Abstract
We have developed a novel assay specific to MraY, which catalyzes the first membrane step in the biosynthesis of bacterial cell wall peptidoglycan. This was accomplished by using UDP-MurNAc-Nɛ-dansylpentapeptide, a fluorescent derivative of the MraY nucleotide substrate, and a partially purified preparation of MraY solubilized from membranes of an Escherichia coli overproducing strain. Two versions of the assay were developed, one consisting of the high-pressure liquid chromatography separation of the substrate and product (dansylated lipid I) and the other, without separation and adapted to the high-throughput format, taking advantage of the different fluorescence properties of the nucleotide and lipid I in the reaction medium. The latter assay was validated with a set of natural and synthetic MraY inhibitors.
The biosynthesis of bacterial peptidoglycan is a complex two-stage process. The first stage consists in the formation of the monomer unit GlcNAc-MurNAc-(pentapeptide), whereas the second one concerns polymerization and maturation reactions (28). In the first stage, two enzymes, MraY and MurG, have in common their attachment to the cytoplasmic membrane and their processing of lipid intermediates: MraY transfers the phospho-MurNAc-pentapeptide motif onto lipid carrier undecaprenyl phosphate, yielding MurNAc-(pentapeptide)-pyrophosphoryl undecaprenol (lipid I), and MurG then adds the GlcNAc residue to yield GlcNAc-MurNAc-(pentapeptide)-pyrophosphoryl undecaprenol (lipid II), which contains the whole monomer unit. In the present study, we have focused our attention to the MraY transferase, which is an integral membrane protein with ten transmembrane segments, five cytoplasmic domains, and six periplasmic domains including the N- and C-terminal ends (4). Besides the synthesis of lipid I, which is fully reversible, MraY catalyzes in vitro an exchange reaction between UMP and UDP-MurNAc-pentapeptide (23). Owing to its exclusive presence in bacteria and to its essential character, which was demonstrated in Escherichia coli (5), MraY is a target of interest for the discovery of novel antibacterial agents. It is inhibited by non-clinically used antibiotics such as tunicamycin, amphomycin, mureidomycin, liposidomycin, and muraymycins (7, 9, 22). Recently, simplified analogues of liposidomycin, named riburamycins, have been shown to be powerful MraY inhibitors and to possess antibacterial activities against gram-positive organisms (13-16) (J. Biton, K. Braham, C. Dini, O. Krebs, P. Lassaigne, F. Monti, T. Stachyra, V. Steier, and J. Zhang, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-361, 2002). To date, several specific assays for the MraY activity have been published (6, 17, 18, 27, 29, 30); however, very few can be adapted to the high-throughput format. In the present work, we took advantage of the fluorescent properties of UDP-MurNAc-Nɛ-dansylpentapeptide and dansylated lipid I (6, 29) to set up a high-throughput screen (HTS) assay for MraY, which was validated with a set of natural and synthetic inhibitors (tunicamycin, mureidomycin B, and riburamycins).
MATERIALS AND METHODS
Materials
Triton X-100, N-laurylsarcosine, and the mixture of tunicamycins were purchased from Sigma (St. Louis, Mo.), and undecaprenyl phosphate was obtained from Larodan Fine Chemicals AB (Malmö, Sweden). Tritiated UDP-MurNAc-pentapeptide was prepared according to the method of Reddy et al. (24) by using meso-[3H]diaminopimelic acid (A2pm). Mureidomycin B was a gift from Hoechst (Frankfurt, Germany). The synthesis of the riburamycin analogues has already been described (13-16).
General procedures
Fermentations were carried out in a fully automated 20-liter bioreactor (Techfors; Infors AG, Bottmingen-Basel, Switzerland). UV-visible spectra were recorded with a UV-1601 spectrophotometer (Shimadzu, Croissy Beaubourg, France). Fluorescence spectra were recorded with a Cary Eclipse apparatus (Varian, Les Ulis, France). Mass spectra were obtained with an electrospray ion-trap spectrometer (model LCQ; Thermo-Finnigan, Les Ulis, France).
Bacterial strains
The Escherichia coli XL1-Blue [recA1 endA1 gyrA96 thi hsdR17 (rK− mK+) supE44 relA1 lac/ F′ proAB+ lacIq Δ(lacZ)M15::Tn10] and K-12 (250HT11) strains were used as the host for plasmids and in the cell-based assay, respectively. The Bacillus cereus ATCC 9634 was used for the accumulation of UDP-MurNAc-pentapeptide.
Construction of plasmids
Standard procedures for molecular cloning (26) and cell transformation (12) were used. The E. coli mraY and Enterobacter cloacae P99 β-lactamase genes were amplified by PCR with O1 and O2 oligonucleotides and with O3 and O4 oligonucleotides, respectively (restriction sites are indicated in boldface in Table 1). The 1,119-bp E. cloacae fragment was cut by PstI and HindIII and inserted into pBAD/Myc-HisB plasmid vector (Invitrogen) opened by the same enzymes. The resulting plasmid was cut by NcoI, treated with Klenow fragment for filling-in of NcoI cohesive ends, and then cut by SacII. The 1,104-bp E. coli mraY fragment restricted by SmaI and SacII was inserted into the latter plasmid. This resulted in plasmid pMP6115, which expresses the MraY-P99 fusion under the control of the strong arabinose-inducible araBAD promoter. For expression of the fusion with a C-terminal His6 extension, pMP6115 was opened by XhoI and XbaI and ligated to two oligonucleotides, O5 and O6 (Table 1), that had been phosphorylated and hybridized together, resulting in plasmid pER139. The internal 1-kb BspHI fragment from pMP6115 carrying the ampicillin resistance gene was deleted and replaced by the 1.3-kb EcoRI kanamycin resistance cartridge (Pharmacia), after filling-in of the restriction sites with the Klenow fragment, resulting in plasmid pER122. For expression of the fusion with a C-terminal His6 extension in a kanamycin-selectable plasmid, the 0.7-kb KpnI-XbaI fragment of pER122 was replaced by the corresponding fragment of pER139, generating plasmid pER145. For introduction of a TEV protease cleavage site between MraY and P99 protein sequences, pER145 was opened by SacII and ligated to two oligonucleotides, O7 and O8 (Table 1), that had been previously phosphorylated and hybridized together. This resulted in the 6.5-kb kanamycin-selectable pER153 plasmid, allowing expression of the mraY gene product (MLVWLAE—-LATLKVR) fused to the His6-tagged mature form of P99 β-lactamase (TPVSEKQ—-HILEALQHHHHHH) via a short intermediate peptide linker (PRENLYFQGR) carrying the TEV protease cleaving site. The sequence of the whole insert cloned into the pER153 plasmid was confirmed by DNA sequencing.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea |
|---|---|
| O1 | 5′-CGCGCGCGCCCCGGGGTTTGGCTGGCCGAACATTTGGTCAAA-3′ |
| O2 | 5′-ACTGGCGTGCCGCGGACGTACCTTCAGCGTTGCCAGACCAATCAG-3′ |
| O3 | 5′-CGCGCGTCTGCAGCTCCGCGGACGCCAGTGTCAGAAAAACAGCTGGC-3′ |
| O4 | 5′-GCGCGCAAGCTTTTACTGTAGCGCCTCGAGGATATGGTATGC-3′ |
| O5 | 5′-TCGAGGCGCTACAGCACCATCACCATCACCATTAATAAT-3′ |
| O6 | 5′-CTAGATTATTAATGGTGATGGTGATGGTGCTGTAGCGCC-3′ |
| O7 | 5′-GTGAAAACCTGTACTTCCAAGGGC-3′ |
| O8 | 5′-CCTTGGAAGTACAGGTTTTCACGC-3′ |
Restriction sites introduced for cloning of mraY and P99 β-lactamase genes as detailed in the text are indicated in bold face.
Preparation of the crude enzyme
E. coli XL1-Blue(pER153) was used for the production of MraY. The culture (12 liters) was performed at 30°C in the medium described by Riesenberg et al. (25). The carbon source was glucose (30 g/liter) and the inoculum size was 2 ml of frozen working cell bank. After total depletion of glucose (A600 = 29), induction by l-arabinose (30 g/liter) for 7 h (final A600 = 30.1) led to a recovery of 493 g of biomass (wet weight). The next steps were performed at 4°C. The pellet (aliquot of 90 g) was resuspended in 100 ml of buffer A (50 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 2 mM 2-mercaptoethanol) containing DNase, RNase, and complete EDTA-free protease inhibitors (cocktail tablets from Boehringer Mannheim), and cells were disrupted with a French press. After centrifugation (40 min at 6,000 × g) and ultracentrifugation (1 h at 80,000 × g), the activity was recovered in the pellet. The enzyme was solubilized by treatment of the pellet in buffer A containing 0.25% N-laurylsarcosine and 20% glycerol for 1 h. After ultracentrifugation (1 h at 80,000 × g), the activity was recovered in the supernatant.
UDP-MurNAc-l-Ala-γ-d-Glu-meso-A2pm-d-Ala-d-Ala (UDP-MurNAc-pentapeptide)
The preparation of this compound (20) was scaled up to the millimole range. Cultures (12 liters) of B. cereus ATCC 9634 in the medium described by Reisenberg et al. (25) supplemented with glucose (30 g/liter) and Casamino Acids (20 g/liter) were performed under the following initial conditions: stirrer speed, 200 rpm; temperature, 30°C; inoculum size, 200 ml of a 17-h old preculture in the same medium (A600 = 14); pH regulated at 6.8; initial aeration rate, 0.33 vvm (air volume/medium volume/min); and dissolved oxygen maintained above 50% by stirrer speed control. After 5 h of growth (A600 = 3.6), chloramphenicol (100 mg/liter), uracil (80 mg/liter), d-glutamic acid (240 mg/liter), l-lysine (1 g/liter), and A2pm (mixture of all isomers, 240 mg/liter) were added, and vancomycin (10 mg/liter) was added 20 min later to ensure UDP-MurNAc-pentapeptide accumulation. Cells were harvested by centrifugation (50 min at 4,650 × g) 60 min after vancomycin addition, providing 120 g of biomass (wet weight). The pellet originating from four cultures was resuspended in 960 ml of buffer B (20 mM potassium phosphate buffer [pH 7.2]), and cells were disrupted with the French press. After centrifugation at 16,000 × g, purification was achieved in four chromatographic steps: (i) Q-Sepharose HP (250 by 200 mm), elution at 260 ml/min with a gradient (22 mM/min) of 0 to 1 M NaCl in buffer B; (ii) Q Hyper D (100 by 50 mm), elution at 30 ml/min with a step of 0.5 M NaCl in buffer B; (iii) Superdex 30 PG (700 by 100 mm) equilibrated in 50 mM Tris-HCl (pH 7.8)-0.5 M NaCl at 40 ml/min; and (iv) the same column equilibrated in water at 40 ml/min. UDP-MurNAc-pentapeptide (1.19 g, 1 mmol) was obtained with >95% purity (analytical high-pressure liquid chromatography [HPLC]). Analytical characteristics include the following: [MH]+, 1,194.4 (calculated) and 1,194.1 (found); and UV (in water), λmax 262 nm (ɛM = 10,000 M−1 cm−1).
UDP-MurNAc-Nɛ-dansylpentapeptide
UDP-MurNAc-pentapeptide (1 mmol) was dissolved in a 1:1 (vol/vol) mixture (175 ml) of 0.25 M sodium hydrogen carbonate and acetone, and dansyl chloride (10 mmol) was added. The solution was stirred for 2 h at room temperature in the dark. Acetone was evaporated off, an insoluble material was removed by centrifugation, and the supernatant was lyophilized. The resulting powder was dissolved in 300 ml of 1:1 (vol/vol) water-acetonitrile, diluted 10 times with buffer B, and subjected to two chromatographic steps: (i) Q Hyper D (100 by 50 mm), elution at 30 ml/min with a gradient (22 mM/min) of 0 to 0.5 M NaCl in buffer B and (ii) Superdex 30 PG (700 by 100 mm) equilibrated in water at 40 ml/min. UDP-MurNAc-Nɛ-dansylpentapeptide (903 mg, 0.63 mmol) was obtained with >95% purity (analytical HPLC). Analytical characteristics included the following: [MH]+, 1,427.2 (calculated) and 1,426.8 (found); UV (in water), λmax 250.5 nm (ɛM = 23,900 M−1 cm−1) and 329 nm (ɛM = 5,270 M−1 cm−1); fluorescence, λmax emission 547.5 nm (excitation at 325 nm) in 20 mM ammonium acetate in 3:1:1 water-methanol-isopropanol; and λmax emission 522 nm (excitation at 325 nm) in 20 mM ammonium acetate in 1:1 methanol-isopropanol.
HPLC assay
Reaction mixtures contained, in a final volume of 50 μl, 50 mM Tris-HCl (pH 7.8), 0.1% Triton X-100, 200 mM KCl, 50 mM MgCl2, 50 μM undecaprenyl phosphate, and 25 μM UDP-MurNAc-Nɛ-dansylpentapeptide. The reaction was initiated by the addition of enzyme (0.2 μg of proteins). After 15 min at 30°C, it was stopped by the addition of 150 μl of eluent A. Dansylated lipid I and residual UDP-MurNAc-Nɛ-dansylpentapeptide were separated on a Vydac C4 column (50 by 4.6 mm; eluent A, 20 mM ammonium acetate in 3:1:1 water-methanol-isopropanol; eluent B, 20 mM ammonium acetate in 1:1 methanol-isopropanol; gradient, 0% eluent A for 3 min, 0 to 100% eluent B for 2 min, 100% eluent B for 5 min, at 0.5 ml/min). Detection and quantitation were performed by monitoring their fluorescence emission at 495 nm (excitation at 325 nm) with a RF-10AXL fluorometer (Shimadzu).
HTS assay
Reactions were carried out as described above in 96- or 384-well plates, and the formation of dansylated lipid I was monitored for 15 min at 30°C by fluorescence enhancement (excitation at 355 nm, emission at 538 nm) by using a Victor2 fluorescence microplate reader (Perkin-Elmer, Courtabœuf, France).
IC50 determinations
The inhibitory effect of the compounds was determined in the HPLC and HTS assays. In both cases, the mixtures contained 5% (vol/vol) dimethyl sulfoxide in order to increase the solubility of the inhibitors. Fifty percent inhibitory concentration (IC50) values were calculated from plots of the percent inhibition versus the inhibitor concentration; data were processed with Grafit software.
Cell-based assay
The method of Maass and Pelzer (21) with permeabilized E. coli cells has been modified recently (13) in order to measure the incorporation of radiolabeled MurNAc-pentapeptide into trichloroacetic acid-precipitable material. The reaction was initiated by the addition of tritiated UDP-MurNAc-pentapeptide to toluenized E. coli K-12(250HT11) cells, in the presence or absence of inhibitors. The reaction was stopped by the addition of trichloroacetic acid, the precipitated material was collected by filtration, and the radioactivity retained on the filter was determined by using a radioactivity counter.
RESULTS AND DISCUSSION
Preparation of solubilized enzyme and of fluorescent substrate
Our first goal was to obtain large amounts of MraY in a solubilized form that would be more suitable for a HTS than a membrane preparation. For this purpose, an overproducing strain was necessary. All previous attempts to overproduce high levels of the MraY protein or to detect it by sodium dodecyl sulfate-polyacrylamide gel electrophoresis have failed. Only a radiolabeled form of this protein has been detected by using in vitro translation experiments (5). We recently analyzed the membrane topology of the E. coli MraY protein. Various fusions between defined fragments of MraY and the mature form of BlaM β-lactamase were generated, and their characterization led to a topological model (4). Interestingly, it was observed that the expression of MraY as a fusion with β-lactamase was an advantage for the stability and/or production of this protein. Indeed, an important increase of MraY activity was detected in cells in which the MraY-BlaM fusion was expressed from the pNF150 plasmid, which is not a very efficient vector for protein expression (19). The activity was significantly lower if only the MraY protein (not fused to BlaM) was expressed (data not shown). Taking these observations into account, we constructed a new plasmid suitable for MraY expression and purification. In this plasmid, pER153, the E. coli mraY and E. cloacae P99 β-lactamase genes were fused together and expressed under the control of the strong arabinose-inducible promoter. The P99 β-lactamase was preferred since it offers the possibility of purification by affinity chromatography (10); a TEV protease cleavage site between MraY and P99 sequences and a His6 tag extension at the C terminus were also introduced in order to facilitate future purification steps. The pER153 plasmid was transformed into wild-type strain XL1-Blue. In crude membrane extracts from the resulting strain, the level of MraY activity after growth in the absence or presence of arabinose was increased 3- and 30-fold, respectively, with respect to the host strain.
Extraction of the enzyme activity contained in the membrane fraction (20 nmol/min/mg of protein) with an N-laurylsarcosine-containing buffer resulted in a 2.5-fold purification: indeed, from 90 g of cells, 200 ml of solubilized membrane proteins at 0.5 mg/ml was obtained, with a specific activity of 50 nmol/min/mg of protein. This value is much higher than those obtained previously: 1 to 2 nmol/min/mg (6) and 2.4 nmol/min/mg (30). The preparation was stored at −80°C for 1 year without significant loss of activity. Neither MurG nor WecA activity was detected: when UDP-[14C]GlcNAc was added to the assay mixture, no radioactive lipid was formed. The preparation was not contaminated with undecaprenyl phosphate since no MraY activity was detected in the assay lacking this substrate. This crude fusion protein was used as source of MraY activity throughout the present study and will be referred to as MraY for sake of simplification.
The second step of the HTS setup was the preparation of a fluorescent substrate for the enzymatic assay. Other investigators had proposed UDP-MurNAc-Nɛ-dansylpentapeptide as a substrate for the MraY activity (6, 23). We decided to reexamine the fluorescence properties of UDP-MurNAc-Nɛ-dansylpentapeptide in the context of an MraY assay, to prepare a millimole amount of the dansylated nucleotide, and to adapt the assay to the high-throughput format.
Development of enzymatic assays
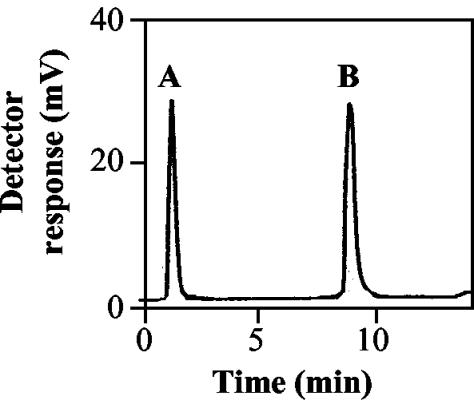
In order to characterize the activity of our partially purified MraY enzyme, we first performed the assay with separation of product and residual substrate. Whereas reaction conditions were similar to those of Brandish et al. (6), separation and quantitation were accomplished by reversed-phase HPLC on a C4 column with a gradient of isopropanol and methanol in water (1, 2) and on-line fluorescence detection. In the chromatograms of the assay mixtures (Fig. 1), the UDP-MurNAc-Nɛ-dansylpentapeptide peak, which eluted early (1.4 min), decreased, whereas a new peak (9.0 min), eluting with 100% organic solvent, appeared. The identity of the new peak as dansylated lipid I was confirmed by on-line mass spectrometry (MS) ([MH]+ = 1,950.4 [calculated] and 1,950.9 [found]) and MS/MS (fragments at m/z 1,201.4 [fragmentation at the allyl phosphate bond] and 1,023.4 [fragmentation at the muramyl phosphate bond plus loss of water]).
FIG. 1.
Chromatogram of an MraY assay mixture. See Materials and Methods for HPLC conditions. (A) UDP-MurNAc-Nɛ-dansylpentapeptide; (B) dansylated lipid I. This example corresponds to a 25% transformation of substrate into product.
Using this assay, some properties of the enzyme were determined and compared to those in the literature. The Km for UDP-MurNAc-Nɛ-dansylpentapeptide and undecaprenyl phosphate were 15 and 10 μM, respectively, by using fixed concentrations of 100 and 50 μM for undecaprenyl phosphate and UDP-MurNAc-Nɛ-dansylpentapeptide, respectively. The Km for the nucleotide substrate was similar to that found by Brandish et al. (19 μM) (6); as far as the natural lipid substrate was concerned, this was the first Km determination. The MraY reaction reached an equilibrium when 40 to 50% UDP-MurNAc-Nɛ-dansylpentapeptide at 25 μM was converted to dansylated lipid I, even with protein amounts up to 1.2 μg; this finding is consistent with the reversibility observed by Neuhaus (23). In contrast to that of Brandish et al. (6), our enzyme preparation was nearly not stimulated by phosphatidylglycerol; this discrepancy may arise from the nature of our enzyme, which is a fusion protein with β-lactamase and, perhaps, more adapted to a water environment.
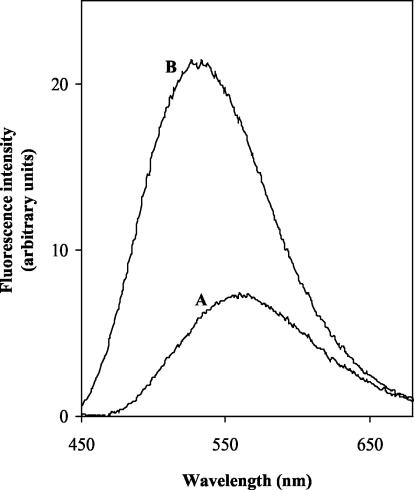
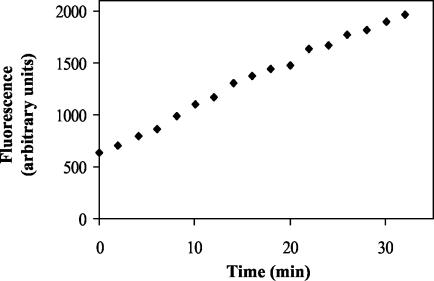
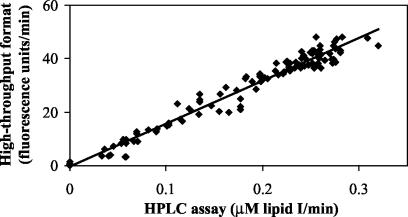
The next step was to perform the assay without separation. For this purpose, the emission fluorescence spectra (excitation at 355 nm) of UDP-MurNAc-Nɛ-dansylpentapeptide and of a synthetic analogue of lipid I, MurNAc(Nɛ-dansylpentapeptide)-pyrophosphoryl (R,S)-α-dihydroundecaprenol (2), were recorded in the reaction buffer. The fluorescence maximum of the dansyl moiety was blue-shifted in the latter compound (531 versus 561 nm), and the fluorescence intensity was increased (Fig. 2). Presumably, this resulted from the passage of the lipid product into detergent micelles. Taking into account the availability of the filters of the fluorescence microplate reader and the excitation/emission couple giving the best signal-to-noise ratio, we chose the following wavelengths: excitation at 355 nm and emission at 538 nm. In these conditions, the fluorescence of the product was sixfold greater than that of the substrate. When recorded in an assay mixture, the fluorescence increased linearly for at least 20 min, allowing an assay time of 15 min (Fig. 3). This assay was clearly adapted to the HTS format, and its reproducibility was attested by the good correlation between the enzymatic rates determined both in the high-throughput format and in the aforementioned HPLC assay (Fig. 4).
FIG. 2.
Fluorescence spectra of UDP-MurNAc-Nɛ-dansylpentapeptide (A [10 μM]) and MurNAc(Nɛ-dansylpentapeptide)-pyrophosphoryl (R,S)-α-dihydroundecaprenol (B [5.35 μM]) in 50 mM Tris-HCl (pH 7.8)-0.1% Triton X-100-200 mM KCl-50 mM MgCl2. The spectrum of the buffer alone was subtracted from both spectra. The excitation wavelength is 355 nm.
FIG. 3.
Time course of the MraY reaction measured in the HTS assay. Fluorescence was recorded every 100 s. The fluorescence enhancement is linear during the first 15 min.
FIG. 4.
Correlation curve between the enzymatic rates of 118 assays determined both with HPLC separation and in the high-throughput format. The variables used were the presence, the nature, and the concentration of inhibitors. The correlation equation is y = 159.99x − 0.25 (r2 = 0.9669).
Validation of the HTS with MraY inhibitors
To further validate the HTS screen, we determined the inhibition of the MraY activity by the natural inhibitors tunicamycins and mureidomycin B and by a series of synthetic compounds named riburamycins (T. Stachyra, J. Biton, C. Dini, S. Feteanu, and D. Le Beller, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-722, 2002). The IC50 values determined (Table 2) were quite similar for both assays. Tunicamycins, mureidomycin B, and the series of riburamycins were also tested in the previously described assay (13) that uses toluene-permeabilized E. coli bacteria. This cell-based assay was usually used to test inhibitors that produced no antibacterial effect on growing cells because of a lack of uptake and access to their targets. Comparison with the screening assay demonstrated (Table 2) a global preservation of the ranking within the riburamycin series in terms of inhibitory potency. One exception was observed with the tunicamycins; this might be due either to their mode of action, which is different from that of mureidomycin B and riburamycins (7), or to their higher affinity for the MraY enzyme within its membrane environment in the cell-based assay.
TABLE 2.
Inhibitory effect on the MraY activity of selected compounds
Assay with HPLC separation of substrate and product; the error on the determination was ≤16%.
Assay in the HTS format; the error on the determination was ≤20%.
Cell-based assay with permeabilized E. coli cells; the error on the determination was ≤30%.
Conclusion
The present study demonstrates progress toward the screening and the characterization of novel MraY inhibitors. As mentioned in the introduction, the existence of naturally occurring inhibitors has recently promoted this enzyme as a potential target for novel antibacterial compounds. The availability of an MraY assay in the HTS format is therefore needed. To date, five HTS assays have been described (3, 8, 11, 17, 30). However, these assays involve one or several additional activities (MurG, transglycosylase, or transpeptidase), and they use a radioactive substrate. Moreover, three of them rely upon the membrane content of undecaprenyl phosphate, which is not known with accuracy and may be variable from one membrane preparation to another. An additional drawback of membrane-based assays is the presence of lipid-synthesizing activities other than MraY and MurG (e.g., WecA [17]); these activities may interfere with an MraY/MurG coupled assay. In the present study, we set up a nonradioactive assay specific to MraY that is much more reliable than the previously utilized cell-based assay. The new assay could be adapted to the HTS format without separation. This HTS was validated by the determination of the IC50 values of a set of natural or synthetic compounds already described as MraY inhibitors: the values obtained at the HTS format were similar to those obtained in the assay with HPLC separation. The large-scale synthesis of the dansylated substrate for MraY and the HTS screening permit the testing of large chemical libraries (up to 200,000 compounds per week) at a reasonable cost. Selected hits can then be thoroughly studied by using the HPLC assay. The cell-based assay remains interesting for an early profiling of MraY inhibitors, to illustrate the role of the bacterial membrane, to predict the behavior of the inhibitors during the MIC measurement steps, and to check their target specificities.
Acknowledgments
This study was supported by grants from the Centre National de la Recherche Scientifique (UMR 8619) and from the Ministère de l'Education Nationale, de la Recherche, et de la Technologie (Biotechnologies).
The important contributions of Cécile Loison and Jacques Dumas are gratefully acknowledged. We also thank Evelyne Réalo and Marie-Thérèse Bocquel for excellent molecular biology support and Fabien Lux and Jacques Winter for fermentation support. We thank Geneviève Auger for the gift of the synthetic analogue of lipid I.
REFERENCES
- 1.Auger, G., M. Crouvoisier, M. Caroff, J. van Heijenoort, and D. Blanot. 1997. Synthesis of an analogue of the lipoglycopeptide membrane intermediate I of peptidoglycan biosynthesis. Lett. Peptide Sci. 4:371-376. [Google Scholar]
- 2.Auger, G., J. van Heijenoort, D. Mengin-Lecreulx, and D. Blanot. 2003. A MurG assay which utilizes a synthetic analogue of lipid I. FEMS Microbiol. Lett. 219:115-119. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa, M. D. F. S., H. O. Ross, M. C. Hillman, R. P. Meade, M. G. Kurilla, and D. L. Pompliano. 2002. A multitarget assay for inhibitors of membrane-associated steps of peptidoglycan biosynthesis. Anal. Biochem. 306:17-22. [DOI] [PubMed] [Google Scholar]
- 4.Bouhss, A., D. Mengin-Lecreulx, D. Le Beller, and J. van Heijenoort. 1999. Topological analysis of the MraY protein catalysing the first membrane step of peptidoglycan synthesis. Mol. Microbiol. 34:576-585. [DOI] [PubMed] [Google Scholar]
- 5.Boyle, D. S., and W. D. Donachie. 1998. mraY is an essential gene for cell growth in Escherichia coli. J. Bacteriol. 180:6429-6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandish, P. E., M. K. Burnham, J. T. Lonsdale, R. Southgate, M. Inukai, and T. D. H. Bugg. 1996. Slow binding inhibition of phospho-N-acetylmuramyl-pentapeptide-translocase (Escherichia coli) by mureidomycin A. J. Biol. Chem. 271:7609-7614. [DOI] [PubMed] [Google Scholar]
- 7.Brandish, P. E., K. I. Kimura, M. Inukai, R. Southgate, J. T. Lonsdale, and T. D. H. Bugg. 1996. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob. Agents Chemother. 40:1640-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandstrom, A. A., S. Midha, C. B. Longley, K. Han, E. R. Baizman, and H. R. Axelrod. 2000. Assay for identification of inhibitors for bacterial MraY translocase or MurG transferase. Anal. Biochem. 280:315-319. [DOI] [PubMed] [Google Scholar]
- 9.Bugg, T. D. H., and P. E. Brandish. 1994. From peptidoglycan to glycoproteins: common features of lipid-linked oligosaccharide biosynthesis. FEMS Microbiol. Lett. 119:255-262. [DOI] [PubMed] [Google Scholar]
- 10.Cartwright, S. J., and S. G. Waley. 1984. Purification of β-lactamases by affinity chromatography on phenylboronic acid-agarose. Biochem. J. 221:505-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandrakala, B., B. C. Elias, U. Mehra, N. S. Umapathy, P. Dwarakanath, T. S. Balganesh, and S. M. deSousa. 2001. Novel scintillation proximity assay for measuring membrane-associated steps of peptidoglycan biosynthesis in Escherichia coli. Antimicrob. Agents Chemother. 45:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagert, M., and S. D. Ehrlich. 1979. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 6:23-28. [DOI] [PubMed] [Google Scholar]
- 13.Dini, C., P. Collette, N. Drochon, J. C. Guillot, G. Lemoine, P. Mauvais, and J. Aszodi. 2000. Synthesis of the nucleoside moiety of liposidomycins: elucidation of the pharmacophore of this family of MraY inhibitors. Bioorg. Med. Chem. Lett. 10:1839-1843. [DOI] [PubMed] [Google Scholar]
- 14.Dini, C., S. Didier-Laurent, N. Drochon, S. Feteanu, J. C. Guillot, F. Monti, E. Uridat, J. Zhang, and J. Aszodi. 2002. Synthesis of sub-micromolar inhibitors of MraY by exploring the region originally occupied by the diazepanone ring in the liposidomycin structure. Bioorg. Med. Chem. Lett. 12:1209-1213. [DOI] [PubMed] [Google Scholar]
- 15.Dini, C., N. Drochon, S. Feteanu, J. C. Guillot, C. Peixoto, and J. Aszodi. 2001. Synthesis of analogues of the O-β-d-ribofuranosyl nucleoside moiety of liposidomycins. Part 1. Contribution of the amino group and the uracil moiety upon the inhibition of MraY. Bioorg. Med. Chem. Lett. 11:529-531. [DOI] [PubMed] [Google Scholar]
- 16.Dini, C., N. Drochon, J. C. Guillot, P. Mauvais, P. Walter, and J. Aszodi. 2001. Synthesis of analogues of the O-β-d-ribofuranosyl nucleoside moiety of liposidomycins. Part 2: role of the hydroxyl groups upon the inhibition of MraY. Bioorg. Med. Chem. Lett. 11:533-536. [DOI] [PubMed] [Google Scholar]
- 17.Hyland, S. A., and M. S. Anderson. 2003. A high-throughput solid-phase extraction assay capable of measuring diverse polyprenyl phosphate: sugar-1-phosphate transferases as exemplified by the WecA, MraY, and MurG proteins. Anal. Biochem. 317:156-164. [DOI] [PubMed] [Google Scholar]
- 18.Inukai, M., F. Ishino, and A. Takaitsuki. 1993. Selective inhibition of the bacterial translocase reaction in peptidoglycan synthesis by mureidomycins. Antimicrob. Agents Chemother. 37:980-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loubens, L., L. Debardieux, A. Bohin, J.-M. Lacroix, and J.-P. Bohin. 1993. Homology between a genetic locus (mdoA) involved in the osmoregulated biosynthesis of periplasmic glucans in Escherichia coli and a genetic locus (hrpM) controlling the pathogenicity of Pseudomonas syringae. Mol. Microbiol. 10:329-340. [DOI] [PubMed] [Google Scholar]
- 20.Lugtenberg, E. J. J., A. van Schijndel-van Dam, and T. H. M. van Bellegem. 1971. In vivo and in vitro action of new antibiotics interfering with the utilization of N-acetyl-glucosamine-N-acetyl-muramyl-pentapeptide. J. Bacteriol. 108:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maass, D., and H. Pelzer. 1981. Murein biosynthesis in ether permeabilized Escherichia coli starting from early peptidoglycan precursors. Arch. Microbiol. 130:301-306. [DOI] [PubMed] [Google Scholar]
- 22.McDonald, L. A., L. R. Barbieri, G. T. Carter, E. Lenoy, J. Lotvin, P. J. Petersen, M. M. Siegel, G. Singh, and R. T. Williamson. 2002. Structures of the muraymycins, novel peptidoglycan biosynthesis inhibitors. J. Am. Chem. Soc. 124:10260-10261. [DOI] [PubMed] [Google Scholar]
- 23.Neuhaus, F. C. 1971. Initial translocation reaction in the biosynthesis of peptidoglycan by bacterial membranes. Acc. Chem. Res. 4:297-303. [Google Scholar]
- 24.Reddy, S. G., S. T. Waddell, D. W. Kuo, K. K. Wong, and D. L. Pompliano. 1999. Preparative enzymatic synthesis and characterization of the cytoplasmic intermediates of murein biosynthesis. J. Am. Chem. Soc. 121:1175-1178. [Google Scholar]
- 25.Riesenberg, D., V. Schulz, W. A. Knorre, H. D. Pohl, D. Korz, E. A. Sanders, A. Ross, and W. D. Deckwer. 1991. High cell density cultivation of Escherichia coli at controlled specific growth rate. J. Biotechnol. 20:17-28. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Somner, E. A., and P. E. Reynolds. 1990. Inhibition of peptidoglycan biosynthesis by ramoplanin. Antimicrob. Agents Chemother. 34:423-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Heijenoort, J. 2001. Recent advances in the formation of bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 18:503-519. [DOI] [PubMed] [Google Scholar]
- 29.Weppner, W. A., and F. C. Neuhaus. 1977. Fluorescent substrate for nascent peptidoglycan synthesis. Uridine diphosphate-N-acetylmuramyl-(Nɛ-5-dimethylaminonaphthalene-1-sulfonyl)pentapeptide. J. Biol. Chem. 252:2296-2303. [PubMed] [Google Scholar]
- 30.Zawadzke, L. E., W. Ping, L. Cook, L. Fan, M. Casperson, M. Kishnani, D. Calambur, S. J. Hofstead, and R. Padmanabha. 2003. Targeting the MraY and MurG bacterial enzymes for antimicrobial therapeutic intervention. Anal. Biochem. 314:243-252. [DOI] [PubMed] [Google Scholar]