Abstract
The HUGO Gene Nomenclature Committee situated at the European Bioinformatics Institute assigns unique symbols and names to human genes. Since 2011, the data within our database has expanded largely owing to an increase in naming pseudogenes and non-coding RNA genes, and we now have >33 500 approved symbols. Our gene families and groups have also increased to nearly 500, with ∼45% of our gene entries associated to at least one family or group. We have also redesigned the HUGO Gene Nomenclature Committee website http://www.genenames.org creating a constant look and feel across the site and improving usability and readability for our users. The site provides a public access portal to our database with no restrictions imposed on access or the use of the data. Within this article, we review our online resources and data with particular emphasis on the updates to our website.
INTRODUCTION
The HUGO Gene Nomenclature Committee (HGNC) maintains a publicly accessible database of unique and approved human names and symbols for protein coding genes and other features found within the human genome (1). The HGNC arose from the Human Gene Mapping community recognizing the need for a single committee with the authority to approve human gene nomenclature in 1977; in 2007, 30 years and >25 000 gene symbols later, the HGNC moved to its current location within the European Bioinformatics Institute on the Wellcome Trust Genome Campus south of Cambridge, UK.
All HGNC entries are manually curated, and the HGNC symbols and names assigned represent a standard, to be used in all publications and databases where a specific gene is discussed or referenced. Our aim is to provide nomenclature that is informative and acceptable to researchers in the field. To accomplish this, we contact researchers working on particular genes and gene families before approving symbols. We also encourage researchers to submit proposals for gene symbols to determine their suitability before publication, and these are considered under strict confidentiality. We also work closely with other nomenclature groups such as those for mouse (2), rat (3) and zebrafish (4) to ensure that orthologous genes are assigned equivalent symbols where possible. Our symbols are used extensively within the human genetics and genomics communities and throughout the databases that concentrate on human genes and proteins, such as Entrez Gene (5), Ensembl (6), Vega (7), ENA/GenBank/DDBJ (8–10), GeneCards (11), UCSC genome browser (12) and UniProt (13), as well as disease-related databases such as Decipher (14), OMIM (15), and COSMIC (16).
DATA
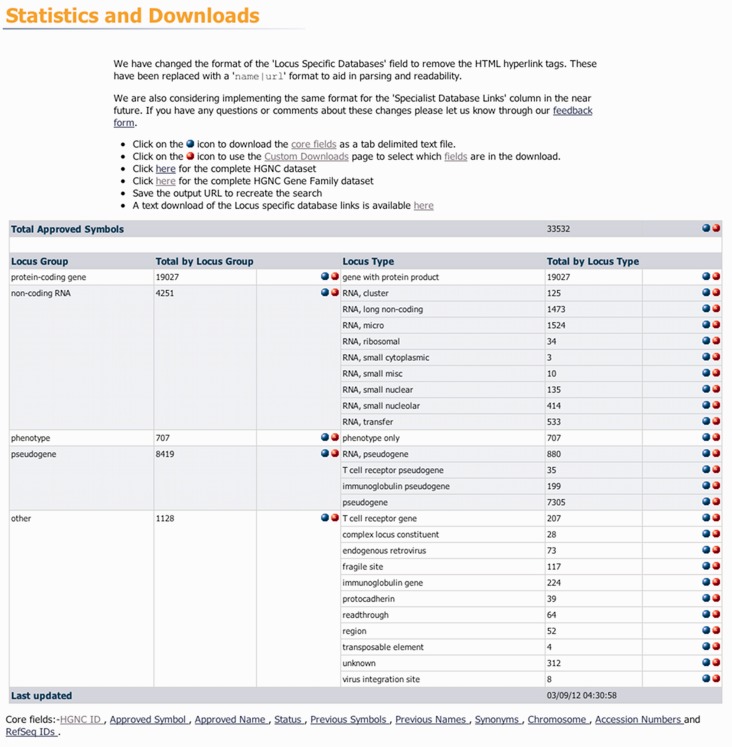
As of the beginning of September 2012, we have 33 532 active entries within our database with 19 027 of the entries being protein coding genes (Figure 1). In May 2003, at Cold Spring Harbor Laboratories, winners were declared for GeneSweep, a competition to estimate how many protein coding genes were present in human, which concluded that <30 000 genes would be found (17). In the intervening years, improvements in genome annotation have led to progressively lower estimates. The Consensus CDS (CCDS) project (18), dedicated to identifying a core set of protein coding regions, contains 18 474 gene IDs as of August 2012. Comparing our figure of 19 027 protein coding genes with that of the CCDS project, we believe we have an almost complete set for the human genome. Our entries are constantly being reviewed and updated with extra information, name and symbol changes and locus type classification changes. We have found that the number of protein coding genes has plateaued, as novel genes have been offset by revision and reclassification of existing entries to non-coding locus types and withdrawal of redundant entries. We also have at present ∼300 entries with a locus type of ‘unknown’. Entries fall in this category when annotation groups disagree on the coding potential of the gene in question. These entries are reviewed regularly, and we are very interested in hearing about new evidence to prove or disprove the coding potential of these genes.
Figure 1.
The statistics and downloads page allows users to retrieve plain text, tab-separated data sets. The user may download the entire gene symbol report data set, gene family sets or download data that belong to a certain locus type. The table on the page shows the total number of entries per locus group and type including all of the non-coding RNA locus types and the number of entries associated to them.
The main areas of growth within our database in the past 2 years have been in naming non-coding RNA genes (19) and pseudogenes, and classifying genes into families and groups. At the start of September 2012, we had 4251 non-coding RNAs, and this figure is expected to increase as more long non-coding RNAs (>200 nucleotides) are annotated (Figure 1). Within our database, the classification of non-coding RNA is a ‘locus group’ that contains several ‘locus types’. Since 2011 (20), we have changed our non-coding RNA locus types to better reflect sequence ontology (21). A full list of current non-coding RNA types can be seen in Figure 1. Pseudogenes account for almost a quarter of our total entries and are the second largest group of genes with approved nomenclature. The vast majority of pseudogenes are non-functional, but the analysis of these genes can be extremely important for insight into the evolution of gene families, and pseudogenes are discussed frequently in literature; so it is important for us to assign meaningful symbols and names to this class of gene. We have introduced a new addition to our locus type classification, ‘readthrough’, in which we currently have 64 entries. This class contains loci such as INS-IGF2 (22) where transcription goes beyond the normal termination sequence of a gene (i.e. INS, insulin), and extends into an adjacent gene [i.e. IGF2, insulin growth factor 2 (somatomedin A)].
WEBSITE
Our website www.genenames.org provides a public access portal to our database. There are no restrictions imposed on access or the use of the data provided by the HGNC. In May 2011, we released a new design for our website to give every page a consistent look and feel throughout the site and to improve user experience and interactions. The new design was mainly built using the Drupal content management system for static pages, and Perl common gateway interface (CGI) for our dynamic content that retrieves data from our MySQL database. Drupal offers us many advantages, the main one being that our curators can add and change content easily and efficiently without needing to learn how to build web pages using HTML and so on. The Perl CGI pages use many comprehensive perl archive network (CPAN) modules especially to connect to the database and to create HTML templates. Using templates separates the design and presentation elements away from the core Perl code, which aids development and maintenance. Owing to the consistent design across the site, users will not notice the difference between Drupal and Perl CGI web pages. We are still looking to update sections of our website and are keen to hear from our users to improve their experience in navigating our site. We shall now explore the new design and the main pages of the site that make genenames.org.
Header and footer
Every page of www.genenames.org has a new header/banner that contains everything you will need to browse through the website. One of the main improvements to the website was the addition of the drop-down menus attached to the tabs (Figure 2) in the header. These allow us to break down the sets of pages that we provide and allow the user to find the page they are looking for without clicking through multiple index pages. To activate the drop-down menus, the user need only hover over a tab, and a column of links will appear. The menus are created using cascading style sheets only, and so if the user prefers to disable JavaScript, the menus will still work. The new footer also uses the same colour scheme as the header and appears on every page. It contains links to our terms of use, our privacy and cookies policy and an email link so that the user can email us with any queries about the site or data contained within (Figure 2).
Figure 2.
A typical gene symbol report showing the shaded box containing the HGNC core data with other information and links below. If the gene is within a gene family, then the gene family field will also be displayed. The new banner featuring a new logo and colour scheme is at the top of the image. The banner contains a simple quick search box to the right and holds tabs that produce drop-down menus, once the mouse cursor hovers over the tab. The website footer containing our affiliations, funding sources and policies is seen at the bottom of the page. The footer also sports an email link so that users can contact us via the link.
Home page
There are four sections to the main body of our new home page. The four sections are: a ‘browse-approved symbols by chromosome’ interactive karyotype image, linking to a ‘Statistics and Downloads’ page specifically for each chromosome (see ‘Downloads’ section); a ‘Quick Gene Search’; a ‘latest news’ section including a link to our new HGNC Twitter feed (@genenames); and a frequently asked questions portal and website search.
Gene search
As discussed in 2011 (20), there are three ways to search for gene symbol reports within genenames.org, all of which can be accessed via the ‘Search Genes’ drop-down menu on the header (Figure 2). The most commonly used search is the ‘Quick Gene Search’, which can also be found on the home page and within the header (Figure 2). The ‘Advanced Gene Search’ tool allows the user to specify which fields in the HGNC data set they would like to search and build more complex queries with multiple search terms. The third search tool we provide is the ‘List Search’, which allows the user to type, paste or upload a list of symbols into a search field.
Gene symbol report
The majority of our users access the genenames.org site to retrieve our gene symbol reports, which are the main interface to our manually curated data and the external database links stored within our database (Figure 2). The gene symbol report page has been completely redesigned to fit our new colour scheme and to make using the page more intuitive. Our core HGNC data are now highlighted at the top of the report within a shaded box. This shaded area features approved nomenclature, a unique HGNC ID, previous nomenclature, synonyms, locus type and the chromosomal location, all of which have been curated manually.
In addition to the HGNC-specific data, we have a wide variety of external links that are relevant to the gene in question. These links are displayed below the shaded box in a table and are grouped together by the type of resource named within the first column. A letter ‘C’ next to the link indicates that the link has been checked and curated by a member of the HGNC. Links that have a ‘D’ placed alongside indicate that the link was derived and downloaded from an external source. Data links are organized into the following sections:
Gene family—Only present if the gene is associated to a family or group. The link will navigate the user to the gene family page.
Specialist database—Appears only if a value is present. Contains specialist external database resource links that are specific to a class of genes. To date, we link out to 14 specialist databases, a list of which can be found at www.genenames.org/useful/symbol-report-documentation, with a brief description.
Homologs—A group of homology-related links, including our own HGNC Comparison of Orthology Predictions (HCOP) orthology data-mining tool and links to orthologous gene entries in mouse genome informatics (MGI) and rat genome database (RGD).
Nucleotide sequences—Links to representative accessions from GenBank/EMBL/DDBJ, RefSeq, the CCDS project and Vega.
Gene resources—Links to the four most popular gene and genome browsers, (Ensembl, Vega, Entrez Gene and UCSC); each resource has two links, one to the gene entry and the other to the genome browser.
Protein resources—Information on proteins encoded by the gene. We include links to UniProt and to InterPro (23), which shows all the domains predicted within the encoded proteins by the InterPro member databases.
Clinical resources—Links to resources for associated phenotypes, diseases and gene mutations.
References—PubMed (24) and CiteXplore (http://www.ebi.ac.uk/citexplore/) hyperlinks, which display the abstracts for references pertinent to the gene. The purpose of the section is not to list all possible published articles for the gene, but to provide links to articles that first describe the gene in question or are particularly relevant to the nomenclature of the gene.
Other database links—New links in this section are to Reactome (25), which contains manually curated, peer-reviewed signalling pathway data, and QuickGo (26) which lists all Gene Ontology terms annotated for the gene product(s).
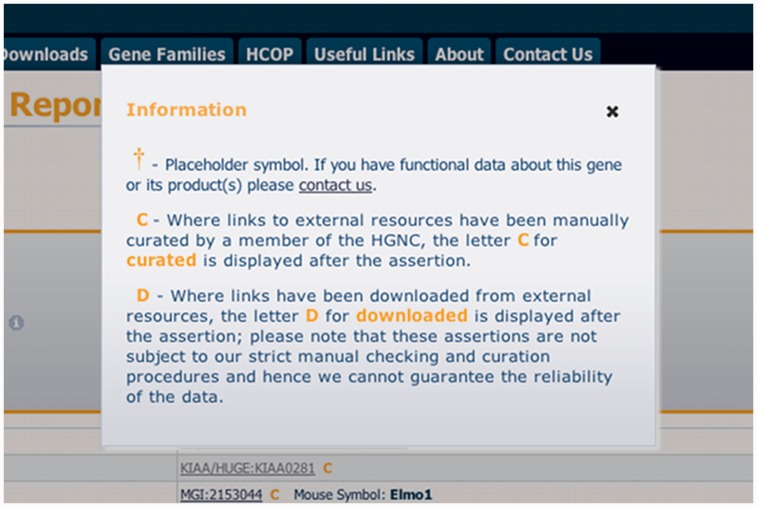
Gene symbol reports include two new icons that are intended to help the user understand the data that we provide. The first is the dagger symbol that can be found next to the gene symbol in certain entries. This informs the user that this gene symbol is a placeholder, and that we are seeking functional data to rename the gene. In the past 2 years, we have made great strides in replacing these placeholder symbols with more informative symbols, reducing the number of C$orf symbols to ∼780 from a peak of 1960, with >200 replaced in 2012 alone. We are always interested in hearing from people who can offer functional information about an entry with a placeholder symbol so that we can reduce these figures further. The second new icon is an ‘i' within a circle that links to information about the field. Both these icons and the ‘C’ and ‘D’ keys once clicked will create a dialog box within the page (Figure 3). The dialog box provides the user with additional information about the field or key. The dialog boxes are created using jQuery UI and retrieve the text using AJAX. The text is retrieved only when asked and is retrieved only once, storing the text so that subsequent calls for the same text will be instant. If the user’s browser has JavaScript switched off, the links will leave the page and fetch the symbol report documentation page that contains all the information about every field (http://www.genenames.org/useful/symbol-report-documentation).
Figure 3.
An example of an information dialog pop up that appears if the keys ‘C’, ‘D’ or the dagger symbol are selected. When the dialog box appears, the background is greyed out, and the dialog comes into focus. The dialog box is scrollable if the text within does not fit into the 400 by 300 px box.
Downloads
The gene symbol report is a good way of browsing individual genes within our database, but many want to retrieve large sets of data. For these users, we offer several ways of downloading data sets. The first of these is via the ‘Statistics and Downloads’, which allows users to download our entire data set, particular locus types and our complete gene families set (Figure 1). It also provides basic statistics of the number of entries we have for each locus group and type and informs the user when the database was most recently updated (UK time). The second is the ‘browse-approved symbols by chromosome’ interface on the homepage, as mentioned earlier. The third tool is the custom downloads, which offers a more advanced way to download our data, and is designed for users that want a specific set of data and/or specific fields within their set. The fourth tool is our BioMart (http://www.genenames.org/biomart) that provides another way of performing complex queries and creating bespoke data sets to download. Both the BioMart Central Portal (27) and our HGNC MartView will allow users to not only retrieve data via the web interface but also via a Perl API, RESTful web service and SOAP web interface.
Gene families/groups
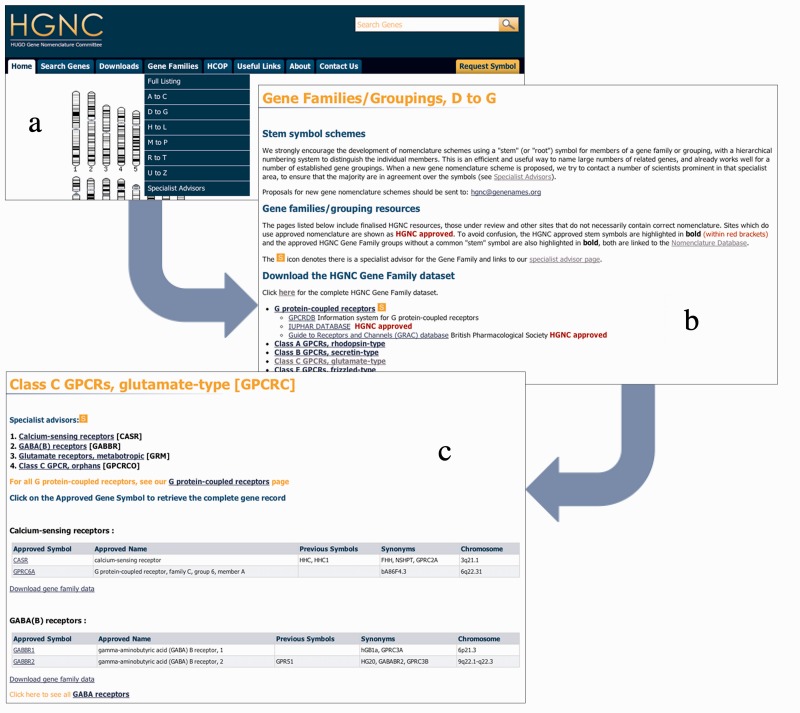
In the past 2 years, we have worked on expanding our resources for gene families and groupings (28). As of September 2012, we have ∼45% of our entries associated with at least one gene family, and we have 475 families, which equate to ∼400 pages of our website dedicated to gene families and sub-families. All families and groups can be found by using the ‘Gene Families’ tab within the header (Figure 4a), which links to a bullet-pointed list of family names and symbols (Figure 4b). To retrieve information about the family, the user must click on a symbol or hyperlinked name to direct them to the selected family page (Figure 4c), which lists all genes and/or subfamilies that belong to the family.
Figure 4.
The most common way of browsing for gene families is shown in the figure. The flow through the pages is seen by following the arrows from image a to image d. From the header, a user would select an option from the drop-down shown (a). This then takes the user to a families list page (b) in which the user selects a family, bringing them to the family page (c). From the family page, the user may then select a gene symbol, taking them to a gene symbol report (see Figure 2).
FUTURE DIRECTION
We are in the process of moving our site and database to two offsite datacentres with multilayer redundancy using virtual machines. Requests to our site will be load balanced between datacentres, which will provide quicker response times during heavy traffic periods. Having a multilayer redundancy infrastructure also allows us to carry out maintenance and repairs without affecting the service for our users, by taking one centre offline while the other serves the content.
We plan to replace our quick gene search with a solr search engine. Many of the world’s largest websites have adopted solr, as it is open source and offers very quick and powerful full-text searches, hit highlighting and faceted searches, as well as many other features. These search engines are also highly scalable, offering efficient replication to other search servers if needed and are optimized for high volumes of web traffic. We hope that by embracing solr, we will improve our search times and will provide a search that will be more intuitive for users. Maintenance of the search will also be easier, as being part of a large community of solr users, we will be able to find help and advice on many forums, published articles and books.
HCOP (HGNC Comparison of Orthology Predictions http://www.genenames.org/hcop) (29) is a tool that searches and displays predicted orthologs of a particular human gene or set of genes made by multiple orthology resources. The reliability of the prediction can be assumed by the number of databases, which concur and by the presence or absence of synteny between the relevant chromosomes, where known. At present HCOP contains data from 14 genomes that can be compared with the human genome. We are planning to expand HCOP by increasing the number of resources and species, and the interface will also be improved for searching and downloading the data. This will aid us in our future work assigning standardized gene names to orthologous genes across vertebrate species. We will also continue to replace placeholder symbols with more informative symbols and introduce novel entries for loci that are only found on alternative assemblies where annotated by the Genome Reference Consortium (30). For any comments or questions concerning our work, please contact us via hgnc@genenames.org.
FUNDING
The Wellcome Trust [081979/Z/07/Z] and [099129/Z/12/Z]; National Human Genome Research Institute [P41 HG03345]. Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank all of their past HGNC colleagues for their invaluable contributions to this project.
REFERENCES
- 1.Wain HM, Lush MJ, Ducluzeau F, Khodiyar VK, Povey S. Genew: the human gene nomenclature database, 2004 updates. Nucleic Acids Res. 2004;32:D255–D257. doi: 10.1093/nar/gkh072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The mouse genome database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40:D881–D886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dwinell MR, Worthey EA, Shimoyama M, Bakir-Gungor B, DePons J, Laulederkind S, Lowry T, Nigram R, Petri V, Smith J, et al. The rat genome database 2009: variation, ontologies and pathways. Nucleic Acids Res. 2009;37:D744–D749. doi: 10.1093/nar/gkn842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SA, et al. ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 2011;39:D822–D829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaminuma E, Kosuge T, Kodama Y, Aono H, Mashima J, Gojobori T, Sugawara H, Ogasawara O, Takagi T, Okubo K, et al. DDBJ progress report. Nucleic Acids Res. 2011;39:D22–D27. doi: 10.1093/nar/gkq1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leinonen R, Akhtar R, Birney E, Bower L, Cerdeno-Tarraga A, Cheng Y, Cleland I, Faruque N, Goodgame N, Gibson R, et al. The European nucleotide archive. Nucleic Acids Res. 2011;39:D28–D31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al. GeneCards Version 3: the human gene integrator. Database. 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s online mendelian inheritance in man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennisi E. Human genome. a low number wins the GeneSweep Pool. Science. 2003;300:1484. doi: 10.1126/science.300.5625.1484b. [DOI] [PubMed] [Google Scholar]
- 18.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright MW, Bruford EA. Naming ‘junk': human non-protein coding RNA (ncRNA) gene nomenclature. Hum. Genomics. 2011;5:90–98. doi: 10.1186/1479-7364-5-2-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The sequence ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monk D, Sanches R, Arnaud P, Apostolidou S, Hills FA, Abu-Amero S, Murrell A, Friess H, Reik W, Stanier P, et al. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum. Mol. Genet. 2006;15:1259–1269. doi: 10.1093/hmg/ddl041. [DOI] [PubMed] [Google Scholar]
- 23.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Eustachio P. Reactome knowledgebase of human biological pathways and processes. Methods Mol. Biol. 2011;694:49–61. doi: 10.1007/978-1-60761-977-2_4. [DOI] [PubMed] [Google Scholar]
- 26.Binns D, Dimmer E, Huntley R, Barrell D, O’Donovan C, Apweiler R. QuickGO: a web-based tool for gene ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guberman JM, Ai J, Arnaiz O, Baran J, Blake A, Baldock R, Chelala C, Croft D, Cros A, Cutts RJ, et al. BioMart central portal: an open database network for the biological community. Database. 2011;2011:bar041. doi: 10.1093/database/bar041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daugherty LC, Seal RL, Wright MW, Bruford EA. Gene family matters: expanding the HGNC resource. Hum. Genomics. 2012;6:4. doi: 10.1186/1479-7364-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eyre TA, Wright MW, Lush MJ, Bruford EA. HCOP: a searchable database of human orthology predictions. Brief. Bioinform. 2007;8:2–5. doi: 10.1093/bib/bbl030. [DOI] [PubMed] [Google Scholar]
- 30.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen HC, Agarwala R, McLaren WM, Ritchie GR, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]