Abstract
Much has changed in the last two years at DGVa (http://www.ebi.ac.uk/dgva) and dbVar (http://www.ncbi.nlm.nih.gov/dbvar). We are now processing direct submissions rather than only curating data from the literature and our joint study catalog includes data from over 100 studies in 11 organisms. Studies from human dominate with data from control and case populations, tumor samples as well as three large curated studies derived from multiple sources. During the processing of these data, we have made improvements to our data model, submission process and data representation. Additionally, we have made significant improvements in providing access to these data via web and FTP interfaces.
INTRODUCTION
Genomic structural variation (GSV) comprises rearrangement events ranging in size from tens to millions of base pairs in size and includes insertions, deletions, inversions, translocations, locus copy number changes and is seen in a diverse class of taxa (2–4). The discovery and characterization of GSV is challenging for a number of reasons (5). A major difficulty in representing these types of variants is obtaining breakpoint resolution of these events. Studies based on microarray technology provide information about sequences involved in variation events, but only a rough estimate of the location of the breakpoints. Current sequencing technology can occasionally provide breakpoint resolution, but often there is a degree of uncertainty about the precise breakpoint location. The variability in the size and type of events that can be detected using a given technology and analysis method underscores the importance of robustly capturing as much experimental information as possible when recording GSV(6).
The European Bioinformatics Institute (EBI) and the National Center for Biotechnology Information (NCBI) maintain permanent public repositories, DGVa (http://www.ebi.ac.uk/dgva) and dbVar (http://www.ncbi.nlm.nih.gov/dbvar), respectively. Both resources provide archival, data accessioning and distribution services for all types of GSV in all species. Together, these archives represent the most comprehensive source of GSV in the world and include data originating from the 1000 Genomes project (estd59 and estd199) (7), The Wellcome Trust Sanger Institute Mouse Genomes (estd118) (8), COSMIC project (estd192) (9) and from numerous clinical genetics studies (e.g. nstd37 and nstd54) (10,11) (Figure 1). Data are submitted to these archives using a standard format that captures the methodology used for calling and validating GSV in individual samples, for aggregating data and representing breakpoint ambiguity. The archives also use Sequence Ontology terms (12) to describe GSV types and associated phenotypic information. The archives exchange data with one another regularly and release them to the scientific community using standard data formats on a monthly basis.
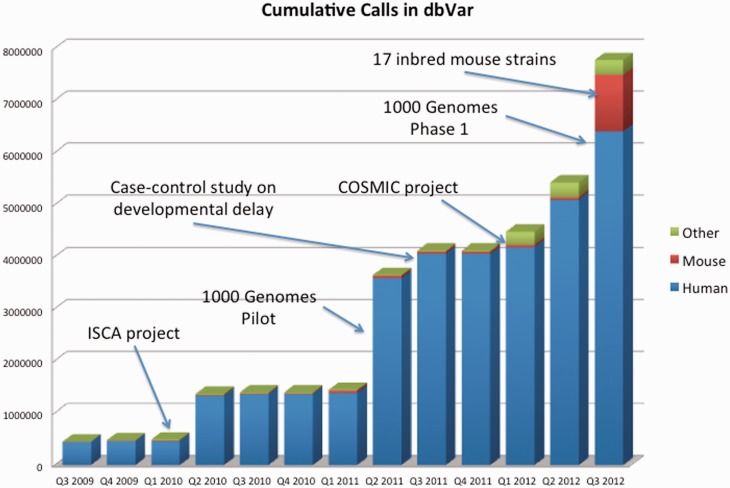
Figure 1.
The data growth since DGVa and dbVar services was launched. The graph shows accumulation of variant calls, stratified by organism. Several large datasets such as the 1000 Genomes project pilot (estd59) and phase I (estd199), structural variation data from 17 in-bred mouse strains (estd118) and the first releases of somatic structural variation from the COSMIC database (estd192), case-control and case-only studies on developmental delay (nstd54) and the International Standard Cytogenetic Array (ISCA) consortium data (nstd37). In addition to human and mouse data the archives include data from dog, pig, fruit fly, macaque, cow, horse, zebrafish, sorghum and chimp.
SHARED DATA REPRESENTATION
The DGVa and dbVar share a data model that is designed to capture and describe the complexity of GSV discovery, validation and genotyping experiments and provides accession numbers for three types of object: the study, the variant region and the variant call. This model allows the representation of a variant region based on the evidence of variation observed in one or more individual samples (the variant calls). The association between calls and regions is made by an assertion method that describes the basis for defining the GSV region. For example, a region might be defined by the set of variant calls overlapping one another by 90% (Figure 2). Variant call and region types are described using Sequence Ontology terms (Table 1).
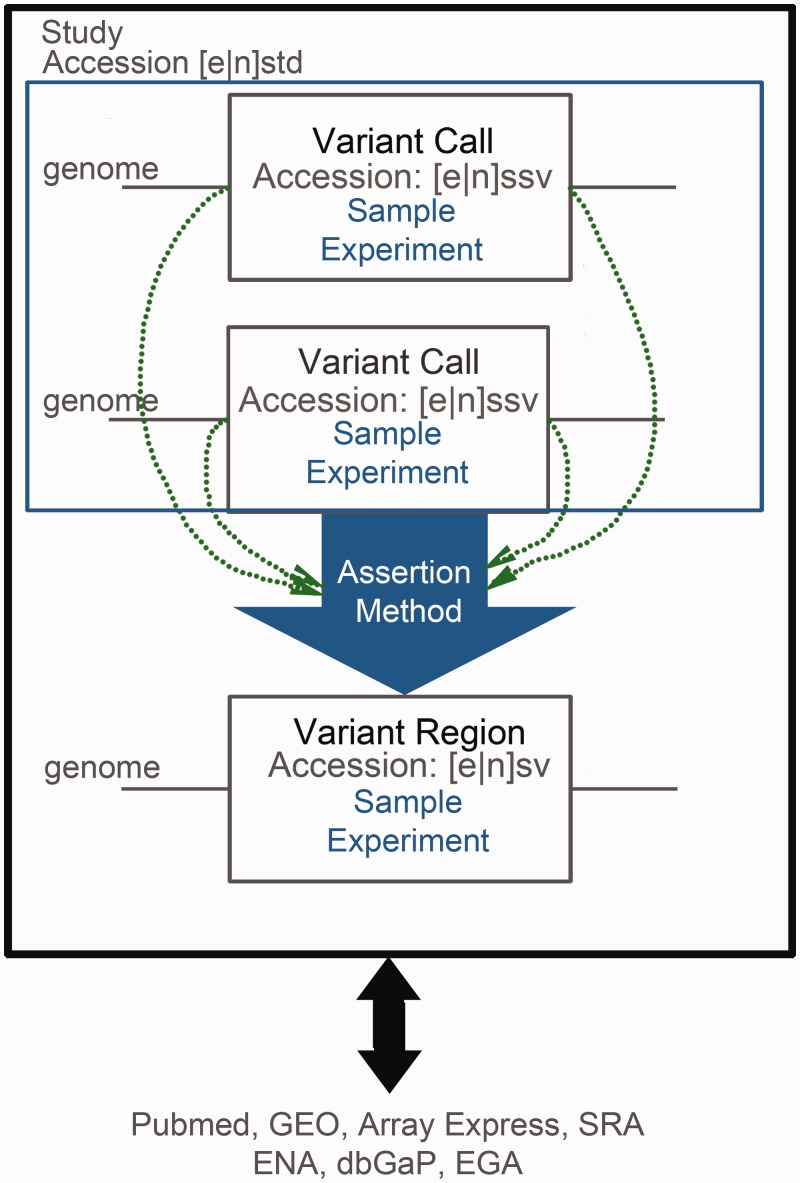
Figure 2.
Graphical representation of the archive data model. The three accessioned objects (studies, calls and regions) are prefixed by an ‘n’ if submitted to dbVar and an ‘e’ if submitted to DGVa. Variation in individual sample genomes is aggregated to a variant region, with respect to a reference genome. Genomic position (indicated by green arrows) does not necessarily overlap completely. Study authors describe the aggregation process in the Assertion method attribute. Discovery and validation methods for each call are stored in the Experiment attribute. This facilitates cross-study analysis of GSV identified using different techniques. Studies point to any external resources that provide access to the raw data used in the experiment or to the publication describing the data.
Table 1.
Variant call types and variant region types
| Variant call type | Associated variant region type |
|---|---|
| Copy number gain | CNV |
| Copy number loss | CNV |
| Deletion | CNV |
| Duplication | CNV |
| Insertion | Insertion |
| Mobile element insertion | Mobile element insertion |
| Novel sequence insertion | Novel sequence insertion |
| Tandem duplication | Tandem duplication |
| Translocation | Translocation |
| Interchromosomal breakpoint | Interchromosomal breakpoint |
| Intrachromosomal breakpoint | Intraschromosomal breakpoint |
| Complex | Complex |
| Unknown | Unknown |
The complex region type can be used for any region where calls of different type (other than CNV) have been called and aggregated into a region by the user. CNV = Copy Number Variation.
Variant calls have a number of associated attributes including the details of the sample(s) or sample set(s) details in which the variation was observed as well as the experimental procedure involved in discovery and/or validation. Combinations of variant call, sample and experiment are unique. Thus, a GSV identified by two different methods, for example, might result in the creation of two separate variant call objects.
The data model accommodates the breakpoint ambiguity associated with a range of experimental and analysis protocols. Three sets of coordinate identifiers are available: start-stop, inner start-stop and outer start-stop. Traditional start and stop coordinates can be used alone to describe variants in which base pair resolution has been achieved. When used in conjunction with the inner and outer coordinate system, the same coordinates allow users to represent an estimated start and stop along with a confidence interval, thus matching the common output of many techniques using next-generation sequencing (NGS) methods. Finally, only inner and/or outer coordinates alone may be used in cases where no start is estimated, as is often the case with array-based techniques, with the inner start and stop defining the region known to be contained within the GSV and the outer start and stop used to define the region likely to contain the breakpoints. All coordinates must be associated with a genome assembly that has been submitted to an International Nucleotide Sequence Database Collaboration (INSDC) database (13). In cases where novel sequence has been identified and genomic coordinates cannot be determined, these novel sequences should be submitted to an INDSC database where it will receive an accession; this identifier can then be referenced by the variant call.
Phenotype information can be associated with samples or sample sets using any of a number of controlled vocabularies, including the Human Phenotype Ontology (14). Our data model also supports assertions of clinical significance to a variant calls to provide explicit links between causative alleles and phenotypes.
DATA SUBMISSION AND RELEASE FROM THE ARCHIVES
Both archives use a common set of well-defined tab delimited files that can be created using Excel to facilitate submission. The submission template collates all the information required to represent the submitter-asserted GSV within the study. The DGVa and dbVar do not store raw data from array-based assays or sequencing experiments; however, submitters are encouraged to pre-submit raw data to a dedicated EBI or NCBI database. Accession numbers from these deposits should be included with the DGVa/dbVar submission. More information about the submission template, including up-to-date guidelines and instructions for accessing the dedicated help-desks, are available on the DGVa and dbVar websites.
Submitted data are processed by the archive that received the initial submission. Processing protocols are shared by both archives and enforce validation rules that aim to ensure data quality and integrity. Once data pass quality control the processing archive issues stable identifiers for the study, all variant calls and regions; these data are then exchanged between archives. Synchronized and timely public release from both databases is the goal and public release can be adjusted to fit with the manuscript publication timelines. The archives support both pre-publication data release, in accordance with the Toronto agreement (15), and data release delayed until publication when requested by the submitters.
Data are made available to the public in Genome Variation Format (GVF) (16) from both archives. A GVF file for each taxonomic name and assembly in a given study can be downloaded; in addition, separate files for germline and somatic mutations, and also for cases where dbVar has remapped submitted data to a more recent version of the assembly are available. dbVar also provides data as tab delimited files and XML format.
ACCESS TO THE DATA THROUGH dbVar WEBSITE
Users can navigate to particular studies using our Study Browser (http://www.ncbi.nlm.nih.gov/dbvar/studies), or they can perform text-based searches using the standard NCBI Entrez search interface (17). Searching for gene symbols or phenotype terms will provide information on studies and variant regions associated with the search query. Users who search by location, either by providing a cytogenetic coordinate or a chromosome location (in the form chr1: start–stop), will be redirected to the dbVar Genome Browser (see below).
Study records provide global information about the study type, variant calls and regions, the samples used, the experimental details as well as any validation experiments performed as part of the study. Publication information for the study is shown as are links to external resources such as OMIM®, dbGaP and submitter resources.
Every submitted variant region is given a dedicated page providing a detailed view of the region. An overview of the variant region is shown at the top, while detailed information is provided below. The detailed information is segregated into labeled tabs. The ‘Genome View’ tab provides a graphical representation of the region in the context of other genome features such as genes. Breakpoint ambiguity, as denoted by endpoint triangles or by translucent color (Figure 3a), and variant call and region type information distinguished by shape and color (Figure 3b), are available in this view. Summary data about overlapping variant regions are available in this tab, with a link to the genome browser that will allow users to browse data from additional studies. Detailed placement information for both the variant calls and regions are shown in the ‘Variant Region Details and Evidence’ tab. Variant calls are also explicitly associated with samples and experimental data in this tab. If there are additional variant calls from a sample, a link is provided so that it is easy to see all calls from a given sample for this study. Additionally, NCBI maps features from submitted assemblies to the current reference assemblies when possible and provides access to all genomic contexts in this tab. Validation information for any calls in this region are available in the ‘Validations’ tab. Detailed information concerning any clinical assertions are in the ‘Clinical Assertions’ tab. While we have a tab reserved for Genotype Information, this is not yet populated. We anticipate adding these data this year, starting with genotype data from the 1000 Genomes project.
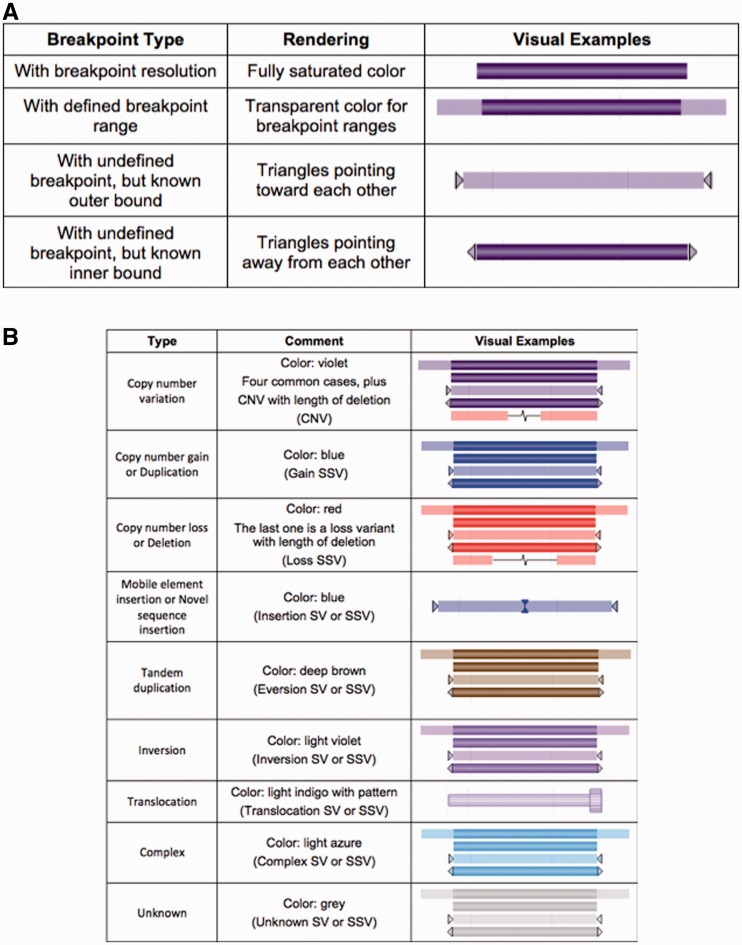
Figure 3.
Rendering of breakpoint ambiguity (A) is shown. Variants with breakpoint resolution are shown with fully saturated color. Breakpoints defining by a range (using inner/outer starts and stops) are shown as fully saturated for the high confidence intervals (the regions defined by the inner start-stop) while the region of breakpoint ambiguity is shown as transparent. In many cases, an undefined breakpoint is submitted, but no likelihood range is provided; in these cases triangles pointing towards each other (when only outer coordinates are provided) or pointing out (when inner coordinates are provided). Rendering call and region type (B) is usually designated by color. SV corresponds to variant region and SSV corresponds to variant calls.
We recently introduced a genome browser to facilitate the graphical view of multiple studies side by side. This viewer also provides access to other genome information such as assembly information, NCBI gene annotation and SNP data, including access to clinically relevant SNPs (in the ‘Clinical Channel’ track) and SNPs that are associated with publications (in the ‘Cited Variants’ track). The top of the page contains information on chromosome location and provides functions for navigating around the genome. A graphical sequence viewer showing annotated features dominates the page. The left-hand column provides a genome overview and navigation widget, a menu for selecting available assemblies, a search function (users can perform term searches or location searches) and information on studies that have data available in the given region. Users can click on the ‘(+)’ or ‘(−)’ to add or remove particular study tracks to the graphical view.
INTEGRATION OF DGVa DATA TO OTHER PUBLIC RESOURCES
The DGVa provides human data to the Database of Genomic Variants (DGV), available from the University of Toronto (18). Utilizing the range of supplied variant properties, DGV merges data of differing qualities, derived using different methodologies to form a high-quality curated reference set of ‘normal’ GSV in humans. The DGV also shows human data from DGVa where samples carry a disease phenotype as separate tracks in the DGV genome browser.
All DGVa archived data are provided to Ensembl, which has developed new ways to visualize GSV data in the genome browser (19). Ensembl uses the same Sequence Ontology terms for the variant classes as DGVa and breakpoint ambiguity is shown using a similar methodology to that applied by dbVar. The GSV can be viewed not only alongside the reference sequence but also against a wealth of other information that includes SNPs and somatic variation, genes and transcripts, mRNA and protein alignments, ncRNAs and regulatory features. The integration of GSV data into such a rich set of genomic annotation provides an extremely powerful tool for elucidating the biological consequences of GSV. All GSV data are integrated as part of the Variant Effect Predictor to provide the variant consequence types for each transcript (20). Ensembl also provides programmatic access to DGVa accessioned variants allowing data from multiple studies to be compared, integrated and analyzed together in novel ways. DGVa data are also made available through Ensembl BioMart to facilitate data mining and integration across all studies and species for researchers without programmatic access.
FUTURE DIRECTIONS
The wealth of GSV information continues to expand both in terms of sheer volume and the nature of associated attributes that are captured. Increasingly these data are accompanied by genotype, phenotype or clinical information, which provides foundation for understanding phenomena such as segregation and variation diversity within populations and in understanding the biological significance of GSV. The data model used by DGVa and dbVar allows for an effective representation of the richness and complexity of GSV information that will be crucial in providing a basis with which to move forward in future integration and analyses.
FUNDING
The Intramural Research Program of the National Institutes of Health, National Library of Medicine for the work on dbVar; the Wellcome Trust (grant number WT084107MA) and by the European Molecular Biology Laboratory for the DGVa. Funding for open access charge: NCBI.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Donna Maglott, Alex Astashyn, Michael DiCuccio, Liangshou Wu and Anatoliy Kuznetsov for internal support for dbVar; Ewan Birney and Jonathan Hinton for early work and support to DGVa; Fiona Cunningham and Laurent Gill for Ensembl collaboration, Simon Forbes for COSMIC collaboration; Steve Scherer, Margie Manker and Lars Feuk for helpful discussions.
REFERENCES
- 1.Church DM, Lappalainen I, Sneddon TP, Hinton J, Maguire M, Lopez J, Garner J, Paschall J, DiCuccio M, Yaschenko E, et al. Public data archives for genomic structural variation. Nat. Genet. 2010;42:813–814. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.She X, Cheng Z, Zöllner S, Church DM, Eichler EE. Mouse segmental duplication and copy number variation. Nat. Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bickhart DM, Hou Y, Schroeder SG, Alkan C, Cardone MF, Matukumalli LK, Song J, Schnabel RD, Ventura M, Taylor JF, et al. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res. 2012;22:778–790. doi: 10.1101/gr.133967.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng L-Y, Guo X-S, He B, Sun L-J, Peng Y, Dong S-S, Liu T-F, Jiang S, Ramachandran S, Liu C-M, et al. Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor) Genome Biol. 2011;12:R114. doi: 10.1186/gb-2011-12-11-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbin R. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yalcin B, Wong K, Bhomra A, Goodson M, Keane TM, Adams DJ, Flint J. The fine-scale architecture of structural variants in 17 mouse genomes. Genome Biol. 2012;13:R18. doi: 10.1186/gb-2012-13-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, Moreno-De-Luca D, Moreno-De-Luca A, Mulle JG, Warren ST, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med. Off. J. Am. Coll. Med. Genet. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karsch-Mizrachi I, Nakamura Y, Cochrane G. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2012;40:D33–D37. doi: 10.1093/nar/gkr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson PN, Mundlos S. The human phenotype ontology. Clinical genetics. 2010;77:525–534. doi: 10.1111/j.1399-0004.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- 15.Birney E, Hudson TJ, Green ED, Gunter C, Eddy S, Rogers J, Harris JR, Ehrlich SD, Apweiler R, Austin CP, et al. Prepublication data sharing. Nature. 2009;461:168–170. doi: 10.1038/461168a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reese MG, Moore B, Batchelor C, Salas F, Cunningham F, Marth GT, Stein L, Flicek P, Yandell M, Eilbeck K. A standard variation file format for human genome sequences. Genome Biol. 2010;11:R88. doi: 10.1186/gb-2010-11-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;40:13–25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet. Genome Res. 2006;115:205–214. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
- 19.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]