Abstract
High-throughput sequencing has revolutionized microbial ecology, but read quality remains a significant barrier to accurate taxonomy assignment and alpha diversity assessment for microbial communities. We demonstrate that high-quality read length and abundance are the primary factors differentiating correct from erroneous reads produced by Illumina GAIIx, HiSeq, and MiSeq instruments. We present guidelines for user-defined quality-filtering strategies, enabling efficient extraction of high-quality data from, and facilitating interpretation of Illumina sequencing results.
Recent advances in high-throughput, short-amplicon sequencing are revolutionizing efforts to describe microbial diversity within and across complex biomes, including the human body1 and Earth’s biosphere2. The greater sequence coverage and lower per-base cost of the Illumina GAIIx, HiSeq, and MiSeq instruments aids this progress over more expensive, lower-coverage platforms. Given unknown sequencing error rates for amplicon data generated by these rapidly evolving instruments and changing chemistries, and the potential for PCR error introduced during short-amplicon sample preparation, quality-filtering is integral to high-throughput sequencing data analysis, removing erroneous reads that otherwise overestimate microbial diversity. “Denoising”3,4, an approach employed to address this issue for amplicon sequencing by the 454 Life Sciences pyrosequencer, is specific to the 454 platform’s error profile, and does not scale to Illumina instruments, which generate tens (MiSeq) to hundreds (GAIIx) to thousands (HiSeq2000) of times more data per run.
Illumina systems provide per-nucleotide Phred quality scores representing the probability that a given base call is erroneous. How best to incorporate these scores in marker-gene-based microbial ecology studies has not been thoroughly investigated, and stringent filtration that discards many reads has been recommended to avoid exaggerated diversity estimates5. Previous investigation into quality-filtering of Illumina data6 focused on whole-genome sequencing applications, where error profiles are expected to differ from those in amplicon-sequencing runs. Additionally, the strategy discussed here differs from Illumina’s quality-filtering software CASAVA, which filters on a per-read basis, while our strategy works on a per-nucleotide basis, truncating reads at the position where their quality begins to drop.
To illuminate the “black box” of Illumina amplicon quality-filtering, we tested the effects of different quality-filtering parameters on taxonomic classification, α– diversity, and β-diversity estimates using the Quantitative Insights into Microbial Ecology (QIIME)7 pipeline (Table S1), outlined in Figure 1. To evaluate the effect of varying parameters in Figure 1, we tested four different ‘mock’ communities sequenced on the GAIIx (n = 1), HiSeq (n = 2), and MiSeq (n = 3) (Table S2). These comprised deliberately combined collections of 12 to 67 bacterial or fungal species whose genomes had been previously sequenced (Tables S3–S6). We also compared free-living and host-associated communities 5,8, representing samples with high β-diversity, and wine9 and spontaneous beer fermentation-associated communities10, representing samples with lower β-diversity, to evaluate the effects of filtering settings on β-diversity comparisons of different community types. Raw read counts and sample counts for all datasets are presented in Table S7.
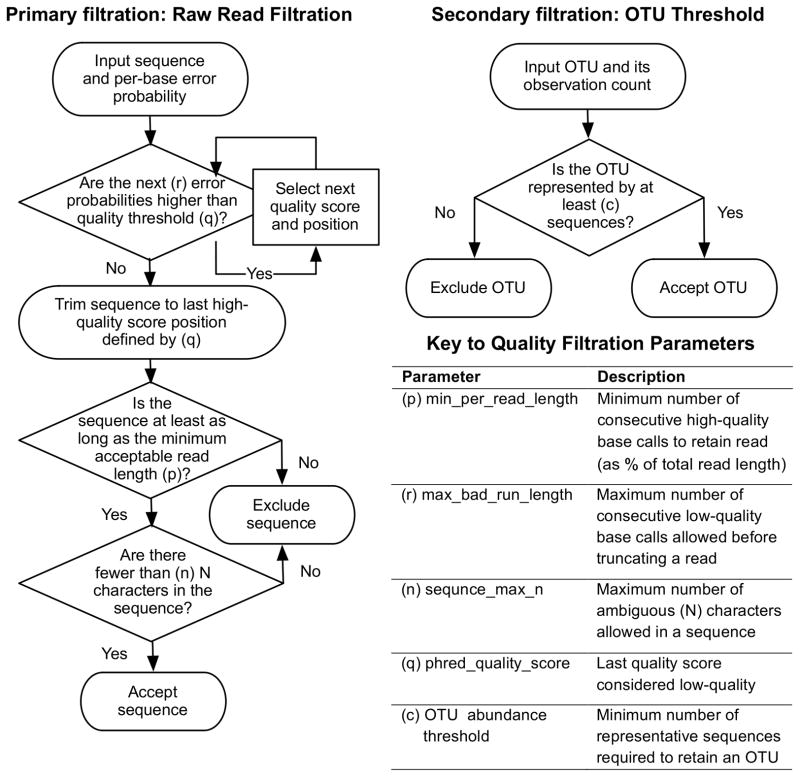
Figure 1.
Quality Filtration Process Flow in QIIME v1.5.0.
We evaluated how primary (p, q, r, and n) and secondary (c; see Figure 1 for definitions) quality-filtering parameters affect analyses using five separate evaluations, defined here.
α– diversity and qualitative taxonomic composition, using mock communities, tests which settings best measure true community composition, minimizing spurious additional OTUs (Figure 2; Figure S1–S7).
quantitative taxonomic composition, using defined mock communities, tests whether different settings introduce biases in specific taxa (Figure S8–S10).
β-diversity, using mock communities, determines whether different settings cause significant differences in phylogenetic composition between identical communities (Table S8).
β-diversity, using real communities, tests whether different settings affect our ability to differentiate sample types in principal coordinates (PCoA) plots (Table S9; Figure 2; Figure S11–S16).
β-diversity, using real communities, tests whether differences detected between communities on different sequencing platforms are consistent with one another.
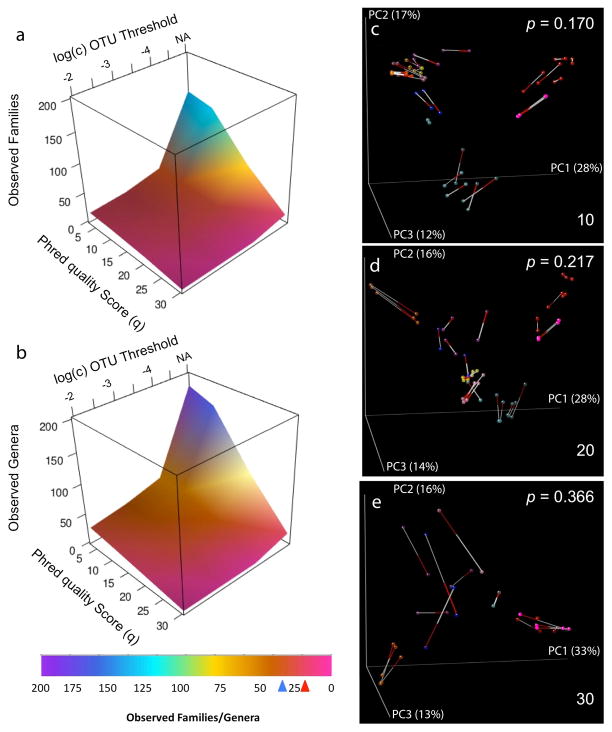
Figure 2.
α- and β-Diversity comparisons of mock community reads filtered using select phred_quality_score (q) settings (dataset 1). A, B: Family-level (A) and genus-level (B) taxon counts for mock communities filtered with variable (q) values at multiple OTU minimum abundance thresholds (c) (as %). Arrows below color key indicate expected genus- (blue) and family-level (red) taxon counts. C, D, E: Procrustes PCoA biplot of GAIIx weighted UniFrac distance comparing variation in (q). Comparison of (q) setting listed in bottom-right corner to (q) = 3. Top-right corner indicates Bonferroni-corrected p-value for Procrustes goodness of fit. Red, human feces; Magenta, mock community; Cyan, human skin; Dark cyan, human tongue; Blue, freshwater; Orange, freshwater creek; Purple, ocean; Yellow, estuary sediment; Pink, soil. All other settings represent defaults in both α- and β-diversity comparisons.
Our results across Evaluations 1–5 reveal general patterns. First, parameters (p), (q) and (c) have a marked effect on α– diversity and estimates of taxonomic composition, but not (n) and (r) (Figure 2A–B; Figure S1–S7). The effects of (p) and (q) were variable across runs in an apparently platform-independent fashion (Figure S4–S5). All settings except high (q) values required secondary filtration with (c) to reach expected taxon counts, but the required level varied between 0.01% to 0.0001% of total sequences, dependant upon (q) and (p) settings.. Increasing (p) also decreased abundance of unassigned sequences and sequences given shallow taxonomic assignment. In all mock data sets studied, extreme settings of (q) and (p), but not (r) and (n), had a marked impacted on taxonomic distribution (Figure S8–S10). These results are described in detail in Supplementary Text Evaluations 1–2. Second, weighted UniFrac11 distances between mock communities (see Supplementary Text Evaluation 3) were more robust to changes in parameter settings than unweighted UniFrac distances at low (c); however, these differences disappear at high (c). Thus as expected, differences in low-abundance OTUs have a larger impact on the unweighted metric. We note that any filtering strategies that remove low-abundance reads make it impossible to apply richness estimation metrics such as ACE and Chao1, which incorporate low-abundance read counts in their calculations. These metrics are unlikely to be accurate, however, if many of these reads actually represent sequencing errors.
Because observations in microbial ecology are often based on PCoA of samples, we applied Procrustes analysis to compare PCoA plots from different parameter settings on both biological and mock communities. We found that conclusions derived from PCoA plots were also robust to differences in parameter settings: the only notable differences occurred at stringent (q), (p), and (c), which result in extreme levels of read filtration that blur the known major distinction between host-associated and free-living communities (Figure 2C–E; Figure S11–S12) and closely related wine and beer fermentation-associated communities (Figure S13–S16). These results are described in detail in Supplementary Text Evaluations 3–4.
Finally, these observations generalize from the GAIIx to the HiSeq2000 and MiSeq platforms. The same β-diversity trends (e.g., separation in host-associated and free-living communities) were observed on all three platforms, and heavily decreased (p) (e.g., p = 0.25) and increased (q) (e.g., q ≥ 20) were the only factors that caused these sample types to erroneously cluster together on the HiSeq2000. These results are described in detail in Supplementary Text Evaluation 5.
Strategic quality-filtering of Illumina sequence data is necessary to retrieve reliable assessments of α- and β-diversity and taxonomic distribution of microbial communities. These results confirm the need to apply appropriate (q), (p), and (c) filters to eliminate erroneous OTU assignments, regulating these parameters depending upon raw sequence length and quality to reach the necessary balance of stringency and sequencing depth. To calibrate optimal filtering settings, we highly recommend including a standardized mock community in each individual sequencing run. We believe this will be necessary for confident comparison of samples from multiple sequencing runs in order to normalize run-to-run PCR and sequencing error, but further work is needed to evaluate which factors (e.g., community composition and complexity) define optimal mock communities for filter calibration under different experimental conditions. For datasets where a mock community is not included for calibration, we recommend the conservative threshold of (c = 0.005%). Further work is also required to address the impact of filtering strategies on the analysis of paired-end reads.
These results also enable users to process sequencing data under specific filtering conditions to support different downstream analyses. For example, users with a majority of high-quality, full-length sequences may wish to increase (q) and (p) in lieu of using (c), thereby retrieving only full-length sequences with low error rates, potentially increasing the discovery rate of rare OTUs (as sequence selection will be based on length and quality, not count). Alternatively, users with shorter reads or reads truncated by early low-quality base calls may wish to increase (r), lower (p), and use a higher (c) threshold. In this way, lower-quality but taxonomically useful reads will be retained, and reliable sequences selected based on abundance rather than error probability. Other users may be more interested in maximizing read count for implementation of machine-learning tools, identifying OTUs with significantly different abundances across metadata categories or treatment regimes, or jackknifing/permutational tests for β-diversity, all of which benefit from increased sample sizes. In this scenario, reads should be filtered using primary filters (q) and (p) instead of (c), which greatly reduces read count. Experienced users can adjust their filtering parameters to control the primary source of filtration (read length, error probability, or abundance) based on the idiosyncrasies of each sequencing run and the demands of the downstream analysis.
These conclusions serve as a benchmark for informed quality-filtering of Illumina amplicon sequence data. With these guidelines, users can confidently extract more, higher-quality sequences and decrease OTU filtration thresholds (c), increasing acuity for rare OTU discrimination and β-diversity comparisons. The Earth Microbiome Project (EMP)2 is adopting these guidelines for routine analysis of all SSU rRNA gene sequencing on the Illumina HiSeq and MiSeq systems, facilitating deeper, more efficient insight into how microbial diversity varies over spatial and temporal scales across our planet. The conclusions drawn from this study are conserved across the HiSeq2000, MiSeq, and GAIIx systems, supporting confident cross-platform data handling. In addition, we recommend new default settings for Illumina processing in QIIME (r = 3; p = 0.75 total read length; q = 3; n = 0; c = 0.005%; see Supplementary Text: Default recommendations for QIIME parameters for additional details), incorporated in the recent release of QIIME 1.5.0. Finally, although quality parameters tested here were evaluated using QIIME, these conclusions are relevant to Illumina amplicon quality-filtering across all bioinformatics pipelines for improved diversity estimates in all taxa and environments.
Supplementary Material
Table S1. Parameter Values and Cross-interactions Tested in This Study. Table S2. Mock Communities Analyzed in This Study. Table S3. Composition of Mock Community A.
Table S4. Composition of Mock Community B. Table S5. Composition of Mock Community C. Table S6. Composition of Mock Community D.
Table S7. Raw Sequence Counts and Sample Counts for Datasets Analyzed in This Study.
Table S8. β-Diversity Comparisons of Mock Communities Between Filtration Settings.
Table S9. β-Diversity Comparisons of Host-associated and Free-living Communities Between Filtration Settings.
Figure S1. α-Diversity comparison of mock community reads filtered using select quality parameter settings (dataset 1). Family-level taxon counts (z-axis) for mock communities filtered with variable split_library_fastq.py parameters, showing all cross-interactions between minimum per-read length as % of total read length (p) (x-axes), OTU abundance threshold (c) (as %; y-axes), maximum bad run length (r) (increasing values left to right), and maximum ambiguous base-call count (n) (increasing values top to bottom) at multiple OTU minimum abundance thresholds (c). Red arrow below color key indicates expected family-level taxon count.
Figure S2. α-Diversity comparison of mock community reads filtered using select quality parameter settings (dataset 1). Genus-level taxon counts (z-axis) for mock communities filtered with variable split_library_fastq.py parameters, showing all cross-interactions between minimum per-read length as % total read length (p) (x-axes), OTU abundance threshold (c) (as %; y-axes), maximum bad run length (r) (increasing values left to right), and maximum ambiguous base-call count (n) (increasing values top to bottom) at multiple OTU minimum abundance thresholds (c). Red arrow below the color key indicates expected genus-level taxon count.
Figure S3. α-Diversity comparison of mock community reads filtered using select quality parameter settings (dataset 1). Observed counts (z-axis) of OTUs clustered at 97% similarity for mock communities filtered with variable split_library_fastq.py parameters, showing all cross-interactions between minimum per-read length as % total read length (p) (x-axes), OTU abundance threshold (c) (as absolute values) (y-axes), maximum bad run length (r) (increasing values left to right), and maximum ambiguous base-call count (n) (increasing values top to bottom) at multiple OTU minimum abundance thresholds (c). Red arrow below the color key indicates expected species count.
Figure S4. Interaction of Phred score threshold (q) and OTU abundance threshold (c) (as %) on observed family and genus-level taxa (datasets 2–6). NA = no OTU abundance threshold was applied. Heading indicates platform type, read length, and expected family-and genus-level counts.
Figure S5. Interaction of minimum per-read length (p) (as % total length) and OTU abundance threshold (c) (as %) on observed family and genus-level taxa (datasets 2–6). NA = no OTU abundance threshold applied. Heading indicates platform type, read length, and expected family- and genus-level counts.
Figure S6. Interaction of maximum ambiguous base-call count (n) and OTU abundance threshold (c) (as %) on observed family and genus-level taxa (datasets 2–6). NA = no OTU abundance threshold applied. Heading indicates platform type, read length, and expected family- and genus-level counts.
Figure S7. Interaction of maximum bad run length (r) and OTU abundance threshold (c) (as %) on observed family- and genus-level taxa (datasets 2–6). NA = no OTU abundance threshold was applied. Heading indicates type of sequencing platform used, read length, and expected family- and genus-level counts.
Figure S8. Family-level taxonomic distribution of mock communities filtered using variable (p) (as % total length), (c), and (q) thresholds (dataset 1). All other parameters were held constant. E, expected values. *default parameters.
Figure S9. Family-level taxonomic Distribution of mock communities filtered using variable (n) and (r) thresholds (dataset 1). All other parameters were held constant. E, expected values.
Figure S10. Taxonomic key to Figures S8 and S9.
Figure S11. Procrustes PCoA of GAIIx weighted UniFrac distance biplot comparing variation in OTU abundance threshold (c), using Illumina GAIIx data (dataset 1). Comparison of (c) setting listed in bottom-right corner to dataset without (c) filtering. Top-right corner indicates Bonferroni-corrected p-value for Procrustes goodness of fit. Red, feces; Magenta, mock community; Cyan, skin; Dark cyan, tongue; Blue, freshwater; Orange, freshwater creek; Purple, ocean; Yellow, estuary sediment; Pink, soil.
Figure S12. Procrustes PCoA weighted UniFrac distance biplot comparing variation in minimum per-read length (p) (as % total read length), using Illumina GAIIx data (dataset 1). Comparison of setting listed in bottom-right corner to (p) = 0.75. Top-right corner indicates Bonferroni-corrected p-value for Procrustes goodness of fit. Red, human feces; Magenta, mock community; Cyan, human skin; Dark cyan, human tongue; Blue, freshwater; Orange, freshwater creek; Purple, ocean; Yellow, estuary sediment; Pink, soil.
Figure S13. Filtering minimally impacts abundance-weighted β-diversity comparisons of gradient communities. Weighted UniFrac PCoA plots comparing bacterial communities of spontaneous beer fermentations (dataset 9) with different filter settings. Row label indicates variable filter setting tested. Labels above plots indicate parameter setting. All other variables held constant to QIIME v1.4.0 defaults (r = 1; n = 0; p = 75 bp; q = 3). Note that (p) represents an absolute value (in nucleotides) in this figure, 150 nt being the maximum read length.
Figure S14. Filtering minimally impacts unweighted β-diversity comparisons of gradient communities. Unweighted UniFrac PCoA plots comparing bacterial communities of spontaneous beer fermentations (dataset 9) with different filter settings. Row label indicates variable filter setting tested. Labels above plots indicate parameter setting. All other variables held constant to QIIME v1.4.0 defaults (r = 1; n = 0; p = 75 bp; q = 3). Note that (p) represents an absolute value (in nucleotides) in this figure, 150 nt being the maximum read length.
Figure S15. Filtering minimally impacts abundance-weighted β-diversity comparisons of cluster communities. Weighted UniFrac PCoA plots comparing bacterial communities of inoculated (blue) and uninoculated (red) wine fermentations (dataset 10) with different filter settings. Row label indicates variable filter setting tested. Labels above plots indicate parameter setting. All other variables were held constant to QIIME v1.4.0 defaults (r = 1; n = 0; p = 75 bp; q = 3). Note that (p) represents an absolute value (in nucleotides) in this figure, 150 nt being the maximum read length.
Figure S16. Filtering minimally impacts unweighted β-diversity comparisons of cluster communities. Unweighted UniFrac PCoA plots comparing bacterial communities of inoculated (blue) and uninoculated (red) wine fermentations (dataset 10) with different filter settings. Row label indicates variable filter setting tested, labels above plots indicate parameter setting. All other variables held constant to QIIME v1.4.0 defaults (r = 1; n = 0; p = 75 bp; q = 3). Note that (p) represents an absolute value (in nucleotides) in this figure, 150 nt being the maximum read length.
Acknowledgments
We thank Georgia Giannoukos, Isabelle Rasolonjatovo, Matthew Gebert, and Laura Wegener Parfrey for contributing mock community sequencing data used in this study. We wish to thank Susan Huse and Antonio Gonzalez for useful feedback and discussions on this manuscript. This work was supported in part by grants from the NIH (NIH DK78669 [JIG], NIH R01HD059127 [DAM], NIH U54HG004969 [DG]), the Juvenile Diabetes Research Fund (DG), the Crohn’s and Colitis Foundation of America (JIG, DG), a Wine Spectator scholarship (NAB), and the Howard Hughes Medical Institute.
Footnotes
Author Contributions
NAB, JGC, DAM, and RK conceived and designed the experiments, and NAB performed the experiments and data analysis. All authors contributed sequencing datasets and wrote the manuscript.
References
- 1.Yatsunenko T, et al. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert JA, Meyer F. ASM Microbe Magazine. Mar, 2012. 2012. [Google Scholar]
- 3.Reeder J, Knight R. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quince C, et al. Nat Methods. 2009;6:639–644. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 5.Caporaso JG, et al. PNAS. 2011;108:4516–4522. [Google Scholar]
- 6.Minoche AE, et al. Genome Biol. 2011;12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caporaso JG, et al. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caporaso JG, et al. ISME. 2012. [DOI] [Google Scholar]
- 9.Bokulich NA, et al. PLoS One. 2012;7(5):e36357. doi: 10.1371/journal.pone.0036357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bokulich NA, Bamforth CW, Mills DA. PLoS One. 2012;7(4):e35507. doi: 10.1371/journal.pone.0035507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozupone CA, Knight R. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, et al. PNAS. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar RC. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Garrity GM, Tiedje JM, Cole JR. Appl Environ Microbiol. 2007;73 doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Santis T, et al. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, et al. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Parameter Values and Cross-interactions Tested in This Study. Table S2. Mock Communities Analyzed in This Study. Table S3. Composition of Mock Community A.
Table S4. Composition of Mock Community B. Table S5. Composition of Mock Community C. Table S6. Composition of Mock Community D.
Table S7. Raw Sequence Counts and Sample Counts for Datasets Analyzed in This Study.
Table S8. β-Diversity Comparisons of Mock Communities Between Filtration Settings.
Table S9. β-Diversity Comparisons of Host-associated and Free-living Communities Between Filtration Settings.
Figure S1. α-Diversity comparison of mock community reads filtered using select quality parameter settings (dataset 1). Family-level taxon counts (z-axis) for mock communities filtered with variable split_library_fastq.py parameters, showing all cross-interactions between minimum per-read length as % of total read length (p) (x-axes), OTU abundance threshold (c) (as %; y-axes), maximum bad run length (r) (increasing values left to right), and maximum ambiguous base-call count (n) (increasing values top to bottom) at multiple OTU minimum abundance thresholds (c). Red arrow below color key indicates expected family-level taxon count.
Figure S2. α-Diversity comparison of mock community reads filtered using select quality parameter settings (dataset 1). Genus-level taxon counts (z-axis) for mock communities filtered with variable split_library_fastq.py parameters, showing all cross-interactions between minimum per-read length as % total read length (p) (x-axes), OTU abundance threshold (c) (as %; y-axes), maximum bad run length (r) (increasing values left to right), and maximum ambiguous base-call count (n) (increasing values top to bottom) at multiple OTU minimum abundance thresholds (c). Red arrow below the color key indicates expected genus-level taxon count.
Figure S3. α-Diversity comparison of mock community reads filtered using select quality parameter settings (dataset 1). Observed counts (z-axis) of OTUs clustered at 97% similarity for mock communities filtered with variable split_library_fastq.py parameters, showing all cross-interactions between minimum per-read length as % total read length (p) (x-axes), OTU abundance threshold (c) (as absolute values) (y-axes), maximum bad run length (r) (increasing values left to right), and maximum ambiguous base-call count (n) (increasing values top to bottom) at multiple OTU minimum abundance thresholds (c). Red arrow below the color key indicates expected species count.
Figure S4. Interaction of Phred score threshold (q) and OTU abundance threshold (c) (as %) on observed family and genus-level taxa (datasets 2–6). NA = no OTU abundance threshold was applied. Heading indicates platform type, read length, and expected family-and genus-level counts.
Figure S5. Interaction of minimum per-read length (p) (as % total length) and OTU abundance threshold (c) (as %) on observed family and genus-level taxa (datasets 2–6). NA = no OTU abundance threshold applied. Heading indicates platform type, read length, and expected family- and genus-level counts.
Figure S6. Interaction of maximum ambiguous base-call count (n) and OTU abundance threshold (c) (as %) on observed family and genus-level taxa (datasets 2–6). NA = no OTU abundance threshold applied. Heading indicates platform type, read length, and expected family- and genus-level counts.
Figure S7. Interaction of maximum bad run length (r) and OTU abundance threshold (c) (as %) on observed family- and genus-level taxa (datasets 2–6). NA = no OTU abundance threshold was applied. Heading indicates type of sequencing platform used, read length, and expected family- and genus-level counts.
Figure S8. Family-level taxonomic distribution of mock communities filtered using variable (p) (as % total length), (c), and (q) thresholds (dataset 1). All other parameters were held constant. E, expected values. *default parameters.
Figure S9. Family-level taxonomic Distribution of mock communities filtered using variable (n) and (r) thresholds (dataset 1). All other parameters were held constant. E, expected values.
Figure S10. Taxonomic key to Figures S8 and S9.
Figure S11. Procrustes PCoA of GAIIx weighted UniFrac distance biplot comparing variation in OTU abundance threshold (c), using Illumina GAIIx data (dataset 1). Comparison of (c) setting listed in bottom-right corner to dataset without (c) filtering. Top-right corner indicates Bonferroni-corrected p-value for Procrustes goodness of fit. Red, feces; Magenta, mock community; Cyan, skin; Dark cyan, tongue; Blue, freshwater; Orange, freshwater creek; Purple, ocean; Yellow, estuary sediment; Pink, soil.
Figure S12. Procrustes PCoA weighted UniFrac distance biplot comparing variation in minimum per-read length (p) (as % total read length), using Illumina GAIIx data (dataset 1). Comparison of setting listed in bottom-right corner to (p) = 0.75. Top-right corner indicates Bonferroni-corrected p-value for Procrustes goodness of fit. Red, human feces; Magenta, mock community; Cyan, human skin; Dark cyan, human tongue; Blue, freshwater; Orange, freshwater creek; Purple, ocean; Yellow, estuary sediment; Pink, soil.
Figure S13. Filtering minimally impacts abundance-weighted β-diversity comparisons of gradient communities. Weighted UniFrac PCoA plots comparing bacterial communities of spontaneous beer fermentations (dataset 9) with different filter settings. Row label indicates variable filter setting tested. Labels above plots indicate parameter setting. All other variables held constant to QIIME v1.4.0 defaults (r = 1; n = 0; p = 75 bp; q = 3). Note that (p) represents an absolute value (in nucleotides) in this figure, 150 nt being the maximum read length.
Figure S14. Filtering minimally impacts unweighted β-diversity comparisons of gradient communities. Unweighted UniFrac PCoA plots comparing bacterial communities of spontaneous beer fermentations (dataset 9) with different filter settings. Row label indicates variable filter setting tested. Labels above plots indicate parameter setting. All other variables held constant to QIIME v1.4.0 defaults (r = 1; n = 0; p = 75 bp; q = 3). Note that (p) represents an absolute value (in nucleotides) in this figure, 150 nt being the maximum read length.
Figure S15. Filtering minimally impacts abundance-weighted β-diversity comparisons of cluster communities. Weighted UniFrac PCoA plots comparing bacterial communities of inoculated (blue) and uninoculated (red) wine fermentations (dataset 10) with different filter settings. Row label indicates variable filter setting tested. Labels above plots indicate parameter setting. All other variables were held constant to QIIME v1.4.0 defaults (r = 1; n = 0; p = 75 bp; q = 3). Note that (p) represents an absolute value (in nucleotides) in this figure, 150 nt being the maximum read length.
Figure S16. Filtering minimally impacts unweighted β-diversity comparisons of cluster communities. Unweighted UniFrac PCoA plots comparing bacterial communities of inoculated (blue) and uninoculated (red) wine fermentations (dataset 10) with different filter settings. Row label indicates variable filter setting tested, labels above plots indicate parameter setting. All other variables held constant to QIIME v1.4.0 defaults (r = 1; n = 0; p = 75 bp; q = 3). Note that (p) represents an absolute value (in nucleotides) in this figure, 150 nt being the maximum read length.