Abstract
Protein O-fucosyltransferase 1 (Pofut1) and protein O-fucosyltransferase 2 (Pofut2) add O-linked fucose at distinct consensus sequences in properly folded epidermal growth factor (EGF)-like repeats and thrombospondin type-1 (TSR) repeats, respectively. Glycan chain elongation past O-fucose can occur to yield a tetrasaccharide on EGF repeats and a disaccharide on TSRs. Elimination of Pofut1 in mice causes embryonic lethality with Notch-like phenotypes demonstrating that O-fucosylation of Notch is essential for its function. Similarly, elimination of Pofut2 results in an early embryonic lethal phenotype in mice, although the molecular mechanism for the lethality is unknown. The recent development of sugar analogs has revolutionized the study of glycans by providing a convenient method for labeling and tracking glycosylation. In order to study O-fucosylation, we took advantage of the recently developed reporter, 6-alkynyl fucose. Using the Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC), or “click” reaction, azido-biotin allows tagging and detection of 6AF-modified proteins. Here we examine whether proteins containing EGF repeats or TSRs with O-fucose consensus sequences are specifically modified with 6AF in cell culture. Using mass spectrometry (MS), we demonstrate that 6AF is efficiently incorporated onto the appropriate consensus sequences on EGF repeats and TSRs. Furthermore, the elongation of the O-fucose monosaccharide on EGF repeats and TSRs is not hampered when 6AF is used. These results show that 6AF is efficiently utilized in a truly bioorthogonal manner by Pofut1, Pofut2 and the enzymes that elongate O-fucose, providing evidence that 6AF is a significant new tool in the study of protein O-fucosylation.
Keywords: 6-Alkynyl fucose, click chemistry, epidermal growth factor-like-repeat, O-fucose, thrombospondin type-1 repeat
Introduction
l-Fucose is a component of many O-linked and N-linked glycan modifications in mammals with physiological and pathophysiological significance in a variety of settings, including the ABO blood group, Lewis-related antigens, core fucosylation of N-linked glycans and O-linked fucose modifications. Thirteen known fucosyltransferases are responsible for addition of fucose to these structures (reviewed in (Becker and Lowe 2003; Ma et al. 2006)), for instance, Fucosyltransferases 1 and 2 (FUT1 and FUT2) add an α(1,2)-fucose to the terminal galactose on the lactosamine-type precursor on a variety of glycoproteins and glycolipids, synthesizing H Antigen, which is expressed in “O” blood group individuals. Sialyl-Lewisb antigen, synthesized by FUT3, allows attachment of Helicobacter pylori to the gastric epithelium, which can lead to peptic ulcer disease and gastric cancer. Sialyl-Lewisx antigen, synthesized by FUT4 and FUT7, plays an essential role in selectin-dependent leukocyte adhesion and lymphocyte homing. Sialyl-Lewisx has also been implicated in mediating human sperm binding to the zona pellucida in fertilization (Pang et al. 2011), and overexpression of sialyl-Lewisx has been implicated in a variety of cancers (Kim and Varki 1997). Core fucosylation of N-linked glycans, catalyzed by FUT8, affects proper functioning of transmembrane signaling receptors. Disruption of core fucosylation of N-linked glycans leads to neonatal lethality and emphysema-like changes in the lungs of mice (Wang et al. 2006), while midline patterning and eye development defects are observed in zebrafish mutants (Seth et al. 2010). Similarly, disruption of O-fucosylation can have devastating effects on development, culminating in embryonic lethality.
O-Fucosylation occurs at consensus sequences on two small cysteine-rich domains: Epidermal growth factor-like (EGF) repeats and Thrombospondin Type 1 Repeats (TSRs). EGF repeats are O-fucosylated by protein O-fucosyltransferase 1 (POFUT1, also known as FUT12) at the consensus, C2xxxx(S/T)C3, where C2 and C3 are the second and third conserved cysteines of the EGF repeat (Wang et al. 2001). While over 100 proteins are predicted to be modified by O-fucose on the basis of the presence of the consensus sequence within EGF repeats (Rampal et al. 2007), the Notch family of receptors have more predicted O-fucose sites than any other protein in the databases (Moloney et al. 2000b; Rana and Haltiwanger 2011). Results from many groups reveal that O-fucosylation is essential for Notch function (Okajima and Irvine 2002; Sasamura et al. 2003; Shi and Stanley 2003; Rampal et al. 2005a), and O-fucose appears to play an important functional role in the ability of agrin to cluster acetylcholine receptors (Kim et al. 2008), but the function of O-fucose on the vast majority of predicted modified proteins is unknown. O-Fucose on EGF repeats in several proteins can be extended by the Fringe family of β3-N-acetylglucosaminyltransferases (Bruckner et al. 2000; Moloney et al. 2000a; Panin et al. 2002) and other enzymes, ultimately to the tetrasaccharide Siaα2-3Galβ1-4GlcNAcβ1-3Fucα1-O-Ser/Thr (Moloney et al. 2000a, b). Fringe-mediated elongation of O-fucose is known to modulate Notch function (Rana and Haltiwanger 2011), but the biological function of elongation on other proteins is not understood.
TSRs are O-fucosylated by protein O-fucosyltransferase 2 (POFUT2, also known as FUT13) at the consensus sequence CX2–3(S/T)CX2G (Hofsteenge et al. 2001; Luo et al. 2006a, b; Ricketts et al. 2007; Wang et al. 2007). Over 50 proteins are predicted to be O-fucosylated on the basis of the presence of this consensus within TSRs, including all the 19 members of the ADAMTS family of matricellular proteases (Du et al. 2010; Leonhard-Melief and Haltiwanger 2010). Disruption of Pofut2 in mice results in embryonic lethality by E10.5 indicating that O-fucosylation of TSRs, like that of EGF repeats, is essential for proper development (Du et al. 2010). Nonetheless, the specific targets responsible for the lethality are not yet known. Several reports demonstrate that O-fucosylation of TSRs is required for proper folding and secretion, suggesting that the lethality may be due to loss of secretion of members of the ADAMTS family (Ricketts et al. 2007; Wang et al. 2007). O-Fucose on TSRs is typically extended by a β3-glucosyltransferase (Kozma et al. 2006; Sato et al. 2006), and mutations in this enzyme lead to Peters'-plus syndrome, an autosomal recessive congenital disorder of glycosylation characterized by a specific malformation of the eye, and associated with developmental defects, cleft lip/palate and mental retardation (Lesnik Oberstein et al. 2006; Reis et al. 2008; Heinonen and Maki 2009). Nonetheless, the biological function of extending O-fucose on any of the proteins predicted to be modified is not known.
To better understand which proteins are actually modified with O-fucose in vivo and the biological function of these modifications, we need to develop better tools for monitoring O-fucosylation. The recent development of sugar analogs has revolutionized the study of glycans by providing a powerful tool for tracking glycosylation in a wide variety of ways. The general strategy involves metabolic labeling cells with a monosaccharide analog bearing an azide (or alkyne) group. The metabolically labeled sugar analog is incorporated onto target glycoproteins and/or glycolipids, allowing visualization after reacting with a reagent bearing the complementary alkyne (or azide) conjugated to an imaging probe. The reaction that covalently couples the azide and alkyne groups is a variation of the Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) (generally referred to as “click chemistry”) (Rostovtsev et al. 2002; Tornoe et al. 2002). This strategy has allowed successful imaging of membrane-associated glycans in developing zebrafish (Laughlin et al. 2008; Dehnert et al. 2012) and Caenorhabditis elegans cell-surface glycans (Laughlin and Bertozzi 2009). Similar strategies using alkynyl monosaccharides have been employed to detect a variety of sugar modifications, including sialylated and fucosylated glycans (Hsu et al. 2007; Besanceney-Webler et al. 2011).
In this report, tracking of fucose modifications is achieved by using the peracetylated fucose analog 6-alkynyl fucose (6AF). 6AF has the structure of l-fucose with an alkyne group incorporated on the C6 carbon (Figure 1A). The related azido-fucose analog has reported toxicity issues and is thus less useful compared with the alkyne analog (Rabuka et al. 2006). 6AF is taken up through the fucose salvage pathway, eventually leading to addition of 6AF onto proteins that normally bear l-fucose (Sawa et al. 2006; Liu et al. 2012). Of the thirteen known fucosyltransferases (FUT1-FUT11; POFUT1/FUT12; POFUT2/FUT13) (Becker and Lowe 2003), the tolerance to fucose analogs has been demonstrated with FUT2-FUT7 (Sawa et al. 2006) and FUT9 (Liu et al. 2012). Most of these studies were done in vitro (Sawa et al. 2006), so little is known about the efficiency of 6AF incorporation in vivo. In addition, no structural analysis of glycans where 6AF has been metabolically incorporated has been reported. Given the variety of fucosyltransferases and the myriad contexts in which fucosylation reactions are involved, the incorporation of 6AF onto its predicted targets must be confirmed before conclusions can be drawn from experiments where 6AF is metabolically incorporated into glycans. Here we address whether proteins containing EGF repeats or TSRs can be metabolically modified by 6AF in cells (by POFUT1 or POFUT2, respectively), and if so, whether enzymes responsible for glycan extension of O-fucose, Lunatic Fringe (Lfng) and β3-glucosyltransferase, also modify 6AF.
Fig. 1.
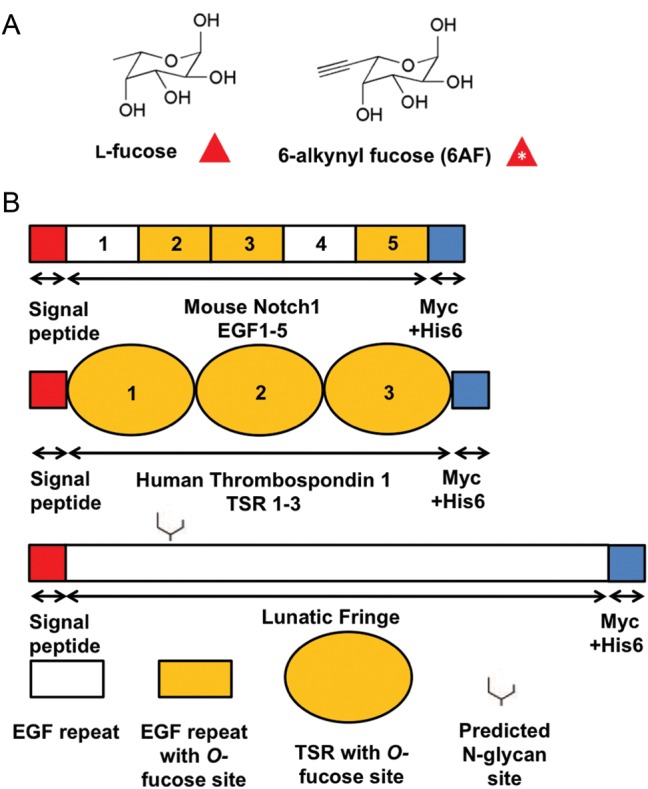
The structure of 6AF and domain map of the constructs used. (A) The structures of l-fucose and 6AF. (B) All expression constructs contained an N-terminal Igk signal peptide (red) and C-terminal Myc-His6 tags (blue) for detection by Western blot and purification. Mouse Notch 1 EGF 1–5 contains three O-fucosylated EGF repeats (indicated by shading) (Shao et al. 2003), and all the three TSRs in Human Thrombospondin 1 TSR 1–3 are O-fucosylated (Hofsteenge et al. 2001), while neither construct contains an N-glycosylation site. Lfng is not predicted to be O-fucosylated and contains one N-glycosylation site at Asn-167.
Results
6AF is successfully incorporated onto EGF repeats and TSRs
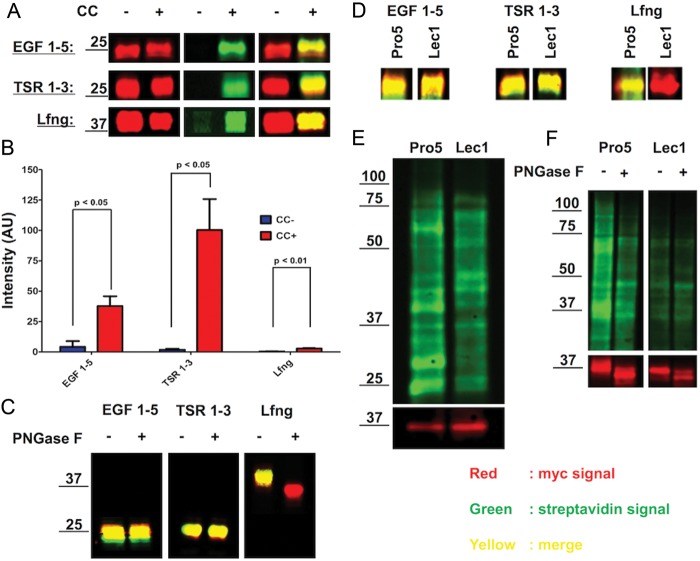
To determine whether 6AF is tolerated by Pofut1 and Pofut2, we examined metabolic incorporation onto several protein fragments: EGF repeats 1–5 from mouse Notch 1 (mN1 EGF1–5), which contains three known Pofut1 modification sites (Rana and Haltiwanger 2011; Rana et al. 2011), and TSRs 1–3 from human Thrombospondin1 (hT1 TSR1-3), which contains three known Pofut2 modification sites (Hofsteenge et al. 2001) (Figure 1B). Constructs encoding secreted versions of mN1 EGF1–5 or hT1 TSR1–3 (or untransfected control) were transfected into wild-type Human Embyronic Kidney 293T (HEK293T) cells, and incubated with 200 µM peracetylated 6AF, or 200 µM peracetylated fucose, for 72 h. Cell lysates and culture media were subjected to CuAAC with azido-biotin (+click chemistry (CC)) or without azido-biotin (−CC), separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), and transferred to nitrocellulose. Both mN1 EGF1–5 and hT1 TSR1–3 could be detected by streptavidin peroxidase indicating the presence of 6AF tagged with biotin on the proteins (Figure 2A). Reactivity with streptavidin peroxidase required 6AF and CC. The anti-myc blot confirmed the expression of mN1 EGF1–5 and hT1 TSR1–3 in transfected media samples. Control media, obtained under identical conditions from untransfected cells, showed no signal with streptavidin or anti-myc. Cell lysate samples, obtained from control cells incubated with 6AF and subjected to the click reaction, showed efficient labeling of a variety of fucosylated glycoproteins. Most of these protein species disappeared in samples without CC or without 6AF incubation, with the exception of two distinct bands, presumably corresponding to endogenously biotinylated proteins (Figure 2A). These results demonstrate that 6AF is successfully incorporated onto EGF repeats and TSRs and selective detection is achieved.
Fig. 2.
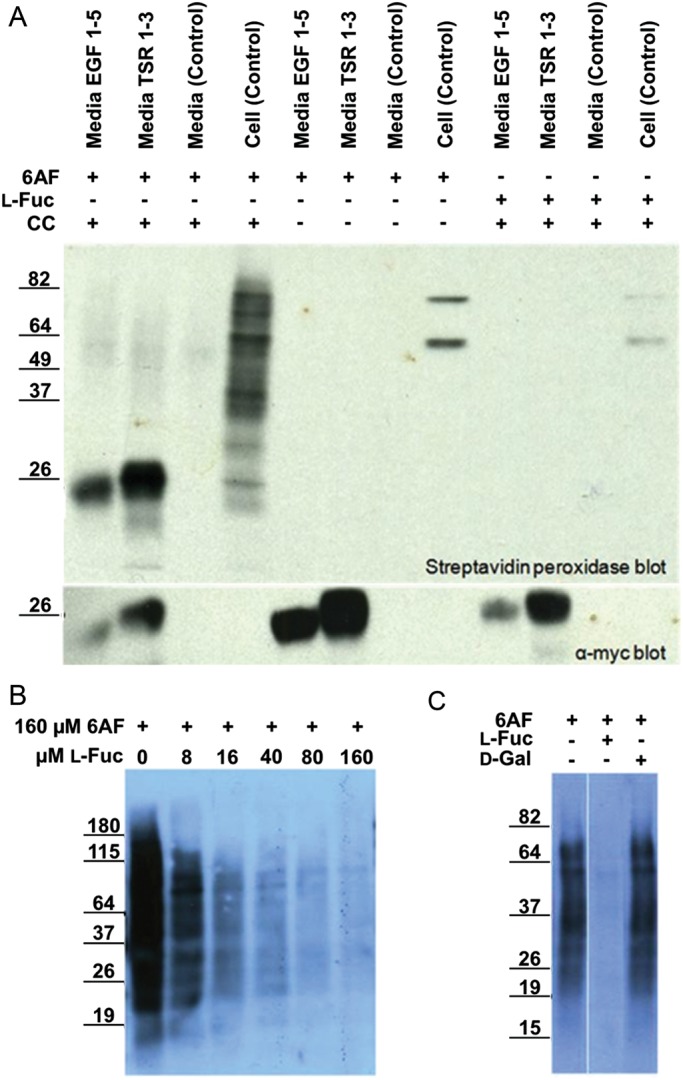
6AF is incorporated onto EGF repeats and TSRs in HEK293T cells. (A) HEK293T cells were either untransfected (Control) or transfected with plasmids encoding mN1 EGF1–5 or hT1 TSR1–3, and incubated with 200 µM peracetylated 6AF (6AF) or peracetylated l-fucose (Fuc) for three days. The media and the lysates were subjected to CuAAC with (+CC) or without (−CC) azido-biotin. The samples were then subjected to SDS–PAGE and Western blotting using Streptavidin-HRP and anti-myc. (B) Cell lysates were prepared from untransfected HEK293T cells incubated with 160 µM peracetylated 6AF and increasing amounts of l-Fucose, subjected to CuAAC with azido-biotin and analyzed as described above. (C) Cell lysates were prepared from untransfected HEK293T cells incubated with 200 µM peracetylated 6AF in the presence or absence of 200 µM l-fucose or d-galactose, subjected to CuAAC with azido-biotin and analyzed as described above.
6AF is specifically incorporated onto fucosylated glycoproteins
To confirm that 6AF is specific to fucosylation within the cell and is not metabolized into other sugars, we examined whether the 6AF signal could be eliminated by excess l-fucose (Figure 2B). Incorporation of 6AF onto endogenous glycoproteins in HEK293T cells decreased in a dose-dependent fashion as the concentration of l-fucose was increased. In contrast, incubation of control cells with 6AF and d-galactose had no effect on 6AF incorporation (Figure 2C). These results establish that 6AF is specifically utilized by fucosylation pathways within cells.
6AF is incorporated into both N-linked and O-linked glycans
The results from Figure 2 suggest that 6AF is incorporated onto EGF repeats and TSRs as well as a large number of unidentified proteins in untransfected cell lysates. As an initial means of characterizing whether the 6AF is incorporated into N- or O-linked glycans in the crude lysates, we took advantage of Pro5 (wild-type) and Lec1 Chinese hamster ovary (CHO) cells. Lec1 cells are unable to synthesize complex N-linked glycans (Stanley et al. 1975) and are predicted to have little or no fucosylated N-glycans (Lin et al. 1994). To confirm that 6AF is incorporated onto EGF repeats, TSRs and N-glycans in this system, we transfected constructs encoding mN1 EGF1–5, hT1 TSR1–3 and Lfng (which contains a single consensus site for N-linked glycan modification on Asparagine (Asn)-167 and no sites of O-fucosylation) (Figure 1), into Pro5 cells grown in the presence of 6AF. All the three proteins were detected in the media of transfected Pro5 cells (anti-myc; red) and incorporated 6AF as measured by streptavidin (green) in a CC-dependent manner (Figure 3A). Although the streptavidin signal for the +CC Lfng is clearly above background (−CC), it is much lower than that for either EGF1–5 or TSR1–3 when normalized to protein levels (Figure 3B). Since Lfng does not contain EGF or TSRs, we predicted that the 6AF signal on Lfng resulted from modification of its N-glycan. To test this, we performed peptide N-glycosidase (PNGase F) digestion on all the three proteins. PNGase F completely removed the streptavidin signal (green) from Lfng and caused a mass shift of several kDa, characteristic of the loss of an N-glycan, but had no effect on the 6AF signal on EGF1–5 or TSR1–3 (Figure 3C). Moreover, when these proteins were expressed in Lec1 cells, 6AF incorporation was lost on Lfng, but not for EGF1–5 or TSR1–3, consistent with 6AF incorporation into the N-glycan on Lfng (Figure 3D).
Fig. 3.
6AF is incorporated into N-glycans found on Lfng and numerous proteins in crude extracts of CHO cells. (A) The Pro5 cells were transfected with plasmids encoding mN1 EGF1–5, hT1 TSR1–3 or Lfng (Figure 1) and grown in the presence of 200 µM peracetylated 6AF. The media samples were subjected to CuAAC with (+ Click Chemistry) or without (−CC) azido-biotin and analyzed as described in Materials and Methods. Protein was monitored using the anti-myc probe (red), and biotin using the streptavidin probe (green), while merging of the two channels results in yellow. (B) Bar graph illustrating the intensity (in arbitrary units (AU) of streptavidin signal in EGF 1–5, TSR 1–3 and Lfng samples, with (+CC) and without (−CC) performing the click chemistry reaction. The experiment was performed in triplicate and the streptavidin intensity values were normalized to anti-myc intensity values. Paired student t-test was performed to determine statistical significance. (C) The samples prepared as in A were subjected to PNGase F digestion as described in Materials and Methods. (D) The plasmids encoding mN1 EGF1–5, hT1 TSR1–3 and Lfng were transfected either into Pro5 or into Lec1 cells in the presence of 200 µM peracetylated 6AF, and the media samples were analyzed as in A. (E) The Pro5 and Lec1 cells transfected with Lfng were grown in the presence of 200 µM peracetylated 6AF and the cell lysates were biotinylated and analyzed as in A. Many proteins are labeled in both cell lines, but the signal is lost on several proteins in Lec1 cells. The bottom panel shows an anti-Myc blot for the Lfng-Myc-His6 to show that similar amounts of extract were loaded in each lane. (F) The Pro5 and Lec1 cells transfected with Lfng were grown in the presence of 200 µM peracetylated 6AF and the cell lysates were biotinylated, PNGase F digested and then analyzed as in A. The intensity of the streptavidin signal in Pro5 cell lysate is decreased after PNGase F digestion, closely resembling the Lec1 cell lysate sample. The bottom panel shows an anti-Myc blot for the transiently transfected Lfng-Myc-His6 to show that similar amounts of extract were loaded in each lane and to confirm that the PNGase F digestion was successful.
Comparison of 6AF incorporation onto endogenous proteins in Pro5 versus Lec1 cells (Figure 3E) revealed that while many proteins are labeled in both CHO cell lines, several are lost in Lec1 cells. These results indicate that some of the 6AF is incorporated into N-glycans on proteins in Pro5 cells. The proteins labeled in Lec1 cells lysates are presumably modified with 6AF on EGF repeats or TSRs, or possibly on mucin-type O-glycans (O-GalNAc modifications). PNGase F digestion of the Pro5 and Lec1 cell lysates (Figure 3F) showed that while the streptavidin signal is very similar in Lec1 lysates in the presence or absence of PNGaseF, the signal in PNGaseF-treated Pro5 cell lysates strongly resembles that in the Lec1 cell lysates. These results confirm that most of the bands detected with streptavidin in the Lec1 cell lysates correspond to proteins modified with O-fucose glycans or mucin-type glycans with terminal fucose.
6AF is efficiently incorporated onto O-fucose sites on EGF repeats and does not perturb glycan elongation by Fringe
Although Figure 2 demonstrates incorporation of 6AF onto EGF repeats and TSRs, it does not address whether the POFUT1 and two consensus sites are modified, measure the efficiency with which the fucose analog is incorporated onto the site or determine whether the presence of the alkyne affects glycan elongation. Specifically, O-fucose on EGF repeats can be elongated by the Fringe family of β3-N-acetylglucosaminyltransferases to the disaccharide GlcNAcβ1-3Fucose, and further elongated by other enzymes to the mature tetrasaccharide Siaα2-3-Galβ1-4GlcNAcβ1-3Fuc (Moloney et al. 2000a). To address the incorporation efficiency and determine whether the presence of the alkyne perturbs glycan elongation beyond the O-fucose on EGF repeats, we used a mass spectral approach. The mass of 6AF is 10 Da greater than that of fucose (accounting for one additional carbon and a net subtraction of two hydrogen atoms, Figure 1A) and thus the mass-to-charge (m/z) ratio will differ according to incorporation of the analog.
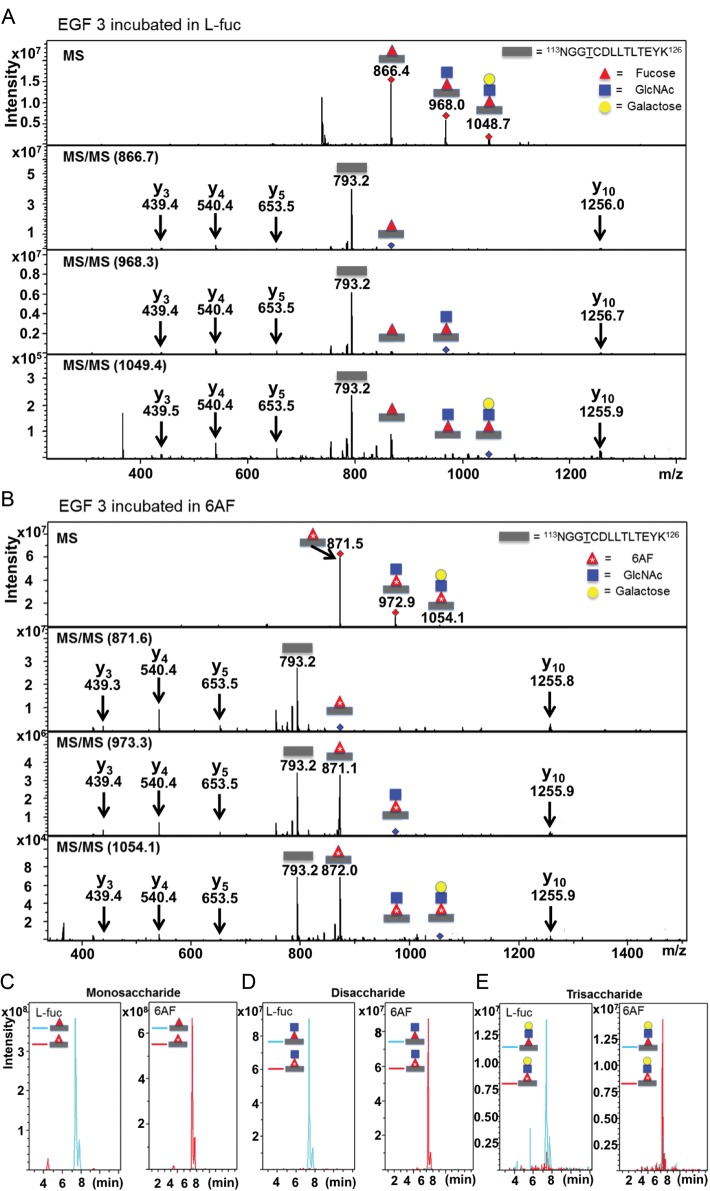
To examine 6AF incorporation and glycan elongation, Pro5 cells were co-transfected with the plasmids encoding mN1 EGF1–5 and Lfng and grown in the presence of either 6AF or l-fucose. Lfng elongates the O-fucose beyond a monosaccharide (Moloney et al. 2000a). The mN1 EGF1-5 protein was purified from medium, digested with trypsin, and the resulting glycopeptides were subjected to nano-liquid chromatography-mass spectrometry (LC-MS)/MS. Mass spectra of a tryptic glycopeptide containing the consensus site for O-fucosylation from EGF3 of mN1 EGF1–5 show ions at an m/z of 866.4, 968.0 and 1048.7 (Figure 4A; top panel). These ions correspond to the predicted masses of doubly charged forms of the mono-, di- and trisaccharide glycoforms of this peptide with natural fucose (note that the tetrasaccharide form of this glycopeptide is not observed under these conditions). The MS/MS spectra of each glycoform (bottom panels) reveal the sequential neutral losses of the modifying glycans as well as several “b” and “y” ions confirming the identity of the peptide. Mass spectra of the same glycopeptide obtained from a sample generated with 6AF show ions at an m/z of 871.5, 972.9 and 1054.1, which correspond to the above masses plus five mass units (Figure 4B). An m/z difference of 5 in the 2+ charged state is consistent with incorporation of 6AF on the peptide containing the O-fucosylation site of EGF3. Fragmentation of each glycoform (Figure 4B, bottom panels) shows that the extra mass is lost at the appropriate position for the fucose. Surprisingly, very little glycopeptide with an m/z corresponding to fucose (e.g. at 866.4, 968.0, or 1048.7) is observed in the MS spectra from the samples generated with 6AF.
Fig. 4.
6AF is efficiently incorporated onto the O-fucosylation site in EGF3 of mouse Notch1 and elongated by Lfng. (A) The Pro5 cells were co-transfected with plasmids encoding mN1 EGF1–5 and Lfng grown in the presence of 200 µM peracetylated l-fucose. mN1 EGF1–5 was purified from the medium, digested with trypsin and subjected to nano-LC-MS/MS analysis as described in Materials and Methods. MS (top panel) and MS/MS (lower panels) spectra of a peptide from EGF3 containing the O-fucose consensus sequence (represented by a gray bar). The ions at m/z 866.4, 968.0 and 1048.7 correspond to the doubly charged forms of the mono-, di- and trisaccharide glycoforms of this peptide. The MS/MS spectra of each glycoform show loss of the modifying glycans to the unglycosylated form of the peptide (m/z 793.2), as well as several “b” and “y” fragmentation ions (indicated in red). (B) mN1 EGF1–5 was prepared as in A except that the cells were grown in 200 µM peracetylated 6AF. MS (top panel) and MS/MS (lower panels) spectra of the same peptide from EGF 3. The ions at m/z 871.5, 972.9 and 1054.1 correspond to the doubly charged forms of the mono-, di- and trisaccharide glycoforms of this peptide with 6AF. These masses closely match the expected increase of m/z of five compared with the l-fuc samples, as the peptides are in the 2+ charge state and as 6AF is 10 Da heavier than l-fucose. The MS/MS spectra confirm loss of the modifying glycans to the unglycosylated form of the peptide (m/z 793.2) and show several “b” and “y” fragmentation ions (in red). EIC for samples obtained from cells incubated in l-fucose (+l-Fuc) and for samples obtained from cells incubated in 6AF (+6AF), for the monosaccharide glycoform (C), the disaccharide glycoform (D) and the trisaccharide glycoform (E) of the peptide from EGF3. Gray rectangle, peptide; red triangle, fucose; red triangle with *, 6AF; blue square, GlcNAc; yellow circle, galactose.
To evaluate the relative amounts of each glycoform in these samples, we performed a semiquantitative mass spectral analysis. Extracted Ion Chromatograms (EIC) were generated for each glycoform of the EGF3 peptide present in the samples, searching for fucose or 6AF forms using the corresponding masses as determined in Figure 4A and B (Figure 4C Monosaccharide glycoform; Figure 4D Disaccharide glycoform; Figure 4E Trisaccharide glycoform). As expected, only the fucose form of the glycopeptides was found in the samples prepared in the presence of l-fucose, while the 6AF form of each glycopeptide was the predominant signal detected in the 6AF samples. These results indicate that 6AF is incorporated at very high efficiencies onto the O-fucose site of EGF repeat 3 under these conditions and that elongation by Fringe is not affected by 6AF.
6AF is efficiently incorporated onto O-fucose sites on TSRs and can be readily elongated to the disaccharide form
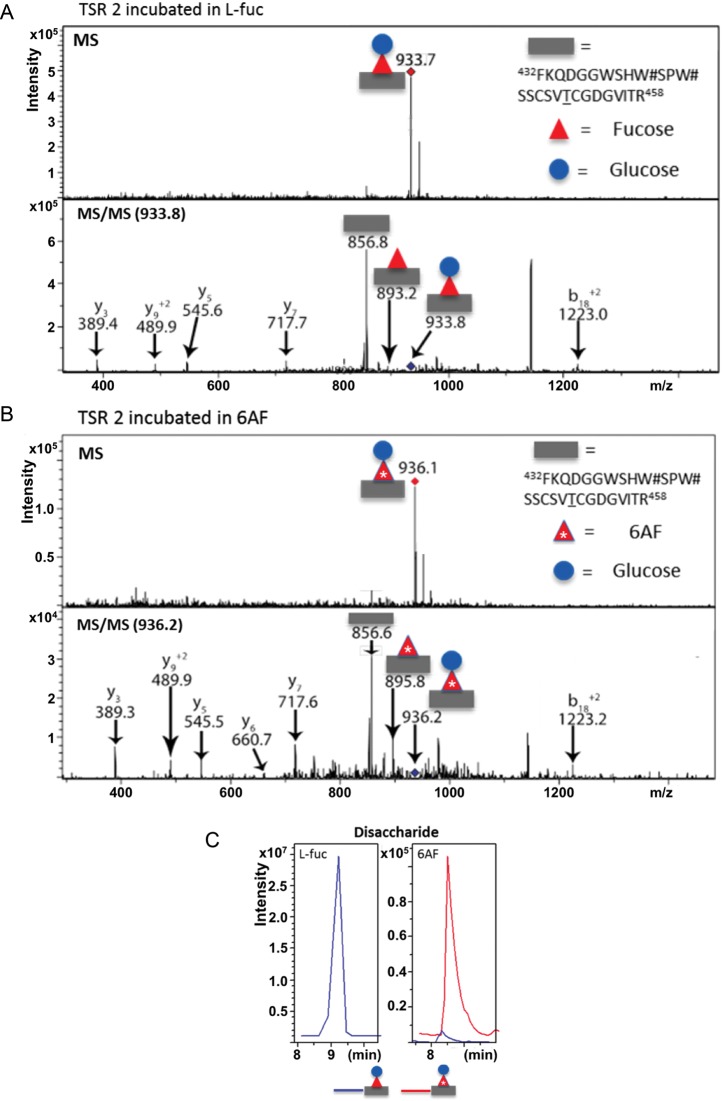
As with the incorporation of 6AF onto EGF repeats, we examined the efficiency of 6AF incorporation onto the glycopeptide containing the O-fucosylation site from TSR2 of TSR 1–3 and whether the β3-glucosyltransferase responsible for elongating O-fucose on TSRs tolerates the analog. Figure 5A shows an MS spectrum (top panel) of the tryptic glycopeptide containing the consensus site for O-fucosylation of TSR 2 obtained from a sample generated in HEK293T cells incubated in l-fucose. The MS spectra contain an ion at an m/z of 933.7, corresponding to the quadruply charged form of the glycopeptide bearing the disaccharide Glcβ1-3Fuc. The MS/MS spectra (Figure 5A, bottom panel) indicate ions at m/z 893.2 and 856.8, corresponding respectively to the monosaccharide and unmodified forms of the peptide. We observed a similar MS spectra obtained from a sample generated as described above, but from cells cultured with 6AF (Figure 5B). The MS spectrum shows a peak at 936.1, which closely corresponds to the mass of the glycopeptide analyzed in Figure 5A plus 2.5 mass units. An m/z difference of 2.5 in the quadruply charged state is consistent with the incorporation of 6AF in place of l-fucose onto the peptide containing the O-fucosylation site from TSR 2. Fragmentation of the glycopeptide (Figure 5B, bottom panel) demonstrates that the extra mass is lost at the appropriate position for the fucose. The EIC for the 6AF sample shows that the glycopeptide containing 6AF was the principal form detected (Figure 5C). These results reveal that 6AF is efficiently incorporated onto the O-fucosylation site of TSR2 and demonstrate that 6AF can be efficiently elongated by the β3-glucosyltransferase.
Fig. 5.
6AF is efficiently incorporated onto TSR2 of Thromospondin1 and elongated by β3-glucosyltransferase. (A) The Pro5 cells were transfected with the plasmid encoding hT1 TSR1–3 and grown in the presence of 200 µM peracetylated fucose. hT1 TSR1–3 was purified from the medium, digested with trypsin and subjected to nano-LC-MS/MS analysis as described in Materials and Methods. MS (top panel) and MS/MS (bottom panel) spectra of a peptide from TSR 2 containing the O-fucose consensus sequence (represented by gray bar). The ion at m/z 933.7 corresponds to the quadruply charged form of this peptide modified with the O-fucose disaccharide. The MS/MS spectra indicate loss of the modifying glycans, first to the monosaccharide form (m/z 893.2) and then to the unglycosylated form of the peptide, (m/z 856.8). Several “b” and “y” fragmentation ions confirming the identity of the peptide are also indicated in black. (B) hT1 TSR1-3 was prepared as in A except that the cells were grown in the presence of 200 µM peracetylated 6AF. MS (top panel) and MS/MS (bottom panel) spectra of the same peptide from TSR 2 are shown. The ion at m/z 936.1 corresponds to the quadruply charged form of this peptide modified with the O-6AF disaccharide. The MS/MS spectra indicate sequential loss of glycans, first to the monosaccharide (m/z 895.8) and then to the unmodified form (m/z 856.6). These masses closely match the expected increase in m/z of 2.5, compared with the l-fucose labeled sample, as the peptides are in the 4+ charge state and as 6AF is 10 Da heavier than l-fucose. Several “b” and “y” ions are also indicated in black. (C) EIC for sample obtained from cells incubated in l-fucose (+l-Fuc) and for sample obtained from cells incubated in 6AF (+6AF), for the disaccharide glycoform of TSR 2. Gray rectangle, peptide; red triangle, fucose; red triangle with*, 6AF; blue circle, glucose.
Discussion
Here we demonstrated that 6AF is efficiently incorporated onto EGF repeats, TSRs, an N-glycan on Lfng and N-glycans on a number of proteins in crude lysates of CHO cells. 6AF is incorporated onto the predicted O-fucosylation sites of the EGF repeats and TSRs, and metabolic labeling at the concentrations used (200 µM) leads to highly efficient incorporation. Glycan elongation of the O-fucose either on EGF repeats or on TSRs does not appear to be affected by the presence of 6AF. 6AF can also be incorporated onto the N-linked glycan of Lfng, although apparently with less efficiency compared with the O-fucosylation of EGF repeats and TSRs. On the basis of these results, 6AF appears to be well tolerated by POFUT1, POFUT2, Lfng and β3-glucosyltransferase during biosynthesis of O-fucose glycans on EGF repeats and TSRs. These data suggest that 6AF is a non-perturbing bioorthogonal analog to l-fucose for the study of O-fucosylation.
The mass spectral data provide strong evidence to indicate that 6AF is incorporated onto the correct O-fucosylation sites on EGF3 and TSR2, as indicated by the increased mass of 10 Da (reflected by an increased m/z of five for the EGF3 peptide in the doubly charged state and an increased m/z of 2.5 for the TSR2 peptide in the quadruply charged state), when compared with the same peptides which utilized l-fucose instead. Interestingly, virtually all of the glycopeptides incorporated 6AF at the concentration we used for metabolic labeling (200 µM). Normally, the fucose salvage pathway is believed to account for <10% of the fucose incorporated onto proteins in cells (Yurchenco and Atkinson 1975). The nearly stoichiometric incorporation of 6AF onto the O-fucose sites of EGF repeats and TSRs indicates that a concentration of 200 µM 6AF is sufficient to shift almost all GDP-fucose production from the de novo pathway to the salvage pathway, as has been suggested elsewhere (Liu et al. 2011). These results also suggest that POFUT1 and POFUT2 utilize GDP-6AF quite efficiently. While no crystal structure of GDP-6AF bound to these enzymes exists, the data suggest that the alkynyl group does not sterically inhibit binding of the nucleotide sugar to the enzyme active site. The recently published structures for POFUT1 from C. elegans and human POFUT2 in complex with GDP-fucose suggest that there is sufficient space in the active site for the alkyne group (Lira-Navarrete et al. 2011; Chen et al. 2012). Future structural studies of both of these enzymes should help us to better understand how GDP-6AF binds to the enzyme active sites.
In contrast to the EGF repeats and TSRs, incorporation of 6AF into N-glycans appears to be sub-stoichiometric under the conditions used in this study. The data in Figure 3A clearly demonstrate some incorporation of 6AF into the N-glycan on Lfng, although this appears to be much lower than incorporation onto the O-fucose sites of EGF or TSR repeats (Figure 3B). There are several potential explanations for this observation. It is possible that only a small fraction of the N-linked glycan on Lfng is normally fucosylated. If so, even if 6AF was incorporated at high efficiency, the signal detected by western blot would be quite low. Alternatively, fucosylation of the N-linked glycan may occur with high frequency, but 6AF itself might be a poorly tolerated substrate for the appropriate fucosyltransferase. Finally, the fucose on the N-glycan may be less accessible for biotinylation during the click reaction. Interestingly, there is a slight difference in the streptavidin intensity between normalized amounts of the EGF repeats and TSRs (Figure 3B), even though the mass spectral analysis shows that 6AF is incorporated at high stoichiometries on both (Figures 4 and 5). This difference suggests that there may be a slight difference in the reactivity of the 6AF on EGF repeats versus TSRs during the click reaction.
While observing the crude lysates of Pro5 and Lec1 cells (Figure 3E), several protein species appear in the Pro5 cell lysates but not in the Lec1 cell lysates, presumably because they bear fucosylated complex-type N-linked glycans, which would not be expected to be synthesized in Lec1 cells. This observation is further confirmed by the data in Figure 3F, which shows that PNGase F treatment causes a decreased streptavidin signal in the Pro5 lysates, resulting in a pattern that closely resembles that from the Lec1 cell lysates. Exhaustive genomic sequencing of CHO cell lines conducted by Xu et al. (2011) indicates that the major N-glycan modifying fucosyltransferase found to be expressed in CHO cells is FUT8. Assuming that this is the case, our results suggest that 6AF is utilized by FUT8, although we cannot comment on how efficiently it is used.
Building on the reports that GDP-6AF is tolerated by FUT2-FUT7 and FUT9, we can now add POFUT1, POFUT2 and most likely FUT8 (though possibly with a lower efficiency) to the growing list of fucosyltransferases that can utilize 6AF. This observation raises the prospect of using 6AF as a means to track fucosylation in a myriad of settings, including on N-glycans, mucin-type O-glycans (O-GalNAc modifications) and O-fucose glycans. A recent study used 6AF, after genetically engineering the cells, in a proteomic study to identify proteins modified on N-glycans specifically by FUT9 (Liu et al. 2012). Similar studies could be done to identify proteins modified with other fucosyltransferases by performing overexpression or RNAi-mediated knockdown of specific enzymes. Because 6AF does not appear to interfere with removal of N-glycans by PNGase F (Figure 3), proteins modified with N-linked versus O-linked 6AF-modified glycans could be separated prior to analysis. In addition to proteomic settings, 6AF can be used to directly track O-fucosylation (and potentially all types of fucosylation) in cell-based studies. This can supplant the use of potentially more expensive radioactive analogs, which also require long autoradiograph development and cumbersome waste disposal, with cheaper, more sensitive and easier to handle 6AF metabolic labeling. Western blots of 6AF-labeled cells can be a convenient readout for the effects on fucosylation in proteins with a mutated fucosylation consensus sequence, in the study of the effects of fucosylation on protein folding and secretion, and a myriad of other potential settings where determination of the fucosylation status of a target protein is desired.
6AF can be a powerful tool in tracking fucosylation, but the very versatility and non-perturbing nature of the fucose analog also demands that caution be exercised in its use. Particularly, imaging studies that utilize 6AF must consider the possibility that the fucose analog will modify a variety of acceptor substrates, perhaps at different levels of efficiency based on the particular fucosyltransferase involved, impacting on the conclusions that can be drawn from such an experiment. Nevertheless, the potential uses of all sugar analogs and 6AF in particular are vast and will likely continue to play a critical role in glycobiology research for the foreseeable future.
Materials and methods
Production of mN 1 EGF 1–5, human TSR 1–3 and human Lfng constructs
The construct for mouse Notch 1 EGF repeats 1–5 was generated as described previously (Shao et al. 2003). The construct for human TSR 1–3 was generated by modifying the procedure in (Luo et al. 2006b) to include the cDNA sequence encoding TSP1- TSR1–3 instead of TSR3. The sequences of the primers used in PCR are available upon request. Successful cloning was confirmed by direct DNA sequencing. The mouse LuFng construct was generated as described previously (Rampal et al. 2005b). All constructs were made in pSecTag2/Hygro C vector (Invitrogen (Life Technologies), Grand Island, NY), which is designed for high-level expression and protein secretion. The vector contains an Igκ chain leader sequence for protein secretion, as well as C-terminal myc epitope and 6-histidine residues, to aid in detection and purification.
Cell culture, metabolic labeling and protein expression
Pro5 and Lec1 CHO cells were grown in integrin and metalloproteinase with thrombospondin motifs (αMEM) (GIBCO (Life Technologies), Grand Island, NY) with 10% fetal calf serum (FCS), while HEK293T cells were grown in Dulbecco's modified eagle medium (DMEM) (GIBCO (Life Technologies), Grand Island, NY) with 10% FCS. Transfections were performed at 50% confluence with pSecTag mNotch 1 EGF 1-5 or pSecTag hTSP1 TSR1-3 or pSecTag Lfng. Briefly, 6 μg of plasmid was suspended in 600 μL 0.15 M NaCl followed by addition of 36 μL Poly(ethylenimine) (PEI) transfection reagent, vortexed and incubated for 10 min at room temperature. The transfection mixture was added to cells grown on 100 mm plates. The cells were then incubated at 37°C for 4 h, after which the media was removed, and the cells were washed once with 2.5 mL of Tris-buffered saline (TBS). The control cells were incubated overnight in 10 mL of αMEM or DMEM, 10% FCS, followed by incubation in 5 mL of OptiMEM for 72 h (GIBCO (Life Technologies), Grand Island, NY), while the experimental cells were additionally supplemented with 200 μM peracetylated 6AF as previously described (Hsu et al. 2007).
Detection of glycoproteins in cell lysate/media
Media was collected and the cells, washed twice in TBS, were lysed in 1 mL of chilled lysis buffer as described (1% Nonidet P-40, 150 mM NaCl, Protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN), 100 mM sodium phosphate, pH 7.5) (Hsu et al. 2007). The cell lysates were incubated on ice and vortexed briefly every five minutes for 30 min. The lysates were centrifuged at 14,000 × g for 10 min at 4°C and the pellet discarded. Click reactions were performed as described (Hsu et al. 2007) on both the labeled media and labeled cell lysate to tag glycoproteins containing 6AF with Biotin (0.1 mM Azido-Biotin (CC Tools, Scottsdale, AZ), 0.1 mM tris-(benzyltriazolylmethyl)amine catalyst (AnaSpec, Fremont, CA), 1 mM copper sulfate, 2 mM sodium ascorbate, in phosphine-buffered saline (PBS), at room temperature for one hour).
The tagged cell lysate and the media were analyzed by immunoblot. Samples from the media and the lysates were separated by 10% SDS-PAGE, transferred to nitrocellulose paper (BioRad, Hercules, CA) and probed with 9E10 α-Myc (1:1000 in phosphine-buffered saline with tween (PBST)), followed by Alexa Fluor 680 Goat Anti-Mouse IgG (Invitrogen (Life Technologies), Grand Island, NY) or HRP-conjugated Goat Anti-Mouse IgG (Jackson ImmunoResearch, West Grove, PA) to detect the transfected proteins. To visualize biotinylated glycoproteins, the tagged media and the cell lysate samples were separated by 10% SDS–PAGE, transferred to nitrocellulose membrane and probed with IRDye800 (Rockland Immunochemicals, Glibertsvile, PA) or Streptavidin-conjugated-peroxidase (1:20,000 in PBST) (Thermo Scientific, Rockford, IL). Western blots using fluorescent probes (Alexa Fluor 680 or IRDye800) were visualized using an Odyssey Imager (LI-COR, Lincoln, NE), while western blots with peroxidase conjugated probes were detected by enhanced chemiluminescence (ECL) blotting substrate (Thermo Scientific, Rockford, IL) and film development.
Specificity/competition experiments
HEK293T cells were grown to 50% confluency in DMEM, 10% FCS and 1% penicillin/streptomycin. The cells were washed one time with 5 mL TBS and resuspended in optimum MEM media supplemented with (0, 8, 16, 40, 80, 160) µM l-fucose and 160 µM peracetylated 6AF or 200 µM galactose and 200 µM peracetylated 6AF. After 72 h at 37°C and 5% CO2, the cells were lysed and biotinylated via CC, and 1 µg per lane was resolved using SDS–PAGE and western blotting as described above. Protein concentration was quantified using the bicinchoninic acid assay protein assay with BSA as standard (Thermo Scientific, Rockford, IL).
Enzymatic cleavage of N-linked glyans by PNGase F
PNGase F digestion was performed as previously described (Lin et al. 1994). Briefly, protein from the cell media or the lysate samples was precipitated in eight volumes of acetone at −20°C overnight. The precipitated proteins were collected by centrifugation at 10,000 × g at 4°C for 15 min. The protein pellet was resuspended in 25 µL of 1% SDS and 1% β-mercaptoethanol by boiling for 5 min. After cooling, 225 µL of 50 mM Tris–HCl (pH 8.6), 0.7% Nonidet P-40, and Protease inhibitor cocktail (1 Protease inhibitor tablet per 10 mL of buffer) was added. The samples were digested with 5 U of PNGase F (produced in-house) for at least eight hours at 37°C.
Glycan analysis via nano-LC-MS/MS
Media from transiently transfected pSecTag Notch 1 EGF1–5 (and co-transfected with pSecTag Lfng, in a 2:1 ratio) Pro5 CHO cells or pSecTag hTSP1 TSR1–3 HEK293T cells supplemented either with 6AF or with l-fucose (above) was collected after 72 h, bound to Ni2+-NTA, washed with 5 mL TBS containing 0.5 M NaCl and 10 mM Imidazole, 5 mL cold RIPA buffer, and eluted with TBS containing 250 mM Imidazole. Protein concentration and purity were assessed by SDS/PAGE and Coomassie Brilliant Blue (Thermo Scientific, Rockford, IL) along BSA standards. Approximately 1 μg EGF1–5 or TSR1–3 was acetone precipitated with four volumes acetone, vortexed and frozen at −80°C overnight. The precipitated protein was reduced, alkylated and typsinized, followed by analysis by nano-LC-MS/MS using a Zorbax 300SB-C18 nano-Chip on an Agilent model 6340 ion trap mass spectrometer using collision-induced dissociation as previously described (Rana et al. 2011).
Funding
This work was supported by the National Institutes of Health [GM61126 and CA0123071 to R.S.H., AI072155 to C.-H.W.].
Conflict of interest
None declared.
Abbreviations
6AF, 6-alkynyl fucose; ADAMTS, a disintegrin and metalloproteinase with thrombospondin motifs; Asn, asparagine CC, click chemistry; CHO, Chinese hamster ovary; CID, collision-induced dissociation; CuAAC, Cu(I)-catalyzed azide-alkyne cycloaddition; DMEM, Dulbecco's modified Eagle medium; EGF, epidermal growth factor; EIC, extracted ion chromatograms; FCS, fetal calf serum; FUC, fucose; FUT, fucosyltransferases; HEK293T, human embryonic kidney; hT1, human thrombospondin; LC-MS, liquid chromatography-mass spectrometry; Lfng, lunatic fringe; MS, mass spectrometry; PBS, phosphine-buffered saline; PBST; phosphine-buffered saline with tween; PNGase F, peptide N-glycosidase F; Pofut, protein O-fucosyltransferase; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; TBS, tris-buffered saline; TSRs, thrombospondin type-1 repeats; αMEM, minimum essential medium alpha.
Acknowledgements
The authors thank Dr. Shinako Kakuda for helping with the mass spectral studies and Dr. Bernadette Holdener, Dr. Isaac Carrico and all the members of the Haltiwanger laboratory for their helpful discussion and critical comments on this manuscript.
References
- Becker DJ, Lowe JB. Fucose: Biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. doi:10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- Besanceney-Webler C, Jiang H, Wang W, Baughn AD, Wu P. Metabolic labeling of fucosylated glycoproteins in Bacteroidales species. Bioorg Med Chem Lett. 2011;21:4989–4992. doi: 10.1016/j.bmcl.2011.05.038. doi:10.1016/j.bmcl.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. doi: 10.1038/35019075. doi:10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- Chen CI, Keusch JJ, Klein D, Hess D, Hofsteenge J, Gut H. Structure of human POFUT2: Insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J. 2012 doi: 10.1038/emboj.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehnert KW, Baskin JM, Laughlin ST, Beahm BJ, Naidu NN, Amacher SL, Bertozzi CR. Imaging the sialome during zebrafish development with copper-free click chemistry. Chembiochem. 2012;13:353–357. doi: 10.1002/cbic.201100649. doi:10.1002/cbic.201100649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Takeuchi H, Leonhard-Melief C, Shroyer KR, Dlugosz M, Haltiwanger RS, Holdener BC. O-fucosylation of thrombospondin type 1 repeats restricts epithelial to mesenchymal transition (EMT) and maintains epiblast pluripotency during mouse gastrulation. Dev Biol. 2010;346:25–38. doi: 10.1016/j.ydbio.2010.07.008. doi:10.1016/j.ydbio.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen TY, Maki M. Peters’-plus syndrome is a congenital disorder of glycosylation caused by a defect in the beta1,3-glucosyltransferase that modifies thrombospondin type 1 repeats. Ann Med. 2009;41:2–10. doi: 10.1080/07853890802301975. doi:10.1080/07853890802301975. [DOI] [PubMed] [Google Scholar]
- Hofsteenge J, Huwiler KG, Macek B, Hess D, Lawler J, Mosher DF, Peter-Katalinic J. C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J Biol Chem. 2001;276:6485–6498. doi: 10.1074/jbc.M008073200. doi:10.1074/jbc.M008073200. [DOI] [PubMed] [Google Scholar]
- Hsu TL, Hanson SR, Kishikawa K, Wang SK, Sawa M, Wong CH. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc Natl Acad Sci USA. 2007;104:2614–2619. doi: 10.1073/pnas.0611307104. doi:10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ML, Chandrasekharan K, Glass M, Shi S, Stahl MC, Kaspar B, Stanley P, Martin PT. O-fucosylation of muscle agrin determines its ability to cluster acetylcholine receptors. Mol Cell Neurosci. 2008;39:452–464. doi: 10.1016/j.mcn.2008.07.026. doi:10.1016/j.mcn.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Varki A. Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconj J. 1997;14:569–576. doi: 10.1023/a:1018580324971. doi:10.1023/A:1018580324971. [DOI] [PubMed] [Google Scholar]
- Kozma K, Keusch JJ, Hegemann B, Luther KB, Klein D, Hess D, Haltiwanger RS, Hofsteenge J. Identification and characterization of abeta1,3-glucosyltransferase that synthesizes the Glc-beta1,3-Fuc disaccharide on thrombospondin type 1 repeats. J Biol Chem. 2006;281:36742–36751. doi: 10.1074/jbc.M605912200. doi:10.1074/jbc.M605912200. [DOI] [PubMed] [Google Scholar]
- Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. doi:10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin ST, Bertozzi CR. In vivo imaging of Caenorhabditis elegans glycans. ACS Chem Biol. 2009;4:1068–1072. doi: 10.1021/cb900254y. doi:10.1021/cb900254y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhard-Melief C, Haltiwanger RS. O-fucosylation of thrombospondin type 1 repeats. Methods Enzymol. 2010;480:401–416. doi: 10.1016/S0076-6879(10)80018-7. doi:10.1016/S0076-6879(10)80018-7. [DOI] [PubMed] [Google Scholar]
- Lesnik Oberstein SA, Kriek M, White SJ, Kalf ME, Szuhai K, den Dunnen JT, Breuning MH, Hennekam RC. Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. Am J Hum Genet. 2006;79:562–566. doi: 10.1086/507567. doi:10.1086/507567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AI, Philipsberg GA, Haltiwanger RS. Core fucosylation of high-mannose-type oligosaccharides in GlcNAc transferase I-deficient (Lec1) CHO cells. Glycobiology. 1994;4:895–901. doi: 10.1093/glycob/4.6.895. doi:10.1093/glycob/4.6.895. [DOI] [PubMed] [Google Scholar]
- Lira-Navarrete E, Valero-Gonzalez J, Villanueva R, Martinez-Julvez M, Tejero T, Merino P, Panjikar S, Hurtado-Guerrero R. Structural insights into the mechanism of protein O-fucosylation. PLoS One. 2011;6:e25365. doi: 10.1371/journal.pone.0025365. doi:10.1371/journal.pone.0025365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T-W, Ito H, Chiba Y, Kubota T, Sato T, Narimatsu H. Functional expression of l-fucokinase/guanosine 5′-diphosphate-l-fucose pyrophosphorylase from Bacteroides fragilis in Saccharomyces cerevisiae for the production of nucleotide sugars from exogenous monosaccharides. Glycobiology. 2011;21:1228–1236. doi: 10.1093/glycob/cwr057. doi:10.1093/glycob/cwr057. [DOI] [PubMed] [Google Scholar]
- Liu TW, Kaji H, Togayachi A, Ito H, Sato T, Narimatsu H. A chemoenzymatic approach toward the identification of fucosylated glycoproteins and mapping of N-glycan sites. Glycobiology. 2012;22:630–637. doi: 10.1093/glycob/cwr189. doi:10.1093/glycob/cwr189. [DOI] [PubMed] [Google Scholar]
- Luo Y, Koles K, Vorndam W, Haltiwanger RS, Panin VM. Protein O-fucosyltransferase 2 adds O-fucose to thrombospondin type 1 repeats. J Biol Chem. 2006a;281:9393–9399. doi: 10.1074/jbc.M511975200. doi:10.1074/jbc.M511975200. [DOI] [PubMed] [Google Scholar]
- Luo Y, Nita-Lazar A, Haltiwanger RS. Two distinct pathways for O-fucosylation of epidermal growth factor-like or thrombospondin type 1 repeats. J Biol Chem. 2006b;281:9385–9392. doi: 10.1074/jbc.M511974200. doi:10.1074/jbc.M511974200. [DOI] [PubMed] [Google Scholar]
- Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158R–184R. doi: 10.1093/glycob/cwl040. doi:10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, Vogt TF. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000a;406:369–375. doi: 10.1038/35019000. doi:10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, Haltiwanger RS. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J Biol Chem. 2000b;275:9604–9611. doi: 10.1074/jbc.275.13.9604. doi:10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- Okajima T, Irvine KD. Regulation of notch signaling by O-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. doi:10.1016/S0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- Pang P-C, Chiu PCN, Lee C-L, Chang L-Y, Panico M, Morris HR, Haslam SM, Khoo K-H, Clark GF, Yeung WSB, Dell A. Human sperm binding is mediated by the Sialyl-Lewisx oligosaccharide on the Zona Pellucida. Science. 2011;333:1761–1764. doi: 10.1126/science.1207438. doi:10.1126/science.1207438. [DOI] [PubMed] [Google Scholar]
- Panin VM, Shao L, Lei L, Moloney DJ, Irvine KD, Haltiwanger RS. Notch ligands are substrates for protein O-fucosyltransferase-1 and Fringe. J Biol Chem. 2002;277:29945–29952. doi: 10.1074/jbc.M204445200. doi:10.1074/jbc.M204445200. [DOI] [PubMed] [Google Scholar]
- Rabuka D, Hubbard SC, Laughlin ST, Argade SP, Bertozzi CR. A chemical reporter strategy to probe glycoprotein fucosylation. J Am Chem Soc. 2006;128:12078–12079. doi: 10.1021/ja064619y. doi:10.1021/ja064619y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R, Arboleda-Velasquez JF, Nita-Lazar A, Kosik KS, Haltiwanger RS. Highly conserved O-fucose sites have distinct effects on Notch1 function. J Biol Chem. 2005a;280:32133–32140. doi: 10.1074/jbc.M506104200. doi:10.1074/jbc.M506104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R, Li AS, Moloney DJ, Georgiou SA, Luther KB, Nita-Lazar A, Haltiwanger RS. Lunatic Fringe, manic Fringe, and radical Fringe recognize similar specificity determinants in O-fucosylated epidermal growth factor-like repeats. J Biol Chem. 2005b;280:42454–42463. doi: 10.1074/jbc.M509552200. doi:10.1074/jbc.M509552200. [DOI] [PubMed] [Google Scholar]
- Rampal R, Luther KB, Haltiwanger RS. Notch signaling in normal and disease states: Possible therapies related to glycosylation. Curr Mol Med. 2007;7:427–445. doi: 10.2174/156652407780831593. doi:10.2174/156652407780831593. [DOI] [PubMed] [Google Scholar]
- Rana NA, Haltiwanger RS. Fringe benefits: Functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr Opin Struct Biol. 2011;21:583–589. doi: 10.1016/j.sbi.2011.08.008. doi:10.1016/j.sbi.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana NA, Nita-Lazar A, Takeuchi H, Kakuda S, Luther KB, Haltiwanger RS. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J Biol Chem. 2011;286:31623–31637. doi: 10.1074/jbc.M111.268243. doi:10.1074/jbc.M111.268243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis LM, Tyler RC, Abdul-Rahman O, Trapane P, Wallerstein R, Broome D, Hoffman J, Khan A, Paradiso C, Ron N, Bergner A, Semina EV. Mutation analysis of B3GALTL in Peters Plus syndrome. Am J Med Genet A. 2008;146A:2603–2610. doi: 10.1002/ajmg.a.32498. doi:10.1002/ajmg.a.32498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts LM, Dlugosz M, Luther KB, Haltiwanger RS, Majerus EM. O-fucosylation is required for ADAMTS13 secretion. J Biol Chem. 2007;282:17014–17023. doi: 10.1074/jbc.M700317200. doi:10.1074/jbc.M700317200. [DOI] [PubMed] [Google Scholar]
- Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. doi:10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Sasamura T, Sasaki N, Miyashita F, Nakao S, Ishikawa HO, Ito M, Kitagawa M, Harigaya K, Spana E, Bilder D, Perrimon N, Matsuno K. Neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions. Development. 2003;130:4785–4795. doi: 10.1242/dev.00679. doi:10.1242/dev.00679. [DOI] [PubMed] [Google Scholar]
- Sato T, Sato M, Kiyohara K, Sogabe M, Shikanai T, Kikuchi N, Togayachi A, Ishida H, Ito H, Kameyama A, Gotoh M, Narimatsu H. Molecular cloning and characterization of a novel human beta1,3-glucosyltransferase, which is localized at the endoplasmic reticulum and glucosylates O-linked fucosylglycan on thrombospondin type 1 repeat domain. Glycobiology. 2006;16:1194–1206. doi: 10.1093/glycob/cwl035. doi:10.1093/glycob/cwl035. [DOI] [PubMed] [Google Scholar]
- Sawa M, Hsu TL, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong CH. Glycoproteomic probes for fluorescent imaging of fucosylated glycans in vivo. Proc Natl Acad Sci USA. 2006;103:12371–12376. doi: 10.1073/pnas.0605418103. doi:10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Machingo QJ, Fritz A, Shur BD. Core fucosylation is required for midline patterning during zebrafish development. Dev Dyn. 2010;239:3380–3390. doi: 10.1002/dvdy.22475. doi:10.1002/dvdy.22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Moloney DJ, Haltiwanger R. Fringe modifies O-fucose on mouse Notch1 at epidermal growth factor-like repeats within the ligand-binding site and the abruptex region. J Biol Chem. 2003;278:7775–7782. doi: 10.1074/jbc.M212221200. doi:10.1074/jbc.M212221200. [DOI] [PubMed] [Google Scholar]
- Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci. 2003;100:5234–5239. doi: 10.1073/pnas.0831126100. doi:10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Caillibot V, Siminovitch L. Selection and characterization of eight phenotypically distinct lines of lectin-resistant Chinese hamster ovary cell. Cell. 1975;6:121–128. doi: 10.1016/0092-8674(75)90002-1. doi:10.1016/0092-8674(75)90002-1. [DOI] [PubMed] [Google Scholar]
- Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. doi:10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- Wang LW, Dlugosz M, Somerville RP, Raed M, Haltiwanger RS, Apte SS. O-fucosylation of thrombospondin type 1 repeats in ADAMTS-like-1/punctin-1 regulates secretion: Implications for the ADAMTS superfamily. J Biol Chem. 2007;282:17024–17031. doi: 10.1074/jbc.M701065200. doi:10.1074/jbc.M701065200. [DOI] [PubMed] [Google Scholar]
- Wang X, Gu J, Miyoshi E, Honke K, Taniguchi N. Phenotype changes of Fut8 knockout mouse: Core fucosylation is crucial for the function of growth factor receptor(s) Methods Enzymol. 2006;417:11–22. doi: 10.1016/S0076-6879(06)17002-0. doi:10.1016/S0076-6879(06)17002-0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P, Haltiwanger RS. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J Biol Chem. 2001;276:40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, Chen W, Xie M, Wang W, Hammond S, Andersen MR, Neff N, Passarelli B, Koh W, Fan HC, Wang J, Gui Y, Lee KH, Betenbaugh MJ, Quake SR, Famili I, Palsson BO. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotechnol. 2011;29:735–741. doi: 10.1038/nbt.1932. doi:10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco PD, Atkinson PH. Fucosyl-glycoprotein and precursor polls in HeLa cells. Biochemistry. 1975;14:3107–3114. doi: 10.1021/bi00685a011. doi:10.1021/bi00685a011. [DOI] [PubMed] [Google Scholar]