database allows users to input new ZFs and ZFAs; (ii) a shadow database temporarily stores user-submitted data, pending approval by the database curator and subsequent loading into the persistent database; (iii) ZiFDB contains 181 Context-Dependent Assembly (CoDA) ZFAs, which were generated by this newly described ZFA engineering platform; and (iv) the database also now contains 319 F1F2 CoDA units and 334 F2F3 CoDA units that can be used to construct CoDA arrays. In total, the new release of ZiFDB contains 1226 ZFs and 1123 ZFAs.
INTRODUCTION
The C2H2 zinc finger (ZF) motif, which was first described in the transcription factor TFIIIA from Xenopus laevis (1), is one of the most abundant DNA-binding motifs in nature. Each ZF comprises about 30 amino acids that fold into a ßßα structure through hydrophobic interactions and binding of a zinc ion by two conserved cysteine and histidine residues. A ZF typically recognizes a continuous 3-bp DNA sequence. Owing to this DNA-binding capacity, ZFs serve as a framework for constructing engineered DNA-binding proteins: ZFs are linked in tandem to form zinc finger arrays (ZFAs), which can recognize extended DNA sequences (2,3).
The methods used to engineer ZFAs to recognize novel target sequences can be classified into two categories: modular assembly and selection-based methods. Modular assembly simply involves linking together ZFs that recognize known target sequences (3). However, modular assembly is not necessarily reliable. For example, in one study, more than half of the ZFAs created by modular assembly showed little or no activity (4). This is because specificity and affinity of a given finger is influenced by context—i.e. the position of a finger in an array and its neighboring fingers. Selection-based methods identify fingers that work well together and are thus more reliable for producing functional ZFAs; however, selection-based methods often require considerable time and a high level of molecular biology expertise to perform. In 2008, the Zinc Finger Consortium implemented a selection-based platform called Oligomerized Pool Engineering (OPEN) (5). Since its debut, >500 ZFAs have been generated by OPEN [558 are housed in Zinc Finger Database (ZiFDB) v2.0].
Despite the high efficacy of OPEN, the labor and expertise required by this method have prevented it from being widely used. With the goal of combining the simplicity of modular assembly with the reliability of OPEN, another publicly available platform, named Context-Dependent Assembly (CoDA), was described in 2011 by the Zinc Finger Consortium (6). To assemble CoDA ZFAs, two, two-finger units, derived through selection, are used that have a common ZF at position two (F2). For example, an F1F2 CoDA unit (recognizing 3′-GAGGGG) is fused to an F2F3 CoDA unit (recognizing 3′-GGGGTG) such that the resulting three-finger array recognizes a novel DNA sequence (3′-GAGGGGGTG). Although Moore et al. (7) showed that the activity of CoDA zinc finger nucleases (ZFNs) is lower than those made by OPEN, CoDA does not require selection steps and is therefore easier for most researchers to use.
Regardless of the method used to construct ZFAs, these engineered DNA-binding domains have become powerful tools for both basic and applied biological research. Engineered zinc finger transcription factors, for example, can be created by fusing transcriptional repressor or activator domains to engineered ZFAs. These artificial transcription factors have been used to repress or activate genes in a variety of species with a high degree of specificity (8,9). Similarly, engineered ZFNs have proven effective as targeted mutagens in diverse eukaryotes (10,11). ZFNs are typically composed of a customized array of ZFs fused to the non-specific Fok1 restriction endonuclease cleavage domain (12). As Fok1 needs to dimerize to be functional, ZFAs are designed in pairs to recognize two unique DNA sequences separated by a short DNA spacer. Binding to the two-target sequences allows Fok1 to dimerize, cut the DNA and introduce a double-stranded break in the spacer. When double-stranded breaks are repaired through non-homologous end-joining or homologous recombination, targeted sequence modifications can be introduced at or near the break site.
ZiFDB is designed to serve as a resource for those interested in engineering custom ZFAs or better understanding how ZF proteins recognize target DNA (13). In addition to housing information on ZFs and engineered ZFAs, ZiFDB is linked to the output from ZiFiT—a software package that assists biologists in finding sites within target genes for engineering ZF proteins (14). Consequently, ZiFDB is particularly valuable for determining whether a given ZFA (or portion thereof) has previously been constructed and whether it has the requisite DNA-binding activity for a given experiment. ZiFDB v2.0 houses an expanded number of ZFAs, from 652 to 1123. Likewise, the number of ZFs has expanded from 716 to 1126. The updated database also allows users to input ZFAs into a shadow database, which are then deposited into the persistent database by the curator on approval. The information in this database will continue to help molecular biologists develop ZF reagents that meet their needs for genome modification.
NEW FEATURES
Shadow database
ZiFDB v2.0 allows users to directly input into the database new information about novel ZFAs. To ensure data quality, a shadow database has been created. The information submitted by the user is collected into the shadow database and held pending approval by the database curator. On approval, the information is then loaded into the persistent database. The information collected into the shadow database includes array name and array type (e.g. if it was derived by OPEN, CoDA or modular assembly), the sequence of the targeted DNA triplets, the sequences of the recognition helices of the ZFs, journal information if the array has been published and information about the submitter (see below for additional details).
New database content
Classes
ZiFDB stores ZF information as a set of objects defined by Java classes. The previous version of ZiFDB had four major classes: Zinc Finger, Zinc Finger Array, Article and Author. Two additional classes were added to this release of ZiFDB to accommodate the two-finger CoDA units (designated as CoDA_F1F2 and CoDA_F2F3). Similar to the Zinc Finger and Zinc Finger Array class designations, both CoDA_F1F2 and CoDA_F2F3 point to the Article class, which provides information about relevant publications. Information about submitters of the new arrays is integrated into ZiFDB’s existing Author class.
Novel three-finger arrays and CoDA units
Since the release of the previous version of ZiFDB, OPEN has been used to generate numerous additional ZFAs. More than 500 OPEN ZFAs are now housed in ZiFDB. CoDA arrays are a new array type in this version of ZiFDB, and the database currently has 181 CoDA arrays. ZiFDB v2.0 also has information about 319 F1F2 CoDA units and 334 F2F3 units.
Updated interface
Search page for CoDA units
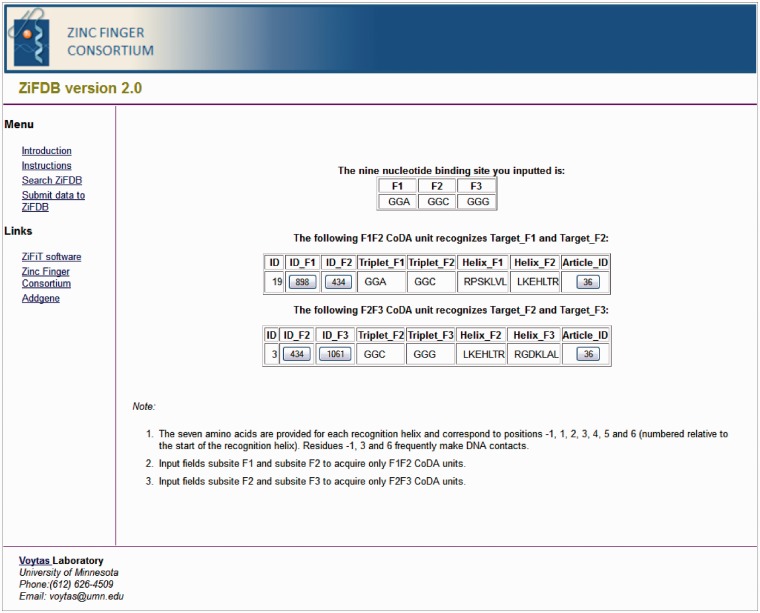
In addition to the ability to search for individual ZFs and three-finger ZFAs, ZiFDB v2.0 allows users to search for all available CoDA units that recognize a given target sequence. By providing the DNA sequence of the F1F2 or F2F3 triplets to be targeted, all of the corresponding CoDA units matching the input sequence are returned. If the user provides the nucleotide triplets for a 9-bp target site, information is provided about both F1F2 and F2F3 units (Figure 1).
Figure 1.
Sample output from the CoDA unit search page when GGA is provided as triplet F1, GGC as triplet F2 and GGG as triplet F3.
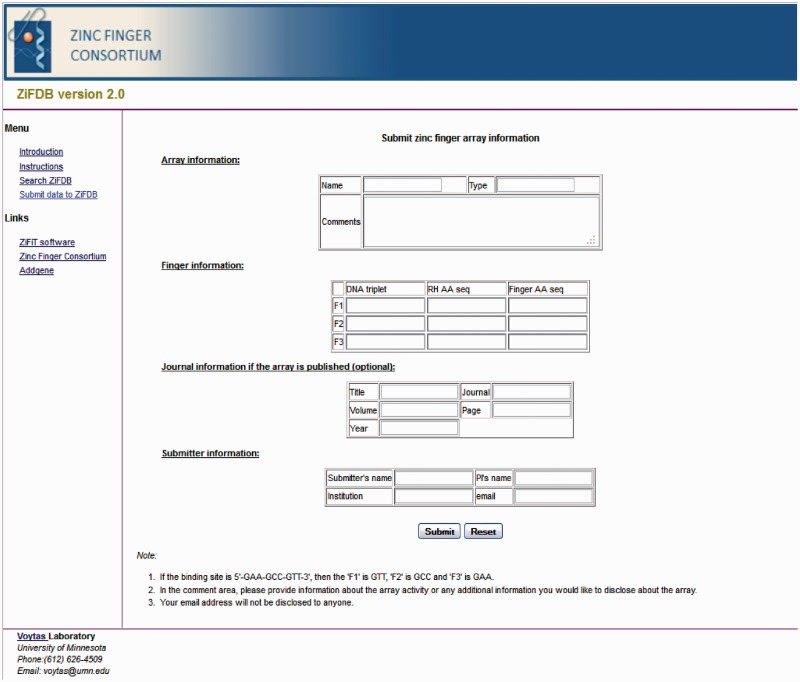
Page for submitting arrays
The array submission page collects four types of information (Figure 2):
Array information. The user can input the name given to a particular ZFA and the array type. As indicated above, the array type specifies the method used to engineer the ZFA (i.e. modular assembly, OPEN or CoDA). It is also possible to designate an array as natural (i.e. found in nature). The option remains to include engineering methods that may be developed at a later date. The user can also provide other useful information such as data concerning array activity.
Finger information. In this section, the DNA triplet, amino acid sequence of the recognition helix and the amino acid sequence of the entire ZF are provided for each finger in a ZFA.
Journal information. If the array has been published, then the citation is provided.
Submitter information. In this section, the name of the submitter and/or principal investigator, the name of the submitter’s institution and an email address are provided. This information is used by the database curator to contact the submitter if there are any questions concerning the data. No information about the submitter is publicly disclosed.
Figure 2.
The array submission page.
Newly added features and attributes in the array query output page
In the output page of the array query, a search link for CoDA units is provided. This makes it convenient for the user to check whether CoDA units are available for assembling ZFAs that target the 9-bp DNA sequence they provided. Another newly added attribute of the array query output is the array type. This currently includes natural, modular assembly, OPEN and CoDA. As the success rate and efficacy of ZFAs engineered by different platforms differ considerably, this information may be valuable for users when choosing previously engineered arrays to use in their experiments.
CONCLUSION
One important new feature in this version of ZiFDB is the added ability of users to input ZFAs and ZFs into the database. This will ensure that the ZiFDB captures new information generated by the scientific community, including unpublished data. In addition, by now housing information about two-finger CoDA units and CoDA arrays, ZiFDB is current with the most recent ZFA engineering practices. Finally, the expanded number of ZFs and ZFAs that are now stored in ZiFDB will provide a rich resource for users interested in either ZFA engineering or better understanding how ZFs recognize their target DNA.
FUNDING
Funding for open access charge: National Science Foundation [DBI-0923827 to D.F.V.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 3.Segal DJ, Beerli RR, Blancafort P, Dreier B, Effertz K, Huber A, Koksch B, Lund CV, Magnenat L, Valente D, et al. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–2148. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 4.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, Khayter C, Ramirez CL, Joung JK, Langenau DM. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urnov FD, Rebar EJ. Designed transcription factors as tools for therapeutics and functional genomics. Biochem. Pharmacol. 2002;64:919–923. doi: 10.1016/s0006-2952(02)01150-4. [DOI] [PubMed] [Google Scholar]
- 9.Blancafort P, Segal DJ, Barbas CF., III Designing transcription factor architectures for drug discovery. Mol. Pharmacol. 2004;66:1361–1371. doi: 10.1124/mol.104.002758. [DOI] [PubMed] [Google Scholar]
- 10.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 12.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu F, Sander JD, Maeder M, Thibodeau-Beganny S, Joung JK, Dobbs D, Miller L, Voytas DF. Zinc Finger Database (ZiFDB): a repository for information on C2H2 zinc fingers and engineered zinc-finger arrays. Nucleic Acids Res. 2009;37:D279–D283. doi: 10.1093/nar/gkn606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]