Abstract
Three-dimensional RNA models fitted into crystallographic density maps exhibit pervasive conformational ambiguities, geometric errors, and steric clashes. To address these problems, we present Enumerative Real-space Refinement ASsisted by Electron density under Rosetta (ERRASER), coupled to PHENIX (Python-based Hierarchical Environment for Integrated Xtallography) diffraction-based refinement. On 24 datasets, ERRASER automatically corrects the majority of MolProbity-assessed errors, improves average Rfree factor, resolves functionally important discrepancies in non-canonical structure, and refines low-resolution models to better match higher resolution models.
Over the last decade, fruitful progress in RNA crystallography has revealed numerous 3D structures of functional RNAs, providing powerful information for understanding their biological functions1-2. Nevertheless, RNA structures are typically solved at low resolution (typically >2.5 Å) compared to protein data. A recent report by the PDB X-ray Validation Task Force noted the ubiquity of bond geometry errors, anomalous sugar puckers, and backbone conformer ambiguities in RNA crystallographic models, and recommended that their assessment be included in PDB validation procedures3. There is thus a critical need for efficient algorithms that can resolve ambiguities in existing and future RNA crystallographic models.
The difficulty of resolving RNA crystallographic errors is underscored by limitations in currently available computational tools. RNABC (RNA Backbone Correction)4 and RCrane (RNA Constructed using RotAmeric NuclEotides)5 can identify and fix backbone conformer errors in some models. However, these methods anchor phosphates and bases to starting positions determined manually and thus only correct a subset of errors. Recent advances in Rosetta RNA de novo modeling6-8 and electron-density-guided protein modeling9-10 have suggested that confident high-accuracy structure prediction may be feasible if guided by experimental data. We have therefore developed a method for Enumerative Real-space Refinement ASsisted by Electron density under Rosetta (ERRASER) and integrated it with the PHENIX tools for diffraction-guided refinement. The protocol is based on exhaustively sampling each nucleotide's possible conformations and scoring by the physically realistic Rosetta energy function supplemented with an electron-density-correlation score (see Online Methods and Supplementary Fig. 1). Based on a benchmark of published crystallographic datasets and newly solved RNA structures, we report that this automated pipeline resolves the majority of geometric errors while retaining or improving correlation to diffraction data.
To measure the effectiveness of the ERRASER-PHENIX pipeline, we collected a test set of 24 RNA-containing crystal structures deposited in the PDB, ranging from small pseudoknots to entire ribosomal subunits (Supplementary Table 1). In parallel, we tested the effectiveness of RNABC and RCrane as alternatives to the ERRASER refinement step in our protocol, as well as PHENIX alone. In the starting PDB-deposited structures, MolProbity tools revealed numerous potential errors in four classes: atom-atom steric clashes, high frequencies of outlier bond lengths or angles, ‘non-rotameric’ backbone conformations, and potentially incorrect sugar puckers11. While not all of these features are necessarily incorrect, their high frequencies in medium-to-low-resolution models (2.5-3.5 Å) compared to high-resolution models (< 2.0 Å) suggest that most are due to inaccurate fits4-5, 11-12.
First, outlier bond lengths and angles (> 4 standard deviations from reference values) in the crystallographic models have mean frequencies of 0.53 % and 1.18 % in the starting PDB coordinates. Some of these outliers are due to different ideal bond geometries used by different refinement packages, and thus PHENIX alone lowered the outlier frequencies substantially. Nevertheless, application of ERRASER-PHENIX gave greater improvement, eliminating all the outlier bond lengths and angles in the benchmark (Table 1 and Supplementary Table 2).
Table1.
Average values for the validation results of the benchmark set.
| Outlier bond(%) | Outlier angle (%) | Clashscore | Outlier backbone rotamer (%) | Potentially incorrect pucker (%) | R | R free | Nucleotide similarity (%)a | Pucker similarity (%)a | |
|---|---|---|---|---|---|---|---|---|---|
| PDB | 0.53 | 1.18 | 18.03 | 18.8 | 5.0 | 0.210 | 0.256 | 64.9 | 91.5 |
| PHENIX | 0.01 | 0.03 | 10.79 | 15.2 | 2.4 | 0.199 | 0.244 | 71.7 | 96.4 |
| RNABC-PHENIX | 0.01 | 0 | 10.03 | 15.3 | 2.4 | 0.200 | 0.244 | 71.9 | 96.3 |
| RCrane-PHENIX | 0.003 | 0.12 | 10.12 | 10.3 | 1.0 | 0.207 | 0.252 | 74.1 | 95.8 |
| ERRASER-PHENIX | 0 | 0 | 7.04 | 7.9 | 0.2 | 0.199 | 0.244 | 80.5 | 97.0 |
Comparison of refined low-resolution models to independent high-resolution models (Supplementary Table 9). Nucleotides in which the differences between all torsion angles were smaller than 40° were denoted ‘similar’. Nucleotides in which torsion angle δ agreed to within 20° were assigned ‘similar’ puckers.
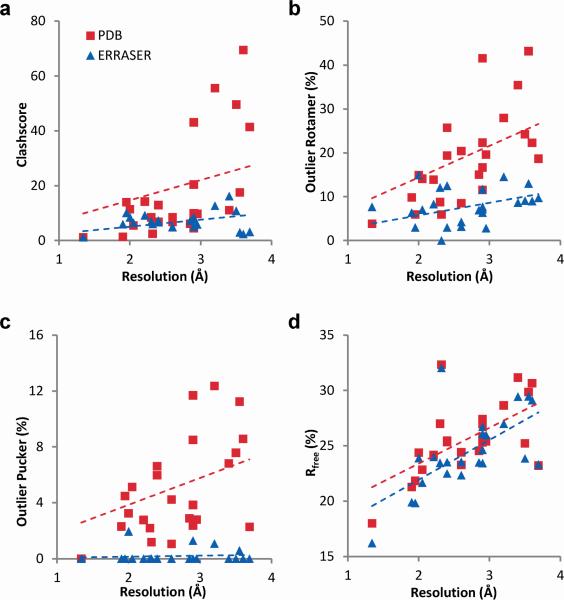
Second, ERRASER-PHENIX substantially reduced the steric clashes in RNA coordinates fitted into low-resolution electron density. In a bacteriophage prohead RNA test case (3R4F), the initial pervasive clashes were reduced by 5-fold with ERRASER-PHENIX (Fig. 1a). Over the entire benchmark, the MolProbity clashscore (number of serious clashes per 1,000 atoms11) was reduced from an average of 18.0 to 7.0 (Fig. 2a). Other refinement approaches that use less stringent or no steric criteria gave higher average clashscores (Table 1 and Supplementary Table 3).
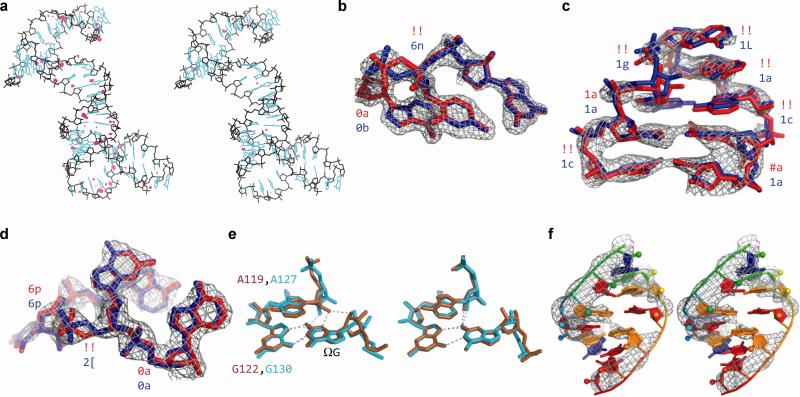
Figure 1.
Examples of geometric improvements by ERRASER-PHENIX. (a) Clash reduction in 3R4F. Red dots: unfavorable clashes. Left: PDB. Right: ERRASER-PHENIX. (b, c, d) Backbone conformation improvement on (b) nucleotides 62-64, chain A of 1U8D, (c) nucleotides 27-34, chain Q of 2OIU and (d) nucleotides 33-36, chain A of 2YGH. Rotamer assignments are shown at each suite. ‘!!’ stands for outlier suites. Red: PDB. Blue: ERRASER-PHENIX. (e) Functionally relevant pucker correction on group I ribozyme models. Brown: 1Y0Q. Cyan: 3BO3. Left: PDB. Right: ERRASER-PHENIX. (f) Base-pair geometry improvement on nucleotides 1-6 and 66-71, chain A of 3P49. Left: PDB. Right: ERRASER-PHENIX.
Figure 2.
Improvements of the crystallographic models by ERRASER-PHENIX across the test cases. (a) clashscore, (b) frequencies of outlier backbone rotamers, (c) frequencies of outlier puckers and (d) Rfree factors (in percentage). The dashed lines give linear fits.
Third, a recent community-consensus analysis indicates that 92.4 % of RNA backbone ‘suites’ (sets of two consecutive sugar puckers with 5 connecting backbone torsions) fall into 54 rotameric classes, many of which are correlated with unique functions12. Non-rotameric suites are thus potential fitting errors. ERRASER-PHENIX reduced the number of such outliers in 22 of 24 cases and the average outlier rate from 19 % to 8 % (Table 1, Supplementary Table 4 and Fig. 2b). This result was particularly striking since the 54-rotamer classification was not used during the Rosetta modeling. In high-resolution cases, the ERRASER-fitted conformer typically agreed better with the electron density by visual inspection (Fig. 1b). For cases with medium-to-low resolution where the starting and remodeled conformer fit the density equally well visually, ERRASER-PHENIX gave substantially more rotameric conformers (Fig. 1c). As an additional test, we applied ERRASER during a recent RNA-puzzles blind trial13 involving a protein-RNA complex. ERRASER-PHENIX changed a suite in the protein-binding kink-turn in starting RNA template (2YGH), from an outlier to the ‘2[’ rotamer consistent with other kink-turn motifs12 (Fig. 1d), which was indeed recovered in the subsequently released crystal structure (3V7E).
Fourth, RNA sugar rings typically exhibit either 2′-endo or 3′-endo conformations, but crystallographic assignments of these puckers can be ambiguous. While sugar pucker errors can be confidently identified using simple geometric criteria, finding alternative error-free solutions remains difficult11. ERRASER-PHENIX reduced the mean pucker error rate by 25-fold, from 5 % to 0.2 %, and gave zero pucker errors in 19 cases (Table 1, Supplementary Table 4 and Fig. 2c). As an example with functional relevance, an adenosine in the active site of the group I ribozyme was fitted with different puckers in independent crystallographic models from bacteriophage Twort (A119 in 1Y0Q) and Azoarcus sp. BH72 (A127 in 3BO3). This discrepancy also led to different hydrogen bonding patterns between the adenosine's 2′-OH group and the guanosine (ΩG) substrate of the ribozyme (Fig. 1e). ERRASER-PHENIX improved agreement between the Twort and Azoarcus models throughout the active site and gave the same 2′-endo pucker conformation and hydrogen-bonding network (Fig. 1e), in agreement with recent double-mutant analyses of group I ribozyme14.
In addition to correcting four classes of MolProbity-identified geometric problems, ERRASER-PHENIX improved other categories of errors. The ERRASER-PHENIX remodeling gave RNA base-pairing patterns with enhanced co-planarity and hydrogen-bonding geometry of interacting bases, as assessed by the automated base-pair assignment program MC-Annotate15 and demonstrated by the a glycine riboswitch example (3P49, Fig. 1f). Furthermore, ERRASER-PHENIX led to remodeling of glycosidic bond torsions (syn vs. anti χ). In cases where higher resolution structures were available, the accuracy of these changes could be confirmed. Complete discussions are given in Supplementary Results and Supplementary Table 5-6.
In addition to the above improvements of geometric features, we also evaluated the fits of our models to the diffraction data using R and Rfree factors. Avoiding increases in Rfree, the correlation to set-aside diffraction data, is critical for preventing overfitting of the experimental data16. The ERRASER-PHENIX pipeline consistently decreased both R and Rfree, lowering Rfree in 22 out of 24 cases. The average R dropped from 0.210 to 0.199 and average Rfree dropped from 0.255 to 0.243 (Table 1, Supplementary Table 7-8 and Fig. 2d). Other methods gave the same or worse average Rfree. As a practical demonstration, we applied ERRASER-PHENIX to a newly solved structure of subdomain IIa from the hepatitis C virus internal ribosome entry site17. The ERRASER-PHENIX model gave fewer errors in all MolProbity criteria and lower R and Rfree, and was therefore deposited into PDB as the final structure (3TZR).
As a separate independent assessment, we compared the similarity of remodeled low-resolution structures to original PDB-deposited models of high-resolution structures with the same sequences. We reasoned that pairs of models with the same sequences should give similar local conformations, and the higher-resolution models could be used as working references. For all 13 such cases (Table 1, Supplementary Table 9-10), ERRASER-PHENIX remodeling gave low-resolution models with increased agreement in backbone torsions and sugar puckers to the deposited high-resolution models. In addition, we evaluated structures related by non-crystallographic symmetry or by internal homology and found that ERRASER improved their agreement in all tested cases (see Supplementary Results and Supplementary Table 11-12).
The quality improvement for lower-resolution models by ERRASER-PHENIX is further illustrated by comparison of the six datasets with worst diffraction resolution (3.20-3.69 Å) with five datasets at high-resolution (1.90-2.21 Å). For the low-resolution datasets, ERRASER-PHENIX improved the mean clashscore from 40.8 to 7.9, lower than the mean clashscore of 9.3 in the original high-resolution models. This value (7.9) is equal to the median clashscore for models solved at 1.8 Å in a recent whole PDB survey3. Similar reductions in outlier bond lengths and angles, outlier backbone rotamers, and anomalous sugar puckers are apparent (Supplementary Table 2-4 and Supplementary Fig. 2).
For RNA crystallographic datasets across a wide range of resolutions and molecular size, ERRASER-PHENIX leads to consistent and substantial reduction of geometric errors, as assessed by independent validation tools and, in some cases, by independent functional evidence. The improved models give similar or better fits to set-aside diffraction data in all cases. For all geometric features, R and Rfree values, the ERRASER-remodeled coordinates are significantly improved compared to starting PDB values (P < 0.02 by Wilcoxon signed-rank test; see Supplementary Table 13). Finally, comparison of remodeled low resolution and independent high resolution datasets indicates that this automated pipeline consistently increases the accuracy of RNA crystallographic models. We therefore expect this algorithm to mark an application of ab initio RNA 3D prediction that will be widely useful in experimental biology.
ONLINE METHODS
Overview of the ERRASER-PHENIX pipeline
The ERRASER-PHENIX pipeline involves three major stages (Supplementary Fig. 1a). The starting model deposited in the PDB was first refined in PHENIX (v. dev-1034), with hydrogen atoms added. The refined model and electron-density map (setting aside the data for Rfree factor calculations; see below) were then passed into Rosetta (v. r50831) for a three-step real-space refinement. First, all torsion angles and all backbone bond lengths and bond angles were subjected to continuous minimization under the Rosetta high-resolution energy function supplemented with electron density correlation score. The Rosetta all-atom energy function models hydrogen bonding, Lennard-Jones packing, solvation, and torsional preferences, and has been successful in the modeling and design of RNA at near-atomic accuracy7-8. The electron density score term is similar to the Rosetta electron density score recently pioneered for application to electron cryomicroscopy and molecular replacement9-10. Second, bond length, bond angle, pucker and suite outliers were identified using phenix.rna_validate. In addition, we also included nucleotides that shifted substantially during the initial Rosetta minimization (evaluated by nucleotide-wise RMSD before and after minimization). These outlier and high-RMSD nucleotides were rebuilt by single-nucleotide StepWise Assembly (SWA) in a one-by-one fashion, where all of a nucleotide's atoms and the atoms up to the previous and next sugar were sampled by an exhaustive grid search of all torsions and a kinematic loop closure at sub-Angstrom resolution (Supplementary Fig. 1b)8, 18. If SWA found a lower-energy alternative structure of the rebuilt nucleotide, this new conformation was accepted. Third, the new model was minimized again in Rosetta. The rebuilding-minimization cycle was iterated three times to obtain the final ERRASER model. This model was again refined in PHENIX against diffraction data to obtain the final ERRASER-PHENIX model. Code is available in the current Rosetta release (3.4), which is freely available to academic users at http://www.rosettacommons.org. ERRASER is also available as a part of the PHENIX package and as a free on-line application through the ROSIE (Rosetta Online Server that Includes Everyone, http://rosie.rosettacommons.org). All the ERRASER-PHENIX remodeled structures discussed in this research are available as Supplementary Data.
The new Rosetta module, ERRASER
The ERRASER protocol consisted of three steps: an initial whole structure minimization, followed by single nucleotide rebuilding, and finally another whole structure minimization. Before passing the models into ERRASER, the PHENIX-generated pdb files were converted to the Rosetta format. Protein components, ligands and modified nucleotides were removed from the model, because current enumerative Rosetta modeling only handles standard RNA nucleotides. To avoid anomalies in refitting, we held fixed the positions of the nucleotides that were bonded or in van der Waals contact with these removed atoms during the next ERRASER step. In 2OIU, a cyclic RNA structure, we also held fixed the first and the last nucleotides in the RNA chain to prevent the bonds from breaking during ERRASER. For structures that have notable interaction through crystal contacts, we manually included the interacting atoms into the ERRASER starting models.
Throughout the ERRASER refinement, an electron density score (unbiased by excluding set-aside Rfree reflections during map creation, see below) was added to the energy function to ensure that the rebuilt structural models retained a reasonable fit to the experimental data. The electron density scoring in our method is slightly different from the one published recently 9-10. Instead of calculating the density profile of the model every time we rescored the model, we pre-calculated the correlation between the density of a single atom and the experimental density in a fine grid. The score was defined as the negative of the sum of the atomic numbers of all the heavy atoms in the model times this rapidly computed real-space correlation coefficient. This new density scoring term, named elec_dens_atomwise, was an order of magnitude faster than the one in the previous Rosetta release, thus reducing the total computational time of our method substantially. To accommodate the change of our energy function caused by the electron density energy constraint, we also modified the weights in the original scoring function. The scoring weights file is included in the Rosetta release named rna_hires_elec_dens.wts.
In addition, we used a new RNA torsional potential for this study. This new potential was obtained by fitting to the logarithm of the histogram of RNA torsions derived from the RNA11 dataset (http://kinemage.biochem.duke.edu/databases/rnadb.php). The RNA11 dataset contains 24,842 RNA suites and 311 different pdb entries, which is much richer and more diverse than the 50S ribosomal subunit model (1JJ2, 2,875 suites) used in deriving the original potential7. This new potential can be turned on by including the tag “-score:rna_torsion_potential RNA11_based_new” in the Rosetta command line (see Supplementary Notes)
During the whole structure minimization, we constrained the phosphate atoms in the RNA to their starting position; this is especially important for low-resolution models where the phosphate positions were not accurately defined by electron density. Errors in phosphate positions were corrected during the latter rebuilding step. We also found that when the molecule was too large, Rosetta was unable to minimize the entire molecule due to slow scoring. Therefore for any molecule larger than 150 nucleotides, we decomposed the RNA into smaller segments with an automated script rna_decompose.py, and minimized each of them sequentially. To retain all interactions, we also included the nucleotides within 5 Å radius of the segment being minimized as fixed nucleotides during the minimization.
After the whole structure minimization, we used phenix.rna_validate to analyze the obtained models. All nucleotides assigned to have outlier bond lengths, bond angles, puckers and/or potentially erroneous backbone rotamers (outliers or regular rotamers with suiteness < 0.1; suiteness is a quality measurement for rotamer assignments12) were identified as problematic and were rebuilt in subsequent Rosetta single nucleotide rebuilding. Furthermore, because the single nucleotide rebuilding region in Rosetta did not match the definition of a “suite”, we rebuilt both the selected nucleotide and the nucleotide preceding it to cover the whole suite for rotameric outliers.
In addition to rebuilding outlier nucleotides, we also computed the nucleotide-wise RMSD between the models before and after minimization. The nucleotides with RMSD larger than 0.05 times the diffraction resolution and within the 20 % of the largest RMSD nucleotide were also selected for rebuilding. We reasoned that because these nucleotides moved substantially after Rosetta minimization, their starting conformations were not favorable in terms of Rosetta energy function and were potentially erroneous.
The single nucleotide rebuilding step used in our method was based on a modified SWA algorithm in which the RNA chain was closed using triaxial kinematic loop closure18. For nucleotides at chain termini, the original SWA sampling was used since no chain closure was required. For rebuilding nucleotides inside the RNA chain, we first created a chain break between O3' and P in the lower suite of the rebuilding nucleotide. Then we sampled all possible torsion angles for εi, ζi, αi, αi+1 in 20° steps, and the two most common conformation of the sugar pucker, 2′-endo and 3′-endo. For each sampled conformation, analytical loop closure was applied to close the chain and determine the remaining 6 torsions (βi, γi, εi, ζi+1, γi+1) which form three pairs of pivot-sharing torsions. The glycosidic torsion χi, and the 2′-OH torsion were sampled after chain closure, and the generated models were further minimized in Rosetta. During the whole rebuilding, we applied a modest constraint to the glycosidic torsion so that it is more stable near the starting conformation, therefore only the base-orientation changes that have substantial Rosetta energy bonus were accepted as the final conformations. To reduce the computational expense, we only searched conformations that were within 3.0 Å RMSD with respect to the starting models.
After the conformational search, 100 lowest energy models were kept and further minimized under the constraint of the Rosetta linear_chainbreak and chainbreak energy term to ensure that the chain break was closed properly in the final model. Finally the best scored model was outputted as the new model for the RNA. If no new low-energy model could be found, then the program kept the starting model of that nucleotide. In the rebuilding process, the candidate nucleotides were rebuilt sequentially from the 5′-end to 3′-end of the RNA sequence. In order to speed up the Rosetta rebuilding process, the nucleotide being rebuilt was cut out from the whole structural model together with all nucleotides within 5 Å radius, rebuilt using SWA, and pasted back to the model.
After all the problematic nucleotides were rebuilt, we minimized the whole model again to further reduce any bond length or angle errors that might have occurred in the rebuilding process, and to improve the overall energy of the model. In this study, the rebuilding-minimization cycle was iterated three times, although single iterations gave nearly equivalent results (not shown). The coordinates of the RNA atoms in the PHENIX model were then substituted by the new coordinates in the Rosetta-rebuilt model to give the final ERRASER output.
The three ERRASER steps discussed above were wrapped into a python script erraser.py and can be performed automatically. The user needs to input a starting pdb file, a ccp4 map file, the resolution of the map and a list of any nucleotides that should be held fixed during refinement due to their interaction with removed atoms.
A sample ERRASER command line used for the refinement of 3IWN is shown below:
erraser.py -pdb 3IWN.pdb –map 3IWN.ccp4 -map_reso 3.2 -fixed_res A37 A58-67 B137 B158-167
Here 3IWN.pdb is the name of PHENIX refined model, 3IWN.ccp4 is the name of ccp4 density map file, -map_reso tag gives the resolution of the density map, and -fixed_res specifies the nucleotides that should remain untouched. “A37” means the 37th nucleotide of chain A in the pdb file.
Examples of the automatically generated Rosetta command lines by the python script are given in Supplementary Notes.
PHENIX refinement
PHENIX19 (v. dev-1034) was used for all the refinements performed in this study. We first prepared the starting models downloaded from the PDB for refinement using phenix.ready_set. This step added missing hydrogen atoms into the models and set up constraint files including ligand constraints and metal coordination constraints. For ligands A23, 1PE and CCC, we substituted the PHENIX-generated ligand constraints with constraint files from the CCP4 monomer library to achieve better geometry. Furthermore, phenix.ready_set did not automatically create bond length and bond angle constraints at the linkage between some modified nucleotides (GDP and GTP) and standard nucleotides, or between the first and the last nucleotide of a cyclic RNA. In such cases these constraints were added manually. Finally, for pdb files with TLS (Translation, Libration, Screw)20 refinement records, the TLS group information was manually extracted from the pdb files and saved in a separate file for further use in PHENIX.
After all the files for the refinement were ready, a four-step PHENIX refinement was performed. In the first step, because PHENIX does not load in TLS records in the pdb files, we performed a one-cycle TLS refinement to recover the TLS information. Second, the models were refined by phenix.refine for three cycles. At this step, ADP (Atomic Displacement Parameters) weight (wxu_scale) was optimized by PHENIX using a grid search, and other parameters were manually determined based on the criteria described below. For higher resolution structures a higher wxc_scale (scale for X-ray vs. Sterochemistry weight) was found to be appropriate. Based on initial tests (on PDB cases 1Q9A and 2HOP, which were not included in this paper's benchmark since they were used to set parameters), we used the following criteria: wxc_scale = 0.5 for Resolution < 2.3 Å, wxc_scale = 0.1 for 2.3 Å ≤ Resolution < 3 Å, wxc_scale = 0.05 for 3 Å ≤ Resolution ≤ 3.6 Å, and wxc_scale = 0.03 for Resolution > 3.6 Å. The ordered_solvent option (automatic water updating) was turned on for all structures. Empirically, we found that the real-space refinement strategy in PHENIX only gave equal or worse R factor, so it was turned off throughout all the refinement steps in this study. TLS refinement was turned on only for structures with TLS record in the deposited PDB files. Third, the models were further refined in phenix.refine for nine cycles using the same parameter set. Fourth, the models were further refined in phenix.refine for three cycles, with all target weights (wxc_scale and wxu_scale) optimized during the run. Other parameters stayed the same as in the first refinement round. Finally, we compared the models by the three different refinement steps and selected the one with best fit to the diffraction data as the final model. For 3OTO, the multi-step PHENIX refinement clearly distorted the starting model and gave worse geometries, so in this case we used the results obtained after the first refinement step. For 3P49, we supplied 1URN as a reference model to improve the protein part of the structure during refinement21.
After the initial refinement, the electron density map was generated from the experimental diffraction data and the PHENIX refined structural model for further ERRASER improvement. We used phenix.maps to create 2mFobs-DFcalc maps in ccp4 format, and diffraction data used for Rfree validation were excluded for the map generation to avoid directly fitting to the Rfree test set during the ERRASER refinement. To avoid Fourier truncation errors due to the missing data, we filled the missing Fobs with Fcalc during the map calculation. The averaged kicked map approach was also used to reduce the noise and model bias of the maps22. An example for the input file used in map creation is given below.
The final PHENIX refinement, after the ERRASER steps, was similar to the starting refinement described above, with small variations. First, there was no need for an initial TLS refinement since the pdb files already had this information at this stage. Second, we ran phenix.ready_set again on the ERRASER model to generate metal coordination constraints for refinements, in case the new model presented different metal coordination patterns than the starting one. The models were then refined using PHENIX in the same multi-step fashion, with the same parameter sets.
Examples of the PHENIX command lines used in this work are given in Supplementary Notes.
Refinement of 3TZR, a new structure of subdomain IIa from the hepatitis C virus IRES domain
The refinement of the 3TZR model currently deposited in the PDB was performed at an earlier stage of this work using an earlier PHENIX version (v1.7.1-743). The initial coordinates for 3TZR were already well-refined in PHENIX, and we therefore maintained the settings from that initial stage. In particular, during the PHENIX refinement, hydrogen atoms were not added to the model, and wxc_scale was set to 0.5. The final PHENIX refinement was performed using the same setting as the initial one.
R and Rfree calculation
For consistency, R and Rfree values of all the models were calculated using phenix.model_vs_data23. For the starting models, the PHENIX-calculated R and Rfree were generally similar to the values shown in the PDB header; both reported in Supplementary Table 7-8. In the main text, we have reported PHENIX-calculated R and Rfree to permit comparisons across the refinement benchmark.
Similarity analysis test
The similarities of the local geometries between similar structural models (Table 2, Supplementary Table 9-12) were evaluated as follows. If differences between the torsion angles (α, β, γ, δ, ε, ζ, χ) of each nucleotide pair were all smaller than 40°, the pair was counted as a similar nucleotide pair. If the difference of the δ angles of a nucleotide pair was smaller than 20°, the pair was assigned as having similar sugar pucker. Finally, RMSDs of all the torsion angles (in degrees) between the model pairs were calculated as an indicator of the model similarity in the torsional space.
Other tools
RNABC4 (v1.11) and RCrane.CLI5 (v1.01) were combined with PHENIX in the same manner as the ERRASER-PHENIX pipeline, by substituting the ERRASER stage with RNABC and RCrane, respectively. Since RNABC rebuilt only one nucleotide per run, a python script was used to achieve automatic rebuilding of all nucleotides. The MolProbity11 analysis was performed using command line tools phenix.clashscore and phenix.rna_validate in the PHENIX package. MC-Annotate15 (v1.6.2) was used to assign base-pairs in starting and refined models. All molecular images in this work were prepared using PyMol, except Figure 1a, which used MolProbity11 and KiNG (Kinemage, Next Generation)24.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. S. Richardson for suggesting this problem and for detailed evaluation of the results which we used to improve the program; C. L. Zirbel and N. B. Leontis for suggestions on base-pair validation; B. Stoner and D. Herschlag for discussions on group I ribozyme active site; T. Terwilliger and J. Headd for aids in integrating ERRASER into PHENIX; S. Lyskov for setting up the ERRASER protocol on the ROSIE Server; the Das lab for comments on the manuscript; and members of the Rosetta and the PHENIX communities for discussions and code sharing. Computations were performed on the BioX2 cluster (NSF CNS-0619926) and XSEDE resources (NSF OCI-1053575). This work is supported by funding from National Institute of Health (R21 GM102716 to R.D. and R01 AI72012 to T.H.), a Burroughs-Wellcome Career Award at Scientific Interface (R.D.), Governmental Scholarship for Study Abroad of Taiwan and Howard Hughes Medical Institute International Student Research Fellowship (F.C.C.), and the C. V. Starr Asia/Pacific Stanford Graduate Fellowship (P.S.).
Footnotes
AUTHOR CONTRIBUTIONS
F.C.C., P.S. and R.D. designed the research. F.C.C. implemented the methods and analyzed the results. P.S. provided code and assisted in data analysis. S.M.D and T.H. provided the starting model and diffraction data of the unreleased 3TZR structure and evaluated its refinement. F.C.C. and R.D. prepared the manuscript. All authors reviewed the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 2.Gesteland RF, Cech T, Atkins JF, editors. The RNA world: the nature of modern RNA suggests a prebiotic RNA world. Edn. 3rd. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2006. [Google Scholar]
- 3.Read Randy J., et al. A New Generation of Crystallographic Validation Tools for the Protein Data Bank. Structure. 2011;19:1395–1412. doi: 10.1016/j.str.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, et al. RNABC: forward kinematics to reduce all-atom steric clashes in RNA backbone. J. Math. Biol. 2008;56:253–278. doi: 10.1007/s00285-007-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keating KS, Pyle AM. Semiautomated model building for RNA crystallography using a directed rotameric approach. Proc. Natl. Acad. Sci. USA. 2010;107:8177–8182. doi: 10.1073/pnas.0911888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das R, Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc. Natl. Acad. Sci. USA. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das R, Karanicolas J, Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat. Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sripakdeevong P, Kladwang W, Das R. An enumerative stepwise ansatz enables atomic-accuracy RNA loop modeling. Proc. Natl. Acad. Sci. USA. 2011;108:20573–20578. doi: 10.1073/pnas.1106516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of Protein Structures into Low-Resolution Density Maps Using Rosetta. J. Mol. Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiMaio F, et al. Improved molecular replacement by density- and energy-guided protein structure optimization. Nature. 2011;473:540–543. doi: 10.1038/nature09964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson JS, et al. RNA backbone: Consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution). RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cruz JA, et al. RNA-Puzzles: A CASP-like evaluation of RNA three-dimensional structure prediction. RNA. 2012 doi: 10.1261/rna.031054.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forconi M, et al. Structure and Function Converge To Identify a Hydrogen Bond in a Group I Ribozyme Active Site. Angew. Chem. Int. Ed. 2009;48:7171–7175. doi: 10.1002/anie.200903006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gendron P, Lemieux S, Major F. Quantitative analysis of nucleic acid three-dimensional structures. J. Mol. Biol. 2001;308:919–936. doi: 10.1006/jmbi.2001.4626. [DOI] [PubMed] [Google Scholar]
- 16.Brunger AT. Free R value: A novel statistical quantity for assessing the accuracy of crystal strucutres. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 17.Dibrov SM, et al. Structure of a hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc. Natl. Acad. Sci. USA. 2012;109:5223–5228. doi: 10.1073/pnas.1118699109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Cryst. D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 21.Headd JJ, et al. Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Cryst. D. 2012;68:381–390. doi: 10.1107/S0907444911047834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Praznikar J, Afonine PV, Guncar G, Adams PD, Turk D. Averaged kick maps: less noise, more signal...and probably less bias. Acta Cryst. D. 2009;65:921–931. doi: 10.1107/S0907444909021933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afonine PV, et al. phenix.model_vs_data: a high-level tool for the calculation of crystallographic model and data statistics. J. Appl. Crystallogr. 2010;43:669–676. doi: 10.1107/S0021889810015608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen VB, Davis IW, Richardson DC. KING (Kinemage, Next Generation): A versatile interactive molecular and scientific visualization program. Protein Sci. 2009;18:2403–2409. doi: 10.1002/pro.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.