Abstract
Since its initial discovery as an atypical PKC-interacting protein, p62 has emerged as a crucial molecule in a myriad of cellular functions. This multifunctional role of p62 is explained by its ability to interact with several key components of various signaling mechanisms. Not surprisingly, p62 is required for tumor transformation owing to its roles as a key molecule in nutrient sensing, as a regulator and substrate of autophagy, as an inducer of oxidative detoxifying proteins, and as a modulator of mitotic transit and genomic stability, all crucial events in the control of cell growth and cancer.
Keywords: p62, Sequestosome1, NF-κB, NRF2, mTORC1, atypical PKCs, Cancer, Autophagy
Regulation of cell growth, survival, and proliferation
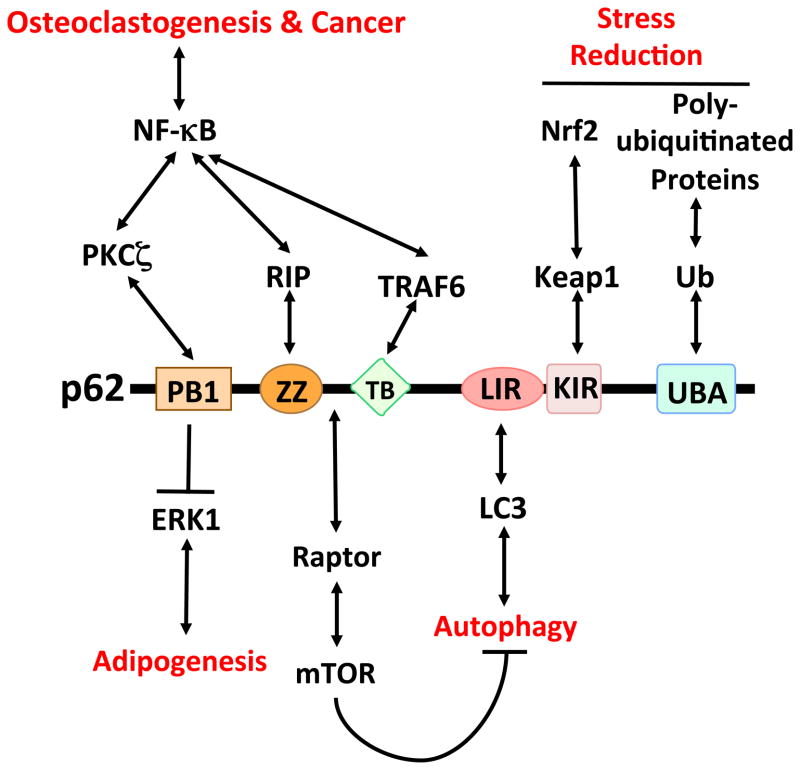
p62 (also known as sequestosome-1) was identified as a novel partner of the atypical protein kinase Cs (aPKCs) in unbiased screens [1,2]. Both the aPKCs and p62 harbor a Phox/Bem 1p (PB1) protein-protein binding domain that governs their interaction and the interactions of aPKC and p62 with the PB1 domains of their respective specific partners, Par-6 and neighbor of BRCA1 gene 1 (NBR1) [3,4]. Initial models suggested that, similar to how Par-6 localizes the aPKCs to the core of the polarity pathways, p62 controls localization of the aPKCs to the nuclear factor kappa B (NF-κB) cascades [3,5,6]. However, further studies demonstrated that, in contrast to the relatively simple structure of Par-6, p62 is rich in protein-interacting sequences, which highlights its essential role as a signaling hub rather than a mere scaffold [7] (Figure 1).
Figure 1.
p62-interacting partners and functions. The PB1 domain of p62 binds and inhibits ERK1, which is crucial for the negative regulation of adipogenesis and obesity and the ensuing inflammation. p62 also interacts with PKCζ through a PB1-PB1 interaction. The ZZ zinc-finger domain binds RIP, which links p62 to NF-κB activation in the TNF pathway. The TB domain binds TRAF6, which is relevant in RANK-induced osteoclastogenesis, as well as in Ras-induced tumorigenesis. p62 also interacts with raptor, which restrains autophagy through mTOR activation. This inhibition of autophagy, because of p62’s ability to interact with LC3, serves to control p62 levels. The interaction with Keap1 might be important for the regulation of Nrf2 and the control of ROS levels. Finally, the UBA domain regulates the interaction of p62 with polyubiquitylated proteins targeted for degradation by the proteasome or autophagy.
Cell proliferation, particularly under tumorigenic conditions, is an intricate process requiring that cells increase in size before undergoing division. In addition, their continued survival depends on overcoming harsh conditions of nutrient and oxygen scarcity. That is, the ability of a cell to activate autophagy, reprogram its metabolism, and control the production of toxic compounds, such as reactive oxygen species (ROS) and misfolded proteins, is another crucial set of parameters that dramatically influences cell proliferation and tumorigenisis [8]. Therefore, growth, mitosis, and survival are tightly controlled through mechanisms that are being progressively unveiled. Cell division must be perfectly orchestrated in a sequential manner, and failure to do so ultimately results in dramatic consequences. For example, dysregulation of progression through the last steps of the cell cycle could influence cell proliferation and promote tumorigenesis through genomic instability [10]. These regulatory mechanisms will likely be rich sources of potential therapeutic targets in cancer. For example, interfering with the nutrient-sensing pathways, which control cell size, will prevent cells from dividing because mitosis only occurs when cells have reached the right size [9].
Previous studies analyzing the phenotype of p62-deficient mice demonstrated that the physiological role of p62 is to control osteoclastogenesis and bone remodeling [11], as well as adipogenesis and obesity [12]. In this article we review the recent literature demonstrating that p62 is also a central regulator of tumorigenesis due to its abilities to modulate (and be a substrate of) autophagy, to control the levels of reactive oxidative species (ROS) and misfolded proteins, and to ensure a timely transit of cell through mitosis, all crucial factors in cancer.
p62 in the control of cell growth and autophagy
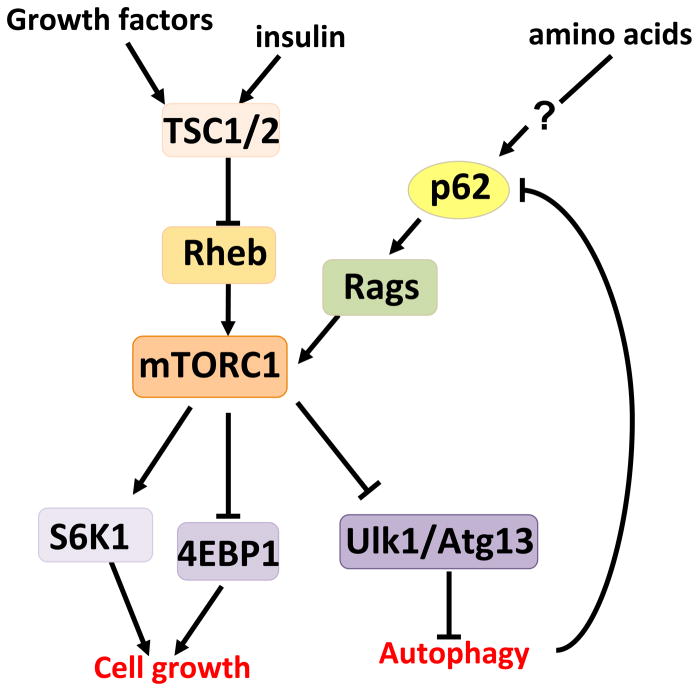
A recent study that used an unbiased proteomic approach uncovered an unanticipated role for p62 in activation of the mammalian target of rapamycin (mTOR) pathway, a central regulator of cell growth and autophagy that integrates nutrient sensing and cell-size control, and is aberrantly activated in many types of cancer [13]. Of the two multiprotein complexes orchestrated by mTOR, mTOR complex (mTORC)1 and mTORC2, p62 specifically associates with mTORC1 through its interaction with raptor, a core component of mTORC1 (Figure 1)[13,14]. mTORC1 is sensitive to inhibition by rapamycin and senses multiple cellular and environmental cues including nutrient availability, energy levels, protein misfolding, quality control, and growth signals [15]. Thus, mTORC1 channels a variety of signals into a coordinated cell growth and proliferation response by promoting protein synthesis and inhibiting autophagy (Figure 2). Most upstream signals converge on TSC1-TSC2, a heterodimeric tumor suppressor that negatively regulates the Ras-related small G protein Rheb, an essential activator of mTORC1 [16]. By contrast, amino acid signaling uses a different GTPase family, the Rag GTPases [17,18]. Rheb, although required for mTORC1 activation by amino acids, might not be directly involved in sensing amino acids [19]. Interestingly, recent findings place p62 specifically in the amino acid-mediated mTORC1 activation pathway, and add a new piece in our understanding of the upstream mechanisms regulating nutrient sensing [13]. Accordingly, in p62-deficient cells amino acid-mediated phosphorylation of the mTORC1 targets S6K and 4EBP1 is severely impaired and, in keeping with decreased mTORC1 activity, autophagy is upregulated [13]. Because p62 is also a substrate of autophagy [20], this creates a feed-forward loop by which p62 activation of mTORC1 increases p62 levels, further promoting mTORC1 activity [21]. This could be essential in conditions of nutrient deprivation, in which a scarcity of nutrients reduces mTORC1 activity and upregulates autophagy. Under these conditions, the p62-mTORC1-autophagy loop might provide a safeguard mechanism to ensure the irreversibility of cell death when nutrients are not available.
Figure 2.
p62 and nutrient sensing. The mTORC1 complex plays an essential role in the control of anabolic responses to nutrient availability, insulin, and growth factors. mTORC1 regulates cell growth through the phosphorylation of S6K and 4E-BP1 and autophagy by targeting Ulk1 and Atg13. Most upstream signals converge on TSC1/2 complex that regulates the mTORC1 activator, Rheb. However, amino acids impinge on a different cascade involving Rags. p62 is selectively required for mTORC1 activation by amino acids, although how p62 senses the signal is not yet fully understood.
How mTORC1 senses nutrients is the key question yet to be resolved. Recent studies in mammals, and others involving genetic screens in yeast and flies, have helped to uncover essential elements in this process [19]. An important step forward is the finding that the Rag GTPases control amino acid-dependent mTORC1 activity by regulating mTORC1 translocation to a lysosomal LAMP2-positive compartment, thus putting mTORC1 in proximity of its activator, Rheb [18]. p62 binds raptor and the Rags, promoting activation of the pathway by favoring formation of the active Rag dimer, through a mechanism that probably involves p62 oligomerization [13]. In addition, p62 is also required for the translocation of mTORC1 to the lysosomal surface [13], which is consistent with the initial observation that p62 is located at Rab7-positive late-endosomal membranes [1]. This is also consistent with recent findings that mTORC1 regulates endocytosis in response to changes in environmental factors such as nutrient availability [22]. Considering the important role of late endosomes and/or lysosomes as factories where amino acids activate mTORC1, it is conceivable that other functional proteins in these intracellular locations play a role in this pathway as well. In fact, recent studies have identified the Ragulator complex (formed by MAP kinase scaffold protein 1 (MP1), p14 and p18) and the vacuolar H+-ATPase as lysosomal anchors of the Rags, as well as crucial components of the lysosome-associated machinery for amino acid sensing [23,24]. Other small GTPases that modulate protein trafficking, such as RalA, Rab5, Rab7, and Arfl, have also been shown to be involved in mTORC1 activation [25–27]. Taken together, these findings point to the intracellular localization of signaling molecules as an important mechanism for controlling signal specificity, and also highlight the crucial role scaffold proteins play as docking platforms to spatially integrate the signal. These observations also highlight the intricate relations between p62 and autophagy through the functional interactions with mTORC1, a central piece in the control of cell survival and growth in cancer cells.
p62 and control of the oxidative stress response in cancer
The interaction of p62 with tumor necrosis factor receptor-associated factor (TRAF) 6 (an important inflammation signaling molecule), and the degradation of p62 by autophagy, are important for the role of p62 in cell survival and tumorigenesis [28]. Several studies have established the importance of controlling p62 levels to prevent cell toxicity and cancer. Investigations on the role of autophagy in normal liver physiology established an intriguing link between the autophagic pathways, inflammation-mediated cell toxicity, and p62. They demonstrated that genetic inactivation of key autophagy molecules, such as Atg7, resulted in p62 accumulation and hepatotoxicity, which led to the generation of liver tumors [29,30]. The simultaneous ablation of p62 in this model reversed the hepatotoxic and carcinogenic effect of autophagy deficiency, implying that p62 overexpression leads to chronic inflammation and cancer in the liver [31]. Collectively, these observations suggest a linear model whereby autophagy deficiency enhances p62 protein levels, which then promotes inflammation, a process that is intimately linked to carcinogenesis in many types of tumors.
However, the connection between autophagy and cancer is complex and in some cases contradictory [32]. As a survival mechanism for cells under nutrient stress, autophagy would be predicted to play a key role as a tumor promoter [33]. However, abundant data support its role as a cancer inhibitor, including the fact that Beclin-1, a protein essential for autophagy, is actually a tumor suppressor [34]. In fact, phosphatase and tensin homolog (PTEN) and the tuberous sclerosis complex (TSC) proteins, which are well-established tumor suppressors, actually promote autophagy through inhibition of mTORC1 [16,35]. Therefore, that Atg7 deficiency results in increased tumorigenesis is consistent with autophagy being a tumor suppressor and establishes p62 as a crucial mediator of that function.
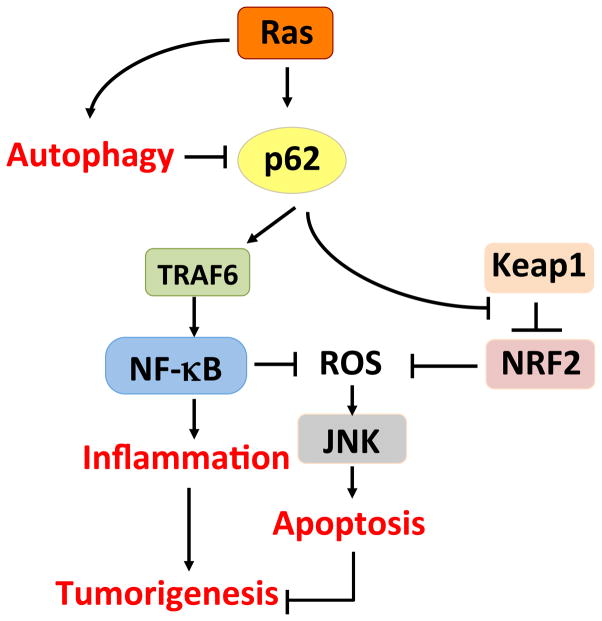
The role of p62 in mediating autophagy-dependent tumor suppression is probably through suppression of ROS. Autophagy deficiency in the liver is associated with enhanced ROS production and activation of Nuclear factor (erythroid-derived 2)-like 2 (NRF2), a transcription factor that controls the expression of key ROS scavengers [36]. Recent studies have linked p62 overexpression to NRF2 activation through the ability of p62 to chelate Keap1, a negative regulator of NRF2 activation [37–39]. Therefore, it is possible to propose a linear model in which reduced autophagy leads to the accumulation of p62, which in turn activates NRF2 to alleviate oxidative stress (Figure 3). This oxidative stress could be caused by the accumulation of, for example, dysfunctional mitochondria that are not cleared because of the impaired autophagic degradation pathways. In keeping with this notion, the lack of p62 in Atg7-deficient livers leads to reduced NRF2 production [29,30].
Figure 3.
p62, Ras, and autophagy. Under conditions of low autophagy, p62 accumulates and activates inflammation via NF-κB, and increases the expression of ROS scavengers via NF-κB and Nrf2. The net effect is enhanced tumorigenesis.
Unfortunately, this simple linear model does not account for all of the available data. For example, the model would predict that the ablation of p62 in autophagy-deficient livers should worsen hepatotoxicity because cells are deprived of the protection against oxidative stress that the p62-NRF2 tandem provides. In marked contrast with this prediction, the hepatotoxicity observed in Atg7-deficient mice is prevented by the simultaneous inactivation of p62 [31]. Moreover, genetic inactivation of NRF2 impairs p62 accumulation in autophagy-deficient hepatocytes [29,30] and in other in vitro cell systems [40]. This complicates the interpretation of the Atg7/p62 doubly deficient phenotype, because it is now unclear whether p62 is upstream or downstream of NRF2 in vivo, and calls into question the relative importance of either protein in liver hepatotoxicity during autophagy deficiency. Taken together, these data make the autophagy-p62 connection in cancer less straightforward.
The linear link between autophagy, p62, and oncogenesis has been further weakened by recent observations that Ras-induced transformation requires p62 [13,41] but also increases autophagy [41]. This suggests a disconnect between autophagy and the Ras-p62 pathway. One potential explanation for these apparently paradoxical observations is that Ras-induced autophagy and Ras-induced p62 are two independent but mutually required arms of the Ras downstream signal. For instance, Ras-induced autophagy would be required for the removal of damaged and dysfunctional mitochondria, which is thought to be associated with metabolic reprogramming and contributes to maintaining low ROS levels. Ras-induced p62 might shuttle the damaged mitochondria to the autophagosome by interacting with the autophagosomal membrane protein Atg8/LC3. That is, p62 would link autophagy and cell transformation not as a linear cascade, but rather as a central hub controlling several survival mechanisms in Ras-transformed cells. In fact, several overexpression and RNAi-mediated knockdown studies have suggested that p62 is important for mitophagy in some systems [42,43]. Unfortunately, however, data from p62 knockout cells do not seem to support such a role for p62 [44,45].
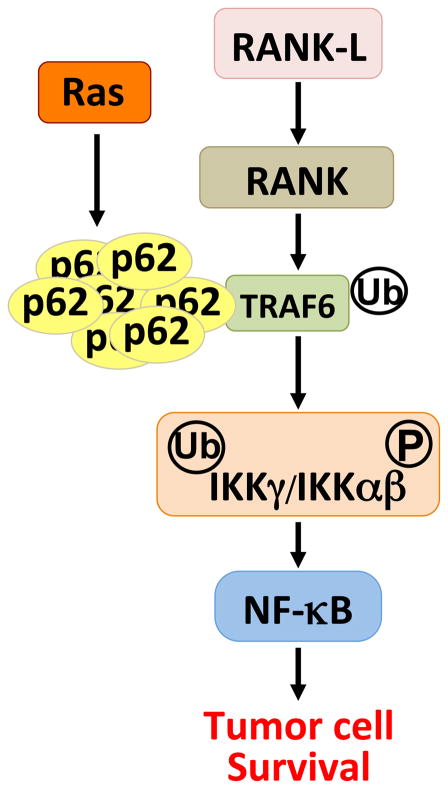
Interestingly, oncogenic Ras modulates p62 levels at a gene transcriptional level through a mechanism involving targeting of an AP-1 enhancer element in the p62 promoter [46]. Importantly, ablation of p62 abolished oncogenic transformation in an in vivo, physiologically relevant endogenous Ras-induced lung cancer model. Thus, whereas wild-type mice efficiently developed lung adenomas and adenocarcinomas upon the expression of oncogenic Ras, the p62 knockout mice were completely resistant. This was the first time that a link between tumor transformation and p62 was revealed, and the first in vivo demonstration that p62 is required for tumorigenesis [46]. Recent data demonstrate that the role of p62 in NF-κB activation might also be crucial to its role in Ras-induced carcinogenesis. In this study, they found that p62 plays an important role in the development of pancreatic ductal adenocarcinomas through a feedforward loop, whereby Ras activates NF-κB; NF-κB then transcriptionally induces p62 [47], and finally, expression of p62 leads likely to the oligomerization of TRAF6, resulting in further NF-κB activation [13]. Thus, the compounded activation of NF-κB by both Ras and the induced p62 causes a p62-dependent pancreatic carcinogenic process. This is consistent with a model whereby Ras-induced p62 overexpression kidnaps a physiological p62-TRAF6 interaction that is important in the activation of NF-κB in response to interleukin- 1 (IL-1) [28], nerve growth factor (NGF) [48], and RANK [11,49–52](Figure 4). Therefore, oncogenic Ras profits from a physiologically important biochemical and genetic link between p62, TRAF6 and NF-κB, to promote tumor cell survival through the ability of this pathway to reduce the production of ROS during transformation in lung cells and tissues [46].
Figure 4.
p62 and tumorigenesis. The interaction of p62 with TRAF6 is important for osteoclastogenesis in response to RANK activation of NF-κB. It is also used at the level of TRAF6 by the Ras oncogene, which promotes the synthesis of p62 to regulate tumor cell survival through the activation of NF-κB.
Induction of the NF-κB signaling cascade is a prominent feature of several tumorigenic processes [53], owing to the ability of NF-κB transcriptional products to act as ROS scavengers [13]. Specifically, the loss of p62 led to the accumulation of Ras-induced ROS above tolerable levels, resulting in oxidative stress and the ensuing JNK activation and apoptosis [46]. These observations, therefore, suggest that p62 is an important molecule in lung carcinogenesis and that TRAF6 is also likely to be relevant in the formation of this type of tumor. Indeed, recent results demonstrate TRAF6 mRNA overexpression and gene amplification in samples from human patients with lung cancer [54]. Consistent with this notion, TRAF6 inactivation in human lung cancer cell lines dramatically reduced NF-κB and inhibited tumorigenesis [54]. Conversely, TRAF6 overexpression in immortalized mouse embryo fibroblasts resulted in NF-κB activation, anchorage-independent growth, and tumor formation [54]. This is an important observation because it shows that the p62-TRAF6 cassette is necessary for Ras-induced transformation during mouse and human lung carcinogenesis. It also suggests that Ras produces oncogenic transformation by hijacking the p62-TRAF6 complex, which under normal circumstances regulates bone remodeling (Figure 4). Consistent with the observation that ectopically overexpressed TRAF6 confers tumor characteristics to normal cells [54], a recent report demonstrated that ectopic overexpression of p62 was also able to promote tumorigenesis [55]. Therefore, p62 regulation of either NF-κB in Ras transformation and/or NRF2 during autophagic inhibition serves to control the toxic levels of ROS and to promote tumorigenesis (Figure 3).
Another twist in the story of p62 and cancer is the role of p62 as a chaperone during the degradation of proteins by autophagy. Under conditions of nutrient starvation, p62 can shuttle Dishevelled (Dvl)2, a crucial component of the pro-tumorigenic Wnt pathway, to autophagomes, leading to the shutdown of this important signaling cascade [56]. This is consistent with the tumor suppressor role of autophagy and might compound with autophagy-dependent p62 degradation to increase cytotoxicity and cell death. Altogether this will produce a reduction in the tumorigenic process. Whatever the link between p62 and autophagy is, it is clear that p62 has emerged as a crucial regulator of oncogenic proteins and ROS levels through at least two different essential transcription factors, NF-κB and NRF2, and that these effects are important for oncogenic transformation in vitro and in vivo [29,30,46,57].
p62 and mitosis
One of the characteristics of tumor cells, in addition to their exceptional ability to survive the harshest conditions, is their uncontrolled capacity to divide [58]. Subversion of the different steps of the cell cycle unquestionably severely alters the proliferative capabilities of cancer cells, their maintenance of chromosome stability and their ability to produce healthy progeny [59–61]. Indeed, subtle alterations in the timing of mitotic entry or exit could have tumorigenic consequences [61,62]. Cyclin-dependent kinases (cdks) regulate progression of mammalian cells through the various phases of the cell cycle [63]. The cdks are heterodimeric protein kinases, each composed of a catalytic subunit known as a cdk and a regulatory subunit known as a cyclin [64]. The mitotic kinase cdk1 controls transit through the late S/G2 phase and early mitosis phases of the cell cycle [64]. Interestingly, while examining p62 levels at different steps of the cell cycle, it was found that p62 is significantly phosphorylated in early mitosis [65]. Pharmacological inhibition or knockdown of cdk1 demonstrated that cdk1 phosphorylates p62 at residues T269 and S272 in mitosis [65]. This strongly suggests that p62 might play a role not only in cell survival, but also in the mitotic control of cancer cells. Indeed, cancer cells expressing a nonphosphorylatable p62 mutant displayed higher tumorigenic properties in vivo and in vitro than the same cells expressing wild-type p62 [65]. This indicates that p62 phosphorylation by cdk1 serves to restrain cell transformation. Of note, another study showed that phosphorylation of p62 at the cdk1 sites might control its abilities to shuttle between the nucleus and cytosol and localize to the promyelocytic leukemia (PML) bodies of asynchronously growing cells [66]. However, if confirmed, this will likely be irrelevant to its role in cell cycle control, because p62 is maximally phosphorylated by cdk1 at pro-metaphase, when the nuclear membrane has already dissolved. Whether the localization of p62 to PML bodies is important for its role as a protumorigenic protein has not been addressed, but it is plausible because the PML proteins are well established tumor suppressors [67]. Further studies are needed to answer this interesting question.
The precise mechanism whereby cdk1-mediated p62 phosphorylation restrains cell transformation involves control of mitotic exit. That is, when tumor cells expressing either wild-type or nonphosphorylatable p62 were released from prometaphase blockade, the cells with nonphosphorylatable p62 exited mitosis and transitioned from mitosis to G1 faster than wild-type cells [65]. Importantly, an enhanced proportion of nonphosphorylatable p62-expressing cells displayed lagging chromosomes and micronuclei, which are indicative of increased genome instability [65]. These are important observations because they indicate that p62 stimulates tumorigenesis not only by controlling ROS levels and therefore promoting cell survival, but also by promoting premature exit from mitosis (unless phosphorylated by cdk1), which leads to an increased proliferation rate and genome instability [65]. p62 promotes premature exit from mitosis through degradation of cyclin B1 via the proteasome, which is also restrained by cdk1-mediated phosphorylation of p62 [65].
Collectively, these results are consistent with a role for p62 in cell cycle control at the level of mitotic exit, which is modulated by cdk1 phosphorylation. Interestingly, another study supports p62 as an important player in late mitotic phases but through a completely different mechanism. That is, it has also been shown that p62 plays an important role in the disposal of the midbody ring, a circular structure that connects two dividing cells, and micronuclei by autophagy [68–70]. Although it remains to be determined how defects in this mechanism of midbody ring disposal and micronuclei removal by p62 influence cell transformation, these results further emphasize the relevance of p62 in the control of cell division.
Concluding remarks
Although there are biochemical details yet to be discerned, it is becoming apparent that p62 is a crucial component of the cell transformation machinery that influences at least three processes: cell growth, survival, and mitosis. The role of p62 as a target of autophagy is very important because it biochemically links nutrient sensing to signaling cascades that regulate inflammation and ROS levels, allowing tumor cells to survive under conditions of autophagy defects. This might be crucial under conditions of autophagy inhibition, for example from Beclin-1 deficiency. However, the role of p62 in cell division is also important, because by controlling cell cycle transit and the levels of cell cycle proteins, which are essential for genome stability, p62 can dramatically impact the tumorigenic potential of cancer cells. Finally, the recent observation that p62 is a ‘nutrient sensor’ for the activation of the mTORC1 pathway unveils its role not only as a target of autophagy, or as a bridge between polyubiquitylated proteins and the autophagosome, but also as an important step in the negative regulation of autophagy in response to nutrient availability.
In this review, we have focused our discussion on the role of p62 in cancer cell biology. However, based on the analysis of p62 knockout mice, it is clear that the role of p62 under non-pathological conditions is to control bone and metabolic homeostasis. Through these physiological functions, p62 might indirectly control cancer as well. First, bone is an important site of cancer metastasis that can be affected by cross-talk between tumor and bone cells. Therefore, p62 could be explored as a potential therapeutic target in metastasis. Second, it is clear that p62 has a role in maintaining metabolic homeostasis by restraining adipogenesis and promoting energy expenditure. Intriguingly, it is becoming clear that obesity-induced inflammation and insulin resistance, in both humans and mice, can promote at least some types of cancer. Therefore, inactivation of p62 at an organismal level could have undesired effects on cancer progression if, as the phenotype p62 knockout mice suggests, it results in enhanced adiposity and obesity. The connections between p62 and obesity have been recently discussed elsewhere [21]. However, it is worth noting that the genetic inactivation of autophagy proteins in mouse adipose tissue results in protection against obesity [71], the opposite phenotype of ablating p62 [12]. This might be because an increase in p62 levels, as a consequence of autophagy inhibition, inhibits ERK1 (Figure 1), which in turn restrains adipogenesis and gives rise to decreased adiposity. Studies in Atg7/p62 double knockout adipose tissue should solve this question. In summary, the multitasking function of p62 makes it a fascinating protein to study from the point of view of cell signaling organization and physiology. Its multifunctional nature make its role in cancer complex, however it could be an attractive target for therapeutic intervention if we can selectively modulate the interactions of p62 with specific signaling molecules, perhaps by aiming at the different interaction modules in its structure.
Acknowledgments
Our research is funded by NIH grants R01AI072581 (J.M.), R01DK088107 (J.M.), R01CA132847 (J.M.), and R01CA134530 (M.T.D-M). We thank Maryellen Daston for editing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sanchez P, et al. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cell Biol. 1998;18 (5):3069–3080. doi: 10.1128/mcb.18.5.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puls A, et al. Interaction of protein kinase C zeta with ZIP, a novel protein kinase C-binding protein. Proc Natl Acad Sci U S A. 1997;94 (12):6191–6196. doi: 10.1073/pnas.94.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moscat J, et al. Cell signaling and function organized by PB1 domain interactions. Mol Cell. 2006;23 (5):631–640. doi: 10.1016/j.molcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Sumimoto H, et al. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE. 2007;2007(401):re6. doi: 10.1126/stke.4012007re6. [DOI] [PubMed] [Google Scholar]
- 5.Ohno S. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr Opin Cell Biol. 2001;13 (5):641–648. doi: 10.1016/s0955-0674(00)00264-7. [DOI] [PubMed] [Google Scholar]
- 6.Macara IG. Par proteins: partners in polarization. Curr Biol. 2004;14 (4):R160–162. [PubMed] [Google Scholar]
- 7.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137 (6):1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeBerardinis RJ, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7 (1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14 (23):R1014–1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 10.Negrini S, et al. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11 (3):220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 11.Duran A, et al. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell. 2004;6 (2):303–309. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez A, et al. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3 (3):211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Duran A, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44 (1):134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110 (2):163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 15.Wullschleger S, et al. TOR signaling in growth and metabolism. Cell. 2006;124 (3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 16.Huang J, et al. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28 (12):4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim E, et al. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10 (8):935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320 (5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, Guan KL. Amino acid signaling in TOR activation. Annu Rev Biochem. 2011;80:1001–1032. doi: 10.1146/annurev-biochem-062209-094414. [DOI] [PubMed] [Google Scholar]
- 20.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171 (4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moscat J, Diaz-Meco MT. Feedback on Fat: p62-mTORC1-Autophagy Connections. Cell. 2011;147 (4):724–727. doi: 10.1016/j.cell.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macgurn JA, et al. TORC1 Regulates Endocytosis via Npr1-Mediated Phosphoinhibition of a Ubiquitin Ligase Adaptor. Cell. 2011;147 (5):1104–1117. doi: 10.1016/j.cell.2011.09.054. [DOI] [PubMed] [Google Scholar]
- 23.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141 (2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H-ATPase. Science. 2011;334 (6056):678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maehama T, et al. RalA functions as an indispensable signal mediator for the nutrient-sensing system. J Biol Chem. 2008;283 (50):35053–35059. doi: 10.1074/jbc.M805822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flinn RJ, et al. The late endosome is essential for mTORC1 signaling. Mol Biol Cell. 2010;21 (5):833–841. doi: 10.1091/mbc.E09-09-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, et al. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010;285 (26):19705–19709. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanz L, et al. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19 (7):1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25 (8):795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inami Y, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193 (2):275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131 (6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 32.Hippert MM, et al. Autophagy in cancer: good, bad, or both? Cancer Res. 2006;66 (19):9349–9351. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- 33.Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev. 2011;21 (1):113–119. doi: 10.1016/j.gde.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue Z, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100 (25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inoki K, et al. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37 (1):19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 36.Taguchi K, et al. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16 (2):123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 37.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12 (3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 38.Lau A, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30 (13):3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Copple IM, et al. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J Biol Chem. 2010;285 (22):16782–16788. doi: 10.1074/jbc.M109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285 (29):22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25 (5):460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12 (2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 43.Geisler S, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6 (7):871–878. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- 44.Okatsu K, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15 (8):887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narendra D, et al. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6 (8):1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duran A, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13 (4):343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Ling J, et al. Kras(G12D)-Induced IKK2/beta/NF-kappaB Activation by IL-1alpha and p62 Feedforward Loops Is Required for Development of Pancreatic Ductal Adenocarcinoma. Cancer cell. 2012;21 (1):105–120. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wooten MW, et al. The p62 scaffold regulates nerve growth factor-induced NF-kappaB activation by influencing TRAF6 polyubiquitination. J Biol Chem. 2005;280 (42):35625–35629. doi: 10.1074/jbc.C500237200. [DOI] [PubMed] [Google Scholar]
- 49.Laurin N, et al. Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in Paget disease of bone. Am J Hum Genet. 2002;70 (6):1582–1588. doi: 10.1086/340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roodman GD. Insights into the pathogenesis of Paget’s disease. Ann N Y Acad Sci. 2010;1192:176–180. doi: 10.1111/j.1749-6632.2009.05214.x. [DOI] [PubMed] [Google Scholar]
- 51.Daroszewska A, et al. A point mutation in the ubiquitin-associated domain of SQSMT1 is sufficient to cause a Paget’s disease-like disorder in mice. Hum Mol Genet. 2011;20 (14):2734–2744. doi: 10.1093/hmg/ddr172. [DOI] [PubMed] [Google Scholar]
- 52.Kurihara N, et al. Contributions of the measles virus nucleocapsid gene and the SQSTM1/p62(P392L) mutation to Paget’s disease. Cell Metab. 2011;13 (1):23–34. doi: 10.1016/j.cmet.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benhar M, et al. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep. 2002;3 (5):420–425. doi: 10.1093/embo-reports/kvf094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Starczynowski DT, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-kappaB pathways in human lung cancer. J Clin Invest. 2011;121 (10):4095–4105. doi: 10.1172/JCI58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137 (6):1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao C, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12 (8):781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 57.Hussain S, et al. Nephrin deficiency activates NF-kappaB and promotes glomerular injury. J Am Soc Nephrol. 2009;20 (8):1733–1743. doi: 10.1681/ASN.2008111219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144 (5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10 (7):478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432 (7015):316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 61.Kops GJ, et al. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol. 2005;169 (1):49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vecchione A, et al. Fez1/Lzts1 absence impairs Cdk1/Cdc25C interaction during mitosis and predisposes mice to cancer development. Cancer cell. 2007;11 (3):275–289. doi: 10.1016/j.ccr.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2 (1):21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 64.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30 (11):630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 65.Linares JF, et al. Phosphorylation of p62 by cdk1 controls the timely transit of cells through mitosis and tumor cell proliferation. Mol Cell Biol. 2011;31 (1):105–117. doi: 10.1128/MCB.00620-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pankiv S, et al. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem. 2010;285 (8):5941–5953. doi: 10.1074/jbc.M109.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108 (2):165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 68.Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat Cell Biol. 2009;11 (1):65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- 69.Kuo TC, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat Cell Biol. 2011;13 (12):1467. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rello-Varona S, et al. Autophagic removal of micronuclei. Cell cycle. 2012;11 (1):170–176. doi: 10.4161/cc.11.1.18564. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y, et al. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106 (47):19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]