Abstract
Plants have large diverse families of small secreted proteins (SSPs) that play critical roles in the processes of development, differentiation, defense, flowering, stress response, symbiosis, etc. Oryza sativa is one of the major crops worldwide and an excellent model for monocotyledonous plants. However, there had not been any effort to systematically analyze rice SSPs. Here, we constructed a comparative platform, OrysPSSP (http://www.genoportal.org/PSSP/index.do), involving >100 000 SSPs from rice and 25 plant species. OrysPSSP is composed of a core SSP database and a dynamic web interface that integrates a variety of user tools and resources. The current release (v0530) of core SSP database contains a total of 101 048 predicted SSPs, which were generated through a rigid computation/curation pipeline. The web interface consists of eight different modules, providing users with rich resources/functions, e.g. browsing SSP by chromosome, searching and filtering SSP, validating SSP with omics data, comparing SSP among multiple species and querying core SSP database with BLAST. Some cases of application are discussed to demonstrate the utility of OrysPSSP. OrysPSSP serves as a comprehensive resource to explore SSP on the genome scale and across the phylogeny of plant species.
INTRODUCTION
It had been known in animals for years that small secreted proteins (SSPs), such as peptide hormones, cytokines/chemokines, digestive enzymes and defensive peptides (antibody, neurotoxin, defensin), played critical roles in development, metabolism, reproduction, differentiation, metamorphosis, predation and other essential aspects of life cycles in animals (1–3). Recently, similarly important functions of SSPs were discovered in plants, when Pearce et al. (4) first identified tomato systemin, an 18-aa peptide, which functions as a signal molecule in the defense-response cascade. Intensive studies in the following two decades unraveled the essential roles of diverse SSP in plants’ physiology throughout their life cycles (5–13).
The initial efforts on identification of plant SSP via biochemical approach made only small progress. They were accelerated lately by the available genomic sequences of increasing number of plant species, including Arabidopsis thaliana, Glycine max and Populus deltoids. To date, attempts were made to take advantage of the genome annotation to predict SSP in A. thaliana (14) and P. deltoids (15), or to profile plant secretome with computational methods (16). Although genomic approach has greatly expanded the list of SSPs in plants, to many plant biologists and bioinformaticians, there are many short-falls and questions remained to be addressed. First, existing genome annotation programs are inadequate to annotate all SSP in plants. As a result, the numbers of small proteins were grossly underestimated in many current genome annotations (14). Lease and Walker (14) tried to recover the missing SSP from A. thaliana by scanning its open reading frame (ORF) encoding short peptides of between 25 and 250 aa in length. A total of 33 809 un-annotated SSPs were predicted in A. thaliana, and 10 247 (30%) were supported by tiling array data. Using the ‘Coding Index’ method, Hanada et al. (16,17) identified 7159 possible SSPs from A. thaliana, with claimed 1% false discovery rate. However, a separate work by Castellana et al. (18) suggested a mere 2% confirmation rates in the above A. thaliana studies. Hoping to avoid false-positive results by starting with trancriptomic data, Yang et al. (15) obtained an initial set of 12 852 ORFs encoding proteins of 10–200 aa in length from P. deltoids.
Oryza sativa, one of the major crops worldwide and an excellent model for monocotyledonous plants, remained open in the study of SSP. Both an important economic crop and a model plant, it is our top priority to explore its SSPs on the whole-genome scale and compare them with other species across the phylogeny of plant species. Here, we present OrysPSSP: a comparative Platform for Small Secreted Proteins from rice and other plants. In the current project, we set out to achieve the following goals:
Building a database of exhaustive SSPs from O. sativa. To make it exhaustive, we created the initial dataset by combining a six-frame translation and an algorithm for gene model prediction. A processing pipeline followed to filter out false data in three steps.
Building flexible and effective validation tools to minimize false discovery rate and enhance usability. We integrated three levels of high-throughput experimental datasets, including gene expression microarray, RNA-seq and tandem mass spectrometry (MS), for the validation of predicted SSPs.
Building a comparative genomics tool for a comprehensive analysis of the conservation of SSPs in 26 plant species. We integrated the genome information from 25 plant species besides O. sativa ssp. japonica. Comparison across the phylogeny would yield insight into the occurrence and evolution of SSPs in plant species.
The present work provides the most comprehensive platform for the study of plant SSP. Its database not only contains SSPs from rice (the best model plant) but also conserves SSPs from 25 other plant species/subspecies. The current official release (v0530) contains a wholly set of 101 048 SSP candidates. About two-thirds of them, 67 559, are located in un-annotated genome regions in rice, while the rest, 33 489, are included in known genes. When validated with dataset at three different levels, 33 350 SSPs were supported by tiling array data, 9431 by RNA-seq data and 18 353 by MS results. When comparing across the phylogeny of 25 plant species, we found the number of conserved SSPs between rice and other plants, in general, was inversely proportional to their evolutionary distance.
DATABASE CONSTRUCTION
Data source
For the reference genome of O. sativa ssp. japonica, we used IRGSP1.0 from the Rice Annotation Project (RAP, http://rapdb.dna.affrc.go.jp/) (19). The annotation of the rice genome was updated by a jointed effort of RAP and the MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) (20). For comparative genomics analysis, the genomes of 25 green plant species were obtained from sources listed in Table 1 (21–30).
Table 1.
Data source for genomes of 25 green plant species
For validation analysis of predicted SSPs, tiling array datasets for seedling root, seedling shoot, panicle and suspension cultured cells of O. sativa ssp. japonica were obtained from the Gene Expression Omnibus (GEO) database (GEO Series accession number: GSE6996, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6996) (31); RNA-seq datasets from root and tip tissues of O. sativa ssp. japonica were downloaded from the Sequence Read Archive (SRA) database (study accession: SRP007395, http://www.ncbi.nlm.nih.gov/sra?term=SRP007365) (32); the proteomics datasets for O. sativa ssp. japonica were retrieved from the PRoteomics IDEntifications database (PRIDE) (experiment accession: 15854–15865, http://www.ebi.ac.uk/pride/) (33).
Data processing pipeline
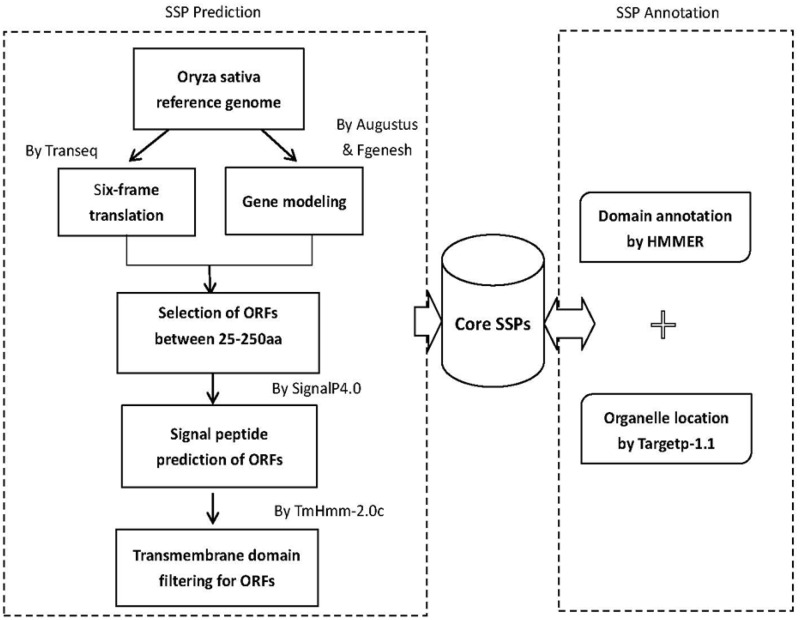
A data pipeline was built to predict and annotate SSP in O. sativa ssp. japonica (Figure 1). To predict SSPs, a core dataset was formed by three steps:
Constructing the starting dataset of small peptides (25–250 aa in length) by combining data from whole-genome screening and from gene modeling using Augustus (v2.5.5) and FGENESH (Softberry, http://www.softberry.com) (34). Whole-genome screening was performed by translating the rice genome in six-frame using the EMBOSS package (35). To recover multiple-exon genes that may be missed from the six-frame translation approach, the gene modeling programs, Augustus (v2.5.5) and FGENESH, were used to predict genes that were to be combined with the previous gene set. For Augustus, a rice gene set supported by full-length cDNAs, expressed sequence tags or proteins in IRGSP1.0 were used for training, and for FGENESH, the gene model of Z. mays, a close neighbor in evolution, was used for gene prediction in O. sativa ssp. japonica var. Nipponbare. The two resulting datasets (90 140 ORFs from six-frame translation and 22 341 from gene modeling) were combined and ORFs encoding peptides of 25–250 aa in length were selected for further analysis.
Screening for N-terminal signaling sequences on the above merged dataset. We used a stand-alone software SignalP 4.0 (36) that uses a combination of artificial neural networks to predict a signal peptide and its cleavage site. As a result, secreted peptides that have N-terminal signaling sequence were retained for further analysis.
Screening for trans-membrane domain. The above dataset was filtered for the presence of trans-membrane helices using TMHMM2.0c (37), which indicates a protein resides in the plasma membrane or an endomembrane.
Figure 1.
Schematic diagram of the data processing pipeline for OrysPSSP.
Using the pipeline, a total of 101 048 putative ORFs for SSP were identified, and about one-third were novel that were located between known genes.
To reveal possible functions of these peptides, the candidates from core SSP dataset were annotated for (i) conserved domains and (ii) organelle location. HMMER(v3.0) was used to scan PfamA database for identifying domains in rice SSP, and 7755 SSP genes were found to have one or more domain matches (38,39). Finally, the organelle location of SSP was predicted using TargetP1.1 (40). Because of ancient origins of secreted proteins in both prokaryotes and eukaryotes, we reasoned that putative SSPs that are conserved in evolution are more likely to be true secreted proteins. So in addition to the validation tool, we provide analytic tool comparing rice SSP to those of other plant species.
Database implementation
The core SSP dataset of O. sativa ssp. japonica var. Nipponbare generated from the data processing pipeline was stored in a MySQL database (v5.5, http://www.mysql.com/) with a relational database design. They include the genome location, pre-protein sequence, signaling peptide sequence, domain annotation, targeting organelle, neighboring genes for a SSP as well as some validation information. All OrysPSSP application tools, including ‘Browse’, ‘Search & Validate’, ‘Compare Genomes’ and ‘BLAST Search’, were built on the MySQL database. They were implemented in JSP and deployed on the Apache Tomcat web server (http://tomcat.apache.org/). OrysPSSP can be accessed through IE 6.0 or higher, Netscape 7.0 or higher and other web browsers such as Safari, Opera, Chrome and Firefox.
WEB INTERFACE AND INTEGRATED TOOLS
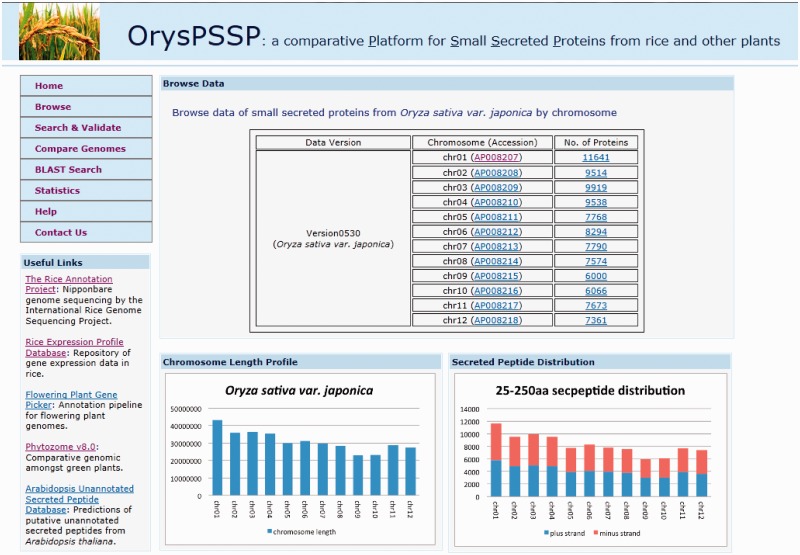
OrysPSSP web interface (Figure 2) consists of eight different modules, integrating the core SSP database and various application tools. The eight modules include (i) ‘Home’: a brief introduction to OrysPSSP; (ii) ‘Browse’: browse SSP from O. sativa ssp. japonica by chromosome; (iii) ‘Search & Validate’: search for SSP using one or multiple search parameters and validate the resulting SSP by applying one or multiple filtering dataset(s); (iv) ‘Compare Genomes’: apply comparative genomics approach to analyze SSPs from O. sativa ssp. japonica that are conserved in other plant species; (v) ‘BLAST search’: use ‘BLAST’ tool to search for SSP contained in the query sequences; (vi) ‘Statistics’: provide basic statistics from the data processing pipelines; (vii) ‘Help’: offer answers to frequently asked questions about OrysPSSP and (viii) ‘Contact us’: include contact information of support for users.
Figure 2.
Snap shot of the ‘Browse’ module from OrysPSSP web interface, which consists of eight modules: (i) ‘Home’; (ii) ‘Browse’; (iii) ‘Search & Validate’; (iv) ‘Compare Genomes’; (v) ‘BLAST search’; (vi) ‘Statistics’; (vii) ‘Help’; and (viii) ‘Contact us’. Details of each module are described in web interface and integrated tools.
OrysPSSPs currently provides users with several flexible tools and integrates related genome resources to search, validate, filter and compare the data. Some useful links are also provided for a number of relevant resources for the same purpose.
Search and validation tool
‘Search & validation’ tool module provides some primary search and filter functions on the database, including text search or filter on chromosome number, strand and/or annotation.
In addition to add value to users’ experience for this informative and applicable platform, we provide validation functions by integrating three levels of experimental datasets: (i) at the transcriptional level, we obtained the tiling array hybridization datasets of seedling root, seedling shoot, panicle and suspension cultured cells from O. sativa ssp. japonica; (ii) with the advancement of more sensitive ‘RNA-seq’ technology in detecting transcripts at low expression levels, we included the O. sativa ssp. japonica RNA-seq dataset (SRP007395) from NCBI SRA database; and (iii) at translation level, we added a peptide spectra dataset (experiment accession: 15854–15865) extracted from the MS of rice tissues from PRIDE (http://www.ebi.ac.uk/pride/) (33). Users can select one, two or three levels of data to perform validation test on small secreted peptides in OrysPSSP. These parameters will be combined as a logical ‘AND’ operation. For validation with the tiling array expression data, a threshold of twice the median hybridization intensity value was used for positive results, and we found evidence for the expression of 18 371 putative small secreted peptides. For validation with the rice RNA-seq data, bases within ORF must be mapped by at least two RNA-seq reads; we identified 3992 putative ORFs supported by RNA-seq. For validation with the MS data, tryptic peptides from each predicted small secreted peptides were screened against the MS data with X-tandem. We obtained supporting evidence for 16 657 SSPs from rice. Furthermore, it is easy for a user to use different combination of search parameters and validation dataset to obtain a subset of SSP that meets his/her research needs.
Comparative analysis to other plant species
‘Compare Genomes’ is an advanced tool module that applies comparative genomics approach to search for SSPs from O. sativa ssp. japonica that are conserved in 25 plant species ranging from moss to angiosperm, including Aquilegia coerulea, Arabidopsis lyrata, A. thaliana, Brachypodium distachyon, Capsella rubella, Carica papaya, Citrus sinensis, Cucumis sativus, Eucalyptus grandis, G. max, Malus domestica, Manihot esculenta, Medicago truncatula, Mimulus guttatus, O. sativa ssp. indica, Phaseolus vulgaris, Physcomitrella patens, Populus trichocarpa, Ricinus communis, Selaginella moellendorffii, Setaria italic, Sorghum bicolor, Thellungiella halophila, Vitis vinifera and Zea mays. It helps users who are interested in studying more conservative SSPs from rice and in looking for model SSPs that have evolutionary root in other plant species.
This module requires users to input a list of rice SSPs to start comparative search. The list of SSPs can be readily generated from the results of other tools or created manually by users. The user input SSPs are used as query to search against the genome sequences of the species users select using BLASTp. While users can select one or multiple species to perform the search, they are treated as logical ‘or’ in the searching operation. The results will show those SSPs that are conserved in any of the selected species.
BLAST search tool
A ‘BLAST’ search tool was integrated into OrysPSSP to help users to search for SSP (of rice source) that map to users’ sequences of interest. Users can either input their queries into query box or upload a file that contains query sequence. The tool was made flexible to allow DNA, mRNA or amino acid sequence type of queries. Users can also modify the common parameters for BLAST tool or can leave them with the default values.
CASES OF APPLICATION
For plant biologists, OrysPSSP would be a valuable resource for study on the functions of novel SSPs in development, signaling, metabolism and reproduction in a model plant like rice. To illustrate the utilities of OrysPSSP, we here describe two application cases inspired by our collaborators.
Mining novel SSPs from O. sativa ssp. japonica
Despite the progress made in the study of SSPs in plants, their number and identities remain largely unknown in rice. To find novel SSP from rice, we used the ‘Search & Validate’ tool by setting the ‘Within known gene’ option to ‘No’ and leaving everything else unchanged. Validation filter was set by checking one of the three check boxes: tilingArray, RNAseq or MS. There were 6213 novel SSPs in rice that passed validation by tilingArray, and 233 and 9788 were validated with RNA-seq data or MS data, respectively. When tilingArray and MS were combined (a moderately stringent filter), 3412 novel SSPs were returned, representing a subset of rice SSPs with relatively high confidence.
Identification of conserved SSPs from O. sativa ssp. japonica
Diverse families of SSP have been identified in plants (41). They include the CLV3/ESR-related (CLE) family, RALF (rapid alkalinization factor), EPF (epidermal patterning factor) family, PDF (plant defensin), DEFL (defensin-like proteins), CEP1 (C-terminally encoded peptide 1), SCR/SP11, LUREs, etc. To find those conserved SSPs in rice and identify new members is of great importance to plant biologists. Using the keyword ‘CLE’ in the ‘Search & Validate’ module (setting ‘Within known gene’ option to ‘No’), we found a new ‘FON2-like CLE protein 2’ (ID: ory_chr06_5621_spd), which had not been known in rice. Using ‘Compare Genome’ module to compare between plant species, ory_chr06_5621_spd was found to be specific to rice. Similarly, seven new members of PDF (plant defensin) (ory_chr11_2644_spd, ory_chr10_5233_spd, ory_chr08_3989_spd, ory_chr08_3386_spd, ory_chr05_4899_spd, ory_chr03_2326_spd and ory_chr01_6059_spd) and 1 new member of RALF (ory_chr11_4864_spd) were discovered in O. sativa ssp. japonica.
SUMMARY AND FUTURE DEVELOPMENT
OrysPSSP serves as a comprehensive resource to explore SSPs from O. sativa and other plant species on the whole-genome scale. It would be beneficial to investigators addressing a variety of questions. For geneticists, they can query the database for SSP located within the target regions of their interest. For plant biologists, the platform is a valuable resource for initiating a study on the functions of novel SSPs in development, signaling or metabolism in a model plant. Alternatively, they can check whether some peptides induced by stress belong to the dataset of un-annotated SSP. Furthermore, they may receive gene and domain information for those matched peptides.
Our current study was improved based on methods from previous works. Multiple-exon ORF genes were processed by our new pipeline, whereas only single-exon gene was included previously (14,16). In addition, three levels of omics date were integrated, which give biologists more dynamic and flexible tools for validation of SSPs. A comparative genomics approach was applied for investigating the diversity of SSP from an evolutionary perspective. Still, there are many aspects that OrysPSSP can be improved. In the future, we plan to enhance OrysPSSP by: (i) integrating tissue-specific omics data from rice for analysis of function and tissue specificity of SSP and (ii) applying our pipeline to more plant species, as well as including high-throughput omics data for validation.
OrysPSSP is a comprehensive platform to explore the full spectrum of SSPs in rice and other plant species. It would help advance our understanding of the essential roles by SSP and yield new insights into the processes of development, differentiation, stress response and symbiosis in plants.
FUNDING
Funding for open access charge: National Key Basic Research Program in China [2013CB127005, 2012CB316501]; Special Program for Transgenic Crops in China [2012ZX08009002, 2011ZX08010002-002]; Shanghai Pujiang Scholarship Program [10PJ1408000].
Conflict of interest statement. None declared.
REFERENCES
- 1.Choo KH, Tan TW, Ranganathan S. SPdb—a signal peptide database. BMC Bioinformatics. 2005;6:249. doi: 10.1186/1471-2105-6-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Zhang Y, Yin Y, Gao G, Li S, Jiang Y, Gu X, Luo J. SPD—a web-based secreted protein database. Nucleic Acids Res. 2005;33:D169–D173. doi: 10.1093/nar/gki093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colombani J, Andersen DS, Leopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. 2012;336:582–585. doi: 10.1126/science.1216689. [DOI] [PubMed] [Google Scholar]
- 4.Pearce G, Strydom D, Johnson S, Ryan CA. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science. 1991;253:895–897. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 6.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 7.Ohyama K, Ogawa M, Matsubayashi Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 2008;55:152–160. doi: 10.1111/j.1365-313X.2008.03464.x. [DOI] [PubMed] [Google Scholar]
- 8.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- 9.Hirakawa Y, Kondo Y, Fukuda H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell. 2010;22:2618–2629. doi: 10.1105/tpc.110.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, et al. Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature. 2009;458:357–361. doi: 10.1038/nature07882. [DOI] [PubMed] [Google Scholar]
- 12.Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463:241–244. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- 13.Fiume E, Fletcher JC. Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell. 2012;24:1000–1012. doi: 10.1105/tpc.111.094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lease KA, Walker JC. The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 2006;142:831–838. doi: 10.1104/pp.106.086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X, Tschaplinski TJ, Hurst GB, Jawdy S, Abraham PE, Lankford PK, Adams RM, Shah MB, Hettich RL, Lindquist E, et al. Discovery and annotation of small proteins using genomics, proteomics, and computational approaches. Genome Res. 2011;21:634–641. doi: 10.1101/gr.109280.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res. 2007;17:632–640. doi: 10.1101/gr.5836207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanada K, Akiyama K, Sakurai T, Toyoda T, Shinozaki K, Shiu SH. sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics. 2010;26:399–400. doi: 10.1093/bioinformatics/btp688. [DOI] [PubMed] [Google Scholar]
- 18.Castellana NE, Payne SH, Shen Z, Stanke M, Bafna V, Briggs SP. Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl Acad. Sci. USA. 2008;105:21034–21038. doi: 10.1073/pnas.0811066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice Annotation Project, Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, Numa H, Sakai H, Wu J, Itoh T, et al. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 2008;36:D1028–d1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao W, Wang J, He X, Huang X, Jiao Y, Dai M, Wei S, Fu J, Chen Y, Ren X, et al. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32:D377–D382. doi: 10.1093/nar/gkh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40:D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. International Brachypodium Initiative. (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature, 463, 763–768. [DOI] [PubMed]
- 26.Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. The genome of the domesticated apple (Malus × domestica Borkh.) Nat. Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 28.Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 30.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 31.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kodama Y, Shumway M, Leinonen R, International Nucleotide Sequence Database Collaboration The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Csordas A, Ovelleiro D, Wang R, Foster JM, Ríos D, Vizcaíno JA, Hermjakob H. PRIDE: quality control in a proteomics data repository. Database. 2012 doi: 10.1093/database/bas004. March 20 (doi: 10.1093/database/bas004; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 36.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 37.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 38.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 39.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 41.Fukuda H, Higashiyama T. Diverse functions of plant peptides: entering a new phase. Plant Cell Physiol. 2011;52:1–4. doi: 10.1093/pcp/pcq193. [DOI] [PubMed] [Google Scholar]