Abstract
Two major challenges in proteomics are the large number of proteins and their broad dynamic range within the cell. We exploited the abundance-dependent Michaelis-Menten kinetics of trypsin digestion to selectively digest and deplete abundant proteins with a method we call DigDeAPr. We validated the depletion mechanism with known yeast protein abundances and observed greater than 3-fold improvement in low abundance human protein identification and quantitation metrics. This methodology should be broadly applicable to many organisms, proteases, and proteomic pipelines.
Shotgun proteomics is a widely used approach for biological discovery.1, 2 An integral part of the process is digestion of complex protein mixtures into peptides using proteases with high sequence specificity. As proteins in cells and tissues often exist in stable higher order structures such as protein complexes or embedded in lipid bilayers, efficient and complete digestion in solution remains a challenge and an area for continuing methodological development. A two-step digestion process for whole cell lysates employing endoproteinase Lys-C digestion in 8 M urea, followed by dilution to 2 M urea and digestion with trypsin facilitated the first comprehensive analysis of the yeast proteome.3 Similarly, the use of multiple proteases either in serial or parallel analyses has improved sequence coverage of proteins.4–7 A chaotrope swap strategy using a molecular weight cutoff spin-filter reduces background chemical noise by removing detergent and undigested material.8 Aggressive strategies to digest membrane proteins for shotgun proteomics are effective for releasing peptides from the lipid bilayer for identification.9, 10 Recently, a new protease was developed and introduced for generating larger peptides for middle-down proteomics.11
The digestion of complex protein mixtures, however, is often biased by the presence of high abundance proteins. High abundance proteins produce a corresponding excess of tryptic peptides, which can also be further digested by trypsin’s endoproteinase activity,12 creating proteolytic background. An excess of high abundance peptides necessitates more chromatographic fractionation, limits dynamic range in the mass spectrometer, and, in turn, biases identification to high abundance proteins in shotgun proteomics.13 Common strategies to address the abundance challenge include affinity depletion and enrichment of proteins with antibody arrays or ligand libraries and prefractionation of proteins and peptides.14 Even with these strategies, the broad dynamic range and the large, varied number of high- and mid-abundance proteins between different sample and cell types present a challenge for the analysis of low abundance proteins. Protease digestion of proteins to peptides can be described by Michaelis-Menten kinetics. The rate of the digestion of a protein (ν) is a function of the substrate concentration ([S]), the maximum rate of reaction under substrate saturated conditions (Vmax), and the substrate concentration (KM) at half Vmax:
| (1) |
Thus, the rate of trypsin digestion of a protein lysate is defined primarily by the protein concentration in relation to the KM of trypsin. For digestion of a single protein these factors affect the digestion time. For a complex protein mixture, these factors also affect the relative rates at which proteins will be digested based on their relative abundances. We derived an equation (Supplementary Note 1) to describe this phenomenon where the digestion rate of an individual low abundance protein (Pi) is efined by that protein’s concentration ([Pi]) and the total protein concentration ([PT]):
| (2) |
Briefly, this equation illustrates that the rate of digestion of proteins is dependent on the concentration of each individual protein within the protein lysate and the relationship between the total protein concentration and KM. In fact, this phenomenon is similar to competitive inhibition of an enzyme. The primary difference is that high abundance “inhibitory” proteins form a peptide product whereas a competitive inhibitor simply dissociates from the enzyme. However, the preference for other proximal tryptic sites on the same high abundance protein likely contributes most to the non-linear inhibitor-like effect.
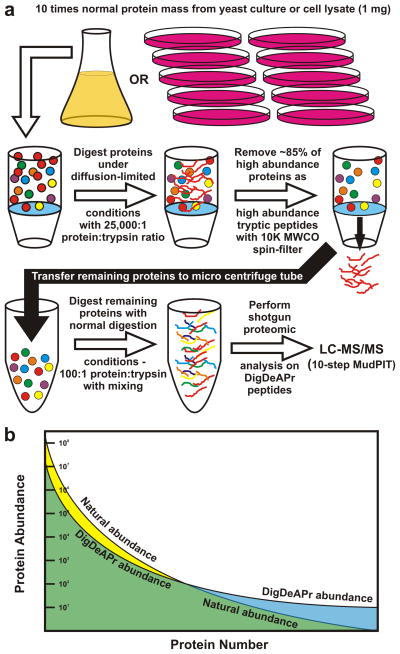
We exploited these digestion phenomena (Supplementary Note 2) to address both the abundance-dependent digestion of proteins and abundance-dependent sampling of peptides by mass spectrometers, with a method we call DigDeAPr. Briefly, 1 mg of proteome (approximately ten times that typically analyzed by Multidimensional Protein Identification Technology (MudPIT) liquid chromatography – tandem mass spectrometry (LC-MS/MS)) is digested to 85 ± 10% completion under trypsin- and diffusion-limited conditions in the presence of 2 M urea (Fig. 1a and Online Methods). High abundance proteins are selectively digested first based on Michelis-Menten kinetics and then removed as peptides using a molecular weight cut-off (MWCO) spin-filter. Although our utilization of a MWCO spin-filter was inspired by chaotrope swapping experiments with spin-filters,8 we did not perform a chaotrope swap. From the mass balance (Online Methods), we routinely found that ~15% of the total protein mass was lost to the spin-filter membrane as either digested and depleted peptides or proteins, consistent with previous claims.15 The residual proteome is then digested using standard trypsin digestion conditions and LC-MS/MS analysis to identify proteins. In this case the digestion method was the same as our Control experiments with 100 μg proteome.
Figure 1.
Schematic and description of the DigDeAPr method. (a) Ten times the normal mass of proteins are digested under trypsin- and diffusion-limited conditions for removal of abundant proteins as abundant peptides with a MWCO spin-filter. The remaining proteins are digested as normal for LC-MS/MS MudPIT analysis. (b) DigDeAPr changes the abundance profile of the proteome by starting with ten times more protein and digesting away ~85% of the most abundant proteins. The higher abundant proteins are preferentially digested by trypsin and depleted as peptides (yellow region), reducing their natural abundance to their DigDeAPr abundance (green region). By using ten times the protein mass to start, the DigDeAPr abundance of low abundance proteins should be ten times higher than their natural abundance (blue region).
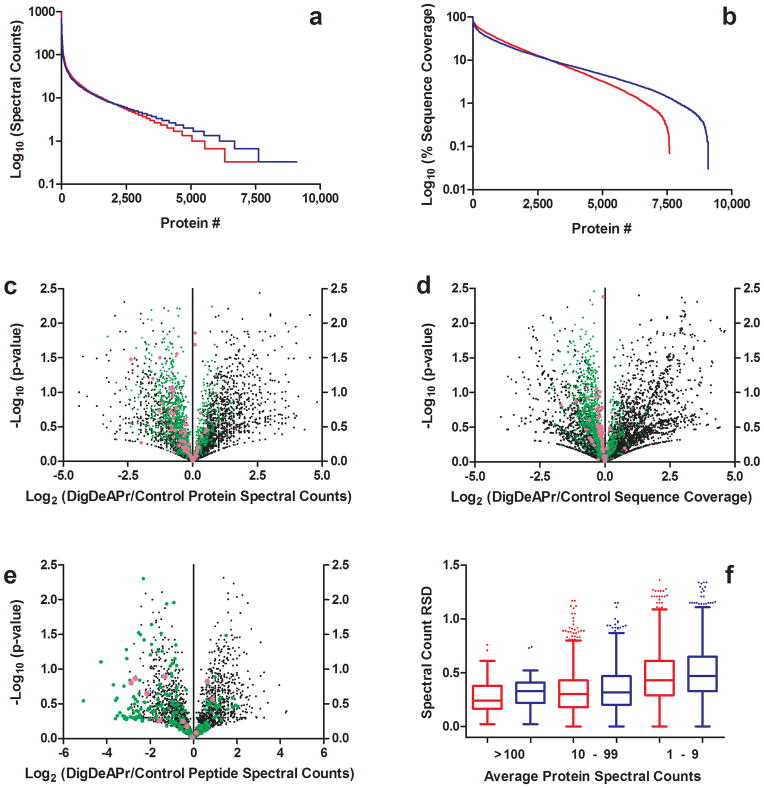
DigDeAPr changes the protein abundance profile of the proteome (Fig. 1b). The most readily identified proteins and peptides are depleted, improving the identification and sequence coverage of lower abundance proteins. With 10-fold higher starting mass of proteome than analyzed in our Control runs, low abundance proteins are 10-fold more abundant within the sample. Similarly, by selective digestion depletion of ~85% of the proteome, the highest abundance proteins should in turn be 10-fold lower. In fact, when we generated rank abundance plots using protein spectral counts and sequence coverage (Fig. 2a–b), both relative measures of protein abundance, we observed this protein abundance profile change between Control and DigDeAPr analyses on human embryonic kidney (HEK) cell lysates.
Figure 2.
Analysis of proteomic metric improvements from DigDeAPr on HEK cell lysates. Rank abundance plots of proteins based on (a) spectral count and (b) sequence coverage from triplicate DigDeAPr (blue) and triplicate Control (red) runs. Volcano plots of the log2 ratio of the average from triplicate DigDeAPr (c) protein spectral counts, (d) protein sequence coverage, and (e) peptide spectral counts and the corresponding average from triplicate Control runs plotted against the p-value. Data points are plotted based on average spectral counts from triplicate Control runs: 1–9 spectral counts (black), 10–99 (green), and more than 100 (magenta). (f) Spectral count reproducibility comparison between Control (red) and DigDeAPr (blue) runs based on average protein spectral counts and their relative standard deviation (RSD).
To further illustrate the changes in protein abundance we performed a statistical comparison of the average spectral count and sequence coverage of proteins identified at least twice in both Control and DigDeAPr triplicate runs (Fig. 2c–d and Supplementary Data). As expected, the spectral count and sequence coverage of high abundance proteins decreased, facilitating increases in the number of identified low abundance proteins, along with their spectral counts and sequence coverages. Specifically, we found ~300 proteins with statistically-significant (p ≤ 0.05) spectral count and sequence coverage changes. Of these proteins, 106 out of 125 proteins with more than five spectral counts in Control runs (ave. 11.3) decreased with an average of 1.95-and 1.78-fold in spectral counts and sequence coverage, respectively. Similarly, 149 out of 175 proteins with less than five spectral counts in Control runs increased with an average of 3.11- and 3.66-fold in spectral counts and sequence coverage, respectively. These statistically-significant changes typify the expected and observed trend for all protein changes found (Fig. 2c–d).
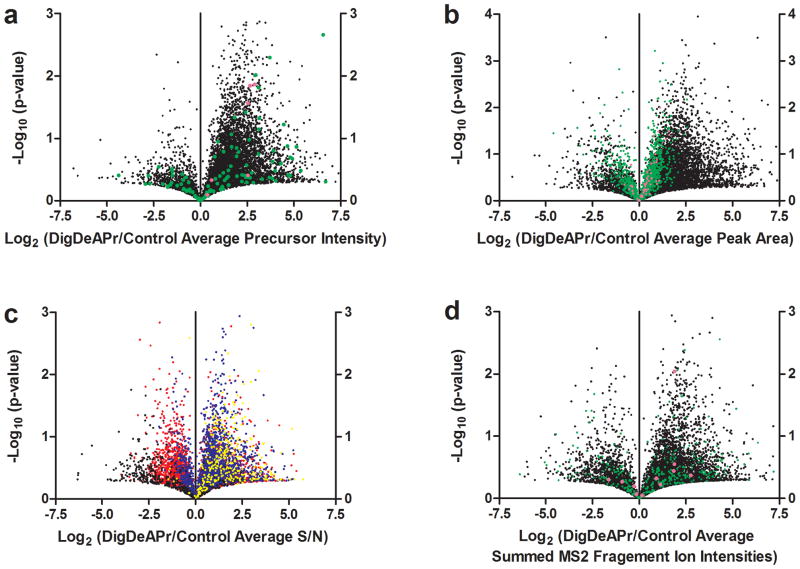
We also performed a comparison at the peptide level and found similar trends (Fig. 2e and Supplementary Data). Peptides identified in Control runs with more than 10 spectral counts had statistically-significant (p ≤ 0.05) reductions in spectral counts from DigDeAPr. Similar analyses of a yeast proteome further validate the abundance-dependent depletion of proteins and peptides (Supplementary Note 3, Supplementary Data, and Supplementary Figs. 1–3). High spectral count, “proteotypic” peptides can suppress other lower abundance less “proteotypic” peptides and typically provide no additional information about protein identity in an experiment. Thus, the protein and peptide spectral count reductions from high abundance proteins and peptides with DigDeAPr led to more protein (7,716 vs. 6,513) and peptide (42,928 vs. 40,592) identifications overall (Fig. 2a and Supplementary Fig. 4a–b) with more protein overlap between runs (Supplementary Fig. 4c–d), from the identification of more new peptides per run (Supplementary Fig. 4e–f). Notably, there were only minor changes to the quality scores between theoretical and experimental peptide spectra for all peptide abundances (Supplementary Fig. 5a–b). These results indicate that improving identification comprehensiveness with DigDeAPr did not adversely affect the quality or confidence of peptides also easily identified in Control runs. Similarly, changes to spectral counts through DigDeAPr did not adversely affect the reproducibility of spectral count quantitation for proteins with fewer than 100 spectral counts in comparison to Control runs (Fig. 2f). Improvements to this and other protein quantitation metrics such as precursor and fragment ion intensities, precursor ion signal-to-noise ratio (S/N), and chromatographic peak area were found (Fig. 3a–d, Supplementary Fig. 6, and Supplementary Data) and are described further (Supplementary Note 4).
Figure 3.
Analysis of proteomic metrics relevant to MS- and MS/MS-based quantitation. Volcano plots of the log2 ratio of the average from triplicate DigDeAPr peptide (a) precursor intensity, (b) chromatographic peak area averaged by protein, (c) precursor S/N, and (d) summed MS/MS fragment ion intensities and the corresponding average from triplicate Control runs plotted against the p-value. Data points in (a), (b), and (d) are plotted based on average spectral counts from triplicate Control runs: 1–9 spectral counts (black), 10–99 (green), and more than 100 (magenta). Data points in (c) are plotted based on average S/N from triplicate Control runs: 1–9 (yellow), 10-–19 (blue), 20–99 (red), and greater than 100 (black).
DigDeAPr directly addresses the main challenges of analyzing whole proteomes by selective digestion based on protein abundance to improve the dynamic range of analysis in an unbiased manner (Supplementary Notes 5–6 and Supplementary Figs. 7–8). Because it relies solely on the KM of a protease and the natural abundance of proteomes, it should be broadly applicable to other organisms, proteases, and proteomic pipelines to improve proteomic sequence coverage. Our method currently uses ten-fold more protein mass than typical comprehensive proteomic analyses, but further optimizations of conditions and the use of higher sensitivity mass spectrometers should make it applicable to mass-limited samples as well. Although we purposely changed the absolute abundance of proteins within a sample using DigDeAPr, the spectral count reproducibility was similar to Control runs, indicating that relative ratios of isotopically labeled protein pairs should remain unchanged as with current protease digestions methodologies. Thus DigDeAPr should also be applicable to quantitative proteomic pipelines using metabolic or chemical labeling strategies.
METHODS
Reagents and Chemicals
Unless otherwise noted all chemicals were purchased from Thermo Fisher Scientific. Deionized water (18.2 MΩ, Barnstead) was used for all preparations.
Growth, isolation, and lysis of log phase yeast
S288C S. cerevisiae strain was obtained from ATCC. 250 mL of log phase cells were grown at 30 °C in YPD media (1% bacto-yeast extract, 2% bacto-peptone, 2% dextrose) to an optical density of 0.6 at 600 nm. The culture was harvested by centrifugation at 3,000 × g for 5 min at 4 °C and washed twice with 10 mL of sterile water. The resulting pellet was snap frozen in liquid nitrogen and placed in −80°C until lysis. The YeastBuster protein extraction reagent (Novagen) was used to lyse cell pellets. The procedure was identical to the manufacturer’s protocol with the addition of 0.5 g of 0.5 mm zirconia beads (RPI Research) per 1 gram of cell pellets. During the 15 min incubation time the lysates were vortexed three times for 30 seconds with one minute rest on ice between cycles. Protein concentration was determined using a non-interfering protein assay kit (Calbiochem).
Cell growth and lysis
Human embryonic kidney cells, HEK 293T, were grown in Dulbecco’s Modified Eagle Medium (Mediatech) supplemented with 10% Fetal Bovine Serum Certified (Invitrogen) to 90% confluency in a 5% CO2 incubator at 37 °C. For collection, plates were washed twice with 20 mL Dulbecco’s Phosphate Buffered Saline (-Mg+, -Ca+) (Invitrogen). Following washing, 1 mL of DPBS containing 1X complete protease inhibitors - EDTA free (Roche) was added to each plate. Cells were lifted from dish surface using Cell Lifter (Corning) and collected into 1.7 mL microcentrifuge tube. Cells were lysed using a probe sonicator at 4 °C, where three cycles of 10 pulses were utilized per sample with 30 seconds on ice between each pulse cycle to offset heating. Lysates were centrifuged at 145,000 × g for 45 minutes. The supernatant was collected as the soluble fraction and used for all subsequent experiments.
Digestion and depletion of abundant proteins
Proteins (~1 mg) were digestion depleted by first denaturing and reducing in 250 μL 8 M urea, 100 mM Tris(hydroxyethylamine) pH 8.5, and 5 mM tris(2-carboxyethyl)phosphine for 30 min. Cysteine residues were acetylated with 10 mM iodoacetamide for 15 min in the dark. The sample was diluted to 1 mL (2 M urea) with 100 mM Tris(hydroxyethylamine) pH 8.5. A 20 μL aliquot was taken for protein quantitation. Trypsin (25 ng, Promega) was added at a 25,000:1 protein:protease mass ratio along with CaCl2 to 1 mM for a 12 hr diffusion-limited digestion at 37 °C. Digests were transferred to regenerated cellulose 10,000 molecular weight cutoff centrifugal filters (Amicon Ultra-4, ULTRACEL 10K, Millipore) and spun at 2.5K × g for 30 min at 4 °C until 100 – 200 μL remained in the filter. A 20 μL aliquot was taken from the flow through for protein quantitation. The cellulose filter was rinsed with 250 μL 8 M urea, 100 mM Tris(hydroxyethylamine) pH 8.5, then diluted to 2 M urea with 750 μL with 100 mM Tris(hydroxyethylamine) pH 8.5. The digest was spun again to 100 – 200 μL. A 20 μL aliquot was taken from the digestion depleted sample for protein quantitation. Protein quantitation was performed in duplicate using BCA analysis (Micro BCA Protein Assay Kit, Pierce) on aliquots taken during digestion and depletion. The protein masses were calculated to ensure mass balance and quantify the extent of digestion depletion using the following equation:
| (1) |
where mpeptide, total is protein mass before digestion depletion, mprotein, depleted is protein mass after digestion depletion, mpeptide, depletion is the peptide mass from the spin-filter flow through, and mpeptide, filter is the peptide mass retained on the spin-filter membrane. Complete protein digestion of digestion depleted samples were continued by transferring the remaining protein solution (100 – 200 μL) to a centrifuge tube, washing the spin-filter membrane twice with 50 μL 8 M urea - 100 mM Tris(hydroxyethylamine) pH 8.5, diluting the protein solution to 2 M urea - 100 mM Tris(hydroxyethylamine) pH 8.5, adding 2 μg trypsin and CaCl2 to 1 mM for an overnight digestion at 37 °C. Peptides were stored at −80 °C until the day of analysis. On the day of analysis peptide samples were acidified to 5% formic acid and spun at 18,000 × g.
Control protein digestion
Proteins (~100 μg) were digested by first denaturing and reducing in 60 μL 8 M urea, 100 mM Tris(hydroxyethylamine) pH 8.5, and 5 mM tris(2- carboxyethyl)phosphine for 30 min. Cysteine residues were acetylated with 10 mM iodoacetamide for 15 min in the dark. The sample was diluted to 2 M urea with 100 mM Tris(hydroxyethylamine) pH 8.5. Trypsin (2 μg as 0.5 μg/μL) was added at a 1:100 protease:protein ratio along with CaCl2 to 1 mM for an overnight digestion at 37 °C. Peptides were stored at −80 °C until the day of analysis. On the day of analysis peptide samples were acidified to 5% formic acid and spun at 18,000 × g.
Multidimensional Protein Identification Technology (MudPIT) analysis
Capillary columns were prepared in-house for LC-MS/MS analysis from particle slurries in methanol. An analytical RPLC column was generated by pulling a 100 μm ID/360 μm OD capillary (Polymicro Technologies, Inc) to 5 μm ID tip. Reverse phase particles (Jupiter C18, 4 μm dia., 90 Å pores, Phenomenex) were packed directly into the pulled column at 800 psi until 15 cm long. The column was further packed, washed, and equilibrated at 100 bar with buffer B followed by buffer A. A MudPIT trapping column was prepared by creating a Kasil frit at one end of an undeactivated 250 μm ID/360 μm OD capillary (Agilent Technologies, Inc.), then successively packed with 2.5 cm strong cation exchange particles (Luna SCX, 5 μm dia., 100 Å pores, Phenomenex) and 2.5 cm reverse phase particles (Aqua C18, 5 μm dia., 125 Å pores, Phenomenex). The Kasil frit was prepared by briefly dipping a 20 cm capillary in well-mixed 300 μL Kasil 1624 (PQ Corporation) and 100 μL formamide, curing at 100 °C for 4 hrs, and cutting the frit to ~2 mm in length. The MudPIT trapping column was equilibrated using buffer A for 15 min at 400 bar. Peptide samples (~100 μg) were loaded onto columns at 400 bar. MudPIT and analytical columns were assembled using a zero-dead volume union (Upchurch Scientific).
LC-MS/MS analysis was performed using an Agilent 1200 HPLC pump and Thermo LTQ-Orbitrap XL using an in-house built electrospray stage. Electrospray was performed directly from the analytical column by applying the ESI voltage at a tee (150 μm ID, Upchurch Scientific) directly downstream of a 1:1000 split flow used to reduce the flow rate to 250 nL/min through the columns. Ten-step MudPIT experiments were performed with consecutive application of 0, 10, 15, 20, 25, 30, 40, 50, 60, 70, 85, and 100% buffer C for 5 min at the beginning of each 2 hr gradient. The repetitive 2 hr gradients were from 100 % buffer A to 60% buffer B over 70 min, up to 100% B over 20 min, held at 100% B for 10 min, then back to 100% A for a 10 min column re-equilibration. HPLC buffers (Honeywell) were 5% acetonitrile 0.1% formic acid (A), 80% acetonitrile 0.1% formic acid (B), and 500 mM ammonium acetate 0.1% formic acid pH 6.0 (C). Precursor scanning in the Orbitrap XL was performed from 300 – 2000 m/z with the following settings, respectively: 5 × 105 target ions, 50 ms maximum ion injection time, and 1 microscan. Data-dependent acquisition of MS/MS spectra with the LTQ on the Orbitrap XL were performed with the following settings: collision-induced dissociation on the 8 most intense ions per precursor scan, 30K automatic gain control target ions, 100 ms maximum injection time, 35% normalized collision energy, and 1 microscan. Dynamic exclusion settings used were as follows: repeat count, 1; repeat duration, 30 second; exclusion list size, 500; and exclusion duration, 60 second. All raw data is available as Thermo. RAW files at http://fields.scripps.edu/published/DigDeAPr2012/
Data analysis
Protein and peptide identification and comparison were done with Integrated Proteomics Pipeline (IP2, http://www.integratedproteomics.com/). Tandem mass spectra were extracted to MS1 and MS2 files from raw files using RawExtract 1.9.9.16 MS/MS spectra were searched against a combined UniProtKB/Swiss-Prot and UniProtKB/VarSplic human database with reversed sequences using ProLuCID.17 Human protein entries were extracted and combined from the complete UniProtKB Swiss-Prot and VarSplic databases downloaded at ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/ on 11/8/2010. The spectral search space included all fully-, half-, and non-tryptic peptide candidates within a 50 ppm window surrounding the peptide candidate precursor mass. Carbamidomethylation (+57.02146) of cysteine was considered as a static modification. Peptide candidates were filtered to 0.1% FDR and proteins candidates to 1% FDR using DTASelect18, 19 with a 10 ppm peptide precursor mass window and statistical consideration of peptide tryptic status and mass accuracy. Spectral count, XCorr, ΔCN and summed fragment ion intensities were extracted from DTASelect results. Precursor intensities and S/N for identified peptides were extracted from MS1 files using in-house software.20 Chromatographic peak areas were extracted with Census.21 Protein physicochemical properties were calculated using an in-house script.22 Calculations and log2 comparisons of protein and peptide spectral counts and peptide XCorr, ΔCN, precursor intensity, S/N, peak area, and fragment ion intensity values were performed using Microsoft Excel (Supplementary Data).
Supplementary Material
Acknowledgments
This project was supported by the National Center for Research Resources (5P41RR011823-17), National Institute of General Medical Sciences (8P41GM103533-17), National Institute of Digestive and Diabetes and Kidney Disease (R01DK074798), National Heart, Lung, and Blood Institute (RFP-NHLBI-HV-10-5), and the National Institute of Mental Health (R01MH067880). We thank Jeffrey N. Savas, Claire M. Delahunty, and Jolene K. Diedrich for comments on the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
B.R.F. designed experiments, performed experiments, analyzed data, and wrote the paper. B.D.S. prepared HEK cell lysates and provided conceptual advice. K.J.W. prepared yeast lysates. T.X., J.C., and S.K.P developed software for data analysis. J.R.Y. wrote the manuscript and provided conceptual guidance.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Cravatt BF, Simon GM, Yates JR., 3rd Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 2.Nilsson T, et al. Nature methods. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- 3.Washburn MP, Wolters D, Yates JR., 3rd Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 4.MacCoss MJ, et al. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choudhary G, Wu SL, Shieh P, Hancock WS. Journal of proteome research. 2003;2:59–67. doi: 10.1021/pr025557n. [DOI] [PubMed] [Google Scholar]
- 6.Swaney DL, Wenger CD, Coon JJ. Journal of proteome research. 2010;9:1323–1329. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran BQ, et al. Journal of proteome research. 2011;10:800–811. doi: 10.1021/pr100951t. [DOI] [PubMed] [Google Scholar]
- 8.Manza LL, Stamer SL, Ham AJ, Codreanu SG, Liebler DC. Proteomics. 2005;5:1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 9.Wu CC, MacCoss MJ, Howell KE, Yates JR., 3rd Nat Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 10.Blonder J, Chan KC, Issaq HJ, Veenstra TD. Nature protocols. 2006;1:2784–2790. doi: 10.1038/nprot.2006.359. [DOI] [PubMed] [Google Scholar]
- 11.Wu C, et al. Nature methods. 2012;9:822–824. doi: 10.1038/nmeth.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picotti P, Aebersold R, Domon B. Mol Cell Proteomics. 2007;6:1589–1598. doi: 10.1074/mcp.M700029-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Sadygov RG, Yates JR., 3rd Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 14.Jmeian Y, El Rassi Z. Electrophoresis. 2009;30:249–261. doi: 10.1002/elps.200800639. [DOI] [PubMed] [Google Scholar]
- 15.Liebler DC, Ham AJ. Nature methods. 2009;6:785. doi: 10.1038/nmeth1109-785a. author reply 785–786. [DOI] [PubMed] [Google Scholar]
- 16.McDonald WH, et al. Rapid Commun Mass Spectrom. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 17.Xu T, et al. Mol Cell Proteomics. 2006;5:S174. [Google Scholar]
- 18.Tabb DL, McDonald WH, Yates JR., 3rd Journal of proteome research. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cociorva D, DLT, Yates JR. Curr Protoc Bioinformatics. Chapter 13(Unit 13–14):2007. doi: 10.1002/0471250953.bi1304s16. [DOI] [PubMed] [Google Scholar]
- 20.Wong CC, Cociorva D, Venable JD, Xu T, Yates JR., 3rd J Am Soc Mass Spectrom. 2009;20:1405–1414. doi: 10.1016/j.jasms.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SK, Venable JD, Xu T, Yates JR., 3rd Nature methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fonslow BR, et al. Journal of proteome research. 2011;10:3690–3700. doi: 10.1021/pr200304u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.