Abstract
AIM: To investigate the suppressive activity of MUTYH variant proteins against mutations caused by oxidative lesion, 8-hydroxyguanine (8OHG), in human cells.
METHODS: p.R154H, p.M255V, p.L360P, and p.P377L MUTYH variants, which were previously found in patients with colorectal polyposis and cancer, were selected for use in this study. Human H1299 cancer cell lines inducibly expressing wild-type (WT) MUTYH (type 2) or one of the 4 above-mentioned MUTYH variants were established using the piggyBac transposon vector system, enabling the genomic integration of the transposon sequence for MUTYH expression. MUTYH expression was examined after cumate induction using Western blotting analysis and immunofluorescence analysis. The intracellular localization of MUTYH variants tagged with FLAG was also immunofluorescently examined. Next, the mutation frequency in the supF of the shuttle plasmid pMY189 containing a single 8OHG residue at position 159 of the supF was compared between empty vector cells and cells expressing WT MUTYH or one of the 4 MUTYH variants using a supF forward mutation assay.
RESULTS: The successful establishment of human cell lines inducibly expressing WT MUTYH or one of the 4 MUTYH variants was concluded based on the detection of MUTYH expression in these cell lines after treatment with cumate. All of the MUTYH variants and WT MUTYH were localized in the nucleus, and nuclear localization was also observed for FLAG-tagged MUTYH. The mutation frequency of supF was 2.2 × 10-2 in the 8OHG-containing pMY189 plasmid and 2.5 × 10-4 in WT pMY189 in empty vector cells, which was an 86-fold increase with the introduction of 8OHG. The mutation frequency (4.7 × 10-3) of supF in the 8OHG-containing pMY189 plasmid in cells overexpressing WT MUTYH was significantly lower than in the empty vector cells (P < 0.01). However, the mutation frequencies of the supF in the 8OHG-containing pMY189 plasmid in cells overexpressing the p.R154H, p.M255V, p.L360P, or p.P377L MUTYH variant were 1.84 × 10-2, 1.55 × 10-2, 1.91 × 10-2, and 1.96 × 10-2, respectively, meaning that no significant difference was observed in the mutation frequency between the empty vector cells and cells overexpressing MUTYH mutants.
CONCLUSION: The suppressive activities of p.R154H, p.M255V, p.L360P, and p.P377L MUTYH variants against mutations caused by 8OHG are thought to be severely impaired in human cells.
Keywords: 8-hydroxyguanine, Mutation, MUTYH, MUTYH-associated polyposis, Oxidative mutagenesis, supF forward mutation assay, piggyBac transposon, Colorectal polyposis
INTRODUCTION
8-hydroxyguanine (8OHG) is an oxidatively damaged form of guanine[1], and because 8OHG can pair with adenine as well as cytosine, the formation of 8OHG in DNA causes a G:C to T:A transversion mutation[2]. To prevent such mutations, excision repair proteins, such as MUTYH (OMIM 604933), that act on 8OHG are present in human cells. The MUTYH protein is a DNA glycosylase that catalyzes the removal of adenine that is mispaired with 8OHG in double-stranded DNA[3-7]. Two major MUTYH proteins, type 1 and type 2, are expressed in human cells as a result of multiple transcription initiation sites and the alternative splicing of mRNA transcripts[4,7]. Because the type 1 protein contains a mitochondrial targeting signal (MTS) in its N-terminal, it is localized in the mitochondria. In contrast, the type 2 protein lacks the N-terminal 14 amino acids of type 1, and this absence leads to the destruction of the MTS; consequently, the type 2 protein is localized in the nucleus[4,7].
Biallelic germline mutations in the MUTYH gene are responsible for MUTYH-associated polyposis (MAP) (OMIM 608456), which is a hereditary disease characterized by multiple colorectal adenomas and carcinomas[8-12]. Most biallelic MUTYH carriers have between 10 and a few hundred colorectal polyps, meaning that MAP shows a phenotypic overlap with two other hereditary colorectal polyposis syndromes: familial adenomatous polyposis (FAP: OMIM 175100) and attenuated FAP (AFAP: OMIM 175100), both of which are caused by inactivation of the APC gene (OMIM 611731)[13,14]. Therefore, screening for germline mutations in MUTYH and APC is important in candidate patients with multiple colorectal polyps. However, even when MUTYH gene variations are detected in the mutation screening, if information regarding the level of the repair activities of the MUTYH variants is lacking, a correct diagnosis of MAP is impossible to make. Thus far, 300 unique DNA variants of the MUTYH gene have been reported in the Leiden Open Variation Database (http://www.lovd.nl/2.0/index_list.php)[15], and the proportion of missense MUTYH variations in the database is larger than nonsense mutations or truncating mutations. For most of the genes, a functional analysis is needed to determine whether the activity of a protein encoded by a missense variant is severely reduced. Thus, the effect of MUTYH variations on repair activity should be examined; however, so far, only a small number of MUTYH variations has been investigated[16-27]. In most of these studies, the DNA glycosylase activities of the variant recombinant proteins were analyzed using a DNA cleavage assay to test the abilities of the variants to cleave double-stranded oligonucleotides containing an A:8OHG mispair in vitro[18,19,21,23-27]. However, because examining the repair activity of MUTYH variant proteins from multiple aspects would lead to a more definitive judgment of the pathogenicity of MUTYH variants and MUTYH has the ability to regulate the mutation frequency in human cells in vivo[28-30], evaluating the mutation frequency in human cells is also valuable. However, at present, the activities of MUTYH variants in the regulation of mutation suppression in human cells in vivo have not been previously reported. Therefore, in this paper, we evaluated the suppressive activities of MUTYH variant proteins against oxidative mutagenesis in human cells. We recently determined the DNA glycosylase activities of 14 type 2 (nuclear form) MUTYH variants using a DNA cleavage assay[27], and based on those results, p.R154H, p.M255V, p.L360P, and p.P377L type 2 proteins were chosen from the tested variants, and their abilities to suppress mutations caused by 8OHG in human cells were analyzed in this study. As far as we know, this is the first report to analyze the suppressive activities of MUTYH variants against oxidative mutagenesis in human cells.
The Human Genome Variation Society (http://www.hgvs.org/) recommends using the transcript variant α5 (NM_001128425.1), which encodes the longest isoform (549 amino acids), as a reference sequence. Therefore, the type 2 proteins p.R154H, p.M255V, p.L360P, and p.P377L used in this study correspond to the reference proteins p.R182H, p.M283V, p.L388P, and p.P405L, respectively.
MATERIALS AND METHODS
Cell line
The human cancer cell line H1299 was obtained from the American Type Culture Collection (Manassas, VA). Cells were maintained at 37 °C in RPMI1640 medium supplemented with 10% fetal bovine serum and penicillin/streptomycin under a 5% CO2 atmosphere. The study design was approved by an institutional review board.
Construction of expression plasmid
Human wild-type (WT) and variant (p.R154H, p.M255V, p.L360P, and p.P377L) MUTYH type 2 cDNAs were polymerase chain reaction-amplified with PfuUltra Hotstart DNA polymerase (Stratagene, La Jolla, CA) and the MUTYH-type 2/pET25b(+) expression vector[27] as a template; the amplified sequence was then inserted into a piggyBac cumate switch inducible vector (System Biosciences, Mountain View, CA) at the NheI and NotI restriction enzyme sites. A WT MUTYH type 2 expression vector with a C-terminal FLAG tag was previously constructed using the pcDNA3.1 expression vector (Invitrogen, Carlsbad, CA)[31]; in this study, the variants were generated using site-directed mutagenesis with a QuikChange Site-directed Mutagenesis kit (Stratagene). All of the vectors were confirmed using DNA sequencing with a BigDye Terminator Cycle Sequencing Reaction Kit (Applied Biosystems, Tokyo, Japan) and an ABI 3100 Genetic Analyzer (Applied Biosystems).
Transfection
A plasmid vector was transfected into H1299 cells using Lipofectamine 2000 reagent (Invitrogen) according to the supplier’s recommendations.
Establishment of stable inducible cell lines
H1299 cells were transfected with the cumate switch inducible vector for MUTYH expression together with the piggyBac transposase vector (System Biosciences). To establish stable inducible cell lines, positively transposed cells were selected using puromycin (1 μg/mL). Because the inducible piggyBac vector features a tight cumate switch combined with the EF1-CymR repressor-T2A-Puro cassette to establish stable cell lines, the addition of cumate solution (System Biosciences) to the puromycin-selected cells led to the induction of MUTYH expression.
Western blotting analysis
Cells were harvested in lysis buffer containing 50 mmol/L HEPES (pH 7.5), 150 mmol/L NaCl, 0.1% sodium dodecyl sulfate (SDS), 1.0% Triton X-100, 0.5% sodium deoxycholate, 100 mmol/L sodium fluoride, 1 mmol/L sodium orthovanadate, and a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO), and the whole-cell extracts were mixed with an equal volume of 2 × SDS sample buffer and boiled. The extract was then subjected to SDS-polyacrylamide gel electrophoresis, and the proteins obtained were electrophoretically transferred to a polyvinylidene fluoride membrane (GE Healthcare Bio-Science, Piscataway, NJ). The membrane was blocked with nonfat milk at room temperature (RT) for 1 h and incubated with an anti-MUTYH monoclonal antibody (clone 4D10; Abnova, Taipei, Taiwan) or an anti-β-tubulin monoclonal antibody (clone 2-28-33; Sigma-Aldrich) at RT for 1 h. After washing with PBS containing 0.05% Tween-20 (PBS-T), the membrane was incubated with an anti-mouse HRP-conjugated secondary antibody (GE Healthcare Bio-Science) at RT for 1 h. The membrane was then washed with PBS-T, and immunoreactivity was visualized using an ECL chemiluminescence system (GE Healthcare Bio-Science).
Indirect immunofluorescence analysis
Cells were fixed with 10% formalin at RT or 4% paraformaldehyde at 4 °C. The cells were permeabilized with 1% Nonidet P-40 in PBS for 5 min and incubated with 10% normal goat serum blocking solution (DAKO, Kyoto, Japan) for 30 min. The cells were then probed with mouse anti-MUTYH monoclonal antibody (4D10) or mouse anti-FLAG M2 monoclonal antibody (Sigma-Aldrich) at RT for 1 h. Indirect immunofluorescence labeling was performed by incubation with an Alexa Fluor 594-conjugated secondary antibody (Molecular Probes, Eugene, OR) at RT for 1 h, and the nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). The immunostained cells were examined under a fluorescence microscope (Olympus BX-51-FL; Olympus, Tokyo, Japan) equipped with epifluorescence filters and a photometric CCD camera (Sensicam; PCO Company, Kelheim, Germany). The captured images were digitized and stored using an image analysis program (MetaMorph; Molecular Devices, Palo Alto, CA).
Shuttle vector plasmid and an indicator bacterial strain
The plasmid pMY189 and the indicator Escherichia coli (E. coli) strain KS40/pKY241 were used for the supF forward mutation assay, as reported previously[30,32]. pMY189 is a shuttle vector containing the bacterial suppressor tRNA (supF) gene. KS40 is a nalidixic acid-resistant (gyrA) derivative of MBM7070 with genotype lacZ (am), CA7070, lacY1, hsdR, hsdM, Δ(araABC-leu)7679, galU, galK, rpsL, thi. The pKY241 plasmid contains a chloramphenicol resistance marker and the gyrA (amber) gene. E. coli KS40/pKY241 cells carrying the active supF gene are sensitive to nalidixic acid and form blue colonies on LB plates containing ampicillin, chloramphenicol, isopropyl-β-D-thiogalactopyranoside (IPTG), and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal), whereas cells carrying the mutated supF gene form white colonies on plates containing nalidixic acid, ampicillin, chloramphenicol, IPTG, and X-gal.
Construction of a shuttle vector plasmid containing an 8OHG residue
pMY189-8OHG, which is the shuttle plasmid pMY189 containing a single 8OHG:cytosine pair at nucleotide position 159 of the supF gene, was prepared according to a previously described method[30]. Briefly, E. coli XL1-Blue MRF’ (Stratagene) and R408 Helper Phage (Stratagene) were used to prepare single-stranded pMY189 DNA, and 30 μg of the single-stranded pMY189 plasmid and a 5-fold molar excess of a 5’-phosphorylated 24-mer oligonucleotide with a single 8OHG at nucleotide position 159 of the supF gene [5’-CGA CTT CGA A (8OHG) G TTC GAA TCC TTC-3’] were annealed in a reaction mixture. Forty units of T4 DNA polymerase (New England Biolabs, Beverly, MA), 600 μmol/L of deoxynucleotide triphosphate (GE Healthcare Bio-Science), 36 Weiss units of T4 DNA ligase (New England Biolabs) and 1 mmol/L of ATP (Nacalai Tesque, Kyoto, Japan) were added to the reaction mixture, and the mixture was incubated at 37 °C for 4 h. Closed circular pMY189-8OHG was isolated using cesium chloride-ethidium bromide density gradient centrifugation.
SupF forward mutation assay
Cells were cultured in the presence of cumate for 3 d for the induction of MUTYH expression, and they were then transfected with the shuttle plasmid pMY189 or pMY189-8OHG. After 48 h, the propagated plasmids were extracted from the cells using a QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA) and digested with DpnI restriction enzyme (New England Biolabs) to eliminate unreplicated plasmids with the bacterial methylation pattern. After purification with Amicon Ultra Centrifugal Filter Units (Millipore, Bedford, MA), the plasmids were introduced into the KS40/pKY241 indicator E. coli strain using electroporation. The transformants were plated onto LB agar plates containing 50 μg/mL of nalidixic acid, 150 μg/mL of ampicillin, and 30 μg/mL of chloramphenicol together with IPTG and X-gal. White colonies on were counted as supF mutants. The mutation frequencies were calculated as the number of supF mutants per the total number of transformants, which were counted on LB plates containing ampicillin, chloramphenicol, IPTG and X-gal.
Statistical analysis
The statistical analysis was performed using an unpaired t-test and JMP software, version 9 (SAS Institute, Cary, NC). P-values less than 0.05 were considered statistically significant.
RESULTS
Establishment of human cells inducibly expressing MUTYH variants
To investigate the ability of MUTYH variants to suppress mutations caused by 8OHG in human cells, we used the piggyBac transposon vector system[33] to establish human cells capable of inducibly expressing MUTYH variants and performed a supF forward mutation assay using the shuttle plasmid pMY189, which contains a single 8OHG in the supF gene. First, H1299 human cancer cells were transfected with a piggyBac cumate switch inducible vector for the expression of WT, p.R154H, p.M255V, p.L360P, or p.P377L MUTYH together with the piggyBac transposase vector; positively transposed cells were then selected with puromycin. We also transfected cells with an empty (parental) piggyBac cumate switch inducible vector and transposase vector. The expression of MUTYH protein after cumate induction was examined using Western blotting analysis using an anti-MUTYH monoclonal antibody (Figure 1A). MUTYH protein was abundantly expressed in cells in which a WT, p.R154H, p.M255V, p.L360P, or p.P377L MUTYH expression vector but not an empty vector was transposed. Immunofluorescence analysis also showed abundant MUTYH protein expression in cells in which a WT, p.R154H, p.M255V, p.L360P, or p.P377L MUTYH expression vector but not an empty vector was transposed (Figure 1B). In accordance with the previous finding that the MUTYH type 2 protein is the nuclear form[4,7], WT MUTYH protein was localized in the nucleus. All of the MUTYH variants were also localized in the nucleus, suggesting that the amino acid changes in p.R154H, p.M255V, p.L360P, and p.P377L did not alter the subcellular localization of these proteins in human cells. With regard to endogenous MUTYH expression, low levels were detected in the immunofluorescence analysis, as shown in the panel of empty vector-transposed cells (Figure 1B). When the intensity of the MUTYH protein signal was enhanced with image-editing software, the signal was observed in both the nucleus and cytoplasm (Figure 1C), which is compatible with the existence of both the type 1 mitochondrial form and the type 2 nuclear form[4,7]. Next, to confirm the nuclear localization of MUTYH type 2 variant forms, we constructed a vector to express MUTYH tagged with a FLAG peptide at the C-terminus and examined the localization of the MUTYH variants using immunofluorescence analysis with an anti-FLAG antibody (Figure 2). All of the variants showed nuclear localization, further suggesting that the amino acid changes in p.R154H, p.M255V, p.L360P, and p.P377L did not alter their subcellular localization in human cells. Together, the above findings indicate that human cells inducibly expressing the MUTYH variants (p.R154H, p.M255V, p.L360P, or p.P377L) and their control cells were properly prepared and were appropriate for evaluating the suppressive activities of MUTYH variants against oxidative mutagenesis in human cells.
Figure 1.
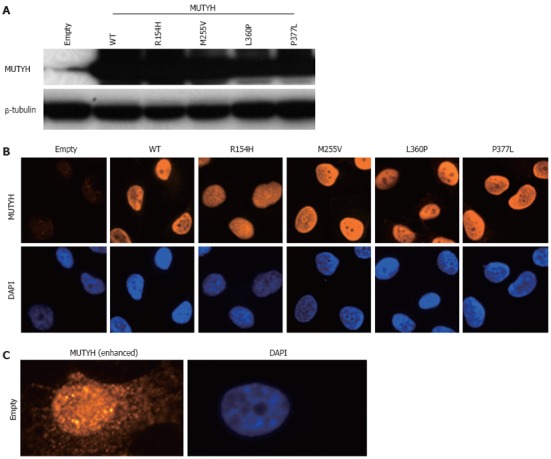
Establishment of H1299 human cell lines inducibly expressing MUTYH variant proteins. A: Detection of MUTYH proteins in cumate-inducible stable cell lines expressing MUTYH in the presence of cumate using Western blotting analysis with an anti-MUTYH antibody. Lysates from empty vector-transposed cells and cells inducibly expressing wild-type (WT) MUTYH or p.R154H, p.M255V, p.L360P, or p.P377L MUTYH variants were analyzed. β-tubulin protein was also analyzed as an internal control; B: Immunofluorescence detection of MUTYH proteins expressed in the cell lines used in (A) in the presence of cumate. The MUTYH protein (red) was stained with anti-MUTYH as the primary antibody and Alexa Fluor 594-conjugated goat anti-mouse IgG as the secondary antibody. The nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) (blue); C: Immunofluorescence detection of endogenous MUTYH proteins in the empty vector-transposed cells as described in (B). The intensity of the signals of MUTYH protein (red) was enhanced with image-editing software to determine the subcellular localization of endogenous MUTYH protein. The nuclei were counterstained with DAPI (blue).
Figure 2.
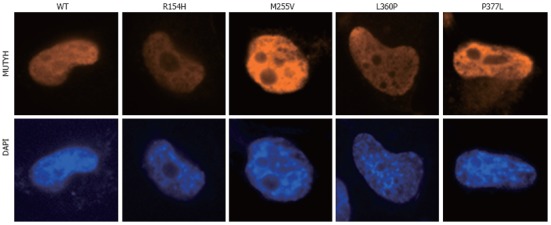
Nuclear localization of MUTYH variant proteins (p.R154H, p.M255V, p.L360P, and p.P377L). H1299 cells were transiently transfected with a vector expressing various types of MUTYH proteins tagged with FLAG, and MUTYH-FLAG protein (red) was stained with anti-FLAG M2 as the primary antibody and Alexa Fluor 594-conjugated goat anti-mouse IgG as the secondary antibody. The nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) (blue). WT: Wild-type.
Impaired activities of MUTYH variants in the suppression of oxidative mutagenesis in human cells in vivo
Next, mutation frequencies were compared for the empty vector-transposed human cells and the cumate-inducible stable cells expressing WT or variant MUTYH using a supF forward mutation assay with the shuttle plasmid pMY189. In this assay, we introduced a single 8OHG residue at position 159 of the supF gene in pMY189. The mutation frequency of supF was 2.2 × 10-2 in the 8OHG-containing pMY189 plasmid and 2.5 × 10-4 in WT pMY189 in empty vector-transposed cells (Figure 3), which was an 86-fold increase in mutation frequency with the introduction of 8OHG. The mutation frequency (4.7 × 10-3) of supF in the 8OHG-containing pMY189 plasmid in cells overexpressing WT MUTYH was significantly lower than in the empty vector-transposed cells. However, the mutation frequencies of supF in the 8OHG-containing pMY189 plasmid in cells overexpressing the p.R154H, p.M255V, p.L360P, or p.P377L MUTYH variant were 1.84 × 10-2, 1.55 × 10-2, 1.91 × 10-2, and 1.96 × 10-2, respectively, meaning that no significant difference was observed in the mutation frequency between the empty vector-transposed cells and the cells overexpressing MUTYH variants. These results suggested that the suppressive activities of p.R154H, p.M255V, p.L360P, and p.P377L MUTYH variants against mutations caused by 8OHG were severely impaired in human cells.
Figure 3.
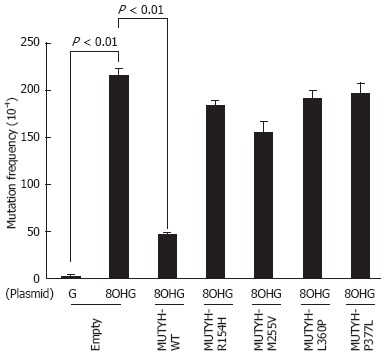
Measurement of the mutation frequency of the supF gene in the pMY189 plasmid using a supF forward mutation assay in H1299 human cell lines inducibly expressing MUTYH variant proteins. Empty vector-transposed cells and cells inducibly expressing wild-type (WT) MUTYH or p.R154H, p.M255V, p.L360P, or p.P377L MUTYH variants in the presence of cumate were transfected with a pMY189 shuttle plasmid, and the mutation frequency of supF in these human cell lines was measured. “8-hydroxyguanine (8OHG)” indicates a pMY189 plasmid containing an 8OHG residue at position 159 of supF, while “G” indicates a pMY189 plasmid containing the WT supF gene. The data are shown as the means ± SE.
DISCUSSION
In this study, human cell lines inducibly expressing MUTYH variants (p.R154H, p.M255V, p.L360P, or p.P377L) were established, and the abilities of these cells to suppress mutations caused by 8OHG were compared using a supF forward mutation assay with a shuttle vector containing an 8OHG residue in the supF gene. The assay showed that the suppressive activities of p.R154H, p.M255V, p.L360P, and p.P377L MUTYH variants against mutations caused by 8OHG were severely impaired in human cells. To the best of our knowledge, this is the first analysis of the suppressive activities of MUTYH variants against oxidative mutagenesis in human cells in vivo.
The type 2 protein is the nuclear form of MUTYH[4,7], and somatic APC and KRAS (OMIM 190070) mutations occur in the nuclear DNA of MAP tumors[8,9,12]; therefore, we believed that it would be more appropriate to investigate type 2 rather than type 1 and we established cell lines expressing the type 2 form in this study. In the supF forward mutation assay using a shuttle vector containing 8OHG, no significant difference was observed in the mutation frequencies between empty vector-transposed cells and cells expressing 1 of the 4 MUTYH variants, indicating the severe impairment of the suppressive activities of the MUTYH variants against mutations caused by 8OHG in human cells in vivo. We previously showed that the adenine DNA glycosylase activity of the p.M255V protein was 10.7% of the level of the WT protein and that the DNA glycosylase activities of the p.R154H, p.L360P, and p.P377L proteins were severely impaired[27]. Thus, the results regarding the regulation of the mutation frequency in the present study are in agreement with DNA glycosylase activity in the previous study. A combination of the results of two distinct analyses, i.e., in vitro and in vivo analyses, would provide more definitive proof of the pathogenicity of the p.R154H, p.M255V, p.L360P, and p.P377L MUTYH variants. Because the diagnosis of MAP depends on whether (1) the clinical phenotypic characteristics of MAP are present in a candidate patient; and (2) the repair activities of the MUTYH variants encoded in the two MUTYH alleles of the patient are decreased, even when gene variations are found in the patient by MUTYH mutation screening, information on the levels of the repair activities of MUTYH variants is indispensable for properly diagnosing MAP. Regarding this point, our results are clinically useful.
Previous studies have provided contradictory results regarding the subcellular localization of MUTYH protein in MAP patients; one paper insisted that MUTYH protein was predominantly localized in the cytoplasm of colorectal tumor cells in MAP patients but not in non-MAP patients, while the other papers denied this localization[34,35]. In the present study, the nuclear localization of the p.R154H, p.M255V, p.L360P, and p.P377L MUTYH type 2 variants was shown using two distinct experiments. Therefore, it seems that the amino acid changes of p.R154H, p.M255V, p.L360P, and p.P377L did not alter the subcellular localization of the MUTYH protein. Similarly, Molatore et al[26] recently reported that 3 missense MUTYH variants other than our variants were all localized in the nucleus.
In this paper, we successfully established cumate-inducible stable human cell lines by utilizing the piggyBac transposon vector system. Transposon technology is an attractive non-viral gene delivery model that allows for efficient genomic integration in a variety of cell types[36]. Among the several transposon systems available, the piggyBac transposon, which was isolated from the genome of the cabbage looper moth (Trichoplusia ni), has been optimized for gene transfer into mammalian cells[36,37]. In practice, the MUTYH expression status in our cell lines after puromycin selection in the presence of cumate clearly demonstrated abundant MUTYH expression in almost all of the cells. Because we performed transient transfection with a shuttle plasmid in the supF forward mutation assay in this study, the genomic integration of the sequence for MUTYH expression using the piggyBac transposon system was well suited to our experiment.
In our experiments, the level of expression of exogenously introduced MUTYH was much higher than the level of expression of endogenous MUTYH. This scenario allowed us to effectively evaluate the activities of MUTYH variants to regulate the mutation frequency, and we believe that such an evaluation was successfully performed. However, we cannot completely exclude the possibility that the functional difference observed under experimental conditions of high MUTYH expression levels does not reflect a true functional difference.
The impaired activity of MUTYH variants was shown using H1299 human lung cancer cells in this study. We used this cell line because we believed that the ability of MUTYH variants to suppress mutations in H1299 cells is not different from their ability in human cells derived from the colorectum. If there are no organ type-specific systems to modulate MUTYH activity, then MUTYH activity is dependent on the MUTYH expression level and MUTYH variation. Moreover, we studied overexpressed and exogenous MUTYH variant proteins in this paper. Therefore, we believe our results can most likely be applied for colorectal cells. However, because it might be possible that the difference in organ type has an effect on the results of functional evaluation, we would like to evaluate this activity in human colorectal cells in the future.
Genetic screening for MUTYH mutations in the diagnosis of colorectal polyposis continues to be performed worldwide, and technological progress in genome sequencing analysis has contributed to efficient and rapid screening protocols. Therefore, increasing MUTYH nucleotide variants are likely to be detected in the future. For appropriate patient management, the levels of the repair activities of MUTYH variant proteins should be evaluated, and our system for determining the abilities of these variants to suppress oxidative mutagenesis in human cells in vivo may be of great use for such evaluations.
COMMENTS
Background
The MUTYH gene is responsible for MUTYH-associated polyposis (MAP), a relatively recently identified hereditary disease. Although 300 MUTYH variants have been found, only a small number of variants has been functionally characterized. Therefore, evaluations of the activities of MUTYH variant proteins are needed for the correct diagnosis of MAP.
Research frontiers
An in vitro DNA cleavage assay was performed to evaluate the repair activities of MUTYH variants. Despite the clinical importance of the multiplicity of functional analytical methods for evaluating the activities of MUTYH variant proteins, until now, the ability of MUTYH variants to suppress oxidative mutagenesis in human cells in vivo has not been previously analyzed.
Innovations and breakthroughs
Human cumate-inducible stable cell lines expressing various MUTYH variants were established using the piggyBac transposon vector system. This is the first report to utilize human cells expressing MUTYH variants encoded by an ectopically transposed gene. Moreover, this is the first report to analyze the suppressive activities of MUTYH variants against oxidative mutagenesis in human cells.
Applications
The results of the present study suggest that the suppressive activities of p.R154H, p.M255V, p.L360P, and p.P377L MUTYH variant proteins against mutations caused by 8-hydroxyguanine (8OHG) are severely impaired in human cells. These in vivo results combined with results from our previous in vitro analysis provide definitive proof of the pathogenicity of p.R154H, p.M255V, p.L360P, and p.P377L MUTYH variants. This conclusion is valuable for the appropriate diagnosis of MAP.
Terminology
The base excision repair protein MUTYH is involved in the repair of the oxidative base lesion 8OHG in DNA. Patients with biallelic inactivating germline mutations in the MUTYH gene are predisposed to MAP, which is characterized by the development of multiple colorectal adenomas and carcinomas.
Peer review
This is a good study in which authors analyze the suppressive activity of MUTYH variant proteins against mutations caused by 8OHG in human cells. Towards understanding the impact of having so many missense mutations among MAP patients, the authors have steadily developed an infrastructure for serving the patients in the future. Through expression of MUTYH WT and 4 variants, subcellular localization, and mutation frequency counting, they suggested that anti-mutation activity of the four MUTYH variants were severely impaired in human cells.
Footnotes
Supported by Grants from the Ministry of Health, Labour and Welfare (21-1); the Japan Society for the Promotion of Science (22590356 and 22790378); the Hamamatsu Foundation for Science and Technology Promotion, the Ministry of Education, Culture, Sports, Science and Technology (221S0001); the Takeda Science Foundation; the Aichi Cancer Research Foundation; and the Smoking Research Foundation
Peer reviewers: Dr. Anthony T Yeung, BS, MS, PhD, Fox Chase Cancer Center, Room R404, 333 Cottman Avenue, Philadelphia, PA 19111-2497, United States; Yeun-Jun Chung, MD, PhD, Professor, Director, Department of Microbiology, Integrated Research Center for Genome Polymorphism, the Catholic University Medical College, 505 Banpo-dong, Socho-gu, Seoul 137-701, South Korea
S- Editor Gou SX L- Editor A E- Editor Zhang DN
References
- 1.Kasai H, Nishimura S. Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res. 1984;12:2137–2145. doi: 10.1093/nar/12.4.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 3.Slupska MM, Luther WM, Chiang JH, Yang H, Miller JH. Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J Bacteriol. 1999;181:6210–6213. doi: 10.1128/jb.181.19.6210-6213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takao M, Zhang QM, Yonei S, Yasui A. Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine: 8-oxoguanine DNA glycosylase. Nucleic Acids Res. 1999;27:3638–3644. doi: 10.1093/nar/27.18.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shinmura K, Yamaguchi S, Saitoh T, Takeuchi-Sasaki M, Kim SR, Nohmi T, Yokota J. Adenine excisional repair function of MYH protein on the adenine: 8-hydroxyguanine base pair in double-stranded DNA. Nucleic Acids Res. 2000;28:4912–4918. doi: 10.1093/nar/28.24.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai-Wu JJ, Su HT, Wu YL, Hsu SM, Wu CH. Nuclear localization of the human mutY homologue hMYH. J Cell Biochem. 2000;77:666–677. [PubMed] [Google Scholar]
- 7.Ohtsubo T, Nishioka K, Imaiso Y, Iwai S, Shimokawa H, Oda H, Fujiwara T, Nakabeppu Y. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, et al. Inherited variants of MYH associated with somatic G: C--> T: A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 9.Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G: C--> T: A mutations. Hum Mol Genet. 2002;11:2961–2967. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- 10.Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, Bisgaard ML, Orntoft TF, Aaltonen LA, Hodgson SV, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 11.Sampson JR, Dolwani S, Jones S, Eccles D, Ellis A, Evans DG, Frayling I, Jordan S, Maher ER, Mak T, et al. Autosomal recessive colorectal adenomatous polyposis due to inherited mutations of MYH. Lancet. 2003;362:39–41. doi: 10.1016/S0140-6736(03)13805-6. [DOI] [PubMed] [Google Scholar]
- 12.Lipton L, Halford SE, Johnson V, Novelli MR, Jones A, Cummings C, Barclay E, Sieber O, Sadat A, Bisgaard ML, et al. Carcinogenesis in MYH-associated polyposis follows a distinct genetic pathway. Cancer Res. 2003;63:7595–7599. [PubMed] [Google Scholar]
- 13.Nielsen M, Morreau H, Vasen HF, Hes FJ. MUTYH-associated polyposis (MAP) Crit Rev Oncol Hematol. 2011;79:1–16. doi: 10.1016/j.critrevonc.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 14.Sheng JQ, Cui WJ, Fu L, Jin P, Han Y, Li SJ, Fan RY, Li AQ, Zhang MZ, Li SR. APC gene mutations in Chinese familial adenomatous polyposis patients. World J Gastroenterol. 2010;16:1522–1526. doi: 10.3748/wjg.v16.i12.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Out AA, Tops CM, Nielsen M, Weiss MM, van Minderhout IJ, Fokkema IF, Buisine MP, Claes K, Colas C, Fodde R, et al. Leiden Open Variation Database of the MUTYH gene. Hum Mutat. 2010;31:1205–1215. doi: 10.1002/humu.21343. [DOI] [PubMed] [Google Scholar]
- 16.Shinmura K, Yamaguchi S, Saitoh T, Kohno T, Yokota J. Somatic mutations and single nucleotide polymorphisms of base excision repair genes involved in the repair of 8-hydroxyguanine in damaged DNA. Cancer Lett. 2001;166:65–69. doi: 10.1016/s0304-3835(01)00435-9. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi S, Shinmura K, Saitoh T, Takenoshita S, Kuwano H, Yokota J. A single nucleotide polymorphism at the splice donor site of the human MYH base excision repair genes results in reduced translation efficiency of its transcripts. Genes Cells. 2002;7:461–474. doi: 10.1046/j.1365-2443.2002.00532.x. [DOI] [PubMed] [Google Scholar]
- 18.Wooden SH, Bassett HM, Wood TG, McCullough AK. Identification of critical residues required for the mutation avoidance function of human MutY (hMYH) and implications in colorectal cancer. Cancer Lett. 2004;205:89–95. doi: 10.1016/j.canlet.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Bai H, Jones S, Guan X, Wilson TM, Sampson JR, Cheadle JP, Lu AL. Functional characterization of two human MutY homolog (hMYH) missense mutations (R227W and V232F) that lie within the putative hMSH6 binding domain and are associated with hMYH polyposis. Nucleic Acids Res. 2005;33:597–604. doi: 10.1093/nar/gki209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker AR, Sieber OM, Shi C, Hua L, Takao M, Tomlinson IP, Eshleman JR. Cells with pathogenic biallelic mutations in the human MUTYH gene are defective in DNA damage binding and repair. Carcinogenesis. 2005;26:2010–2018. doi: 10.1093/carcin/bgi166. [DOI] [PubMed] [Google Scholar]
- 21.Bai H, Grist S, Gardner J, Suthers G, Wilson TM, Lu AL. Functional characterization of human MutY homolog (hMYH) missense mutation (R231L) that is linked with hMYH-associated polyposis. Cancer Lett. 2007;250:74–81. doi: 10.1016/j.canlet.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanaru-Fujisawa R, Matsumoto T, Ushijima Y, Esaki M, Hirahashi M, Gushima M, Yao T, Nakabeppu Y, Iida M. Genomic and functional analyses of MUTYH in Japanese patients with adenomatous polyposis. Clin Genet. 2008;73:545–553. doi: 10.1111/j.1399-0004.2008.00998.x. [DOI] [PubMed] [Google Scholar]
- 23.Ali M, Kim H, Cleary S, Cupples C, Gallinger S, Bristow R. Characterization of mutant MUTYH proteins associated with familial colorectal cancer. Gastroenterology. 2008;135:499–507. doi: 10.1053/j.gastro.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kundu S, Brinkmeyer MK, Livingston AL, David SS. Adenine removal activity and bacterial complementation with the human MutY homologue (MUTYH) and Y165C, G382D, P391L and Q324R variants associated with colorectal cancer. DNA Repair (Amst) 2009;8:1400–1410. doi: 10.1016/j.dnarep.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsbring M, Vik ES, Dalhus B, Karlsen TH, Bergquist A, Schrumpf E, Bjørås M, Boberg KM, Alseth I. Catalytically impaired hMYH and NEIL1 mutant proteins identified in patients with primary sclerosing cholangitis and cholangiocarcinoma. Carcinogenesis. 2009;30:1147–1154. doi: 10.1093/carcin/bgp118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molatore S, Russo MT, D’Agostino VG, Barone F, Matsumoto Y, Albertini AM, Minoprio A, Degan P, Mazzei F, Bignami M, et al. MUTYH mutations associated with familial adenomatous polyposis: functional characterization by a mammalian cell-based assay. Hum Mutat. 2010;31:159–166. doi: 10.1002/humu.21158. [DOI] [PubMed] [Google Scholar]
- 27.Goto M, Shinmura K, Nakabeppu Y, Tao H, Yamada H, Tsuneyoshi T, Sugimura H. Adenine DNA glycosylase activity of 14 human MutY homolog (MUTYH) variant proteins found in patients with colorectal polyposis and cancer. Hum Mutat. 2010;31:E1861–E1874. doi: 10.1002/humu.21363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamane A, Shinmura K, Sunaga N, Saitoh T, Yamaguchi S, Shinmura Y, Yoshimura K, Murakami H, Nojima Y, Kohno T, et al. Suppressive activities of OGG1 and MYH proteins against G: C to T: A mutations caused by 8-hydroxyguanine but not by benzo[a]pyrene diol epoxide in human cells in vivo. Carcinogenesis. 2003;24:1031–1037. doi: 10.1093/carcin/bgg056. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki T, Harashima H, Kamiya H. Effects of base excision repair proteins on mutagenesis by 8-oxo-7,8-dihydroguanine (8-hydroxyguanine) paired with cytosine and adenine. DNA Repair (Amst) 2010;9:542–550. doi: 10.1016/j.dnarep.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Shinmura K, Goto M, Suzuki M, Tao H, Yamada H, Igarashi H, Matsuura S, Maeda M, Konno H, Matsuda T, et al. Reduced expression of MUTYH with suppressive activity against mutations caused by 8-hydroxyguanine is a novel predictor of a poor prognosis in human gastric cancer. J Pathol. 2011;225:414–423. doi: 10.1002/path.2953. [DOI] [PubMed] [Google Scholar]
- 31.Tao H, Shinmura K, Hanaoka T, Natsukawa S, Shaura K, Koizumi Y, Kasuga Y, Ozawa T, Tsujinaka T, Li Z, et al. A novel splice-site variant of the base excision repair gene MYH is associated with production of an aberrant mRNA transcript encoding a truncated MYH protein not localized in the nucleus. Carcinogenesis. 2004;25:1859–1866. doi: 10.1093/carcin/bgh206. [DOI] [PubMed] [Google Scholar]
- 32.Matsuda T, Yagi T, Kawanishi M, Matsui S, Takebe H. Molecular analysis of mutations induced by 2-chloroacetaldehyde, the ultimate carcinogenic form of vinyl chloride, in human cells using shuttle vectors. Carcinogenesis. 1995;16:2389–2394. doi: 10.1093/carcin/16.10.2389. [DOI] [PubMed] [Google Scholar]
- 33.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 34.Di Gregorio C, Frattini M, Maffei S, Ponti G, Losi L, Pedroni M, Venesio T, Bertario L, Varesco L, Risio M, et al. Immunohistochemical expression of MYH protein can be used to identify patients with MYH-associated polyposis. Gastroenterology. 2006;131:439–444. doi: 10.1053/j.gastro.2006.05.049. [DOI] [PubMed] [Google Scholar]
- 35.van der Post RS, Kets CM, Ligtenberg MJ, van Krieken JH, Hoogerbrugge N. Immunohistochemistry is not an accurate first step towards the molecular diagnosis of MUTYH-associated polyposis. Virchows Arch. 2009;454:25–29. doi: 10.1007/s00428-008-0701-y. [DOI] [PubMed] [Google Scholar]
- 36.Macdonald J, Taylor L, Sherman A, Kawakami K, Takahashi Y, Sang HM, McGrew MJ. Efficient genetic modification and germ-line transmission of primordial germ cells using piggyBac and Tol2 transposons. Proc Natl Acad Sci USA. 2012;109:E1466–E1472. doi: 10.1073/pnas.1118715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cary LC, Goebel M, Corsaro BG, Wang HG, Rosen E, Fraser MJ. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]


