Abstract
It is 14 years since the IMGT/HLA database was first released, providing the HLA community with a searchable repository of highly curated HLA sequences. The HLA complex is located within the 6p21.3 region of human chromosome 6 and contains more than 220 genes of diverse function. Of these, 21 genes encode proteins of the immune system that are highly polymorphic. The naming of these HLA genes and alleles and their quality control is the responsibility of the World Health Organization Nomenclature Committee for Factors of the HLA System. Through the work of the HLA Informatics Group and in collaboration with the European Bioinformatics Institute, we are able to provide public access to these data through the website http://www.ebi.ac.uk/imgt/hla/. Regular updates to the website ensure that new and confirmatory sequences are dispersed to the HLA community and the wider research and clinical communities. This article describes the latest updates and additional tools added to the IMGT/HLA project.
INTRODUCTION
The IMGT/HLA database was established to provide a locus-specific database (LSDB) for the allelic sequences of the genes in the HLA system, also known as the human major histocompatibility complex (MHC). The MHC is one of the most complex and polymorphic regions of the human genome, with excess of 220 genes (1). The core genes of interest in the HLA system are 21 highly polymorphic HLA genes, found within the 6p21.3 region of the short arm of human chromosome 6, whose protein products mediate human responses to infectious disease and influence the outcome of cell and organ transplants. Three distinct regions have been identified within the MHC. The class I region is located at the telomeric end of the MHC and encodes the genes for the HLA class I molecules, HLA-A, -B and -C. These are co-dominantly expressed on the cell surface and responsible for presenting intracellularly derived peptides to CD8-positive T cells. The class II region lies at the centromeric end of the MHC and encodes HLA class genes HLA-DRA, -DRB1, -DRB3, -DRB4, -DRB5, -DQA1, -DQB1, -DPA1 and -DPB1. HLA class II expression is limited to cells involved in immune responses, where these molecules present extracellularly derived peptides to CD4-positive T cells. Located between the class I and class II regions lies the class III region where a number of non-HLA genes with immune function are located. With a nomenclature covering more than 50 genes and 8000 alleles, there is an obvious need for a curated LSDB to manage these highly polymorphic variants. The first public release of the IMGT/HLA database was made on the 16 December 1998 (2). Since then the database has been updated every 3 months, in a total of 55 releases, to include all the publicly available sequences officially named by the World Health Organization (WHO) Nomenclature Committee at the time of release.
The naming of new HLA genes and allele sequences and their quality control is the responsibility of the WHO Nomenclature Committee for Factors of the HLA System, which first met in 1968. This committee meets regularly to discuss the issues of nomenclature and has published 19 major reports (3–21) initially documenting the serologically defined HLA antigens and more recently the genes and alleles defined by nucleotide sequences. The IMGT/HLA database provides the nomenclature committee with the online tools necessary for its task. The dissemination of new allele names and sequences is of paramount importance in the clinical transplant setting, because the variation that distinguishes HLA alleles can have a critical impact on the outcome of a haematopoietic stem cell transplant (22,23). The identification, verification and publication of the sequences of these variants through a centralized resource are necessary for accurate identification of HLA alleles in a clinical setting. Sequencing of HLA alleles began in the late 1970’s, predominantly using protein-based techniques to determine the sequences of HLA class I allotypes. The first complete HLA class I allotype sequence, B7.2, now known as B*07:02:01, was published in 1979 (24). The first HLA class II allele, DRA*01:01, was defined by protein sequencing and later in 1982 by DNA sequencing (25–27). The first HLA DNA sequences or alleles were named by the WHO Nomenclature Committee for Factors of the HLA System (10) in 1987. At that time, 12 class I alleles and 9 class II alleles were named: in the first 8 months of 2012, the WHO Nomenclature Committee was able to assign names to 1163 alleles (Figure 1).
Figure 1.
The number of HLA alleles named each year and included in the IMGT/HLA Database. The recent surge in the number of submissions received by the database is clearly shown.
IMGT/HLA DATA SOURCES
The IMGT/HLA database receives submissions from laboratories across the world. These submissions are curated and analysed, and if they meet the strict requirements, an official allele designation is assigned. The IMGT/HLA database is the official repository for the WHO Nomenclature Committee for factors of the HLA system and is the only way of receiving an official allele designation for a sequence. The sequence is then incorporated into the next 3-monthly release of the database. Since its release in December 1998, the database has received over 14 000 submissions. These submissions come from a variety of sources; the majority are from laboratories involved in clinical HLA typing, for hospitals or donor registries, or commercial organizations performing contract HLA typing for large haematopoietic stem cell donor registries. Further data have been submitted following large-scale genome sequencing projects (1,28). All submissions must meet strict acceptance criteria before the sequence receives an official designation. These minimum standards cover the methodologies used to define the sequence, the length of sequence submitted and the source of the sequence; the full list of the minimum criteria can be seen at http://www.ebi.ac.uk/imgt/hla/subs/submit.html. Around 3% of the submissions received fail to meet these criteria and are rejected. In addition, all the submissions received by the IMGT/HLA database are also available from the International Nucleotide Sequence Database Collaboration (INSDC) (29). The INSDC consists of DNA DataBank of Japan (Japan), GenBank (USA) and the EMBL-European Nucleotide Archive (ENA) (UK) (30–32). The ENA entries also contain database cross-references to the IMGT/HLA entries. The cross-references to the IMGT/HLA database are also included in ENSEMBL (33) and vertebrate genome annotation (VEGA) entries (34).
TOOLS AVAILABLE AT IMGT/HLA
The IMGT/HLA database provides a diversity of tools for the analysis of HLA sequences. Some of these tools were custom written for the IMGT/HLA database, and others were incorporated from the existing set of tools provided on the European Bioinformatics Institute’s (EBI) website (35,36). The website (Figure 2) includes tools for producing user-defined sequence alignments at the protein, cDNA and gDNA level. The user is also able to perform queries for particular HLA alleles; the output provides access to detailed information on any HLA allele, including information on the ethnic origin of the source, database cross-references and seminal publications. This information is also available through integration with the Sequence Retrieval System (SRS) service at EBI (37).
Figure 2.
The IMGT/HLA homepage, which acts as a portal to the different tools provided on the website.
Tools have also been developed to support the laboratories that sequence HLA. The use of sequence-based typing (SBT) as a method for defining the HLA type is well documented (38,39); most SBT typing strategies currently employed use the exon 2 and exon 3 sequences for HLA class I analysis and exon 2 alone for HLA class II analysis. Because of the heterozygous nature of the SBT analysis, the combinations of many pairs of alleles may give an ambiguous typing result; currently, there are over 60 000 recognized ambiguous combinations. The IMGT/HLA maintains and regularly updates a listing of these ambiguous allele combinations. The document also includes a list of all alleles that are identical over exons 2 + 3 for HLA class I and exon 2 for HLA class II.
Where possible, sequence data, both nucleotide and protein, from the IMGT/HLA database is incorporated into the EBI’s suite of search tools including FASTA (40) and BLAST (41) and downloadable from the EBI’s File Transfer Protocol (FTP) directory in a variety of commonly used formats like FASTA, MSF and PIR.
LATEST DEVELOPMENTS
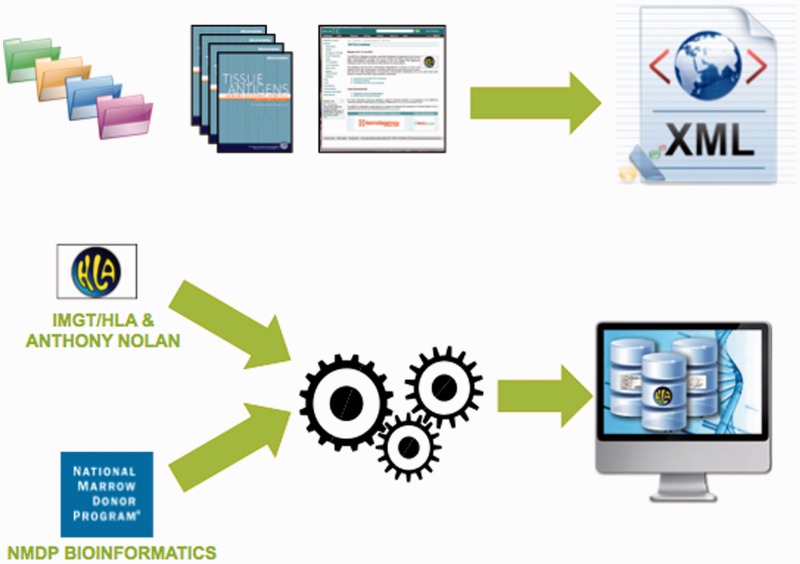
In 2012, the IMGT/HLA database added an Extensible Markup Language (XML) export to the data formats available. XML is a simple but flexible language that defines a set of rules for encoding documents in a format that is both human and machine readable. Designed to meet the challenges of large-scale electronic publishing, XML is playing an increasingly important role in the exchange of scientific data. The data format has been developed in a collaborative project between the HLA Informatics Group of the Anthony Nolan Research Institute and the Bioinformatics Department of the National Marrow Donor Programme (NMDP). The NMDP Bioinformatics group has previous success in developing an XML format for electronically communicating HLA typing data, the Histoimmunogenetics Markup Language file format (42). This experience facilitated the collaboration to develop a similar project for publishing the data contained within each release of the IMGT/HLA database. The new format combines the data present in the multiple files of each quarterly IMGT/HLA release into a single file. The IMGT/HLA database provides an FTP site for the retrieval of sequences in a number of pre-formatted files. The sequences are provided as FASTA, PIR and MSF formats, as well as an archive of the sequence alignments and an ENA flat file like formatted copy of the database. The NMDP Bioinformatics Department has also developed a suite of tools for importing data into different database schema, both open source and proprietary, allowing incorporation into different laboratory systems (Figure 3). Additional XML exports are being developed for other sections of the IMGT/HLA database. Further developmental work on a suite of tools for integrating the XML into laboratory systems used by HLA-typing laboratories is underway.
Figure 3.
The IMGT/HLA export combines a number of existing file formats and data source into a single format. The data are available from the IMGT/HLA database. The tool set is available from the Bioinformatics Group, of the National Marrow Donor Program. Together these allow the user to import the HLA data directly into their local database structure.
HLA matching is a critical factor when considering potential donors for patients receiving allogeneic transplants for haematological disorders (22,23). The most recent development on the IMGT/HLA website is an online tool to implement the T-cell epitope matching algorithm described by Zino et al. (43–45) and updated by Fleischhauer and Shaw (46). This algorithm classifies the HLA-DPB1 alleles into a number of groups based on functional studies and protein motifs. Predictive analysis of the HLA-DPB1 mismatches between patient and donor based on T-cell epitope (TCE) groups has the potential to distinguish between mismatches that are tolerated (permissive) from those that increase the risks of poor clinical outcome (non-permissive). This tool allows the user to enter the HLA-DPB1 of a prospective patient and donor pair and view the predicted TCEs and resulting prediction of the effect of mismatching when selecting appropriate donors for HSCT recipients. Any allele that does not have a TCE group ‘protein’ is analysed for a motif match to particular protein motifs of those alleles with known TCE group. If the tool needs to predict the TCE group for an allele, then a warning is issued within the output to the user, to ensure that the lack of functional studies is acknowledged. The implementation of an easy to use online tool makes it simple for all those staff involved with selecting donors for transplantation to factor in DPB1 mismatches into their own search algorithms and procedures.
FUTURE DEVELOPMENTS
A major challenge for the database is to keep up with the increasing number of allele sequences that are being submitted. In recent years, the number of sequences in the database increased on average by 29% each year. The database must develop new tools for the visualization of sequences while maintaining the high standards set in the presentation and quality of the HLA sequences and nomenclature to the research community. The database aims to continually develop new tools and refine existing tools to meet this challenge.
CONCLUSIONS
The IMGT/HLA database provides a centralized resource for everybody interested, clinically or scientifically, in the HLA system. The database and accompanying tools allow the study of HLA alleles from a single site on the World Wide Web. It aids in the management and development of HLA nomenclature, providing a continuing and updated resource for the WHO Nomenclature Committee. The challenges for the database are to keep up with this increase in submitted sequences, keep pace with the increasing difficulties in performing analyses on the larger datasets and develop new tools for the visualization of the sequences while maintaining the high standards set in the presentation and quality of the HLA sequences and nomenclature to the research community.
LICENSING
The IMGT/HLA database is covered by the Creative Commons Attribution-NoDerivs Licence, which is applicable to all copyrightable parts of the database, which includes the sequence alignments. This means that users are free to copy, distribute, display and make commercial use of the databases in all legislations, provided they give the appropriate credit (47,48). If users intend to distribute a modified version of the data in any form, then they must ask us for permission; this can be done by contacting hla@alleles.org for further details of how modified data can be reproduced.
FUNDING
Histogenetics; One Lambda Inc.; Conexio; Abbott Molecular Laboratories Inc.; European Federation for Immunogenetics; Gen-Probe; LabCorp; Life Technologies; Olersup SSP; 454 Sequencing; American Society for Histocompatibility and Immunogenetics; Anthony Nolan; Asia-Pacific Histocompatibility and Immunogetics Association; BAG Healthcare; Be the Match Foundation; DKMS, Inno-train Diagnostik GMBH; National Marrow Donor Program; Rose and Zentrum Knochenmarkspender-Register Deutschland; Imperial Cancer Research Fund (now Cancer Research UK) and a EU Biotech grant [BIO4CT960037]. Funding for open access charge: The publication costs will be met by the Anthony Nolan Research Institute.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Angie Dahl of the Be The Match Foundation, for her work in securing ongoing funding for the database. They also thank all the individuals and organizations that support the work financially. The authors thank Martin Maiers, Jane Pollack, Adrienne Walts, Joel Schneider, Read Fritsch, Anthony Barber and John Freeman of the Bioinformatics Department of the National Marrow Donor Program for their assistance in developing the XML format.
APPENDIX
ACCESS AND CONTACT
IMGT/HLA Homepage: http://www.ebi.ac.uk/imgt/hla/
-
IMGT/HLA FTP Site: ftp://ftp.ebi.ac.uk/pub/databases/imgt/hla/
Contact: hla@alleles.org
REFERENCES
- 1.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Jr, Wright MW, et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 2.Robinson J, Bodmer JG, Malik A, Marsh SGE. Development of the international immunogenetics HLA database. Hum. Immunol. 1998;59:17. [Google Scholar]
- 3.WHO Nomenclature Committee. Nomenclature for factors of the HL-a system. Bull. World Health Organ. 1968;39:483–486. [PMC free article] [PubMed] [Google Scholar]
- 4. WHO Nomenclature Committee. (1970) WHO Terminology Report. In: Terasaki PI (ed). Histocompatibility Testing, 1970. Munksgaard, Copenhagen, pp. 49.
- 5.WHO Nomenclature Committee. Nomenclature for factors of the HL-A system. Bull. World Health Organ. 1972;47:659–662. [PMC free article] [PubMed] [Google Scholar]
- 6.WHO IUIS Terminology Committee. Nomenclature for factors of the HLA system. Bull. World Health Organ. 1975;52:261–265. [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Nomenclature Committee. Nomenclature for factors of the HLA system, 1977. Tissue Antigens. 1978;11:81–86. doi: 10.1111/j.1399-0039.1978.tb01231.x. [DOI] [PubMed] [Google Scholar]
- 8. WHO Nomenclature Committee. (1980) Nomenclature for factors of the HLA system, 1980. In: Terasaki PI (ed). Histocompatibility Testing, 1980. UCL Tissue Typing Laboratory, Los Angeles, pp. 18–20.
- 9.WHO Nomenclature Committee. Nomenclature for factors of the HLA system 1984. Tissue Antigens. 1984;24:73–80. [PubMed] [Google Scholar]
- 10.WHO Nomenclature Committee. Nomenclature for factors of the HLA system, 1987. Tissue Antigens. 1988;32:177–187. doi: 10.1111/j.1399-0039.1988.tb01655.x. [DOI] [PubMed] [Google Scholar]
- 11.Bodmer JG, Marsh SGE, Parham P, Erlich HA, Albert E, Bodmer WF, Dupont B, Mach B, Mayr WR, Sasasuki T, et al. Nomenclature for factors of the HLA system, 1989. Tissue Antigens. 1990;35:1–8. doi: 10.1111/j.1399-0039.1990.tb01749.x. [DOI] [PubMed] [Google Scholar]
- 12.Bodmer JG, Marsh SGE, Albert ED, Bodmer WF, Dupont B, Erlich HA, Mach B, Mayr WR, Parham P, Sasazuki T, et al. Nomenclature for factors of the HLA system, 1990. Tissue Antigens. 1991;37:97–104. doi: 10.1111/j.1399-0039.1991.tb01853.x. [DOI] [PubMed] [Google Scholar]
- 13.Bodmer JG, Marsh SGE, Albert ED, Bodmer WF, Dupont B, Erlich HA, Mach B, Mayr WR, Parham P, Sasazuki T. Nomenclature for factors of the HLA system, 1991. Hum. Immunol. 1992;34:4–18. doi: 10.1016/0198-8859(92)90079-3. [DOI] [PubMed] [Google Scholar]
- 14.Bodmer JG, Marsh SGE, Albert ED, Bodmer WF, Dupont B, Erlich HA, Mach B, Mayr WR, Parham P, Sasazuki T. Nomenclature for factors of the HLA system, 1994. Tissue Antigens. 1994;44:1–18. doi: 10.1111/j.1399-0039.1994.tb02351.x. [DOI] [PubMed] [Google Scholar]
- 15.Bodmer JG, Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Charron D, Dupont B, Erlich HA, Mach B, Mayr WR. Nomenclature for factors of the HLA system, 1995. Tissue Antigens. 1995;46:1–18. doi: 10.1111/j.1399-0039.1995.tb02470.x. [DOI] [PubMed] [Google Scholar]
- 16.Bodmer JG, Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Charron D, Dupont B, Erlich HA, Fauchet R, Mach B, et al. Nomenclature for factors of the HLA system, 1996. Tissue Antigens. 1997;49:297–321. doi: 10.1111/j.1399-0039.1997.tb02759.x. [DOI] [PubMed] [Google Scholar]
- 17.Bodmer JG, Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Hansen JA, Mach B, Mayr WR, et al. Nomenclature for factors of the HLA system, 1998. Tissue Antigens. 1999;53:407–446. doi: 10.1034/j.1399-0039.1999.530421.x. [DOI] [PubMed] [Google Scholar]
- 18.Marsh SGE, Bodmer JG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Hansen JA, Mach B, Mayr WR, et al. Nomenclature for factors of the HLA system, 2000. Tissue Antigens. 2001;57:236–283. doi: 10.1034/j.1399-0039.2001.057003236.x. [DOI] [PubMed] [Google Scholar]
- 19.Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Mach B, Mayr WR, et al. Nomenclature for factors of the HLA system, 2002. Tissue Antigens. 2002;60:407–464. doi: 10.1034/j.1399-0039.2002.600509.x. [DOI] [PubMed] [Google Scholar]
- 20.Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Hurley CK, Mach B, et al. Nomenclature for factors of the HLA system, 2004. Tissue Antigens. 2005;65:301–369. doi: 10.1111/j.1399-0039.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- 21.Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernandez-Vina M, Geraghty DE, Holdsworth R, Hurley CK, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, Fernandez-Vina M, Flomenberg N, Horowitz M, Hurley CK, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 23.Shaw BE, Mayor NP, Russell NH, Apperley JF, Clark RE, Cornish J, Darbyshire P, Ethell ME, Goldman JM, Little AM, et al. Diverging effects of HLA-DPB1 matching status on outcome following unrelated donor transplantation depending on disease stage and the degree of matching for other HLA alleles. Leukemia. 2010;24:58–65. doi: 10.1038/leu.2009.239. [DOI] [PubMed] [Google Scholar]
- 24.Orr HT, Lopez de Castro JA, Lancet D, Strominger JL. Complete amino acid sequence of a papain-solubilized human histocompatibility antigen, HLA-B7. 2. Sequence determination and search for homologies. Biochemistry. 1979;18:5711–5720. doi: 10.1021/bi00592a030. [DOI] [PubMed] [Google Scholar]
- 25.Lee JS, Trowsdale J, Travers PJ, Carey J, Grosveld F, Jenkins J, Bodmer WF. Sequence of an HLA-DR alpha-chain cDNA clone and intron-exon organization of the corresponding gene. Nature. 1982;299:750–752. doi: 10.1038/299750a0. [DOI] [PubMed] [Google Scholar]
- 26.Wake CT, Long EO, Strubin M, Gross N, Accolla R, Carrel S, Mach B. Isolation of cDNA clones encoding HLA-DR alpha chains. Proc. Natl Acad. Sci. USA. 1982;79:6979–6983. doi: 10.1073/pnas.79.22.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang C, Kratzin H, Gotz H, Thinnes FP, Kruse T, Egert G, Pauly E, Kolbel S, Wernet P, Hilschmann N. [Primary structure of class II human histocompatibility antigens. 2nd Communication. Amino acid sequence of the N-terminal 179 residues of the alpha-chain of an HLA-Dw2/DR2 alloantigen (author's transl)] Hoppe-Seyler's Zeitschrift fur Physiologische Chemie. 1982;363:671–676. [PubMed] [Google Scholar]
- 28.Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L, Jones MC, Horton R, Hunt SE, Scott CE, et al. The DNA sequence and analysis of human chromosome 6. Nature. 2003;425:805–811. doi: 10.1038/nature02055. [DOI] [PubMed] [Google Scholar]
- 29.Karsch-Mizrachi I, Nakamura Y, Cochrane G. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2012;40:D33–D37. doi: 10.1093/nar/gkr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kodama Y, Mashima J, Kaminuma E, Gojobori T, Ogasawara O, Takagi T, Okubo K, Nakamura Y. The DNA Data Bank of Japan launches a new resource, the DDBJ Omics Archive of functional genomics experiments. Nucleic Acids Res. 2012;40:D38–D42. doi: 10.1093/nar/gkr994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amid C, Birney E, Bower L, Cerdeno-Tarraga A, Cheng Y, Cleland I, Faruque N, Gibson R, Goodgame N, Hunter C, et al. Major submissions tool developments at the European Nucleotide Archive. Nucleic Acids Res. 2012;40:D43–D47. doi: 10.1093/nar/gkr946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38(Suppl.):W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McWilliam H, Valentin F, Goujon M, Li W, Narayanasamy M, Martin J, Miyar T, Lopez R. Web services at the European Bioinformatics Institute-2009. Nucleic Acids Res. 2009;37:W6–W10. doi: 10.1093/nar/gkp302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harte N, Silventoinen V, Quevillon E, Robinson S, Kallio K, Fustero X, Patel P, Jokinen P, Lopez R. Public web-based services from the European Bioinformatics Institute. Nucleic Acids Res. 2004;32:W3–W9. doi: 10.1093/nar/gkh405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santamaria P, Lindstrom AL, Boyce-Jacino MT, Myster SH, Barbosa JJ, Faras AJ, Rich SS. HLA class I sequence-based typing. Hum. Immunol. 1993;37:39–50. doi: 10.1016/0198-8859(93)90141-m. [DOI] [PubMed] [Google Scholar]
- 39.Rozemuller EH, Bouwens AG, van Oort E, Versluis LF, Marsh SGE, Bodmer JG, Tilanus MG. Sequencing-based typing reveals new insight in HLA-DPA1 polymorphism. Tissue Antigens. 1995;45:57–62. doi: 10.1111/j.1399-0039.1995.tb02415.x. [DOI] [PubMed] [Google Scholar]
- 40.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Maiers M. A community standard XML message format for sequencing-based typing data. Tissue Antigens. 2007;69:69–71. doi: 10.1111/j.1399-0039.2006.76061.x. [DOI] [PubMed] [Google Scholar]
- 43.Zino E, Vago L, Di Terlizzi S, Mazzi B, Zito L, Sironi E, Rossini S, Bonini C, Ciceri F, Roncarolo MG, et al. Frequency and targeted detection of HLA-DPB1 T cell epitope disparities relevant in unrelated hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2007;13:1031–1040. doi: 10.1016/j.bbmt.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 44.Crocchiolo R, Zino E, Vago L, Oneto R, Bruno B, Pollichieni S, Sacchi N, Sormani MP, Marcon J, Lamparelli T, et al. Nonpermissive HLA-DPB1 disparity is a significant independent risk factor for mortality after unrelated hematopoietic stem cell transplantation. Blood. 2009;114:1437–1444. doi: 10.1182/blood-2009-01-200378. [DOI] [PubMed] [Google Scholar]
- 45.Zino E, Frumento G, Marktel S, Sormani MP, Ficara F, Di Terlizzi S, Parodi AM, Sergeant R, Martinetti M, Bontadini A, et al. A T-cell epitope encoded by a subset of HLA-DPB1 alleles determines nonpermissive mismatches for hematologic stem cell transplantation. Blood. 2004;103:1417–1424. doi: 10.1182/blood-2003-04-1279. [DOI] [PubMed] [Google Scholar]
- 46.Fleischhauer K, Shaw BE, Gooley T, Malkki M, Bardy P, Bignon JD, Dubois V, Horowitz MM, Madrigal JA, Morishima Y, et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13:366–374. doi: 10.1016/S1470-2045(12)70004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson J, Malik A, Parham P, Bodmer JG, Marsh SGE. IMGT/HLA database—a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- 48.Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SGE. The IMGT/HLA database. Nucleic Acids Res. 2011;39:D1171–D1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]