Abstract
Background
Reconstructing the evolutionary history of organisms using traditional phylogenetic methods may suffer from inaccurate sequence alignment. An alternative approach, particularly effective when whole genome sequences are available, is to employ methods that don’t use explicit sequence alignments. We extend a novel phylogenetic method based on Singular Value Decomposition (SVD) to reconstruct the phylogeny of 12 sequenced Drosophila species. SVD analysis provides accurate comparisons for a high fraction of sequences within whole genomes without the prior identification of orthologs or homologous sites. With this method all protein sequences are converted to peptide frequency vectors within a matrix that is decomposed to provide simplified vector representations for each protein of the genome in a reduced dimensional space. These vectors are summed together to provide a vector representation for each species, and the angle between these vectors provides distance measures that are used to construct species trees.
Results
An unfiltered whole genome analysis (193,622 predicted proteins) strongly supports the currently accepted phylogeny for 12 Drosophila species at higher dimensions except for the generally accepted but difficult to discern sister relationship between D. erecta and D. yakuba. Also, in accordance with previous studies, many sequences appear to support alternative phylogenies. In this case, we observed grouping of D. erecta with D. sechellia when approximately 55% to 95% of the proteins were removed using a filter based on projection values or by reducing resolution by using fewer dimensions. Similar results were obtained when just the melanogaster subgroup was analyzed.
Conclusions
These results indicate that using our novel phylogenetic method, it is possible to consult and interpret all predicted protein sequences within multiple whole genomes to produce accurate phylogenetic estimations of relatedness between Drosophila species. Furthermore, protein filtering can be effectively applied to reduce incongruence in the dataset as well as to generate alternative phylogenies.
Keywords: Singular value decomposition, Phylogenomics, Comparative genomics, Drosophila phylogeny
Background
Methods that determine phylogenies based on a restricted number of genes can be negatively affected by horizontal gene transfers, incomplete lineage-sorting, introgression, and the unrecognized comparison of paralogous genes. The recent explosive increase in the number of completely sequenced genomes allows us to consider inferring gene and/or organismal relationships using complete sequence data. Several methods for generating phylogenies based on whole genome information have been explored, and many of these have been applied to re-examine the phylogeny of Drosophila. These include methods based primarily or exclusively on gene content [1], gene order [2], and detailed comparisons of operationally defined orthologs [3]. However, these methods often fail to provide detailed and unbiased comparisons of a high fraction of sequences and instead produce phylogenies based on greatly filtered, preselected datasets. We developed a phylogenetic method that provides accurate comparisons for a high fraction of sequences within whole genomes without the prior identification of orthologous or homologous sites [4]. Our approach allows a relatively comprehensive comparison of complete genome protein sequence, thereby taking into account a higher fraction of total sequence information and providing comprehensive definitions for the various species of interest. This method has been successfully applied to a number of diverse species including vertebrate mitochondrial genomes, plant viral genomes, and eukaryotic nuclear genomes [4-7].
Complete genome sequences for 10 additional species of Drosophila were added to the sequences already available for D. melanogaster and D. pseudoobscura in order to improve the precision and sensitivity of evolutionary inference regarding these organisms [8]. As a result, the currently accepted species phylogeny for these organisms has been further refined and resolved. However, these methods generally continue to utilize greatly filtered data sets primarily comprised of selected single copy orthologous sequences [9-14].
Many such studies have resulted in what is largely considered to be a fully resolved phylogeny for the 12 sequenced species of Drosophila. However, some doubts remain with respect to the placement of certain members of the melanogaster group: D. erecta, D. yakuba and D. melanogaster, placement of the Hawaiian species: D. grimshawi, and to some extent virilisrepleta group: D. virilis and D. mojavenis[15-19]. Among these, the placement of D. erecta and D. yakuba with respect to D. melanogaster is perhaps least certain. Though evidence has been presented to support all the possible phylogenies with respect to D. melanogaster, D. erecta, and D. yakuba, support for each of these phylogenies is not uniformly strong [12]. In this study we apply our more inclusive whole genome phylogenetic method on the 12 genomes of Drosophila to further investigate and validate our current understanding of their phylogenetic relationships.
Results and discussion
Preliminary studies were conducted using a small dataset comprising only 6 genomes of the melanogaster group (D. melanogaster, D. sechellia, D. simulans, D. erecta, D. yakuba and D. ananassae) with a total of 100,851 predicted proteins. Further studies were conducted using a large dataset consisting of all the 12 Drosophila spp. genomes with a total of 193,622 proteins (Table 1). Additional 11 genome datasets excluding one of the melanogaster group species were also constructed for the detailed analysis of the phylogenies. Although there were large similarities in the total number of genes among the Drosophila species, there were large variations in the total number of predicted proteins (Table 1). It seems likely that the melanogaster genome is more fully annotated with a larger number of alternatively spliced transcripts producing multiple (but perhaps slightly different) protein products relative to other Drosophila genomes. Among the 12 species, D. melanogaster had the highest number of predicted proteins (22,765) and D. virilis had the lowest (14,491). Each species’ contribution to the dataset was in the range of 7.48% to 8.51% except for D. melanogaster which contributed about 11.76% for the total. In previous studies, we noted that a modest size difference in genomes has little effect on the final outcome of the tree [4,6].
Table 1.
List of 12 Drosophila spp used in the analysis, along with the number of predicted proteins
| # | Species | Genes | Proteins |
|---|---|---|---|
| 1 |
Drosophila simulans |
16117 |
15415 |
| 2 |
Drosophila sechellia |
17286 |
16471 |
| 3 |
Drosophila melanaogaster |
15431 |
22765 |
| 4 |
Drosophila erecta |
15810 |
15048 |
| 5 |
Drosophila ananassae |
15978 |
15070 |
| 6 |
Drosophila yakuba |
16904 |
16082 |
| 7 |
Drosophila pseudoobscura |
16712 |
16308 |
| 8 |
Drosophila persimilis |
17573 |
16878 |
| 9 |
Drosophila willistoni |
16385 |
15513 |
| 10 |
Drosophila mojavensis |
15179 |
14595 |
| 11 |
Drosophila virilis |
15343 |
14491 |
| 12 | Drosophila grimshawi | 15885 | 14986 |
Higher dimension SVD analysis
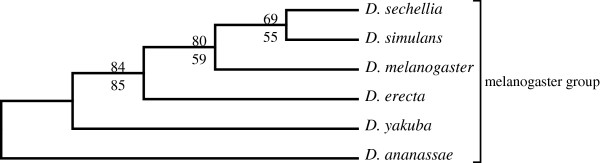
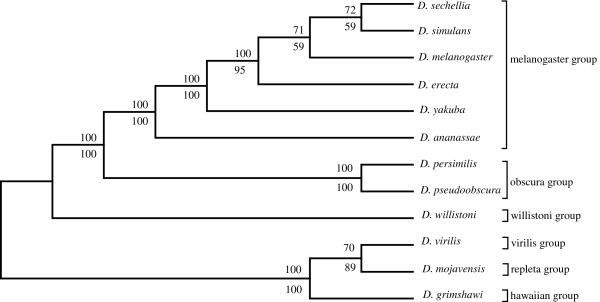
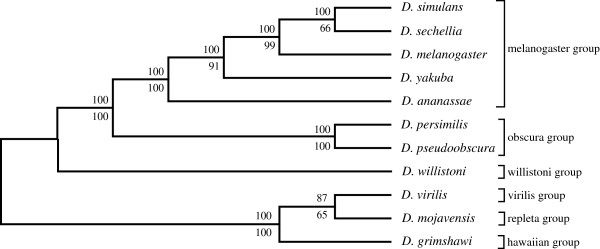
Figure 1 and 2 shows the SVD-based topology obtained via Neighbor-joining for the 6 and 12 genome Drosophila species data sets respectively. Two types of resampling methods were used to estimate branch statistics for this tree. The bottom value on each branch was generated using a traditional bootstrap procedure [4] by sampling 800 singular triplets to construct 700 species trees. The top value on each branch was generated using a successive, delete-one jackknife procedure [4] wherein the least dominant singular vector was removed successively (from 800 to 100 vectors) to generate 700 ordered sets of singular vectors, and a new tree was estimated following each removal. Most of the branches were well supported following application of either the modified jackknife procedure or the bootstrap procedure. Bootstrap yielded a slightly lower branch support for the D. sechellia, D. simulans, and D. melanogaster branch but all other branches were strongly supported by both procedures. The observed difference was likely due to the uniform use of the 700 most dominant vectors in our modified jackknife procedure, while in contrast, the standard bootstrap samples randomly over all 800 vectors generated. The end result is a phylogeny that corresponds well to the currently accepted phylogeny [12,20-22], except for D. erecta and D. yakuba, which remain adjacent in the tree, but fail to cluster as sister species.
Figure 1.
The higher dimension SVD tree for the 6 Drosophila spp., using all 700 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
Figure 2.
The higher dimension SVD tree for the 12 Drosophila spp., using all 700 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
In order to further examine the robustness of the data supporting the correct tree, we performed a series of analyses by systematically excluding protein sequences that were poorly described by their corresponding singular vectors in terms of projection values. The theoretical projection values for a given protein range from −1 to +1. In the first step, all protein sequences having projection value less than or equal +0.001 and more than or equal to −0.001 were removed (about 9,500 sequences). The filter was increased stepwise with an increment of 0.001 and each corresponding dataset was used in turn to construct a tree. When about 54.54% (105,596 sequences) of the original dataset was removed (projection value less than or equal to +0.003 and more than or equal to −0.003), a unique clustering of D. erecta with D. schelliea was observed (Additional file 1). Continued successive increases in stringency to remove poorly described proteins failed to alter this novel cluster until more than 95% (185,039) of the total protein sequences were removed. This resulted in a re-clustering of D. erecta with D. yakuba as sister species, but this was accompanied by the movement of D. melanogaster to a novel position (Additional file 2). Removing a high fraction of poorly described proteins (those with smaller projections on any singular vector) would presumably tend to produce a more highly correlated data set consisting of smaller sets of highly conserved proteins. The tree generated using the modified jack knife procedure, rather than the bootstrap, showed a similar branching pattern. Branch support values for the tree exceeded 80% in all cases, and only 60% for the D. yakuba and D. erecta cluster.
Lower dimension SVD analysis
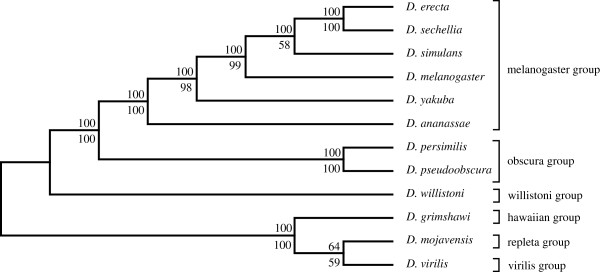
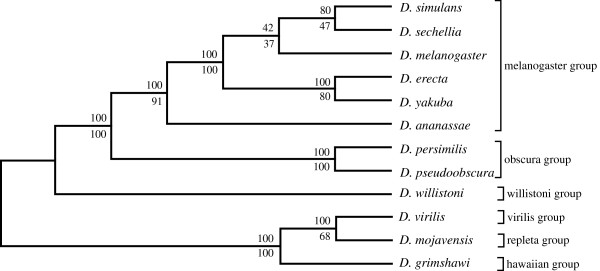
A corresponding lower dimension analyses of the Drosophila spp. was also conducted using the same procedure but with fewer (500) singular triplets. Here the bootstrap branch statistics were generated by sampling 100 random sets of 150 singular triplets to construct 100 species trees. The delete-one jackknife values were generated using 400 ordered sets of singular vectors. Trees were estimated following each successive removal of a least dominant vector from 500 to 100 vectors. The SVD phylogeny obtained for the unfiltered 12 Drosophila species dataset (Figure 3) corresponds well to the currently accepted phylogeny, except for D. erecta, which shows a novel affinity with D. sechellia. It proved possible to disrupt this novel affinity after reducing the number of proteins used in the summation step by 97.57% (Figure 4) by applying a relatively severe filter (projection value less than or equal to +0.035 and more than or equal to −0.035) and thus using only the remaining highly correlated data set consisting of smaller sets of highly conserved proteins. Branch support values for the tree exceeded 70% in all cases, and more than 80% except for the D. melanogaster, D. yakuba and D. erecta cluster.
Figure 3.
The lower dimension SVD tree for the 12 Drosophila spp., using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
Figure 4.
The lower dimension SVD tree for the 12 Drosophila spp., using 300 vectors, with heavy filtering of proteins with projection values ≤ ±0.035. A total of 4430 (2.43%) proteins were used for constructing trees (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
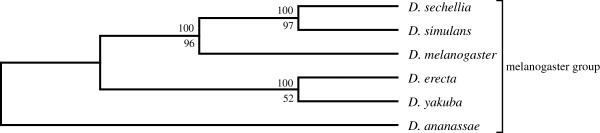
In order to study the relationships among members of the melanogaster group without the influence of D. erecta, a slightly smaller dataset of 11 Drosophila species (178,574 total predicted proteins) was used for analysis. This data set produced the currently accepted phylogeny with strong branch support (Figure 5) [12,20-22]. The observed relationship was consistent across different levels of protein filtering. Both the bootstrap and the modified jackknife produced strong branch support values for most branches.
Figure 5.
The lower dimension SVD tree for the 11 Drosophila species (excluding D. erecta) using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
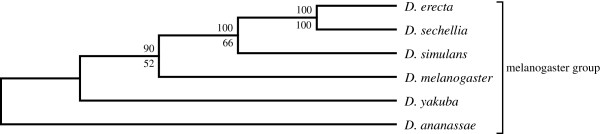
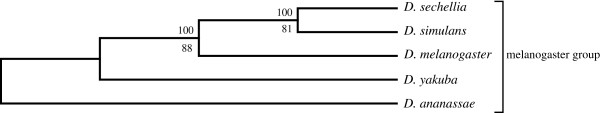
A similar result was obtained with an even smaller dataset that included only 6 genomes with 100,851 predicted proteins (Figure 6 and 7). When subjected to SVD analysis, this produced the currently accepted phylogeny for all 6 members of the melanogaster group, but only under stringent protein filtering (Figure 7). The effect of including more proteins using a less severe protein filter was similar for both the 12 genome tree and the 6 genome tree: D. erecta fails to cluster with D. yakuba and instead clusters with D. sechellia. However, just like in the 11 Drosophila dataset, exclusion of D. erecta from the melanogaster group produced the currently accepted phylogeny with strong branch support (Figure 8) without filtering any proteins. The effect of other genomes on the phylogeny was systematically studied by excluding one of the melanogaster group species from the original 12 genome dataset. All these analyses showed the novel D. sechellia and D. erecta clustering (Additional file 3, Additional file 4, Additional file 5 and Additional file 6) except for the dataset from which D. sechellia was excluded which produced the currently accepted phylogeny (Additional file 7). But, all datasets produced the currently accepted phylogeny under stringent filtering conditions (Additional files 8, 9, 10, 11, and 12).
Figure 6.
The lower dimension SVD tree for the 6 Drosophila species (melanogaster group) using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
Figure 7.
The lower dimension SVD tree for the 6 Drosophila spp., using 300 vectors, with heavy filtering of proteins with projection values ≤ ±0.035. A total of 4048 (4.06%) proteins were used for constructing trees (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
Figure 8.
The lower dimension SVD tree for the 5 Drosophila species (melanogaster group, excluding D. erecta) using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
Conclusions
Our results indicate that it is possible to consult and interpret all predicted protein sequences within multiple whole genomes to produce accurate phylogenetic estimations of relatedness between Drosophila species. Unlike our approach, the most recent independent standard analyses based on whole genome sequence information depend upon filtered data sets in which a restricted number of highly conserved and putatively orthologous genes were compared. In addition, unlike standard methods which use sequence alignments, our method uses angles between high dimensional vectors to estimate evolutionary distance. Despite these novelties in method, the phylogenetic tree derived for the 6 species of the melanogaster group, as well as all 12 species of Drosophila, exhibits strong branch support values and corresponds almost exactly to the currently accepted phylogeny. We conclude that it is possible to include the entire dataset for a more inclusive and potentially more robust analysis using a novel method to produce equivalent results.
This greatly expanded data set appears to contain a strong component of conflicting sequence information that specifically causes D. erecta and D. sechellia to cluster, but this was observed only when more than 55% (105,596) of the proteins are removed. However, this cluster disappears again when 95% (185,039) of poorly described proteins are removed. At lower dimensions, the D. erecta and D. sechellia cluster appears to be stable under various filter settings. Only under stringent filtering conditions could the correct phylogeny be restored. Additionally exclusion of either D. sechellia or D. erecta from the 12 species dataset could produce the currently accepted phylogeny.
The relative placement of D. erecta and D. yakuba with respect to D. melanogaster was largely uncertain until multigene analyses tended to support the same standard tree [9-14,23]. This standard tree is well supported in multiple distinct analyses and is essentially non-controversial, representing the currently accepted statement concerning the relatedness of the first twelve fully sequenced Drosophila genomes. However, previous single gene analyses supported a variety of distinct trees [15,16,24-30], and more comprehensive surveys of putative orthologs revealed a high frequency of conflicting trees [11-13]. Even though the currently accepted phylogeny had the strongest support, depending on the evolutionary model applied, roughly 40% of all orthologous genes examined supported alternative phylogenies within the melanogaster subgroup [12]. In this case, the standard D. erecta/D. yakuba cluster was specifically examined, and only two alternatives, those in which either of these species specifically clustered instead with D. melanogaster, were considered. Two reasons are commonly offered to explain the conflicts observed in these surveys of single gene phylogenies: incomplete lineage sorting, and introgression. Either of these processes could potentially be at least partly responsible for the novel grouping of D. erecta and D. sechellia we observed under the special mid-range filtering conditions reported here.
An alternative but not mutually exclusive explanation for the conditional novel clustering observed in this work is that the sequence signal causing this exists primarily outside of a reasonably complete list of identifiable orthologs (Additional file 2). Although not a necessity, this signal could easily be interpreted as homoplasious. This interpretation is supported by the fact that the standard clustering of D. yakuba and D. erecta was observed again when using only protein sequences with the highest projection values, which includes a small subset of proteins that are more likely to represent close homologs or orthologs. It is also possible that the sequence signal responsible might not be exclusively located outside identifiable orthologs, but might also be partly embedded within orthologs as similar subsets of specific sequence changes within these genes. In either case, it would still be interesting to further investigate the source and strength of these presumed homoplasies, given that they specifically and consistently support a single alternative placement for a single species within a complex tree.
Regardless of their location relative to orthologs, if the sequence characteristics within our all-inclusive analysis that consistently result in the association of D.erecta with D. sechellia represent homoplasious molecular responses to one or more environmental conditions, then this represents a third widely recognized mechanism for generating phylogenetic conflict within sequence data: adaptive convergence. Hence the affinity observed here between erecta and sechellia could result from non-random homoplasy with evolutionary significance. As an example for illustration, consider that D. sechellia and D. erecta are two of only three “specialist” species in the phylogeny that have adapted to specific food sources, and unlike the third species (virilis), they are closely related members of the melanogaster subgroup and have both adapted to particular fruits [31]. Although this single proposed adaptation might seem unlikely to be the sole source of a homoplasious signal capable of clustering D. sechillia and D. erecta in our analysis, multiple similar undiscovered or undescribed convergences could produce a sufficiently robust signal.
Methods
Datasets
Complete predicted protein sequences for 12 Drosophila species were downloaded from the ‘Assembly, Alignment and Annotation of 12 Drosophila species’ website (http://rana.lbl.gov/drosophila/) and were compiled into a single dataset. Various distinct subsets of this larger dataset were also constructed. The number of protein sequences found within the genome of each species of Drosophila is summarized in Table 1.
Peptide frequencies and SVD
The twenty amino acids provide 160,000 possible tetrapeptides, defining each row of the peptide frequency matrix. For every protein, the frequency of each of these tetrapeptides formed the columns of the matrix. The resulting matrix is thus a peptide frequency matrix (A), with each column providing protein vector definitions using 160,000 separate tetrapeptide frequency elements. In our previous studies, using tripeptides we were able to estimate similarities between highly divergent, small set of proteins [32]. It was also shown that tetrapeptides work better for larger data sets derived from vertebrate mitochondrial genomes or whole bacterial genomes [4]. Since, pentapeptides did not add any resolution for estimating similarities on our simulated datasets (unpublished); we chose tetrapeptides for constructing frequency matrix. A peptide frequency matrix was generated for all the three datasets, separately. The resulting matrix was then subjected to a truncated SVD analysis that generates three component matrices: the “left” matrix or “peptide” matrix (U), the “right” matrix or “protein” matrix (V) and the central matrix (∑). The original matrix can be reformed using the relation A = U ∑ VT. The “protein” vectors provided in the “right” factor matrix are known to provide reduced dimensional definitions for all proteins in the dataset as linear combinations of the orthogonal “right” singular vectors [6]. The dataset could produce a total of 910 singular vectors with the reduced dimensional space. An examination of the contribution provided by the less dominant singular vectors showed that these vectors tended to decrease the resolution of the resulting phylogenetic tree (not shown). Using the first 800 vectors was thus determined to be sufficient. The current phylogenetic studies were conducted under two different SVD settings, one referred as “higher dimension,” where we used a total of 800 singular triplets as output and the other referred as “lower dimension” using only 400 singular triplets as output. The SVD was then applied to the 12, 11 and 6 species datasets of Drosophila separately. Three output matrices were obtained consisting of 800 (for higher dimension analysis) and 500 (for lower dimension analysis) singular triplets (left and right singular vectors and their corresponding singular value). Higher the value of vector elements, most dominant is the singular vector and these singular vectors define one or two conserved gene families (or subfamilies) as particular linear combinations of proteins. The detailed comparative information contained within the hundreds of singular vectors and their corresponding motifs and gene families was subsequently used to build a species phylogeny by summing all the SVD-derived right protein vectors separately for each organism and then comparing the relative orientation of the resulting species vectors [6].
Filtering proteins
A systematic exclusion of protein sequences, based on their projection values were done to filter poorly described proteins. The projection value represent a given protein range from −1 to +1. In the first step, all protein sequences having projection value less than or equal +0.001 and more than or equal to −0.001 were removed (about 9,500 sequences). The filter was increased stepwise with an increment of 0.001 and each corresponding dataset was used in turn to construct a tree.
Species trees and branch support
Distance matrices were derived by summing all the SVD derived right protein vectors for a given organism and then comparing the relative orientation of the resulting species vectors using the program cosdist. Species trees were subsequently derived from distance matrices using Phylip-Neighbor. Two distinct resampling methods were used to provide branch support: a traditional bootstrap procedure and a modified jackknife procedure. For the bootstrap, a fixed number of singular vectors were randomly sampled from the total singular vectors generated and were used to construct 100 species trees. For the successive delete-one jackknife procedure [4-7], the least dominant singular vector was removed successively (from the total vectors generated, down to 100 vectors) to generate ordered sets of singular vectors, and a new tree was estimated following each removal.
Competing interests
The authors declare that there are no competing interests.
Authors’ contributions
GS established the overall concept and approach, and AS completed the phylogenetic analysis, as well as producing all tables, figures, and writing early drafts of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
SVD (higher dimension) tree for the 12 Drosophila spp., using all 700 vectors, with filtering cut off value of ±0.003, retaining 88,026 (45.46%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (higher dimension) tree for the 12 Drosophila spp., using all 700 vectors, with filtering cut off value of ±0.032, retaining 8,583 (4.43%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. melanogaster), using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. simulans using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. ananassae) using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. yakuba) using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11Drosophila species (excluding D. sechellia) using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11Drosophila species (excluding D. melanogaster), using 300 vectors, with filtering cut off value of ±0.035, retaining 4146 (2.43%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. sechellia), using 300 vectors, with filtering cut off value of ±0.035, retaining 4271 (2.43%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. simulans), using 300 vectors, with filtering cut off value of ±0.035, retaining 4611 (2.61%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. ananassae), using 300 vectors, with filtering cut off value of ±0.035, retaining 4343 (2.45%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. yakuba), using 300 vectors, with filtering cut off value of ±0.035, retaining of 4628 (2.63%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
Contributor Information
Arun Seetharam, Email: aseetharam@sycamores.indstate.edu.
Gary W Stuart, Email: gstuart@indstate.edu.
Acknowledgements
Support for this work was provided in part by the Biology Department and the School of Graduate Studies at Indiana State University. In addition, help with software development and modification was provided by Yihua Bai from the Center for Instructional and Research Technology at ISU.
References
- Dutilh BE, Snel B, Ettema TJ, Huynen MA. Signature genes as a phylogenomic tool. Mol Biol Evol. 2008;25(8):1659–1667. doi: 10.1093/molbev/msn115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda E, Moya A, Silva FJ. Genome rearrangement distances and gene order phylogeny in gamma-Proteobacteria. Mol Biol Evol. 2005;22(6):1456–1467. doi: 10.1093/molbev/msi134. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311(5765):1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Berry MW. An SVD-based comparison of nine whole eukaryotic genomes supports a coelomate rather than ecdysozoan lineage. BMC Bioinformatics. 2004;5:204. doi: 10.1186/1471-2105-5-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart GW, Moffett K, Baker S. Integrated gene and species phylogenies from unaligned whole genome protein sequences. Bioinformatics. 2002;18(1):100–108. doi: 10.1093/bioinformatics/18.1.100. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Moffett K, Leader JJ. A comprehensive vertebrate phylogeny using vector representations of protein sequences from whole genomes. Mol Biol Evol. 2002;19(4):554–562. doi: 10.1093/oxfordjournals.molbev.a004111. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Moffett PK, Bozarth RF. A comprehensive open reading frame phylogenetic analysis of isometric positive strand ssRNA plant viruses. Arch Virol. 2006;151(6):1159–1177. doi: 10.1007/s00705-005-0692-9. [DOI] [PubMed] [Google Scholar]
- Drosophila.12.Genomes.Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450(7167):203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Bhutkar A, Gelbart WM, Smith TF. Inferring genome-scale rearrangement phylogeny and ancestral gene order: a Drosophila case study. Genome Biol. 2007;8(11):R236. doi: 10.1186/gb-2007-8-11-r236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN. et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450(7167):203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Machado CA, Hey J. The causes of phylogenetic conflict in a classic Drosophila species group. Proc Biol Sci. 2003;270(1520):1193–1202. doi: 10.1098/rspb.2003.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard DA, Iyer VN, Moses AM, Eisen MB. Widespread discordance of gene trees with species tree in Drosophila: evidence for incomplete lineage sorting. PLoS Genet. 2006;2(10):e173. doi: 10.1371/journal.pgen.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Jensen JD, Pool JE, Aquadro CF. Phylogenetic incongruence in the Drosophila melanogaster species group. Mol Phylogenet Evol. 2007;43(3):1138–1150. doi: 10.1016/j.ympev.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, DeSalle R, Lee EK, O'Grady P. Using whole genome presence/absence data to untangle function in 12 Drosophila genomes. Fly (Austin) 2008;2(6):291–299. doi: 10.4161/fly.7481. [DOI] [PubMed] [Google Scholar]
- Russo CA, Takezaki N, Nei M. Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol. 1995;12(3):391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- Lewis RL, Beckenbach AT, Mooers AO. The phylogeny of the subgroups within the melanogaster species group: likelihood tests on COI and COII sequences and a Bayesian estimate of phylogeny. Mol Phylogenet Evol. 2005;37(1):15–24. doi: 10.1016/j.ympev.2005.02.018. [DOI] [PubMed] [Google Scholar]
- O'Grady PM, Kidwell MG. Phylogeny of the subgenus Sophophora (Diptera: Drosophilidae) based on combined analysis of nuclear and mitochondrial sequences. Mol Phylogenet Evol. 2002;22(3):442–453. doi: 10.1006/mpev.2001.1053. [DOI] [PubMed] [Google Scholar]
- Remsen J, O'Grady P. Phylogeny of Drosophilinae (Diptera: Drosophilidae), with comments on combined analysis and character support. Mol Phylogenet Evol. 2002;24(2):249–264. doi: 10.1016/S1055-7903(02)00226-9. [DOI] [PubMed] [Google Scholar]
- Powell JR. Progress and prospects in evolutionary biology: the Drosophila model. New York: Oxford University Press; 1997. [Google Scholar]
- Singh ND, Larracuente AM, Sackton TB, Clark AG. Comparative Genomics on the Drosophila Phylogenetic Tree. Annu Rev Ecol Evol S. 2009;40:459–480. doi: 10.1146/annurev.ecolsys.110308.120214. [DOI] [Google Scholar]
- Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN. et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450(7167):219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han SG. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 2007;3(11):e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A, True JR. Phylogeny of the Oriental Drosophila melanogaster species group: a multilocus reconstruction. Syst Biol. 2002;51(5):786–805. doi: 10.1080/10635150290102410. [DOI] [PubMed] [Google Scholar]
- Arhontaki K, Eliopoulos E, Goulielmos G, Kastanis P, Tsakas S, Loukas M, Ayala F. Functional constraints of the Cu, Zn superoxide dismutase in species of the Drosophila melanogaster subgroup and phylogenetic analysis. J Mol Evol. 2002;55(6):745–756. doi: 10.1007/s00239-002-2370-9. [DOI] [PubMed] [Google Scholar]
- Gailey DA, Ho SK, Ohshima S, Liu JH, Eyassu M, Washington MA, Yamamoto D, Davis T. A phylogeny of the Drosophilidae using the sex-behaviour gene fruitless. Hereditas. 2000;133(1):81–83. doi: 10.1111/j.1601-5223.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- Jeffs PS, Holmes EC, Ashburner M. The molecular evolution of the alcohol dehydrogenase and alcohol dehydrogenase-related genes in the Drosophila melanogaster species subgroup. Mol Biol Evol. 1994;11(2):287–304. doi: 10.1093/oxfordjournals.molbev.a040110. [DOI] [PubMed] [Google Scholar]
- Ko WY, David RM, Akashi H. Molecular phylogeny of the Drosophila melanogaster species subgroup. J Mol Evol. 2003;57(5):562–573. doi: 10.1007/s00239-003-2510-x. [DOI] [PubMed] [Google Scholar]
- Matsuo Y. Molecular evolution of the histone 3 multigene family in the Drosophila melanogaster species subgroup. Mol Phylogenet Evol. 2000;16(3):339–343. doi: 10.1006/mpev.2000.0810. [DOI] [PubMed] [Google Scholar]
- Schlotterer C, Hauser MT, von Haeseler A, Tautz D. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol Biol Evol. 1994;11(3):513–522. doi: 10.1093/oxfordjournals.molbev.a040131. [DOI] [PubMed] [Google Scholar]
- Shibata H, Yamazaki T. Molecular evolution of the duplicated Amy locus in the Drosophila melanogaster species subgroup: concerted evolution only in the coding region and an excess of nonsynonymous substitutions in speciation. Genetics. 1995;141(1):223–236. doi: 10.1093/genetics/141.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow TA, O’Grady PM. Drosophila biology in the genomic age. Genetics. 2007;177(3):1269–1276. doi: 10.1534/genetics.107.074112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart GW, Berry MW. A comprehensive whole genome bacterial phylogeny using correlated peptide motifs defined in a high dimensional vector space. J Bioinform Comput Biol. 2003;1(3):475–493. doi: 10.1142/S0219720003000265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SVD (higher dimension) tree for the 12 Drosophila spp., using all 700 vectors, with filtering cut off value of ±0.003, retaining 88,026 (45.46%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (higher dimension) tree for the 12 Drosophila spp., using all 700 vectors, with filtering cut off value of ±0.032, retaining 8,583 (4.43%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. melanogaster), using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. simulans using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. ananassae) using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. yakuba) using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11Drosophila species (excluding D. sechellia) using 300 vectors, without filtering any proteins (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11Drosophila species (excluding D. melanogaster), using 300 vectors, with filtering cut off value of ±0.035, retaining 4146 (2.43%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. sechellia), using 300 vectors, with filtering cut off value of ±0.035, retaining 4271 (2.43%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. simulans), using 300 vectors, with filtering cut off value of ±0.035, retaining 4611 (2.61%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. ananassae), using 300 vectors, with filtering cut off value of ±0.035, retaining 4343 (2.45%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).
SVD (lower dimension) tree for the 11 Drosophila species (excluding D. yakuba), using 300 vectors, with filtering cut off value of ±0.035, retaining of 4628 (2.63%) protein sequences (upper branch values, modified jackknife and lower branch values, bootstrap procedure for tree generation).