Abstract
Lipidomics is a critical part of metabolomics and aims to study all the lipids within a living system. We present here the development and evaluation of a sensitive capillary UPLC-MS method for comprehensive top-down/bottom-up lipid profiling. Three different stationary phases were evaluated in terms of peak capacity, linearity, reproducibility, and limit of quantification (LOQ) using a mixture of lipid standards representative of the lipidome. The relative standard deviations of the retention times and peak abundances of the lipid standards were 0.29% and 7.7%, respectively, when using the optimized method. The linearity was acceptable at >0.99 over 3 orders of magnitude, and the LOQs were sub-fmol. To demonstrate the performance of the method in the analysis of complex samples, we analyzed lipids extracted from a human cell line, rat plasma, and a model human skin tissue, identifying 446, 444, and 370 unique lipids, respectively. Overall, the method provided either higher coverage of the lipidome, greater measurement sensitivity, or both, when compared to other approaches of global, untargeted lipid profiling based on chromatography coupled with MS.
Keywords: ultra-performance liquid chromatography (UPLC), tandem mass spectrometry (MS/MS), electrospray ionization (ESI), top-down/bottom-up lipid profiling
Like genes and proteins, lipids are essential cellular components with functions ranging from support of membrane architecture [1] and maintenance of energy homeostasis [2–4] to modulation of enzyme activities and transduction of cellular events (e.g., proliferation and death) [5]. Dysregulated lipid metabolism is associated with a variety of pathological conditions, including diabetes mellitus [6–8], neurological disorders [9–11], cancers [12,13], and infectious diseases [14–16].
Lipids have been classified into 8 categories by the International Committee for the Classification and Nomenclature of Lipids in conjunction with the LIPID MAPS Consortium, based on their chemically distinct functional backbones and hydrophobic and hydrophilic elements [17–19]. However, due to the diversity of constituent fatty acids (e.g., various chain lengths, degrees of unsaturation, positions of double bonds, etc) and the combinatorial possibilities of backbone substitution, it is estimated that there are hundreds of molecular species within each of these categories. In addition, there are dramatic concentration differences between different classes of lipids, depending on cell type and cellular location [20].
For global quantitative analyses of lipid molecular species in complex mixtures, mass spectrometry (MS)-based techniques have become the method of choice since the initial use of electrospray ionization (ESI)-MS in the characterization of diacylglycerols [21]. There are two fundamentally different MS-based approaches commonly employed for lipid analysis [22]. The infusion-MS based approach, often referred to as “shotgun lipidomics”, subjects a total lipid extract to direct MS analysis without any prior chromatographic separation [9,20]. In general, this approach offers the most rapid and efficient analysis of individual molecular species from crude lipid extracts and has achieved success in analyzing well-studied lipid classes [9,20]; however, shotgun lipidomics is limited for discovery of novel or unexpected molecular species because only the species with known MS/MS fragmentation patterns are considered. In addition, because there are no chromatographic separation steps, ion suppression effects and lipid–lipid interactions can affect the ionization process, which makes the analysis of less concentrated species or those with low ionization efficiency more difficult [20]. Further, it is challenging to distinguish and quantify isobaric and isomeric (typically only one isobar or isomer is biologically important) species due to the low resolution mass analyzers (e.g., triple quadrupole MS) typically used in shotgun lipidomics studies [23].
The other approach commonly used in lipidomics analyses employs a high performance liquid chromatography (HPLC) step to separate lipids. The LC eluate is then coupled directly to a mass spectrometer for further online analysis. The chromatographic focusing provided by HPLC in combination with ESI-MS makes this approach the most sensitive method for analysis of lipids [24]. Normal- and reversed-phase HPLC are the two most commonly used separation methods. Normal-phase HPLC employs silica columns, and the separation is based on the physical properties of the lipid head groups such that lipids belonging to the same class or subclass co-elute without resolution of individual lipid molecular species, as a result it is used primarily to analyze non-polar, neutral lipids [25] or in off-line or online fractioning strategies prior to targeted or multidimensional analyses [26]. Conversely, reversed-phase HPLC relies on the hydrophobic interactions between the alkyl chains on the stationary phase and those of the lipid fatty acids to separate molecular species in the same lipid class or subclass [20,24,5]. However, co-elution of molecular species from different lipid classes can occur [27,28], and in some cases, large or highly hydrophobic lipid species cannot completely elute from the column, making the quantification of these species difficult [20].
Compared with wide bore analytical columns, capillary HPLC columns offer higher sensitivity and smaller sample requirements [29]. Due to continuous innovations in surface chemistry and decreases in particle sizes of column packing materials, we evaluated the performance of three reversed-phase packing materials [27,30–33] – C8 (1.7 μm), C18 (3.0 μm) and HSST3-C18 (1.8 μm) – in capillary column format as the front end method for separation of a representative lipid standard mixture comprised of 55 species from 6 lipid classes. Reproducible retention times and peak areas were obtained with these reversed-phase capillary columns, and the limits of quantification for most of the lipid standards were sub-fmol. The use of high resolution LTQ-Orbitrap and LTQ-Orbitrap Velos mass spectrometers employing higher energy collision dissociation (HCD) fragmentation provides relative quantification and confident identification of lipid molecular species based on accurate mass measurements of precursor ions and diagnostic fragment ions obtained from data-dependent MS/MS events (i.e. combined top-down and bottom-up lipidomics) [34]. The utility of our method was further demonstrated in comprehensive analyses of lipids from three representative biological samples, a human lung epithelial cell line (Calu-3), rat plasma, and a model human skin tissue. Overall, the method provided either higher coverage of the lipidome, greater measurement sensitivity, or both, when compared to other approaches of global, untargeted lipid profiling based on chromatography coupled with MS.
MATERIALS AND METHODS
Chemicals
HPLC-grade acetonitrile and isopropanol were purchased from Fisher Scientific (Fair Lawn, NJ), while MS-grade ammonium acetate was obtained from Fluka (St. Louis, NJ).
Lipid standards
Lipid standards (Table S1) were purchased from Avanti Polar Lipids (Alabaster, AL), and equimolar standard mixtures were prepared in the following concentrations: 0.5 nM, 1 nM, 5 nM, 10 nM, 50 nM, 100 nM and 500 nM. Deuterated 1,2-dimyristoyl-sn-glycero-3-phosphocholine (14:0 PC D4) was spiked into the unlabeled standard mixtures at a fixed concentration of 50 nM in order to generate calibration curves for evaluating the dynamic range and sensitivity of the LC-MS method and not for absolute quantification of lipids.
Lipid extraction
Lipids were extracted from biological samples (see Supplemental Methods) using cold (−20 °C) chloroform/methanol (2:1, v/v) in a 5:1 ratio over the sample volume. The mixtures were vortexed for 10 s and allowed to stand on ice for 10 min, followed by vortexing for an additional 10 s. After centrifugation at 13,523g for 10 min, 200 μL of the chloroform layers were removed by pipetting and dried in vacuo. Prior to LC-MS analysis, the lipid extracts were reconstituted in 200 μL of isopropanol containing 10 mM ammonium acetate. This sample processing approach resulted in an extraction recovery efficiency of ≥70% for eight lipid standards representative of the major lipid classes present in mammalian samples and evaluated according to the method of Matuszewski et al [35] (see Supplemental Methods).
Capillary ultra-performance liquid chromatography-mass spectrometry (UPLC-MS)
A small trapping column (180 μm i.d. × 2 cm) packed with reversed-phase particles (Symmetry C18, 5 μm; Waters, Milford, MA) was used prior to the analytical column for fast (within 1.5 min at a flow rate of 10 μL/min) loading of lipid samples, followed by washing of the column-bound lipids to remove chemical impurities and non-lipid sample components. Reconstituted lipids (4 μL) were loaded onto the trapping column under the following isocratic conditions: 93% acetonitrile/water (40:60) containing 10 mM ammonium acetate (solvent A) and 7% acetonitrile/isopropanol (10:90) containing 10 mM ammonium acetate (solvent B). The lipids retained on the trapping column were then forward-flushed to the analytical column using gradient elution (Table S1).
Three different analytical columns (Table 1) were slurry packed, as previously described [27], and evaluated in separations of the lipid standard mixture and biological sample extracts using a Waters NanoAcquity LC system. The columns (150 μm × 20 cm) were maintained at 40°C in a column oven, and gradient elution was performed as shown in Table S1 over 90 min.
Table 1.
LC packing material, column characteristics, and calculated peak capacities for the capillary columns. The pressure ranges listed indicate the LC operating pressures at the beginning and end of the analyses. All the three columns (150 μm i.d. × 20 cm) run with flow rate at 1 μL/min. The overall peak capacities were calculated as the average of the peak capacities from the 11 selected lipids species from Table 1.
| Column | Manufacturer | Particle size (μm) | Pressure (psi) | Starting time (min) | Ending time(min) | Peak capacity |
|---|---|---|---|---|---|---|
| HSS T3 C18 | Waters | 1.8 | 3000–8000 | 11.4 | 76.7 | 121 |
| Jupiter C18 | Phenomenex | 3 | 900–2000 | 11.2 | 72.2 | 78 |
| C8 | Waters | 1.7 | 3000–8000 | 10.7 | 73.0 | 84 |
The LC system was interfaced to a LTQ-Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA) using a chemically etched ESI emitter [36], and the ESI emitter and MS inlet capillary potentials were 2.2 kV and 12 V, respectively. Data-dependent MS/MS scan events were performed in the ion-trap (collision-induced dissociation; CID) or Orbitrap (HCD) using a normalized collision energy (NCE) of 35 and 40 arbitrary units, respectively. Both CID and HCD were set with a maximum charge state of 2 and an isolation width of 2 m/z units. An activation Q value of 0.18 was used for CID. The full scan mass ranges for positive and negative ESI modes were 300–2000 and 200–2000 m/z, respectively. In other analyses, a LTQ-Orbitrap Velos (Thermo Scientific) mass spectrometer was used with a NCE of 35 and 30 arbitrary units for CID and HCD, respectively. Each sample was analyzed in triplicate in both ESI modes.
Data processing
Data were acquired under the control of Thermo Xcalibur software (Thermo Scientific). Chromatographic peaks of lipid standards were integrated using Xcalibur Quan Browser in order to construct calibration curves. The PRISM Data Analysis system [37], a series of software tools freely available at http://ncrr.pnl.gov/software/ and developed in-house, was used to process and analyze the LC-MS data generated from the biological sample lipid extracts. The first step involved deisotoping of the raw MS data to give the monoisotopic mass, charge state, and intensity of the major peaks in each mass spectrum using Decon2LS [38]. The data were next examined in a 2-D fashion using MultiAlign to identify groups of mass spectral peaks that were observed in sequential spectra (i.e., “features”) using an algorithm [39] that computes a Euclidean distance in n-dimensional space for combinations of peaks. LC-MS features were then chromatographically aligned across all replicates for each sample using the LCMSWARP algorithm [40] in MultiAlign, and the identities of detected lipids were determined using the in-house software LipidMiner (see Supplemental Methods).
RESULTS AND DISCUSSION
Total lipid extracts of cell, tissue, and biofluid samples are complex, with sample components often numbering in the hundreds to thousands and present in a wide range of concentration. For example, participants in the LIPID MAPS consortium recently performed a quantitative assessment of the human plasma lipidome and measured 588 individual molecular species of fatty acyls (107), glycerolipids (73), glycerophospholipids (160), sphingolipids (204), sterol lipids (36), and prenol lipids (8) using targeted molecular analysis approaches [41]. Importantly, these species were present in concentrations ranging from 0.009 pmol/mL (the eicosanoid 11-trans-LTC4) to 0.820 μmol/mL (free cholesterol; total cholesterol was measured as 3.76 μmol/mL) [41], almost 10 orders of magnitude difference and mirroring the concentration range reported for human plasma proteins [42]. The quantitative measurement of the range of lipids in biological samples is not yet possible in a single analysis with current technology. However, useful information for assessing biological status can be obtained from untargeted lipid profiling experiments, and the goal of the current study was to develop a sensitive capillary LC-MS method for such applications.
LC-MS method development and separation characteristics
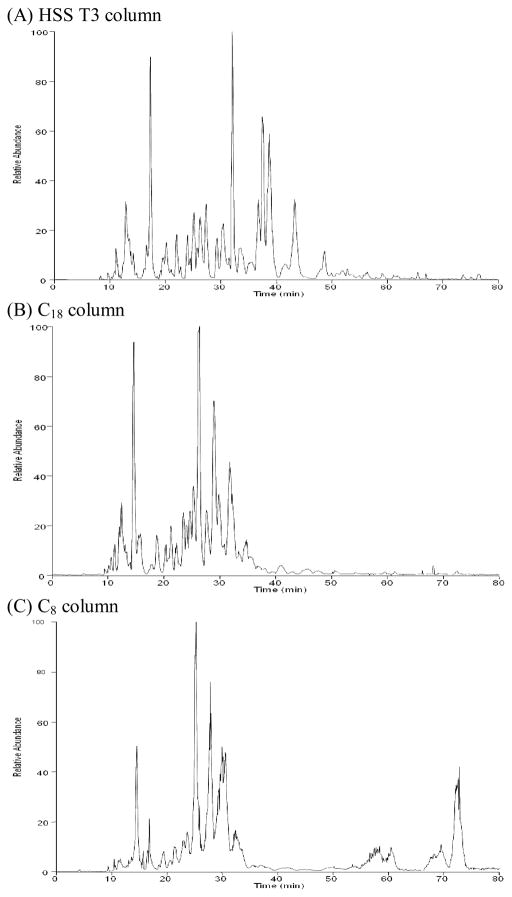
The aim of untargeted lipid profiling experiments is the detection and relative quantification of as many sample components as reasonably possible in order to identify those lipids that can be used to describe a disease, growth condition, or some external perturbation. In the current study, we used a capillary column and associated low flow rate in order to exploit the increased ESI-MS efficiency (defined as the number of analyte ions recorded at the MS detector divided by the number of analyte molecules delivered to the ESI emitter [29]) and, therefore, overall sensitivity of the measurements obtained from this format [43–47]. Similarly, we chose to compare lipid separations obtained using sub-2μm particles to that obtained from a more conventional LC packing material (Table 1), since such UPLC [48] particles provide increased LC separation efficiency through a decrease in the height equivalent to a theoretical plate, resulting in an increase in the number of theoretical plates per column [49]. To ensure that the method would be broadly applicable in lipid profiling experiments, we utilized a mixture comprised of 54 representative lipid standards, including glycerophospholipids (glycerophosphocholine, PC; lysoglycerophosphocholine, LPC glycerophosphoethanolamine, PE; lysoglycerophosphoethanolamine, LPE; glycerophosphoserine, PS; lysoglycerophosphoserine, LPS; glycerophosphoglycerol, PG; lysoglycerophosphoglycerol, LPG; and cardiolipins), glycerolipids (monoradylglycerols, MG; diradylglycerols, DG; triradylglycerols, TG; glycosyldiradylglycerols, MGDG and DGDG), sterol lipids (cholesterol and its esters), sphingolipids (sphingomyelin, SM and ceramides, Cer), fatty acyls (free fatty acids, FA) and one isotope labeled standard 1,2-dimyristoyl-sn-glycero-3-phosphocholine-1,1,2,2-d4 (14:0 PC D4). Based on commercial availability, at least three different molecular species from each class were chosen when possible, including one lipid with an unsaturated acyl chain, one lipid with a short acyl chain, and one lipid with a long acyl chain (Table S1). The mobile phase composition and gradient were then optimized to obtain the best separation possible of the standard mixture. The separation of lipid standards obtained using capillary columns packed with the three materials is shown in Figure 1 and is similar to recent reports of reversed-phase LC separations of lipids [28,50,27] in the sense that species from more polar classes such as lysoglycerophospholipids and FA eluted during the beginning of the separation, followed by more hydrophobic classes such as glycerophospholipids, sphingolipids, and smaller glycerolipids (MG and DG) in the middle of the separation, and followed finally by the most hydrophobic classes such as sterol esters and larger glycerolipids (TG) at the end of the separation (Table S1 and Figure S1). In addition, molecular species in a given class with more carbon atoms in the acyl chains and fewer or no double bonds tended to retain on the reversed-phase columns for a longer time compared to those with fewer carbon atoms or more double bonds. Some classes, such as TG with long fatty acyl chains (e.g., 24:1/24:1/24:1 TG in the lipid standard mixture), can be difficult to completely elute from a traditional C18 stationary phase, resulting in peak broadening and sample carryover. Indeed, we observed carryover of TG species during capillary LC-MS analyses of lipid extracts from blood plasma samples when using our previously published method [27], which relied on a solvent system based on methanol. Importantly, the incorporation of isopropanol into the current solvent system resulted in complete elution of the 24:1/24:1/24:1 TG with no carry-over detected in subsequent blank samples. In addition, we added a flushing procedure at the end of the gradient by ramping down from 99.5% solvent B after the end of the separation to 60%, followed by a return to 99.5% (Table S2) to further clean the column.
Figure 1.
Representative positive ESI chromatograms produced from capillary LC separations of the lipid standard using the HSS T3, C18 and C8 materials.
To provide an estimate of LC separation quality, we calculated the peak capacity for each capillary column. LC column peak capacity is a measure of the column efficiency and is generally defined as the number of fully resolved peaks that can fit shoulder-to-shoulder within the separation space [51]. In our analyses, peak elution began ~10 min after sample injection, and the last lipid standard eluted at ~76 min (Figure 1, Tables 1 & S1). So as not to bias the calculation of peak capacity (e.g., by selecting a single narrow chromatographic peak), eleven lipid molecular species from 11 different lipid classes (marked in bold in Table S1) were selected for calculating peak capacities as the elution time range (t) divided by 4σ, where 2σ corresponds to the width of a LC peak at 60% height [52]. The LC peak capacities were then estimated based on the average of the peak capacities calculated for the 11 selected standards. As shown in Table 1, the HSS T3 column provided the highest peak capacity in separations of the lipid standard mixture. The peak capacities could be potentially improved by using longer columns or longer separation times, as reported in a previous study [53], which showed that chromatographic peak capacities of 1000–1500 were possible for complex metabolite mixtures.
The intraday and interday variation was assessed by performing repeated analyses of the lipid standards on the same day or over three consecutive days using all three capillary columns. The median relative standard deviation (RSD) of the retention times of the standards was 0.29%, 0.64%, and 0.51% for the capillary columns packed with the HSS T3, traditional C18, and C8 materials, respectively. Similarly, the median RSD of the peak areas of the standards was 7.7%, 5.4%, and 8.2% for HSS T3, traditional C18, and C8 materials, respectively. These values are comparable to those in recent reports of LC-MS analyses of lipids [50,26,28].
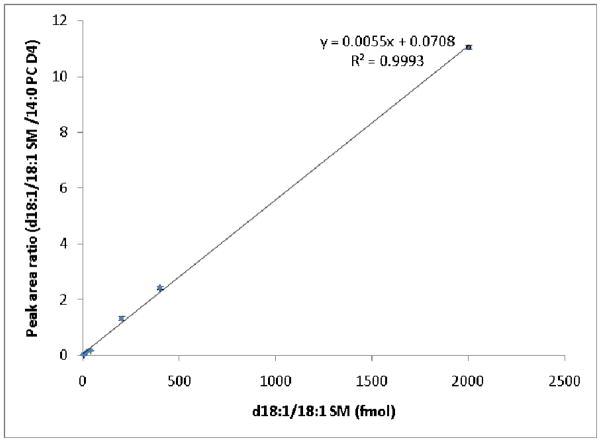
To estimate the sensitivity of the capillary LC-MS method when using columns packed with the three materials shown in Table 1, a dilution series of the lipid standard mixture was analyzed in triplicate. Table 2 shows the limits of quantification (LOQ; defined here as the lowest amount of analyte that can be accurately measured at a signal-to-noise ratio of ≥10) for columns containing each material and the associated signal-to-noise (S/N) ratios for the lowest amount of lipid standard mixture injected. For the 11 lipid molecular species shown in Table 2, the LOQs were generally ≤2 fmol for each column. The variations in LOQs for the 11 different lipid standards can be explained by differences in their ionization efficiencies. For example, glycerophospholipids such as PC, PE, and PS and sphingolipids such as SM and Cer are more easily ionized than neutral lipids such as CE and TG because they form protonated rather than adducted (e.g., NH4) ions. In particular, the cardiolipin in our study generated both protonated and ammoniated ions, with about 5- to 10-fold higher intensities from ammoniated ions. It is important to note the relatively large S/N associated with the LOQs in Table 2, indicating that these values could be extended to the hundreds of amol and that the associated limits of detection (LOD; defined here as the minimum amount of analyte that can be accurately measured with a S/N of 3) may be much lower. With a few exceptions, the capillary column packed with the HSS T3 material provided the highest S/N for the 11 molecular species shown in Table 2. To evaluate the linearity of the method when using the three columns, standard curves were constructed. The HSS T3 column in particular showed R2 values > 0.99 over 3 orders of magnitude in concentration (Figure S2) for all lipids in Table 2 with the exception of 18:0 MGDG (R2 = 0.9841). Figure 3 shows the standard curve of d18:1/18:1 SM with an R2 of 0.9993. These values are comparable with those of recent reports of LC-MS analyses of lipids [50,26,28].
Table 2.
Limits of quantification (LOQ) and signal-to-noise (S/N) ratios obtained for representative lipids using the capillary LC columns evaluated in this study.
| Lipid molecular species | Waters HSS T3 | Jupiter C18 | Waters C8 | |||
|---|---|---|---|---|---|---|
| LOQ | S/N | LOQ | S/N | LOQ | S/N | |
| 14:0 LPC | 2 fmol | 59 | 2 fmol | 15 | 2 fmol | 43 |
| 18:0/18:0 PE | 2 fmol | 58 | 2 fmol | 27 | 2 fmol | 26 |
| 18:1 LPS | 2 fmol | 70 | 2 fmol | 50 | 4 fmol | 106 |
| 14:0/14:0 PG | 2 fmol | 51 | 2 fmol | 44 | 2 fmol | 35 |
| 19:0 DG | 2 fmol | 24 | 2 fmol | 25 | 2 fmol | 30 |
| 24:1 CE | 2 fmol | 78 | 2 fmol | 48 | 2 fmol | 159 |
| d18:1/18:1 SM | 2 fmol | 41 | 2 fmol | 33 | 2 fmol | 15 |
| d18:1/24:0 Cer | 2 fmol | 20 | 2 fmol | 32 | 4 fmol | 11 |
| 18:1/18:1/18:1/18:1 CL | 2 fmol | 21 | 2 fmol | 24 | 2 fmol | 46 |
| C20:1 Δ11 Cis FA | 2 fmol | 77 | 2 fmol | 55 | 2 fmol | 30 |
| 18:0/18:0 MGDG | 2 fmol | 22 | 2 fmol | 18 | 20 fmol | 44 |
Figure 3.
Standard curve of SM d18:1/18:1 generated from triplicate UPLC-MS analysis of the serially diluted lipid standard mixture using the HSS T3 capillary column. Deuterated PC 14:0 was added as an internal standard in a constant amount (200 fmol). The amounts of SM d18:1/18:1 injected were 2 fmol, 4 fmol, 20 fmol, 40 fmol, 200 fmol, 400 fmol and 2000 fmol.
Analysis of lipid extracts from biological samples
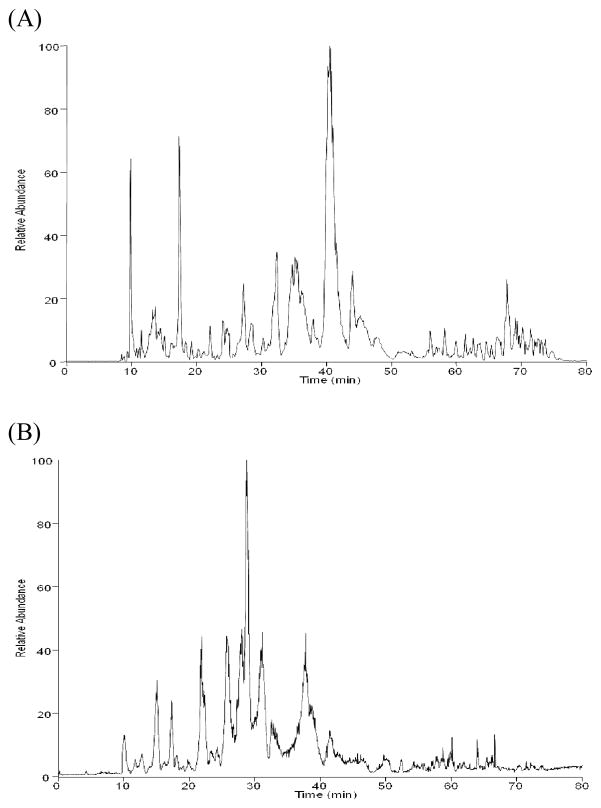
To evaluate the performance of the three packing materials in lipid profiling experiments, we analyzed lipid extracts from Calu-3 cells, rat blood plasma, and human skin tissue in triplicate using each of the capillary columns described above. Importantly, the reproducibility of detected lipid abundances was excellent for all three capillary columns, with Pearson correlation coefficients >0.98 between technical replicates. Consistent with our observations in the analysis of the lipid standard mixture, the HSS T3 column showed superior performance compared to the traditional C18 capillary column and the C8 capillary UPLC column in terms of the number of lipid features reproducibly (n = 3) detected. For example, 2230 lipid features were reproducibly detected from positive ESI analysis of Calu-3 cell lipids using the HSS T3 capillary UPLC column as compared to the traditional C18 capillary column (1784 lipid features reproducibly detected) and the C8 capillary UPLC column (1599 lipid features reproducibly detected). This is due to the higher separation efficiency (Table 1) obtained using the sub-2 μm particles compared to the 3 μm particles used in the traditional C18 column and to the greater retention of lipids by the C18 stationary phase relative to the C8 stationary phase used in the other capillary UPLC column (Figure 2 shows a representative chromatogram obtained from analysis of Calu-3 cell lipids using the HSS T3 capillary UPLC column). Therefore, we focused our analysis on the data generated from the capillary UPLC-MS method using the HSS T3 column.
Figure 2.
Representative chromatograms produced from UPLC separations of Calu-3 cell lipids using the capillary HSS T3 column. (A) positive ESI mode and (B) negative ESI mode.
The variation in the analysis of complex lipid extracts using the HSS T3 capillary column was evaluated by calculating the RSDs of the retention times and peak areas of lipid features detected in triplicate analysis of Calu-3 cell lipids. As in the analysis of the lipid standards, the median RSDs of the retention times and peak areas of the 2230 lipid features reproducibly detected in positive ESI analysis of Calu-3 cell lipids were 0.33% and 11%, respectively. The median RSD of the lipid feature peak areas is higher than those (e.g., 5% [28], 4.3–6.2% [50], and 2.4–6.4% [26]) in recent reports of LC-MS analyses of lipids; however, this slight increase may be attributed to an increase in the variability of the “open” ESI source used in our laboratory and the lack of sheath gas.
To determine if the method could provide comprehensive analysis of lipids from biological samples, we structurally identified the lipid features detected in Calu-3 cells, rat blood plasma, and human skin tissue. Lipids detected in lipid profiling or lipidomics analyses are typically identified using one of two approaches. In top-down lipidomics, the masses or m/z values of detected lipid features are compared directly to known lipid masses or m/z values in custom or publicly available databases, or the observed masses or m/z values are used to calculate candidate empirical formulae for the detected lipids, which are then compared to database entries [54,55,28]. In bottom-up lipidomics, lipids are fragmented in targeted or data-dependent MS/MS analyses, and diagnostic and other ions are used to assign lipid class and fatty acid composition [56,27,57,58]. In this work, we used a LTQ-Orbitrap Velos instrument and combined top-down and bottom-up lipidomics in the same analysis to structurally identify detected lipids in the current study. Because the fragment ion low mass cut-off limits the identification of lipids during CID MS/MS in ion-trap instruments [27], we also used alternating data-dependent CID and HCD fragmentation during analyses of lipid extracts from biological samples in order to obtain as much useful MS/MS information as possible (representative positive and negative ion MS/MS spectra of lipid molecular species from the major lipid classes detected in our study are shown in Supplemental Figure S3).. The latter mode provides triple quadrupole-like fragmentation, and Schuhmann et al. recently demonstrated that HCD on a LTQ-Orbitrap Velos improved the confidence of lipid identification in shotgun lipidomics (i.e., direct infusion-MS/MS) studies compared to CID [34]. Recent work by Bushee and Argikar suggested that LC-MS methods relying on data-dependent MS/MS characterizations of small molecules should incorporate both HCD and CID for comprehensive structural information [59]. The data produced from the combined top-down/bottom-up approach was processed using the in-house software LipidMiner for lipid identification (See Supplemental Methods). The software first generates a list of all fragment ions produced from each data-dependent MS/MS event along with the associated precursor ions. The software then applies an in silico “triple quadrupole-like” filter to identify the diagnostic (i.e., fragment or neutral-loss) ions [57,60,61,20] (Table S3) specific to each lipid class. The software then groups the results of this filtering process by lipid class, and the accurate masses of the precursor ions are matched by class to entries in the Lipid Maps (www.lipidmaps.org) database. Finally, comparison of the retention times of identified lipids to the retentions times or ranges of the lipid standards was used as an additional metric of identification confidence. From this process, 446, 444, and 370 lipid molecular species were identified in the Calu-3, rat blood plasma, and human skin tissue samples, respectively (Tables S4–S7).
Comparison of the capillary UPLC-MS method with other LC-MS approaches
In order to benchmark our approach, we compared our results to those obtained from recently reported chromatography-MS-based lipidomics and lipid profiling methods. While some of these methods are also based on the use of sub-2 μm LC particles, the columns were large bore (2.1 mm i.d.) and the associated flow rates relatively high at 300–500 μL/min, which leads to decreased ESI efficiency and overall decreased sensitivity [43–47]. Indeed, 2 out of the 4 methods [28,50] reported fewer lipid features detected and fewer lipids structurally identified than the method described in this paper, although the software and general methodologies used for feature detection and lipid identification vary and can influence the final numbers. In addition, these metrics depend on the dynamic range of the mass spectrometer used and the LC sample loading.
The findings of Castro-Perez et al. [50] offer the fairest comparison to the method reported herein. Their method utilized a large bore (2.1 mm i.d.) column containing the same packing material (Waters HSS T3 C18) and same solvent system as used in our capillary UPLC method. The total separation time for their method was 15 min (making the method useful for rapid screening of samples) compared to the 90 min separation in our method, and they reported fewer detected lipid features (1500) and structurally identified lipids (284) in the analysis of human plasma lipids, compared to the 2000+ lipid features reproducibly (n = 3) detected and 444 lipids structurally identified in our analysis of rat plasma lipids. The fewer numbers of detected and structurally identified lipids in their study is likely a result of the relatively fast gradient and high flow rate used (400 μL/min), which may have resulted in reduced separation peak capacity and decreased sensitivity. Indeed, the lowest amounts of measured lipid standards reported by Castro-Perez and colleagues ranged from 14–73 pmol compared to the LOQs of 2 fmol (almost 4 orders of magnitude difference) in our study.
Sandra et al. [28] and Nie et al. [26] published recent papers detailing LC methods of comparable separation times (89 and 114 min, respectively) to the method reported here (90 min). Again, both groups utilized large bore (2.1 mm i.d.) columns and relatively high flow rates (300 or 500 μL/min). The method reported by Sandra and colleagues resulted in the detection of 1528 features and at least 55 structurally identified human plasma lipids (presumably more species were structurally identified, but the authors only provide information for 55). Nie et al. reported the structural identification of 721 lipids from rat peritonea – nearly double the number of lipids structurally identified using our method in the analysis of a similar sample type (i.e. 370 lipids reproducibly identified in human skin tissue). However, this method was based on an online 2-D separation of lipids, with the first dimension consisting of normal-phase separation of lipids by class, followed by reversed-phase separation of each normal-phase fraction. Thus, the peak capacity, although not reported by the authors, should be high due to the orthogonality of the two separation dimensions. Despite the decrease in sample complexity of each normal-phase fraction, Nie and colleagues report LODs of 55–65 fmol [26] – at least 27-fold higher than the typical LOQs reported in the current work.
Quehenberger et al. [41] also reported higher numbers of structurally identified human plasma lipids (588) compared to the 444 rat plasma lipids identified in our study. Importantly, the authors chose to use targeted approaches to obtain absolute quantitative data for all of the lipids identified in their study. The advantage to such an approach is the specificity and sensitivity offered by the analytical methodology. However, this approach may be difficult to reproduce by most researchers, considering that 7 different laboratories participated in the measurement of the plasma lipids and that the total separation time of all the methods employed was >290 min (if sample preparation time is factored in, then the total analysis time far exceeds any of the other methods discussed above). Nevertheless, the methods of Quehenberger and colleagues resulted in an impressive, quantitative characterization of the human plasma lipidome and can be repeated when necessary, particularly if stoichiometric information is required.
A special acknowledgement must be made to the work of Moon [62–65] and Taguchi [66] and colleagues. While we have previously reported the use of capillary LC coupled with MS for the analysis of lipids [27], their laboratories have exploited the sensitivity gained with capillary LC since 2005. Moon and colleagues, in particular, have excelled in using nanoflow-scale (e.g., 300 nL/min) separations in the analysis of lipids. However, because their methods are centered on the analysis of phospholipids, we assume that the chromatography has been optimized for these classes of lipids only and therefore have not included them with the other more global methods discussed above.
CONCLUSIONS
In summary, we have developed a capillary UPLC-MS lipidomics approach that provides either higher coverage of the lipidome, greater measurement sensitivity, or both, when compared to other approaches of global, untargeted lipid profiling based on chromatography coupled with MS. In addition, the linearity and reproducibility of the method are comparable to those of recent reports [50,26,28]. We anticipate that the method will be particularly valuable in the analysis of samples of limited quantity, e.g., needle biopsies, aspirate fluids, and exhaled breath condensates.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Department of Health and Human Services, under Contract Number HHSN272200800060C. Additional support was provided by the NIAID under Award Number U54AI081680 and by the Office of Science, U.S. Department of Energy (DOE), under the Low Dose Radiation Research Program. Work was performed at the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RLO 1830.
References
- 1.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. nrm2330 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astrup A, Dyerberg J, Selleck M, Stender S. Nutrition transition and its relationship to the development of obesity and related chronic diseases. Obes Rev. 2008;9(Suppl 1):48–52. doi: 10.1111/j.1467-789X.2007.00438.x. OBR438 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Russo GL. Dietary n-6 and n-3 polyunsaturated fatty acids: from biochemistry to clinical implications in cardiovascular prevention. Biochem Pharmacol. 2009;77(6):937–946. doi: 10.1016/j.bcp.2008.10.020. S0006-2952(08)00777-6 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48(12):2547–2559. doi: 10.1194/jlr.R700014-JLR200. R700014-JLR200 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4(7):594–610. doi: 10.1038/nrd1776. nrd1776 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Oresic M, Simell S, Sysi-Aho M, Nanto-Salonen K, Seppanen-Laakso T, Parikka V, Katajamaa M, Hekkala A, Mattila I, Keskinen P, Yetukuri L, Reinikainen A, Lahde J, Suortti T, Hakalax J, Simell T, Hyoty H, Veijola R, Ilonen J, Lahesmaa R, Knip M, Simell O. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med. 2008;205(13):2975–2984. doi: 10.1084/jem.20081800. jem.20081800 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorensen CM, Ding J, Zhang Q, Alquier T, Zhao R, Mueller PW, Smith RD, Metz TO. Perturbations in the lipid profile of individuals with newly diagnosed type 1 diabetes mellitus: lipidomics analysis of a Diabetes Antibody Standardization Program sample subset. Clin Biochem. 2010;43(12):948–956. doi: 10.1016/j.clinbiochem.2010.04.075. S0009-9120(10)00211-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross RW, Han X. Lipidomics in diabetes and the metabolic syndrome. Methods Enzymol. 2007;433:73–90. doi: 10.1016/S0076-6879(07)33004-8. S0076-6879(07)33004-8 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Han X. Multi-dimensional mass spectrometry-based shotgun lipidomics and the altered lipids at the mild cognitive impairment stage of Alzheimer’s disease. Biochim Biophys Acta. 2010;1801(8):774–783. doi: 10.1016/j.bbalip.2010.01.010. S1388-1981(10)00023-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao L, Spassieva SD, Jucius TJ, Shultz LD, Shick HE, Macklin WB, Hannun YA, Obeid LM, Ackerman SL. A deficiency of ceramide biosynthesis causes cerebellar purkinje cell neurodegeneration and lipofuscin accumulation. PLoS Genet. 2011;7(5):e1002063. doi: 10.1371/journal.pgen.1002063. PGENETICS-D-11-00180 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, Troncoso JC, Mattson MP. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. Proc Natl Acad Sci U S A. 2004;101(7):2070–2075. doi: 10.1073/pnas.0305799101. 0305799101 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandis AZ, Wenk MR. Lipid-based biomarkers for cancer. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(26):2830–2835. doi: 10.1016/j.jchromb.2009.06.015. S1570-0232(09)00422-X [pii] [DOI] [PubMed] [Google Scholar]
- 13.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59 (1):80–84. [PubMed] [Google Scholar]
- 14.Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, Metz TO, Camp DG, 2nd, Waters KM, Smith RD, Rice CM, Katze MG. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6(1):e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain M, Petzold CJ, Schelle MW, Leavell MD, Mougous JD, Bertozzi CR, Leary JA, Cox JS. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc Natl Acad Sci U S A. 2007;104(12):5133–5138. doi: 10.1073/pnas.0610634104. 0610634104 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wenk MR. Lipidomics of host-pathogen interactions. FEBS Lett. 2006;580(23):5541–5551. doi: 10.1016/j.febslet.2006.07.007. S0014-5793(06)00840-4 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. A comprehensive classification system for lipids. J Lipid Res. 2005;46(5):839–861. doi: 10.1194/jlr.E400004-JLR200. E400004-JLR200 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50(Suppl):S9–14. doi: 10.1194/jlr.R800095-JLR200. R800095-JLR200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harkewicz R, Dennis EA. Applications of mass spectrometry to lipids and membranes. Annu Rev Biochem. 2011;80:301–325. doi: 10.1146/annurev-biochem-060409-092612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24(3):367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 21.Duffin KL, Henion JD, Shieh JJ. Electrospray and tandem mass spectrometric characterization of acylglycerol mixtures that are dissolved in nonpolar solvents. Anal Chem. 1991;63 (17):1781–1788. doi: 10.1021/ac00017a023. [DOI] [PubMed] [Google Scholar]
- 22.Dennis EA. Lipidomics joins the omics evolution. Proc Natl Acad Sci U S A. 2009;106(7):2089–2090. doi: 10.1073/pnas.0812636106. 0812636106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell TW, Pham H, Thomas MC, Blanksby SJ. Identification of double bond position in lipids: from GC to OzID. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(26):2722–2735. doi: 10.1016/j.jchromb.2009.01.017. S1570-0232(09)00036-1 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Li M, Zhou Z, Nie H, Bai Y, Liu H. Recent advances of chromatography and mass spectrometry in lipidomics. Anal Bioanal Chem. 2011;399(1):243–249. doi: 10.1007/s00216-010-4327-y. [DOI] [PubMed] [Google Scholar]
- 25.Hutchins PM, Barkley RM, Murphy RC. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J Lipid Res. 2008;49(4):804–813. doi: 10.1194/jlr.M700521-JLR200. M700521-JLR200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie H, Liu R, Yang Y, Bai Y, Guan Y, Qian D, Wang T, Liu H. Lipid profiling of rat peritoneal surface layers by online normal- and reversed-phase 2D LC QToF-MS. J Lipid Res. 2010;51(9):2833–2844. doi: 10.1194/jlr.D007567. jlr.D007567 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding J, Sorensen CM, Jaitly N, Jiang H, Orton DJ, Monroe ME, Moore RJ, Smith RD, Metz TO. Application of the accurate mass and time tag approach in studies of the human blood lipidome. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871(2):243–252. doi: 10.1016/j.jchromb.2008.04.040. S1570-0232(08)00286-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandra K, Pereira Ados S, Vanhoenacker G, David F, Sandra P. Comprehensive blood plasma lipidomics by liquid chromatography/quadrupole time-of-flight mass spectrometry. J Chromatogr A. 2010;1217(25):4087–4099. doi: 10.1016/j.chroma.2010.02.039. S0021-9673(10)00258-X [pii] [DOI] [PubMed] [Google Scholar]
- 29.Metz TO, Zhang Q, Page JS, Shen Y, Callister SJ, Jacobs JM, Smith RD. The future of liquid chromatography-mass spectrometry (LC-MS) in metabolic profiling and metabolomic studies for biomarker discovery. Biomark Med. 2007;1(1):159–185. doi: 10.2217/17520363.1.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nygren H, Seppanen-Laakso T, Castillo S, Hyotylainen T, Oresic M. Liquid chromatography-mass spectrometry (LC-MS)-based lipidomics for studies of body fluids and tissues. Methods Mol Biol. 2011;708:247–257. doi: 10.1007/978-1-61737-985-7_15. [DOI] [PubMed] [Google Scholar]
- 31.Masoodi M, Eiden M, Koulman A, Spaner D, Volmer DA. Comprehensive lipidomics analysis of bioactive lipids in complex regulatory networks. Anal Chem. 2010;82(19):8176–8185. doi: 10.1021/ac1015563. [DOI] [PubMed] [Google Scholar]
- 32.Rainville PD, Stumpf CL, Shockcor JP, Plumb RS, Nicholson JK. Novel application of reversed-phase UPLC-oaTOF-MS for lipid analysis in complex biological mixtures: a new tool for lipidomics. J Proteome Res. 2007;6(2):552–558. doi: 10.1021/pr060611b. [DOI] [PubMed] [Google Scholar]
- 33.Shockcor J, Crowe H, Yu K, Shion H. Application Note. Waters Corporation; Milford, MA: 2010. Analysis of Intact Lipids from Biologics Matrices by UPLC/High Definition MS. [Google Scholar]
- 34.Schuhmann K, Herzog R, Schwudke D, Metelmann-Strupat W, Bornstein SR, Shevchenko A. Bottom-up Shotgun Lipidomics by Higher Energy Collisional Dissociation (HCD) on LTQ Orbitrap Mass Spectrometers. Anal Chem. 2011 doi: 10.1021/ac102505f. [DOI] [PubMed] [Google Scholar]
- 35.Matuszewski BK, Constanzer ML, Chavez-Eng CM. Strategies for the assessment of matrix effect in quantitative bioanalytical methods based on HPLC-MS/MS. Anal Chem. 2003;75 (13):3019–3030. doi: 10.1021/ac020361s. [DOI] [PubMed] [Google Scholar]
- 36.Kelly RT, Page JS, Luo Q, Moore RJ, Orton DJ, Tang K, Smith RD. Chemically etched open tubular and monolithic emitters for nanoelectrospray ionization mass spectrometry. Anal Chem. 2006;78(22):7796–7801. doi: 10.1021/ac061133r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiebel GR, Auberry KJ, Jaitly N, Clark DA, Monroe ME, Peterson ES, Tolic N, Anderson GA, Smith RD. PRISM: a data management system for high-throughput proteomics. Proteomics. 2006;6(6):1783–1790. doi: 10.1002/pmic.200500500. [DOI] [PubMed] [Google Scholar]
- 38.Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. Decon2LS: An open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics. 2009;10:87. doi: 10.1186/1471-2105-10-87. 1471-2105-10-87 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics. 2007;23(15):2021–2023. doi: 10.1093/bioinformatics/btm281. btm281 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Jaitly N, Monroe ME, Petyuk VA, Clauss TR, Adkins JN, Smith RD. Robust algorithm for alignment of liquid chromatography-mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal Chem. 2006;78(21):7397–7409. doi: 10.1021/ac052197p. [DOI] [PubMed] [Google Scholar]
- 41.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CR, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51(11):3299–3305. doi: 10.1194/jlr.M009449. jlr.M009449 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1 (11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt A, Karas M, Dulcks T. Effect of different solution flow rates on analyte ion signals in nano-ESI MS, or: when does ESI turn into nano-ESI? J Am Soc Mass Spectrom. 2003;14(5):492–500. doi: 10.1016/S1044-0305(03)00128-4. S1044030503001284 [pii] [DOI] [PubMed] [Google Scholar]
- 44.Smith RD, Tang KQ, Page JS. Charge competition and the linear dynamic range of detection in electrospray ionization mass spectrometry. Journal of the American Society for Mass Spectrometry. 2004;15(10):1416–1423. doi: 10.1016/j.jasms.2004.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen Y, Jacobs JM, Camp DG, 2nd, Fang R, Moore RJ, Smith RD, Xiao W, Davis RW, Tompkins RG. Ultra-high-efficiency strong cation exchange LC/RPLC/MS/MS for high dynamic range characterization of the human plasma proteome. Anal Chem. 2004;76(4):1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- 46.Wilm MS, Mann M. Electrospray and Taylor-Cone Theory, Doles Beam of Macromolecules at Last. International Journal of Mass Spectrometry. 1994;136 (2–3):167–180. [Google Scholar]
- 47.Delamora JF, Loscertales IG. The Current Emitted by Highly Conducting Taylor Cones. Journal of Fluid Mechanics. 1994;260:155–184. [Google Scholar]
- 48.Plumb R, Castro-Perez J, Granger J, Beattie I, Joncour K, Wright A. Ultra-performance liquid chromatography coupled to quadrupole-orthogonal time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2004;18(19):2331–2337. doi: 10.1002/rcm.1627. [DOI] [PubMed] [Google Scholar]
- 49.Vandeemter JJ, Zuiderweg FJ, Klinkenberg A. Longitudinal Diffusion and Resistance to Mass Transfer as Causes of Nonideality in Chromatography. Chemical Engineering Science. 1956;5 (6):271–289. [Google Scholar]
- 50.Castro-Perez JM, Kamphorst J, DeGroot J, Lafeber F, Goshawk J, Yu K, Shockcor JP, Vreeken RJ, Hankemeier T. Comprehensive LC-MS E lipidomic analysis using a shotgun approach and its application to biomarker detection and identification in osteoarthritis patients. J Proteome Res. 2010;9(5):2377–2389. doi: 10.1021/pr901094j. [DOI] [PubMed] [Google Scholar]
- 51.Shen Y, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. High-efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics. Anal Chem. 2002;74 (16):4235–4249. doi: 10.1021/ac0202280. [DOI] [PubMed] [Google Scholar]
- 52.Giddings JC. United Separation Science. John Wiley & Sons; New York: 1991. [Google Scholar]
- 53.Shen Y, Zhang R, Moore RJ, Kim J, Metz TO, Hixson KK, Zhao R, Livesay EA, Udseth HR, Smith RD. Automated 20 kpsi RPLC-MS and MS/MS with chromatographic peak capacities of 1000–1500 and capabilities in proteomics and metabolomics. Anal Chem. 2005;77(10):3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- 54.Graessler J, Schwudke D, Schwarz PE, Herzog R, Shevchenko A, Bornstein SR. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS One. 2009;4(7):e6261. doi: 10.1371/journal.pone.0006261. 1371/journal.pone.0006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwudke D, Hannich JT, Surendranath V, Grimard V, Moehring T, Burton L, Kurzchalia T, Shevchenko A. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal Chem. 2007;79(11):4083–4093. doi: 10.1021/ac062455y. [DOI] [PubMed] [Google Scholar]
- 56.Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom Rev. 2003;22(5):332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 57.Yang K, Cheng H, Gross RW, Han X. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal Chem. 2009;81(11):4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang JS, Barcelona MJ. Structural determination and quantitative analysis of bacterial phospholipids using liquid chromatography electrospray ionization mass spectrometry. Journal of Microbiological Methods. 1998;33 (1):23–35. [Google Scholar]
- 59.Bushee JL, Argikar UA. An experimental approach to enhance precursor ion fragmentation for metabolite identification studies: application of dual collision cells in an orbital trap. Rapid Commun Mass Spectrom. 2011;25(10):1356–1362. doi: 10.1002/rcm.4996. [DOI] [PubMed] [Google Scholar]
- 60.Quehenberger O, Armando A, Dumlao D, Stephens DL, Dennis EA. Lipidomics analysis of essential fatty acids in macrophages. Prostaglandins Leukot Essent Fatty Acids. 2008;79(3–5):123–129. doi: 10.1016/j.plefa.2008.09.021. S0952-3278(08)00134-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomas A, Deglon J, Lenglet S, Mach F, Mangin P, Wolfender JL, Steffens S, Staub C. High-Throughput Phospholipidic Fingerprinting by Online Desorption of Dried Spots and Quadrupole-Linear Ion Trap Mass Spectrometry: Evaluation of Atherosclerosis Biomarkers in Mouse Plasma. Anal Chem. 2010;82(15):6687–6694. doi: 10.1021/ac101421b. [DOI] [PubMed] [Google Scholar]
- 62.Ahn EJ, Kim H, Chung BC, Kong G, Moon MH. Quantitative profiling of phosphatidylcholine and phosphatidylethanolamine in a steatosis/fibrosis model of rat liver by nanoflow liquid chromatography/tandem mass spectrometry. J Chromatogr A. 2008;1194(1):96–102. doi: 10.1016/j.chroma.2008.04.031. S0021-9673(08)00709-7 [pii] [DOI] [PubMed] [Google Scholar]
- 63.Kim H, Ahn E, Moon MH. Profiling of human urinary phospholipids by nanoflow liquid chromatography/tandem mass spectrometry. Analyst. 2008;133(12):1656–1663. doi: 10.1039/b804715d. [DOI] [PubMed] [Google Scholar]
- 64.Kim H, Min HK, Kong G, Moon MH. Quantitative analysis of phosphatidylcholines and phosphatidylethanolamines in urine of patients with breast cancer by nanoflow liquid chromatography/tandem mass spectrometry. Anal Bioanal Chem. 2009;393(6–7):1649–1656. doi: 10.1007/s00216-009-2621-3. [DOI] [PubMed] [Google Scholar]
- 65.Lee JY, Min HK, Moon MH. Simultaneous profiling of lysophospholipids and phospholipids from human plasma by nanoflow liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2011;400(9):2953–2961. doi: 10.1007/s00216-011-4958-7. [DOI] [PubMed] [Google Scholar]
- 66.Taguchi R, Houjou T, Nakanishi H, Yamazaki T, Ishida M, Imagawa M, Shimizu T. Focused lipidomics by tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;823(1):26–36. doi: 10.1016/j.jchromb.2005.06.005. S1570-0232(05)00408-3 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.