Abstract
The Human Ageing Genomic Resources (HAGR, http://genomics.senescence.info) is a freely available online collection of research databases and tools for the biology and genetics of ageing. HAGR features now several databases with high-quality manually curated data: (i) GenAge, a database of genes associated with ageing in humans and model organisms; (ii) AnAge, an extensive collection of longevity records and complementary traits for >4000 vertebrate species; and (iii) GenDR, a newly incorporated database, containing both gene mutations that interfere with dietary restriction-mediated lifespan extension and consistent gene expression changes induced by dietary restriction. Since its creation about 10 years ago, major efforts have been undertaken to maintain the quality of data in HAGR, while further continuing to develop, improve and extend it. This article briefly describes the content of HAGR and details the major updates since its previous publications, in terms of both structure and content. The completely redesigned interface, more intuitive and more integrative of HAGR resources, is also presented. Altogether, we hope that through its improvements, the current version of HAGR will continue to provide users with the most comprehensive and accessible resources available today in the field of biogerontology.
INTRODUCTION
The Human Ageing Genomic Resources (HAGR, http://genomics.senescence.info) is a web portal encompassing several online databases and tools, aiming to organize the increasing amount of data and information relevant to the biology of ageing, and make them accessible to the research community. Since its first publication in 2005 (1), HAGR has been the leading online resource for biogerontologists, acting as a reference point for various studies and in particular for genetic and evolutionary studies of longevity and ageing.
Initially, HAGR was established around two manually curated databases: GenAge, a database of genes potentially associated with human ageing, and AnAge, a database of ageing, longevity and life history traits in animals. While the goal of GenAge is to function as a compilation of genetic observations reflecting our current knowledge about human ageing, AnAge contains an extensive collection of longevity records, developmental, reproductive and metabolic traits and other key observations related to ageing in >4000 vertebrate species. Since then, the GenAge and AnAge databases have been continuously curated, and new data have been incorporated. Moreover, entire new data sets have also been included. In the update published in 2009 (2), a new list of human genes tested for their possible association with human longevity and a data set of genes associated with longevity and/or ageing in the most studied model organisms were added to GenAge. Although the latter does not include human genes, it was included to serve as a tool for researchers studying ageing in model organisms and because many ageing-related discoveries in model organisms could provide important insights into human ageing (3,4).
With the emergence of new high-throughput technologies, many genes associated with ageing and longevity are still being identified, in particular in model organisms (5–10). Although the field of biogerontology is rapidly evolving, we are still relatively far from having a complete picture of the human ageing process (11), and the need to continue collecting and systematically organizing what we know about the genetics and comparative biology of ageing is more important than ever. In particular, extracting quantitative data from the scientific literature is especially important for performing bioinformatics and systems biology analyses and for guiding experiments. Of note, some genes related to ageing are already being targeted for drug discovery, and translating findings from the genetics of ageing into clinical interventions is an emerging prospect (12).
This article describes the major updates in HAGR since its previous publication in 2009. After a brief introduction to each database, general statistics on content and descriptions of the new types of annotations introduced in HAGR are presented, and a newly added database—GenDR, which focuses on dietary restriction (DR)-associated genes—is then described. Finally, the new HAGR interface, completely redesigned to be more intuitive and user friendly, with an improved cross-integration of our different databases and tools, is presented.
DATABASE CONTENT
New content and features in GenAge
The main aim of the GenAge database (http://genomics.senescence.info/genes/) is to host high-quality curated gene-centric information relevant to human ageing. Although initially GenAge was designed to include only human genes potentially associated with ageing, the database has significantly grown since, and several new gene sets have been added to it. For example, GenAge includes, since 2008, a list of genes from model organisms based on genetic manipulation experiments (2).
Currently, the database is divided into three main sections intertwined through weblinks and cross-references: (i) the set of human ageing-associated genes, which includes the few genes directly related to ageing in humans, and the best candidate human genes, supported by evidence from model organisms, cellular experiments and functional analyses; (ii) the set of longevity-associated genes in model organisms, based on lifespan-modulating genetic interventions; and (iii) a list of genes whose expression is commonly altered during ageing in multiple tissues of mammalian species, inferred from microarray data (Table 1). In addition to these data sets, a work-in-progress list of genes analysed for their possible association with human longevity in population studies is also available in GenAge (http://genomics.senescence.info/genes/longevity.html).
Table 1.
Gene data sets in GenAge
| Gene set | Description |
|---|---|
| Human | A comprehensive set of genes potentially associated with human ageing. The list contains genes that have been directly linked to ageing in humans, as well as the best candidate genes, supported by different types of evidence in model organisms, cells and/or functional analyses. |
| Models | A set of genes in model organisms (predominantly from Mus musculus, Drosophila melanogaster, Caenorhabditis elegans and Saccharomyces cerevisiae) shown to significantly affect lifespan through genetic manipulations. |
| Microarray | A list of genes commonly altered during mammalian ageing from a meta-analysis of microarray studies (13). |
Since our last update in 2009, we have made numerous improvements in terms of both the quantity and the quality of the content in GenAge. The data set of human genes in GenAge has grown only slightly, accounting now for 288 genes, an increase of 27 genes. As mentioned before, the data set contains a list of genes potentially associated with human ageing. For each gene, a description compiled from the studies that link the gene to ageing is provided. It should be noted that our focus is on genes that might affect the ageing process, rather than individual age-related pathologies; genes affecting multiple, even if not all, age-related processes or pathologies may be selected. Besides containing genes directly linked to human ageing (mostly those genes in which mutations result in segmental progeroid syndromes), the homologous genes with the strongest evidence from model organisms, especially from mammals, will typically be found in the human data set. More formally, genes were included in the human gene data set, and annotated accordingly, if one or more of the following criteria were met, the gene was directly linked: (i) to ageing in humans; (ii) to ageing in a mammalian model organism; (iii) to human longevity and/or multiple age-related phenotypes (the only new criteria since our last update); (iv) to ageing in a non-mammalian model organism; (v) to ageing in a cellular model system; (vi) to the regulation or control of genes previously linked to ageing; (vii) to a pathway or mechanism linked to ageing; or (viii) if the gene was acting downstream of a pathway, mechanism or other gene product linked to ageing. As these data are under continuous curation, and new observations are being actively added to existing entries, the quality of the data set has also been improved since our previous update; for example, the bibliography for this data set has increased significantly, and it currently comprises >2200 references.
In contrast to the human data set, whose improvements were mostly qualitative, the latest GenAge build hosts now >1700 genes associated with longevity in model organisms, a drastic boost compared with the 2009 update (a 2.17-fold increase, in fact, in the total number of entries). This rapid increase in the volume of data in the model organisms data set could be mainly attributed to the significant advancements in high-throughput technologies in recent years. For example, in yeast, >750 of the total number of longevity observations (73%) come from several recent large-scale screens (6–10).
All the model organism gene entries in GenAge are based on experimentally validated results from the peer-reviewed scientific literature and are manually extracted by our database curators. Genes are considered for inclusion if genetic manipulations (including knockout, mutations, overexpression or RNA interference) result in noticeable changes in the ageing phenotype and/or lifespan. In the cases where a reduction in lifespan is observed, we include only studies in which the authors are linking the gene interventions to ageing (either mechanistically or by checking various signs and markers of ageing). Exceptions are genes from large-scale experiments in yeast, though as detailed below, these have a different classification. Although we try to be as objective as possible, our selection process is still to a certain degree subjective. Our long-established policy is to have an inclusive rather than exclusive policy, but providing the evidence and links to the relevant literature to allow users to reach their own opinions about each featured gene.
In addition to the increase in the number of entries, in the current update, we have altered the database schema allowing us to host for each longevity-associated gene multiple observations from the same or different studies. By doing this, we are aligning our policy for the data set of genes in model organisms with that for the human data set, namely, reporting all supporting and/or conflicting results instead of only the first study. As of writing (build 16), GenAge provides 1708 genes and 2121 lifespan observations (on average 1.24 observations per gene), of which only a small number (4% of the genes) represent conflicting results (Table 2). Moreover, most (83%) of these ‘conflicting’ results were found for yeast genes.
Table 2.
Summary of the genes and longevity observations for model organisms
| Entries in GenAge | M. musculus | D. melanogaster | C. elegans | S. cerevisiae | Total |
|---|---|---|---|---|---|
| Number of genes | 91 | 128 | 680 | 809 | 1708 |
| Pro-longevity | 64 | 85 | 221 | 43 | 413 |
| Anti-longevity | 24 | 41 | 439 | 217 | 721 |
| Necessary for fitness | – | – | – | 485 | 485 |
| Number of observations | 119 | 151 | 801 | 1050 | 2121 |
| Observations per gene | 1.31 | 1.17 | 1.18 | 1.30 | 1.24 |
| Greatest lifespan increase | 50% | 92% | 10-fold | 6-fold | – |
One of the reasons GenAge has been so popular since its creation is because it has allowed computational gerontologists to access and directly use its data. In this update, we have gone one step further and acknowledged that GenAge can be even more useful if more quantitative data is included. As such, the structure and content of the database have been updated, and the new version of GenAge includes extensive quantitative data. Namely, we have extracted for each experiment, where data was available, the effect (relative change) that a certain genetic intervention has on the mean and/or maximal lifespan. In total, >1250 observations, accounting for 1057 genes for model organisms, have been annotated this way.
Additionally, we have recorded the type of intervention and categorized longevity-associated genes either as pro- or anti-longevity. The criteria for the division of longevity-associated genes into pro- and anti-longevity are based on the type of intervention (loss-of-function or gain-of-function) and its impact on lifespan, and were described previously (3). Briefly, pro-longevity genes are defined as the genes whose overexpression extends lifespan, or whose decreased activity (e.g. because of knockout or RNA interference) reduces lifespan. As aforementioned, in the latter case, we focus on genes linked to ageing processes, such as genes in which mutations result in signs of premature ageing. As this can be problematic for yeast large-scale lifespan screens, yeast genes derived from screens or for which a link to ageing processes has not been observed have been annotated as ‘necessary for fitness’ genes instead of ‘pro-longevity’ genes. Anti-longevity genes are those for which the aforementioned interventions have the opposite effects. In cases where conflicting results were observed, or the data were not sufficient to draw a definite conclusion, our policy has been to keep all observation and annotate the genes as ‘Unclear’ and ‘Unannotated’, respectively. Not taking into account genes with conflicting results, GenAge catalogues a total of 721 anti-longevity genes, 413 pro-longevity genes and 485 genes necessary for fitness (Table 2). It should be, however, noted that this does not necessarily reflect the real genome-wide distribution of pro- and anti-longevity genes, as our data does not account for any biases introduced by the type of experimental design most commonly used by gerontologists.
As in previous versions (1,2), additional external information (including homologues, cytogenetic information, gene ontology annotation, protein–protein interaction data and sequence information) and links are also incorporated in GenAge.
GenDR—genomics of DR
DR, of which caloric restriction is the most widely studied regimen, is the most robust non-genetic intervention shown to extend lifespan in a multitude of species, from yeast to mammals (12,14). However, the exact mechanisms of how DR extends lifespan remain unknown. To decipher the mechanisms of DR in a systematic fashion, we established GenDR (http://genomics.senescence.info/diet/), the first database of DR-associated genes. Because GenDR and related analysis of DR networks have been recently described elsewhere (15), they will only be briefly described herein. To create GenDR, we compiled from the literature a list of DR-essential genes from model organisms. DR-essential genes were defined as those which, if genetically modified, interfere with DR-mediated lifespan extension and, ideally, do not affect the lifespan of animals on an ad libitum diet (or at least do not appear to be merely causing disease). A subset of these genes act as genetic DR mimetics, as their manipulation leads to an increased lifespan for ad libitum fed animals, which is not further extended by DR. One such example is the growth hormone receptor gene in mice (16), in fact the only mouse gene currently in GenDR. In GenDR, the respective homologues of DR-essential genes are included for all the common model organisms, as well as for humans (15).
A complementary data set in GenDR is a list of genes consistently differentially expressed in mammals under DR. In a recent meta-analysis, a common signature of genes differentially expressed in DR across different mammalian species, strains, tissues and experiments was derived. This signature provides a set of genes that are most robustly responding to DR (17).
Presently, build 1 of GenDR features 158 DR-essential genes plus 173 genes that are part of the conserved molecular signature of DR in mammals. We hope that GenDR will help decipher DR-mediated life extension and promote the development of pharmacological DR mimetics with clinical applications (12). Importantly, GenDR has been fully integrated in HAGR with abundant cross-links between GenDR and GenAge.
Enhancing data quality in AnAge, the database of animal ageing and longevity
As previously mentioned, the AnAge database (http://genomics.senescence.info/species/) hosts ageing-related observations, longevity records and a multitude of additional data (including developmental and reproductive traits, taxonomic information and basic metabolic characteristics) for >4000 animal species. Since its inception (1), the main focus of AnAge has been on longevity data and in particular on the quality of data on maximum longevity, rather than merely taking the highest value that is error prone (18). AnAge includes a qualifier of confidence in the data, an estimate of sample size to aid the use of longevity data in comparative studies of ageing, whether maximum longevity comes from a specimen kept in captivity or from the wild and the specific source of the longevity record. Longevity records are manually curated and, if necessary, evaluated by contacting experts (veterinarians, zoologists, etc.) with first-hand experience in a given taxa to assess the reliability of the data, as previously described (18). Anecdotes, unverified longevity claims and controversial issues are mentioned as additional observations. As such, AnAge is arguably the ‘gold standard’ longevity data in animals.
To assist and facilitate comparative studies of ageing, quantitative data on other traits that often correlate with longevity are also included in AnAge, but these are often taken from other data compilations (19–25). Nonetheless, a variety of automated quality control procedures are in place to detect potential errors in all data in AnAge (18).
The focus of AnAge has always been on vertebrates and on mammals in particular. AnAge features higher quality data from mammals, also because they tend to be subjected to more studies than other taxa. At present (build 12), AnAge features data for 4205 species, of which >1000 species are mammals, but also, >1000 species of birds, >500 reptiles, nearly 200 amphibians and nearly 1000 fish species (Table 3) are present. Seventeen other classes, each represented by only a few species in AnAge (maximum lifespan ranging from 0.16 to 15 000 years) have been excluded from Table 3.
Table 3.
Summary of AnAge species according to taxonomic class
| Class | Number of speciesa | Range of MLS (years) | Average and STDEV of MLS (years) |
|---|---|---|---|
| Aves | 1088 (1186) | 3–79 | 19.2 ± 16.4 |
| Mammalia | 989 (1330) | 2.1–211 | 19.0 ± 15.6 |
| Actinopterygii | 811 (822) | 0.16–205 | 17.9 ± 22.8 |
| Reptilia | 508 (542) | 0.4–177 | 21.3 ± 17.0 |
| Amphibia | 149 (173) | 4.1–102 | 15.6 ± 10.8 |
| Chondrichthyes | 115 (116) | 6–75 | 22.3 ± 14.3 |
aIncluded in the table are the species with data quality annotated as acceptable or above. In brackets the total number of species present in AnAge is given.
MLS, maximum lifespan; STDEV, standard deviation.
As AnAge also serves as an informational website for research on ageing, traditional biomedical model organisms, including yeast and invertebrates, are featured. In fact, all species for which there are ageing- or DR-associated genes are present in AnAge with various cross-links between AnAge and HAGR’s gene-centric databases.
Although in terms of content, AnAge has grown only slightly, we have consistently continued to curate its data and enhance its quality. Additionally, new information and references have also been added to AnAge entries. Specifically, we added a modest 109 new species (mostly birds: 91), suggesting that AnAge may be reaching saturation in terms of reflecting the available quality data on longevity of vertebrates and of mammals in particular. Indeed, of the hundreds of longevity records and entries updated, the vast majority (87%) were from birds. This is mostly because of the continually breaking of longevity records in the wild from various banding studies (26,27). In addition, ample information and >200 references have been added, and >1000 references are now cited in AnAge. The information provided for the most important organisms for research on ageing has also been expanded. For example, in the case of the traditional biomedical model organisms, additional information on ageing phenotypes and/or age-related changes and pathologies has been added, and AnAge thus serves as a reference for the organisms mentioned in GenAge and GenDR.
Overall, AnAge continues to be the reference longevity database for comparative and evolutionary studies of ageing. Although its coverage appears to be reaching saturation, at least for mammals, data quality continues to be improved with more information added and more studies cited (in total, HAGR now includes observations for >1950 species). Like for all HAGR databases, AnAge’s interface has been greatly improved, while maintaining a familiar functionality, and cross-links with other databases has been expanded. Lastly, AnAge has also proven valuable for a variety of studies in other areas, from conservation studies to various evolutionary analyses that use the comparative method and/or benefit from longevity data.
Other informational resources
Associated with HAGR is senescence.info (http://www.senescence.info), which aims to provide an informational repository on the science of ageing. Since its creation, senescence.info has continually grown and has been recently updated. We hope that it provides a comprehensive introduction to biogerontology, for both scientists andnon-scientists alike, covering a wide range of aspectsof ageing research and its social implications. The senescence.info website is also an important resource for students and educators and is a source of teaching materials for various courses on ageing. However, contrary to HAGR, which involves several curators and experts, senescence.info is developed by only one of us (J.P.M.).
A complementary resource to HAGR is the Who’s Who in Gerontology website (http://whoswho.senescence.info/), based around the WhosAge database, which contains information on individuals, mainly researchers, and companies working on ageing and lifespan extension. At the time of writing, WhosAge contains information for 262 researchers and 22 companies working on ageing and related fields. During the development of HAGR, a series of software programs (mostly Perl and SPSS scripts) have been also created to help our team in a variety of bioinformatic analyses, such as demographic analysis of age-related mortality (28), a repository of which is available to users as well (http://genomics.senescence.info/software/).
NEW AND IMPROVED INTERFACE
HAGR has grown extensively since it was first published, encompassing more data on ageing and incorporating new databases. One of the key problems that sites encounter when they have grown so rapidly over time is a steady decline in their usability and navigation: sections become harder to discover and navigational features become overloaded. To address and significantly improve these issues, we have completely redesigned the HAGR interface and added many new features.
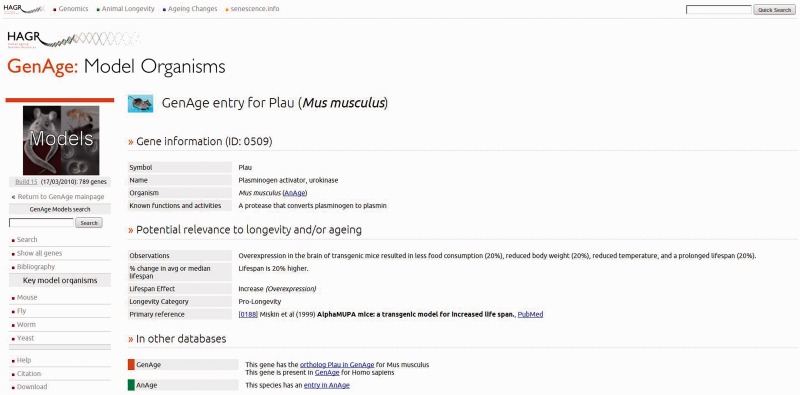
The redesign has overhauled the interface, giving it a new visual look that is more consistent and helps tie each of the resources into a single cohesive piece, as well as improving the navigation to enhance the discoverability of sections. There are now two parts to the navigation, the first being the global navigation bar, which is present across the top of each page in HAGR. This provides quick and consistent navigation to each section, as well as integration for the database-specific searches into a single location. It also provides links to external ageing resources that are relevant, including the Digital Ageing Atlas—a portal of ageing-related changes, which is currently under development (http://ageing-map.org)— senescence.info and the WhosAge database. Finally, the new design provides a global search function that can query each database in HAGR, providing a simple way of searching the data in HAGR without knowing exactly which database is relevant. To complement the global navigation bar, each page contains a left navigation bar that contains context-specific links and information related to the current page. For instance, while using the model organisms' gene section in GenAge, the navigation contains search functions and links to tools related to this data set (Figure 1). This, combined with the greater clarity and structure that the visual refresh provides, makes it much easier for users to find the relevant section and information.
Figure 1.
An entry to the model organisms in the GenAge database for the Plau gene.
All data sets in HAGR have been better integrated and linked to each other, and each gene entry now contains direct links to all other relevant entries in HAGR’s data sets. For example, researchers can quickly see from GenAge if homologues, from the InParanoid database (29), for a model organism gene are present in the human ageing gene set as well or if the gene is present in GenDR. Species information can also be easily accessed through links to AnAge.
While each database was adapted to the new visual style, GenAge was given some further enhancements to help improve the quality of the information within, as mentioned earlier. To support these changes, the search function has gained the ability to filter and sort by organisms and gene annotations (observations of ‘increased’ or ‘decreased’ lifespan effect, and pro- and anti-longevity designations), allowing for a quicker identification of the gene(s) of interest. When presenting search results, if a gene has multiple observations, all suggesting the same effect on lifespan, that effect and the highest observed value will be shown (and implicitly used for filtering).
Another resource that has been completely redesigned is Who’s Who in Gerontology, which has seen changes both to the interface and to its available features. The section focused on individuals has gained a density map indicating the number of researchers working on ageing in each country, as well as a better indication for each person of which country they are currently working in. Each company also now lists their approximate location on a map. These new features combined with a new cleaner design make the Who’s Who resource much easier to use, as well as providing more information than previously.
AVAILABILITY
Same as with our previous access policy, the collection of databases in HAGR is freely available at http://genomics.senescence.info. For all databases, we also provide users with the possibility to export, download and reuse the data for their own analyses, under a Creative Commons Attribution license. Feedback via email is heartily welcome, and we encourage the subscription to the HAGR mailing list to be informed of major updates and changes in HAGR.
CONCLUDING REMARKS
Since its creation, HAGR has proved to be a widely used science of ageing portal with much needed resources for biogerontologists. Both GenAge and AnAge have been featured in a number of other databases and resources, and HAGR has been cited >200 times (Table 4). For example, GenAge has been used as the basis for all the longevity networks hosted in the NetAge database [(30); http://netage-project.org], is featured in JenaLib [(31); http://www.fli-leibniz.de/IMAGE.html] and participates in LinkOut from NCBI resources like OMIM and Entrez Gene (32). AnAge is a content partner of the Encyclopedia of Life (http://eol.org), and its data have also been incorporated into the Biology of Ageing (http://biologyofaging.org) portal, the Animal Diversity Web (http://animaldiversity.ummz.umich.edu) and the Comparative Cellular and Molecular Biology of Longevity Database [(33); http://genomics.brocku.ca/ccmbl/].
Table 4.
Summary of third-party works using and/or citing HAGR
| Year | 2004a | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012b |
|---|---|---|---|---|---|---|---|---|---|
| HAGR citations | 1 | 6 | 7 | 11 | 25 | 33 | 44 | 46 | 23 |
| User statisticsc | 3 | 6.6 | 10.6 | 11.9 | 15.7 | 16.1 | 14.8 | 16.5 | 15.1 |
aStarting in mid-2004.
bBy mid-2012.
cUser statistics include the number of unique visitors per month (in thousands) of all resources combined.
Overall, the current version of the HAGR provides users with a new intuitive interface, significantly augmented information content in the rapidly evolving field of ageing and a better integration of its resources. It is fitting that as a resource for the systems biology of ageing, HAGR has become a resource in which the whole is greater than the sum of its parts. Developments in high-throughput approaches, such as recent advances in next-generation sequencing (34), promise to continue generating ever greater amounts of data in biogerontology that we expect HAGR to continue to accommodate, and thus, we anticipate HAGR to steadily grow. We hope that these and other ongoing improvements will further enhance the use of HAGR’s collection of databases and allow HAGR to continue to be the leading online resource for biogerontology.
FUNDING
Wellcome Trust [ME050495MES to J.P.M.]; Ellison Medical Foundation (to J.P.M.); European Commission FP7 Health Research [HEALTH-F4-2008-202047 to V.E.F., in part]. Funding for open access charge: Wellcome Trust [ME050495MES].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank the numerous contributors and experts who have provided valuable advice and suggestions concerning our resources, and in particular Steve Austad for helping curate AnAge.
REFERENCES
- 1.de Magalhães JP, Costa J, Toussaint O. HAGR: the Human Ageing Genomic Resources. Nucleic Acids Res. 2005;33:D537–D543. doi: 10.1093/nar/gki017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Magalhães JP, Budovsky A, Lehmann G, Costa J, Li Y, Fraifeld V, Church GM. The Human Ageing Genomic Resources: online databases and tools for biogerontologists. Aging Cell. 2009;8:65–72. doi: 10.1111/j.1474-9726.2008.00442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budovsky A, Tacutu R, Yanai H, Abramovich A, Wolfson M, Fraifeld V. Common gene signature of cancer and longevity. Mech. Ageing Dev. 2009;130:33–39. doi: 10.1016/j.mad.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Wolfson M, Budovsky A, Tacutu R, Fraifeld V. The signaling hubs at the crossroad of longevity and age-related disease networks. Int. J. Biochem. Cell Biol. 2009;41:516–520. doi: 10.1016/j.biocel.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 5.Curran S, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Managbanag JR, Witten TM, Bonchev D, Fox LA, Tsuchiya M, Kennedy BK, Kaeberlein M. Shortest-path network analysis is a useful approach toward identifying genetic determinants of longevity. PLoS One. 2008;3:e3802. doi: 10.1371/journal.pone.0003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18:564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabrizio P, Hoon S, Shamalnasab M, Galbani A, Wei M, Giaever G, Nislow C, Longo VD. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 2010;6:e1001024. doi: 10.1371/journal.pgen.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, Boeke JD, Smith JS. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet. 2010;6:e1000921. doi: 10.1371/journal.pgen.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laschober GT, Ruli D, Hofer E, Muck C, Carmona-Gutierrez D, Ring J, Hutter E, Ruckenstuhl C, Micutkova L, Brunauer R, et al. Identification of evolutionarily conserved genetic regulators of cellular aging. Aging Cell. 2010;9:1084–1097. doi: 10.1111/j.1474-9726.2010.00637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Magalhães JP. The biology of ageing: a primer. In: Stuart-Hamilton I, editor. An Introduction to Gerontology. Cambridge, UK: Cambridge University Press; 2011. pp. 21–47. [Google Scholar]
- 12.de Magalhães JP, Wuttke D, Wood SH, Plank M, Vora C. Genome-environment interactions that modulate aging: powerful targets for drug discovery. Pharmacol. Rev. 2012;64:88–101. doi: 10.1124/pr.110.004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Magalhães JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontana L, Partridge L, Longo VD. Extending healthy life span—from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wuttke D, Connor R, Vora C, Craig T, Li Y, Wood S, Vasieva O, Reis RS, Tang F, de Magalhães JP. Dissecting the gene network of dietary restriction to identify evolutionarily conserved pathways and new functional genes. PLoS Genet. 2012;8:e1002834. doi: 10.1371/journal.pgen.1002834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonkowski MS, Rocha JS, Masternak MM, Al Regaiey KA, Bartke A. Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc. Natl Acad. Sci. USA. 2006;103:7901–7905. doi: 10.1073/pnas.0600161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plank M, Wuttke D, van Dam S, Clarke SA, de Magalhães JP. A meta-analysis of caloric restriction gene expression profiles to infer common signatures and regulatory mechanisms. Mol. Biosyst. 2012;8:1339–1349. doi: 10.1039/c2mb05255e. [DOI] [PubMed] [Google Scholar]
- 18.de Magalhães JP, Costa J. A database of vertebrate longevity records and their relation to other life-history traits. J. Evol. Biol. 2009;22:1770–1774. doi: 10.1111/j.1420-9101.2009.01783.x. [DOI] [PubMed] [Google Scholar]
- 19.Hayssen VD, Van Tienhoven A, Van Tienhoven A. Asdell’s Patterns of Mammalian Reproduction: A Compendium of Species-Specific Data. Ithaca, NY: Comstock Publishing Associates; 1993. [Google Scholar]
- 20.Dunning J. CRC Handbook of Avian Body Masses. Boca Raton: CRC Press; 2008. [Google Scholar]
- 21.Nowak R. Walker’s Mammals of the World. Baltimore: Johns Hopkins University Press; 1999. [Google Scholar]
- 22.Jones KE, Bielby J, Cardillo M, Fritz SA, O’Dell J, Orme CDL, Safi K, Sechrest W, Boakes EH, Carbone C, et al. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology. 2009;90:2648. [Google Scholar]
- 23.Froese R, Pauly D, editors. FishBase. World Wide Web electronic publication; 2012. www.fishbase.org, version (August 2012, date last accessed) [Google Scholar]
- 24.Ernest SKM. Life history characteristics of placental non-volant mammals. Ecology. 2003;84:3402–3402. [Google Scholar]
- 25.Poole A, editor. The Birds of North America Online. Ithaca, NY: Cornell Laboratory of Ornithology; 2005. http://bna.birds.cornell.edu/BNA/ (August 2012, date last accessed) [Google Scholar]
- 26.Fransson T, Kolehmainen T, Kroon C, Jansson L, Wenninger T. EURING List of Longevity Records for European Birds. 2010. [Google Scholar]
- 27.Lutmerding JA, Love AS. Longevity Records of North American Birds. 2012. Version 2012.2. Bird Banding Laboratory, Patuxent Wildlife Research Center, Laurel, MD. [Google Scholar]
- 28.de Magalhães JP, Cabral JA, Magalhães D. The influence of genes on the aging process of mice: a statistical assessment of the genetics of aging. Genetics. 2005;169:265–274. doi: 10.1534/genetics.104.032292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer ELL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2009;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tacutu R, Budovsky A, Fraifeld VE. The NetAge database: a compendium of networks for longevity, age-related diseases and associated processes. Biogerontology. 2010;11:513–522. doi: 10.1007/s10522-010-9265-8. [DOI] [PubMed] [Google Scholar]
- 31.Hühne R, Koch FT, Sühnel J. A comparative view at comprehensive information resources on three-dimensional structures of biological macro-molecules. Brief. Funct. Genomic. Proteomic. 2007;6:220–239. doi: 10.1093/bfgp/elm020. [DOI] [PubMed] [Google Scholar]
- 32.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuart JA, Liang P, Luo X, Page MM, Gallagher EJ, Christoff CA, Robb EL. A comparative cellular and molecular biology of longevity database. Age (Dordr) 2012 doi: 10.1007/s11357-012-9458-y. 27 July (doi:10.1007/s11357-012-9458-y; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Magalhães JP, Finch CE, Janssens G. Next-generation sequencing in aging research: emerging applications, problems, pitfalls and possible solutions. Ageing Res. Rev. 2010;9:315–323. doi: 10.1016/j.arr.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]