Abstract
PlantRNA database (http://plantrna.ibmp.cnrs.fr/) compiles transfer RNA (tRNA) gene sequences retrieved from fully annotated plant nuclear, plastidial and mitochondrial genomes. The set of annotated tRNA gene sequences has been manually curated for maximum quality and confidence. The novelty of this database resides in the inclusion of biological information relevant to the function of all the tRNAs entered in the library. This includes 5′- and 3′-flanking sequences, A and B box sequences, region of transcription initiation and poly(T) transcription termination stretches, tRNA intron sequences, aminoacyl-tRNA synthetases and enzymes responsible for tRNA maturation and modification. Finally, data on mitochondrial import of nuclear-encoded tRNAs as well as the bibliome for the respective tRNAs and tRNA-binding proteins are also included. The current annotation concerns complete genomes from 11 organisms: five flowering plants (Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, Medicago truncatula and Brachypodium distachyon), a moss (Physcomitrella patens), two green algae (Chlamydomonas reinhardtii and Ostreococcus tauri), one glaucophyte (Cyanophora paradoxa), one brown alga (Ectocarpus siliculosus) and a pennate diatom (Phaeodactylum tricornutum). The database will be regularly updated and implemented with new plant genome annotations so as to provide extensive information on tRNA biology to the research community.
INTRODUCTION
Transfer RNAs (tRNAs) play essential roles in cell viability. Beyond their major function in translating the genetic code, tRNAs are implicated in many other processes such as viral replication, amino acid biosynthesis or cell wall remodeling (1–3). Eukaryotic genomes encode a complex population of tRNA genes and the expression of tRNA species is subject to a tight regulation in particular at the transcriptional level. The development and cell differentiation of tissues may be affected by the steady-state levels of certain tRNAs [e.g. (4,5)]. The existence of tRNA isodecoders, i.e. tRNAs sharing the same anticodon but having distinct body sequences, has also the potential to play important regulatory roles (6,7). Another major observation is that subcellular tRNA trafficking pathways within the cell, for instance between the cytosol and the nucleus or the cytosol and mitochondria, play a role in RNA quality control, stress response or organelle biogenesis. Finally, over the past 5 years, deep sequencing approaches have revealed the existence of new small non-coding RNAs originating from tRNAs and are thus called tRFs (for tRNA-derived RNA fragments). Some of these tRFs were shown to be induced in response to stress, during aging or to be involved in translation inhibition (8). The number of novel functions played by tRNAs or tRNA-derived fragments is increasing rapidly and the above list is not exhaustive. This is likely the visible part of the iceberg and further studies will necessitate easy access to the most accurate and complete set of data concerning tRNA gene populations of given eukaryotic organisms, as well as to relevant biological information such as upstream promoter regions, intron sequences, mitochondrial import or tRNA-related enzymes.
As photosynthetic organisms possess three compartments encoding genetic information (i.e. the nucleus, the chloroplast and the mitochondrion), they represent the most intricate and thus interesting models to obtain an integrative view of the tRNA gene population present within a eukaryotic cell in terms of gene content, organization, expression and function. In addition, the primary endosymbiosis of a cyanobacteria in a heterotrophic protist leading to the present-day chloroplast and the existence of secondary and tertiary endosymbiosis events leading to the great diversity of photosynthetic eukaryotes from algae to land plants represent landmark evolutionary events (9,10) that are worth to be analyzed at the tRNA gene level.
We thus decided to focus on tRNA genes from photosynthetic organisms. In 2000, the first complete sequence of a plant genome, that of Arabidopsis thaliana, was published (11). Since then, with the completion of several plant and algal genomes, we now have the opportunity to study their complete tRNA gene sets. Indeed, such an analysis was recently achieved for five flowering plants (12), but the data were not accessible online. So far, three major tRNA databases are available: (i) the Transfer RNA database (tRNAdb; http://trnadb.bioinf.uni-leipzig) (13), (ii) the tRNA Gene DataBase Curated by Experts (tRNADB-CE; http://trna.nagahama-i-bio.ac.jp) (14) and (iii) the Genomic tRNA Database (GtRNAdb; http://gtrnadb.ucsc.edu/) (15). The first database, tRNAdb, is a restructured version of the first compilation of tRNA and tRNA gene sequences (16) and contains >12 000 tRNA gene sequences from 577 organisms. This database is the only one that also provides 623 tRNA sequences from 104 organisms and thus offers interesting information on nucleotide modifications. However, concerning higher plants, only few sequences are available. For example, only 247 nuclear tRNA gene sequences mostly from A. thaliana are annotated and no full nuclear genome has been analyzed. In the second database, tRNADB-CE, the authors provide >287 000 tRNA gene sequences from various evolutionary divergent organisms. In particular, more than half of the sequences come from metagenome analyses of microorganisms from different environmental samples. This reliable database represents a nice tool to use tRNA gene sequences as genus-specific markers and study microbial population. tRNADB-CE also includes tRNA genes retrieved from 121 complete plastidial genomes, but nuclear tRNA genes from only two higher plant species (A. thaliana and Oryza sativa) are given. Neither mitochondrial tRNA genes nor nuclear tRNA genes from other photosynthetic eukaryotes can be retrieved. The third database, GtRNAdb, compiles tRNA gene sequences from various complete genomes thanks to the powerful tRNAscan-SE program (17) and 11 land plant genomes were analyzed. As mentioned by the authors, the database is not curated which results in the occurrence of high numbers of errors (e.g. it provides 639 tRNA gene for the Brachypodium distachyon or 738 for O. sativa while the accurate numbers are 479 and 516, respectively) and no tRNA genes from other types of photosynthetic eukaryotes are available. Thus, each of these tRNA databases offers complementary information. However, none of them is dedicated to photosynthetic organisms and only very partial sets of data on plant or algal tRNA genes are available through the three web interfaces. Here, the PlantRNA database brings together the information from 11 eukaryotes representative of evolutionary distinct branches of the photosynthetic lineage including brown and green algae, glaucophytes, bryophytes, flowering plants and diatoms. More than 4350 manually curated sequences of tRNA genes encoded by the nuclear, plastidial or mitochondrial genomes of these 11 species are accessible through the website. Biological information relevant to tRNA biology (e.g. intron sequences, flanking sequences controlling tRNA gene expression, mitochondrial tRNA import or tRNA-related enzyme genes) is also provided and will be presented below.
DATABASE CONTENT AND WEB INTERFACE
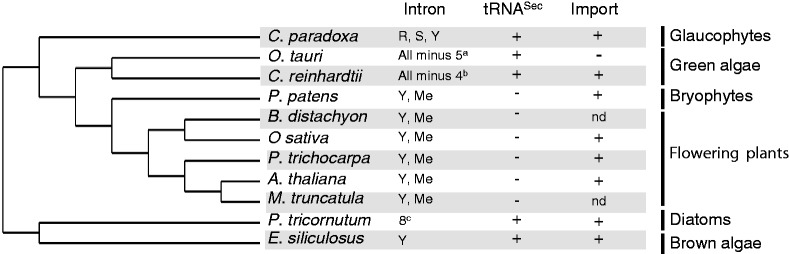
The organisms were selected based on two criteria: (i) the quality of their genomes annotation and (ii) their representativeness of evolutionary divergent branches of the photosynthetic lineage (Figure 1). From the manually curated lists of tRNA genes that we recently extracted from the genomes of five angiosperms (A. thaliana, O. sativa, Populus trichocarpa, Medicago truncatula and B. distachyon) and one green alga (Chlamydomonas reinhardtii) (12), we retrieved tRNA genes from the genomes of six other photosynthetic organisms, namely, another green alga (Ostreococcus tauri), a glaucophyte (Cyanophora paradoxa), a brown alga (Ectocarpus siliculosus) and a pennate diatom (Phaeodactylum tricornutum). Recently, the release of assembly v5 of the C. reinhardtii nuclear genome was available and the tRNA gene data were updated. Most sources of genome sequences are cited in (12) or available at http://bioinformatics.psb.ugent.be/webtools/bogas/ or http://www.phytozome.net/ (18). Whole nuclear, plastidial and mitochondrial genomes were scanned by tRNAscan-SE (17) and then manually annotated as described in (12). For each tRNA gene, the linear secondary structure as well as biological information are given. This includes (i) 5′- and 3′-flanking sequences that are involved in the control of gene expression or on polyT termination sequences, (ii) A and B boxes involved in TFIIIC transcription factor binding and (iii) intron sequences. In addition, two other levels of information are provided. The first concerns the subcellular localization of each tRNA. Mitochondrial tRNA import is a widespread process, in particular in the plant kingdom (19,20). It is thus relevant, either based on experimental data or on prediction to provide the scientific community with the mitochondrial import status of nuclear-encoded tRNA species. Second, information on the population of enzymes related to tRNA biogenesis and function is given. This is particularly true for A. thaliana where >150 enzymes were identified [e.g. (21–23)]. In addition, even though the annotation of whole genomes is still largely incomplete, we also provide information on tRNA-related enzyme genes for six of the other photosynthetic species (including bryophyte, green and brown algae and diatom) present in the PlantRNA database. This information includes accession numbers corresponding to genes encoding enzymes involved in maturation steps such as 5′ and 3′ processing, CCA addition and genes coding for aminoacyl-tRNA synthetases. Due to the importance of the dual-targeting phenomenon for proteins involved in translation in plants, we also put strong effort in providing the subcellular localization of these enzymes.
Figure 1.
Phylogenetic tree of photosynthetic organisms found in the PlantRNA database. The phylogenetic tree was constructed with the full-length 18S rRNA gene sequences using the neighbor-joining method. The presence of intron-containing tRNA genes, of genomes encoding a tRNASec and the occurrence of tRNA mitochondrial import are reported. aMinus 5 = A, E, Mi, V, W; bminus 4 = Q, H, Mi, F; c8 = R, E, Q, Me, P, S, T, W, Y. The one-letter code for amino acids is used.
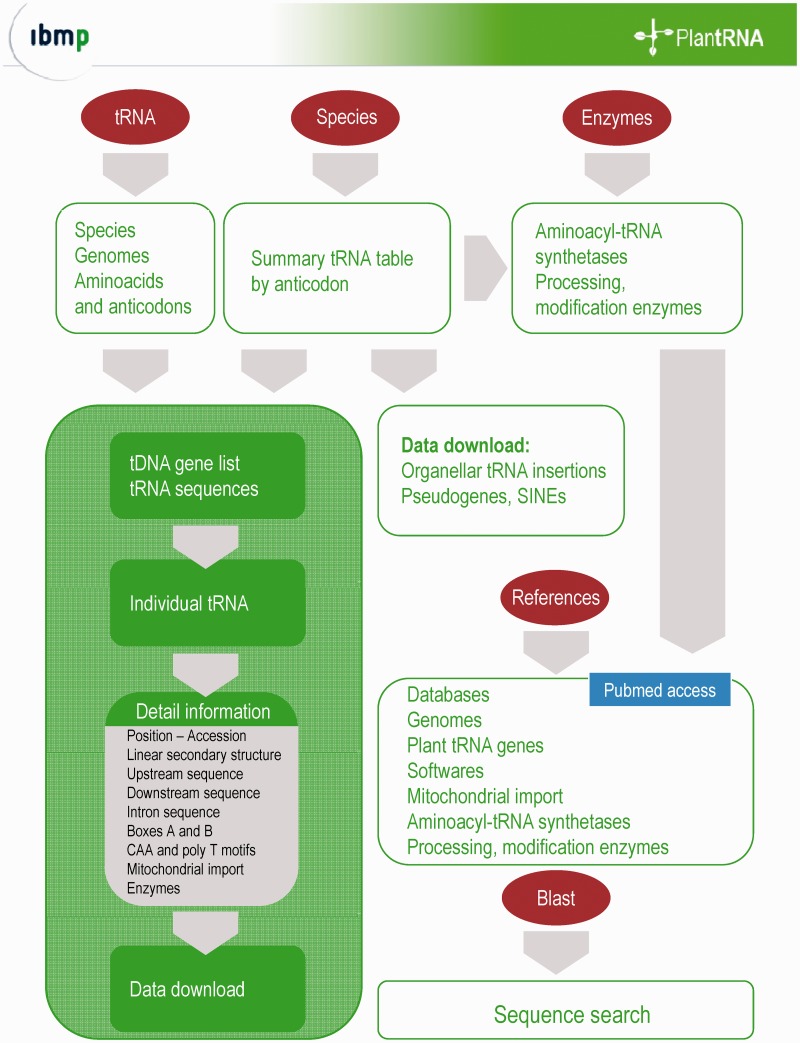
All sequences and biological datasets are stored in a database implemented in MySQL version 5 (http://dev.mysql.com). The MySQL database is structured into 32 normalized tables. The querying of underlying SQL database is implemented using Java servlets running on Apache Tomcat server. As shown in Figure 2, different search forms are available through the homepage. The entry point ‘tRNA’ allows searching by organism, genome (nuclear, plastidial and/or mitochondrial), amino acid and anticodon and gives access to tRNA gene lists. The entry point ‘Species’ allows access to global information (including number of tRNA genes, presence of suppressor or selenocysteine tRNA genes). A summary tRNA table provides access to tRNA gene lists and individual biological data. For each tRNA gene of a tRNA gene list, detailed information is available. This includes organism, chromosome, position, tRNA type, anticodon, upstream and downstream sequences, intron sequences and mitochondrial import. tRNA gene sequences and optional information can be downloaded in xls or FASTA file formats. In addition, a SQL dump of the database is available upon request. Although the database is focused on true tRNA genes (by means of manual curation), we annotated some of the sequences as pseudogenes. We also annotated numerous non-expressed mitochondrial or plastidial tRNA gene sequences inserted into nuclear genomes and inadequately recognized by tRNAscan-SE as true tRNA genes. These sequences as well as the sequences of previously identified short interspersed tRNA-related elements (12,24) can also be downloaded in xls or FASTA file formats. The entry point ‘Enzymes’ allows access to a list of aminoacyl-tRNA synthetases and processing or modification enzymes alongside with their accession numbers, subcellular localization (experimentally validated or predicted by appropriate organellar targeting prediction programs such as Predotar or TargetP (25,26)). Finally, a blast search entry is available and key references are provided.
Figure 2.
PlantRNA database: overview of the web interface and basic functions.
RESULTS AND DISCUSSION
In total, 3821, 368 and 186 tRNA genes were registered from the nuclear, plastidial and mitochondrial genomes, respectively, of the 11 photosynthetic organisms (note that the mitochondrial DNA sequences of P. trichocarpa, M. truncatula and B. distachyon are not available). The usefulness of PlantRNA implementation is illustrated below by a non-exhaustive list of data that can be retrieved from its web interface.
First, the selection of intron-containing tRNA genes shows that all photosynthetic organisms studied here possess tRNA genes with introns but their number and identity greatly vary (Figure 1). The same two families of nuclear tRNA genes (tRNATyr gene and elongator tRNAMet gene) contain intronic sequences (between positions 37 and 38 of their tRNA sequences) in flowering plants and in the bryophyte Physcomitrella patens, while many more tRNA genes contain introns in the two green algae. In the two stramenopiles (diatom and brown alga), tRNA genes corresponding to eight amino acids possess introns in P. tricornutum, while only the family of tRNATyr gene contains intron sequences in E. siliculosus, thus demonstrating the independent acquisition of intron sequences among phylogenetically related species. Interestingly, it is worth to note that in all photosynthetic organisms, intron sequences are always found in tRNATyr gene. This intron acquisition thus very likely occurred before the divergence between plants and metazoans. In human, tRNATyr belongs to the very rare human intron-containing tRNAs (27) and is essential for the presence of a pseudouridine residue at position 35 of the anticodon (28).
Second, another evolutionary interesting aspect is the presence of a tRNASec among a eukaryotic genome. The presence of selenoproteins is not restricted to the animal kingdom and the occurrence of Sec-containing proteins was reported in algae such as Chlamydomonas or in diatoms (29,30) while higher plants lost the ability to synthesize seleno-containing proteins and no tRNASec is present (Figure 1). However, due to its unusual secondary structure, tRNASec is not often retrieved from genome sequences during genome annotation by tRNAscan-SE using default parameters. For example, while selenoproteins are found in stramenopiles (30–32), no tRNASec gene sequence had yet been annotated either in the diatom P. tricornutum or in the brown alga E. siliculosus nuclear genomes. Here, the tRNASec sequences from these two organisms were identified and added to the database, thus confirming the maintenance of a selenocysteine pathway in evolutionary divergent algae and glaucophytes.
Third, mitochondria from different eukaryotic organisms either have a limited set of tRNA genes or no tRNA genes at all. To compensate this deficiency, mitochondrial import of a variable number of nucleus-encoded tRNAs has been demonstrated in several organisms (19,20). The number and identity of mitochondria-imported tRNAs greatly vary between species and this is especially true in the plant kingdom. Here, based on experimental evidence or on the insufficient number of mitochondrial tRNA genes, the PlantRNA database provides information on tRNA mitochondrial import (Figure 1). While the green microalga O. tauri does not apparently need to import nucleus-encoded tRNAs, the green alga C. reinhardtii imports most of its mitochondrial tRNAs. Annotating the other mitochondrial tRNA genes revealed that both stramenopiles, the brown alga E. siliculosus and the diatom P. tricornutum, lack essential tRNA genes (including tRNAThr), thus implying the need to import nucleus-encoded tRNAs. In the glaucophyte C. paradoxa, mitochondrial tRNAThr genes are also missing. Very interestingly, this is reminiscent of the absence of tRNAThr gene in the mitochondrial genome of the protozoan Reclinomonas americana. This mitochondrial genome more closely resembles the genome of the bacterial ancestor at the origin of the present-day mitochondria than do any other mitochondrial DNA (33). For a yet unknown reason, mitochondrial import of nucleus-encoded tRNAThr seems to be the most conserved and the easiest tRNA import event.
Finally, eukaryotic nuclear tRNA genes are usually transcribed by RNA polymerase III (Pol III) thanks to highly conserved internal promoters, called A and B boxes. However, upstream elements were also found to greatly contribute to transcription efficiencies of many tRNA genes. This is particularly true in higher plants where highly conserved TATA-like elements in the region between −25 and −35 upstream tRNA gene sequences followed by CAA triplets in the −1 to −10 regions are particularly frequent (12). Analyzing the upstream sequences of nuclear tRNA gene in all photosynthetic organisms registered in the PlantRNA database using WebLogo (34,35) has confirmed the existence of these conserved sequence signatures for the tRNA genes of the five flowering plants (Supplementary Figure S1). Interestingly and in contrast, TATA and CAA motifs are considerably less frequent in the glaucophyte C. paradoxa and the green alga O. tauri and are very rare in the other green alga C. reinhardtii, the brown alga E. siliculosus and the bryophyte P. patens. It is to note that in the diatom, P. tricornutum, while there is no obvious conserved CAA sequence, an AT-rich region is present between −30 and −40 of the upstream tRNA gene sequences. The lack of conserved motif in the upstream tRNA gene sequences of several photosynthetic organisms resembles the situation found in animal genomes where the presence of TATA and CAA motifs does not occur frequently (5). It thus suggests that evolutionary divergent pathways for tRNA gene expression regulation exist in the photosynthetic kingdom. At the other extremity of nuclear tRNA gene sequences, pol III transcription termination is triggered by short runs of T residues. As shown in Supplementary Figure S1, such stretches of T residues can be found downstream of the majority of tRNA genes. Nevertheless, two exceptions do exist. In the green alga C. reinhardtii, as previously observed (24), many downstream tRNA gene sequences lack such a polyT tail because of the presence of polycistronic tRNAs, a situation usually not found in eukaryotes. Quite strikingly, we also show here that in the moss P. patens, only 40% of the tRNA genes possess a polyT stretch of at least 4 Ts within their 25 nt downstream sequences. This observation suggests a peculiar genomic organization of the tRNA genes in this organism and hints that an alternative tRNA transcription termination process might be operative in P. patens.
FUTURE DIRECTIONS
The PlantRNA will be updated continuously. First, information on tRNA-related enzymes, up to now included for 7 out of the 11 photosynthetic organisms will be implemented as soon as appropriate high-quality genomic annotations will be accessible. Second, we will continue to upgrade the quality of the web interface and offer new search possibilities. Third, as the number of completed nuclear genomes from other photosynthetic organisms is increasing rapidly, the database will be implemented with the new tRNA gene sequences and their related biological information on a regular basis. From an evolutionary point of view, this must be achieved not only for whole genome sequences of flowering plants such as potato (36), grapevine (37) or apple (38) but also for lower photosynthetic organisms such as lycophytes, haptophytes or cryptophytes (39). From an environmental point of view, it will be interesting to analyze and compare tRNA gene content and organization of extremophile photosynthetic organisms such as Thellungiella salsuginea, a close relative of Arabidopsis but highly resistant to abiotic stresses (40). tRNA genes from these organisms will be accurately annotated and incorporated in the database. Finally, a long-term objective will be to enrich the biological information content of the database, e.g. through the implementation of tRNA gene expression profiles, the description of occurring tRFs and 3D structure models of plant tRNAs.
DATABASE ACCESS
PlantRNA can be accessed freely at http://PlantRNA.ibmp.cnrs.fr. All published data performed with the help of the PlantRNA database should refer to this article.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
Centre National de la Recherche Scientifique; University of Strasbourg; French Agence Nationale de la Recherche (ANR) [ANR-09-BLAN-0240-01, ANR-11-BSV8 008 01]; French National Program ‘Investissement d’Avenir’ (Labex MitoCross); University of Strasbourg and the French Ministère de l’Education et de la Recherche [to B.G. and M.M.]. Funding for open access charge: CNRS, IBMP.
Conflict of interest statement. None declared.
REFERENCES
- 1.Banerjee R, Chen S, Dare K, Gilreath M, Praetorius-Ibba M, Raina M, Reynolds NM, Rogers T, Roy H, Yadavalli SS, et al. tRNAs: cellular barcodes for amino acids. FEBS Lett. 2010;584:387–395. doi: 10.1016/j.febslet.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurto RL. Unexpected functions of tRNA and tRNA processing enzymes. Adv. Exp. Med. Biol. 2011;722:137–155. doi: 10.1007/978-1-4614-0332-6_9. [DOI] [PubMed] [Google Scholar]
- 4.Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 5.Zhang G, Lukoszek R, Mueller-Roeber B, Ignatova Z. Different sequence signatures in the upstream regions of plant and animal tRNA genes shape distinct modes of regulation. Nucleic Acids Res. 2011;39:3331–3339. doi: 10.1093/nar/gkq1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geslain R, Pan T. Functional analysis of human tRNA isodecoders. J. Mol. Biol. 2010;396:821–831. doi: 10.1016/j.jmb.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudinger-Thirion J, Lescure A, Paulus C, Frugier M. Misfolded human tRNA isodecoder binds and neutralizes a 3′ UTR-embedded Alu element. Proc. Natl Acad. Sci. USA. 2011;108:E794–E802. doi: 10.1073/pnas.1103698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuck AC, Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Bowman JL, Floyd SK, Sakakibara K. Green genes-comparative genomics of the green branch of life. Cell. 2007;129:229–234. doi: 10.1016/j.cell.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Reyes-Prieto A, Weber AP, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- 11.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 12.Michaud M, Cognat V, Duchêne AM, Maréchal-Drouard L. A global picture of tRNA genes in plant genomes. Plant J. 2011;66:80–93. doi: 10.1111/j.1365-313X.2011.04490.x. [DOI] [PubMed] [Google Scholar]
- 13.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abe T, Ikemura T, Sugahara J, Kanai A, Ohara Y, Uehara H, Kinouchi M, Kanaya S, Yamada Y, Muto A, et al. tRNADB-CE 2011: tRNA gene database curated manually by experts. Nucleic Acids Res. 2011;39:D210–D213. doi: 10.1093/nar/gkq1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:w686–w689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salinas T, Duchêne AM, Maréchal-Drouard L. Recent advances in tRNA mitochondrial import. Trends Biochem. Sci. 2008;33:320–329. doi: 10.1016/j.tibs.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Duchêne AM, Pujol C, Maréchal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr. Genet. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- 21.Duchêne A, Giritch A, Hoffmann B, Cognat V, Lancelin D, Peeters N, Zaepfel M, Maréchal-Drouard L, Small ID. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen P, Jager G, Zheng B. Transfer RNA modifications and genes for modifying enzymes in Arabidopsis thaliana. BMC Plant Biol. 2010;10:201. doi: 10.1186/1471-2229-10-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gobert A, Gutmann B, Taschner A, Gossringer M, Holzmann J, Hartmann RK, Rossmanith W, Giegé P. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 2010;17:740–744. doi: 10.1038/nsmb.1812. [DOI] [PubMed] [Google Scholar]
- 24.Cognat V, Deragon JM, Vinogradova E, Salinas T, Remacle C, Maréchal-Drouard L. On the evolution and expression of Chlamydomonas reinhardtii nucleus-encoded transfer RNA genes. Genetics. 2008;179:113–123. doi: 10.1534/genetics.107.085688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 26.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 27.Green CJ, Sohel I, Vold BS. The discovery of new intron-containing human tRNA genes using the polymerase chain reaction. J. Biol. Chem. 1990;265:12139–12142. [PubMed] [Google Scholar]
- 28.Van Tol H, Beier H. All human tRNATyr genes contain introns as a prerequisite for pseudouridine biosynthesis in the anticodon. Nucleic Acids Res. 1988;16:1951–1196. doi: 10.1093/nar/16.5.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao M, Carlson BA, Novoselov SV, Weeks DP, Gladyshev VN, Hatfield DL. Chlamydomonas reinhardtii selenocysteine tRNA[Ser]Sec. RNA. 2003;9:923–930. doi: 10.1261/rna.5510503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castellano S, Novoselov SV, Kryukov GV, Lescure A, Blanco E, Krol A, Gladyshev VN, Guigo R. Reconsidering the evolution of eukaryotic selenoproteins: a novel nonmammalian family with scattered phylogenetic distribution. EMBO Rep. 2004;5:71–77. doi: 10.1038/sj.embor.7400036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lobanov AV, Fomenko DE, Zhang Y, Sengupta A, Hatfield DL, Gladyshev VN. Evolutionary dynamics of eukaryotic selenoproteomes: large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol. 2007;8:R198. doi: 10.1186/gb-2007-8-9-r198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dudkiewicz M, Szczepinska T, Grynberg M, Pawlowski K. A novel protein kinase-like domain in a selenoprotein, widespread in the tree of life. PLoS One. 2012;7:e32138. doi: 10.1371/journal.pone.0032138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider A, Maréchal-Drouard L. Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- 34.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X, Pan S, Cheng S, Zhang B, Mu D, Ni P, Zhang G, Yang S, Li R, Wang J, et al. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- 37.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 38.Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. The genome of the domesticated apple (Malus × domestica Borkh.) Nat. Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 39.Janouskovec J, Horak A, Obornik M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc. Natl Acad. Sci. USA. 2010;107:10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H-J, Zhang DY, Oh DH, Dassanayake M, Liu B, Huang Q, Sun HX, Xia R, Wu Y, Wang YN, et al. Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl Acad. Sci. USA. 2012;109:12219–12224. doi: 10.1073/pnas.1209954109. [DOI] [PMC free article] [PubMed] [Google Scholar]