Background: Lipoglycans modulate the initiation of immune responses by interacting with TLR2.
Results: A hypermannosylated lipomannan variant, once recognized by the immune system, enhances both innate responses and Th17 differentiation.
Conclusion: Altered lipoglycan structures are differently recognized by innate immune cells with an impact on the adaptive immune response.
Significance: Specific lipoglycan structures may be useful to modulate immune responses.
Keywords: Cytokine, Dendritic Cells, Glycosyltransferases, T Cell, Toll-like Receptors (TLR)
Abstract
Toll-like receptors (TLRs) recognize pathogens by interacting with pathogen-associated molecular patterns, such as the phosphatidylinositol-based lipoglycans, lipomannan (LM) and lipoarabinomannan (LAM). Such structures are present in several pathogens, including Mycobacterium tuberculosis, being important for the initiation of immune responses. It is well established that the interaction of LM and LAM with TLR2 is a process dependent on the structure of the ligands. However, the implications of structural variations on TLR2 ligands for the development of T helper (Th) cell responses or in the context of in vivo responses are less studied. Herein, we used Corynebacterium glutamicum as a source of lipoglycan intermediates for host interaction studies. In this study, we have deleted a putative glycosyltransferase, NCgl2096, from C. glutamicum and found that it encodes for a novel α(1→2)arabinofuranosyltransferase, AftE. Biochemical analysis of the lipoglycans obtained in the presence (wild type) or absence of NCgl2096 showed that AftE is involved in the biosynthesis of singular arabinans of LAM. In its absence, the resulting molecule is a hypermannosylated (hLM) form of LAM. Both LAM and hLM were recognized by dendritic cells, mainly via TLR2, and triggered the production of several cytokines. hLM was a stronger stimulus for in vitro cytokine production and, as a result, a more potent inducer of Th17 responses. In vivo data confirmed hLM as a stronger inducer of cytokine responses and suggested the involvement of pattern recognition receptors other than TLR2 as sensors for lipoglycans.
Introduction
The immune system has evolved several mechanisms of defense against pathogens, favoring pathogen clearance while ensuring host integrity. The innate immune system is the first line of host defense against invading pathogens, being mediated by phagocytes including macrophages and dendritic cells (DC)6 (1). These cells express host germ line-encoded pattern recognition receptors (PRRs) that recognize specific pathogen-associated molecular patterns present in microorganisms, resulting in the initiation of innate immune responses (2). To date, the best studied PRRs are toll-like receptors (TLRs) that sense components of bacteria, fungi, protozoa and viruses, thus playing a major role in innate immunity (2).
TLR2 binds a wider range of ligands than other members of the TLR family. TLR2 in association with TLR1 or TLR6 is essential for recognizing bacterial lipoproteins and lipopeptides as well as a variety of compounds, including lipoteichoic acids, lipoarabinomannan (LAM), and zymosan (2). Structural studies have revealed that the recognition of lipopeptide ligands by TLR2 induces the formation of TLR2-TLR1 or TLR2-TLR6 dimers, bringing together the intracellular domains of these TLRs, which is needed to initiate signaling cascades (3, 4). However, the structure-function relationship between TLR2 and ligands other than lipoproteins, particularly LAMs, is not so well understood. This issue is relevant in the field of mycobacteria, because pathogens of the Mycobacterium genus are well known for their unique and complex cell wall, composed by several molecules, including lipoglycans, able to activate TLR2 (5). Lipoglycans are surface-exposed macromolecules (6) and can be found as lipomannan (LM) and LAM. In mycobacteria, LM is a linear 20–25-sugar residue oligopolymer of α(1→6) mannose units decorated with 7–10 singular α(1→2) mannose units. This mature LM is proposed to be further glycosylated by a large arabinan domain (∼70 arabinose units) and capped by 2–3 mannose residues, resulting in mannose-capped LAM (Man-LAM) (7, 8). The structure of LM and LAM differs between virulent and non-virulent species of mycobacteria (7–10), suggesting that these molecules play a key role in the outcome of host-pathogen interactions. Importantly, recent evidence points to the fact that subtle structural variations on LM and LAM determine their biological activity and affect immunological responses (10–14). It has been suggested that LM is a stronger inducer of TLR2 responses than LAM (15), and the ability of LM to activate TLR2 is determined by the mannan chain length (14). Furthermore, truncation of the arabinan domain of LAM increases its ability to activate TLR2 (13). In addition, truncated LAM leads to altered phagocytosis of mycobacteria (16), and, more recently, LM from avirulent mycobacteria were shown to be less branched and to have shorter mannose domains, being more efficiently presented to T cells in the context of CD1b (10). The acylation state of mycobacterial LM was also shown to modulate TLR2 responses (12).
The study of TLR2 in infection and immunity has been well addressed (17). In the case of Mycobacterium tuberculosis, its in vitro recognition by macrophages and DC strongly depends on TLR2, with decreased expression of several cytokines in its absence (18–21). In vivo, lack of TLR2 did not compromise bacterial control (18, 19, 22–24) but impacted the maintenance of T helper (Th) 17 cells in the lungs of infected animals (25) and granuloma formation (18, 19, 25). Finally, in humans, the association of tlr2 polymorphisms with susceptibility to tuberculosis remains controversial (26) and appears to greatly depend on the genetics of the host/bacteria interplay (27).
In this study, we sought to further understand the impact of the arabinan branching in the phenotype of Th cell responses generated in vitro. Because most of the genetic loci encoding for cell wall biogenesis are essential in the context of M. tuberculosis, we have used Corynebacterium glutamicum as a model to obtain LAM variants (8, 28, 29). We have generated a C. glutamicum mutant deficient in a putative glycosyltransferase (NCgl2096), which resulted in abrogation of LAM and led to the accumulation of a novel form of hypermannosylated lipomannan (hLM). Both LAM and hLM were recognized by DC, mainly via TLR2, and triggered the production of both pro- and anti-inflammatory cytokines. hLM was a stronger stimulus for in vitro cytokine production and also a more potent inducer of Th17 responses. In vivo responses were also increased upon hLM recognition. Importantly, our in vivo data point to the involvement of PRRs other than TLR2 as sensors for lipoglycans.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
C. glutamicum and Escherichia coli DH5αmcr were grown in BHI, CGXII, and LB at 30 and 37 °C with kanamycin and ampicillin (50 μg/ml), wherever appropriate. All chemicals were of reagent grade and obtained from Sigma-Aldrich.
Construction of Plasmids, Mutant, and Complemented Strain
To delete aftE, the deletion vector pK19mobsacBΔ-aftE was made, and crossover PCR was used with genomic DNA of C. glutamicum as template and two different PCRs with primer pairs AB and CD (supplemental Table 1). The resulting PCR product served as template for primer pairs AD. The PCR product contained 18 nucleotides of the 3′-end of the respective gene together with genomic upstream sequences and 36 nucleotides of the 5′-end together with genomic downstream sequences. All plasmids used in this work were confirmed by sequencing. AftE was deleted by electroporating pK19mobsacBΔ-aftE into C. glutamicum and then selecting for sucrose resistance (30). Chromosomal deletions were confirmed using primer pairs AD as well as the additional new primer pairs EF (overhangs required for hybridization are given in italics in supplemental Table 1) hybridizing outside of the regions used for plasmid constructions. To construct pEKEx2-Cg-aftE, Cg-aftE was amplified using primer pairs AD, with the former providing the sequence CTGCAG as a ribosome binding site. The amplified product was ligated between PstI and EcoRI in pEKEx2. Plasmid pEKEx2-Cg-aftE was electroporated in C. glutamicum ΔaftE, resulting in C. glutamicum ΔaftE pEKEx2-Cg-aftE.
Extraction and Biochemical Characterization of Lipoglycans
Lipoglycans were extracted from delipidated cells as described previously (31, 32) and treated with H2O2 before immunological assays to remove any endotoxin contamination (13) and to yield a negative Limulus amebocyte lysate test using E-TOXATETM (Sigma-Aldrich). For MALDI-TOF-MS, 2,5-dihydroxybenzoic acid was used as a matrix, and samples (0.5 μl; 10 μg/μl) were analyzed on a Voyager DE-STR MALDI-TOF instrument (PerSeptive Biosystems) using linear mode detection. Furthermore, lipoglycans were derivatized for glycosyl and linkage analysis as described previously (31). All of the data were collected and analyzed using Xcaliber (version 1.2).
HEK293 Cell Culture and Stimulation Assay
Human HEK293 cells transfected with TLR2 (33) were cultured as described previously (13). For stimulation assays, trypsine-released HEK293 cells were washed with and resuspended in culture medium at 5.55 × 105 cells/ml, after which 90 μl of cell suspension (5 × 104 cells) was transferred to a 96-well U-bottom plate (Greiner) and left for 2 h to let the cells re-adhere. After this, cells were stimulated (in triplicate) with 50 μg/ml LAM or hLM in a final stimulation volume of 100 μl. Non-stimulated cells and cells stimulated with 50 ng/ml Pam3CSK4 served as negative and positive controls, respectively. Culture supernatants were harvested after 24 h by centrifugation and analyzed for IL-8 content using ELISA (Invitrogen).
Animals
Eight-week-old female C57BL/6 (BL/6) and BALB/c mice were obtained from Charles River (Barcelona, Spain); TLR2−/− mice in a C57BL/6 background and DO11.10 mice in a BALB/c background were maintained at the Life and Health and Sciences Research Institute. All protocols were performed according to the European Union Directive 86/609/EEC and previously approved by the Portuguese authority Direção Geral de Veterinária.
Culture of Bone Marrow-derived DC
5 × 106 bone marrow cells were seeded per well in 6-well plates in DMEM supplemented with 10% FBS, 1% HEPES, 1% sodium pyruvate (Invitrogen), and 20% of GM-CSF and incubated at 37 °C in 5% CO2. On days 2 and 4, the medium was renewed. On day 6, non-adherent cells were collected and resuspended in fresh medium with 20% GM-CSF, and 2.5 × 106 cells/well were cultured overnight. On day 7, cultures were stimulated with different doses of LAM or hLM (0–100 μg/ml), with LPS (25 ng/ml) or left unstimulated as controls, and RNA and supernatants were harvested 6 and 24 h thereafter, respectively. EC50 was calculated using a non-linear fit regression in the dose-response curves.
Co-cultures of CD4+ T Cells and DC
Naive CD4+ T cells were purified from spleens of OVA(323–339)-TCR transgenic DO.11.10 mice using magnetic CD4+ beads (GK1.5, Miltenyi Biotec). These cells (1 × 106 cells/ml) were co-cultured with BALB/c DC (5 × 105 cells/ml) in the presence of LAM or hLM (10 μg/ml) and peptide OVA(323–339) (2.5 μm) for 7 days at 37 °C in 5% CO2. CD4+ T cells (1 × 106 cells/ml) were recovered on day 7 and co-cultured with DC for an additional round of differentiation with fresh BALB/c DC (5 × 105 cells/ml) and LAM or hLM for an additional 3 days at 37 °C in 5% CO2. At the end of the culture, CD4+ T cells were recovered and restimulated in the presence of PMA (50 ng/ml) and ionomycin (500 ng/ml) for 4 h and in the presence of brefeldin A (40 μg/ml) for the last 2 h. The frequency of OVA(323–339)-specific, IFN-γ- or IL-17-producing CD4+ T cells was determined by flow cytometry.
In Vivo Model of Peritoneal Challenge
Groups of five mice were injected intraperitoneally with 50 or 400 μg/mouse of LAM or hLM and sacrificed at specific time points postchallenge. Peritoneal cells were harvested with cold PBS, counted, and processed for flow cytometry or RNA extraction and real-time PCR analysis. Resident PECs were harvested from unstimulated BL/6 or TLR2−/− mice and stimulated in vitro for 3 or 6 h with LAM and hLM (10 μg/ml). Cytokine mRNA quantification was performed by real-time PCR.
Real-time PCR Analysis
Total RNA was extracted with TRIzol® (Invitrogen), and reverse transcription was performed using the ReverteAidTM H Minus First Strand cDNA synthesis kit (Fermentas). The cDNA was then subjected to real-time PCR for quantification of IL-12p40, IL-12p35, IL23p19, TNF-α, IL-6, IL-10, CXCL2, and ubiquitin (supplemental Table 1) with SYBR Green supermix (Bio-Rad) in a CFX 96 real-time (Bio-Rad) system. The quantification of the target genes was analyzed using the CFX 96 real-time software and normalized against mouse ubiquitin mRNA.
Flow Cytometry Analysis
Cell surface markers were analyzed using specific antibodies against Ly6G (APC, 1A8; BioLegend), GR1 (PercP-Cy5.5, RB6–8C5; BioLegend), and CD11b (phycoerythrin, M1/70; eBioscience) for identification of neutrophils or against CD4 (eFluor450, RM4–5; eBioscience) for identification of CD4+ T cells. For intracellular detection of cytokines, specific antibodies for IL-17 (phycoerythrin, eBio17B7; eBioscience) and IFN-γ (FITC, XMG1.2; eBioscience) or isotype control antibodies (BioLegend) were used. Fluorescence was analyzed using an LSRII flow cytometer and FACSDiva Software (BD Bioscience) and data with FlowJo software (TreeStar).
ELISA
Supernatants from LAM- or hLM-stimulated DC were screened for IL-12p40, IL-23, TNF-α, IL-6, IL-12p70, and IL-10 by Ready Set-Go ELISA kits (eBioscience).
Statistical Analysis
One-way ANOVA post hoc Tukey's t test was used with a confidence interval of 95% when multiple comparisons were evaluated. Two-way ANOVA was used in kinetic evaluations. Data were expressed as means ± S.E.; p < 0.05 was considered statistically significant.
RESULTS
Construction and Growth of C. glutamicum ΔaftE
In Corynebacterineae, like M. tuberculosis and C. glutamicum, α(1→6) mannosyltransferases MptA (Rv2174/NCgl2093) and MptB (Rv1459c/NCgl1505) catalyze the α(1→6)-Manp transfer to distal and proximal parts of the LM backbone (32, 34), and MptC (Rv2181/NCgl2100) adds singular α(1→2)-Manp on linear LM (35, 36). In C. glutamicum, two other glycosyltransferases are present in tandem with MptC; whereas the role of MptD (NCgl2097) has been established as a second α(1→2) mannopyranosyltransferase involved in mannan branching of LM and LAM (35), the role of the third glycosyltransferase, NCgl2096, remains to be identified (supplemental Fig. 1a). The NCgl2096-encoded protein (referred to as AftE hereafter) consists of eight transmembrane helices and two negatively charged loops (6 Asp, 2 Glu) (37), probably required for glycosyltransferase activity as observed for the arabinosyltransferase Emb (supplemental Fig. 1b) (38). Furthermore, BLAST analysis suggests close similarity to mycobacterial mannopyranosyltransferases MptC and PimE and arabinosyltransferases EmbA, EmbB, and EmbC, suggesting it to be a glycosyltransferase.
To further understand the function of AftE, a non-replicative plasmid pK19mobsacBΔaftE was constructed consisting of 13 nucleotides of the 3′-end of aftE together with genomic upstream sequences and 36 nucleotides of the 5′-end together with genomic downstream sequences. This vector was used to transform C. glutamicum to kanamycin resistance (Kanr), indicating chromosomal integration. Subsequently, sucrose resistance (Sucr) clones were selected in a second round of positive selection, indicating loss of the vector-bound sacB function (30). From 12 Kans and Sucr clones analyzed via PCR, seven had lost aftE, whereas in the remaining five, the wild type phenotype was restored. One of the representative clones with deleted aftE was selected and is referred to here as C. glutamicum ΔaftE (supplemental Fig. 2a). In addition, aftE was cloned in a shuttle vector pEKEx2 and electroporated into C. glutamicum ΔaftE to achieve the complemented strain, C. glutamicum ΔaftE pEKEx2-Cg-aftE. C. glutamicum ΔaftE was grown in liquid CGXII and BHI media, and its growth pattern was studied. C. glutamicum ΔaftE had a slightly reduced growth rate of 0.32 ± 0.01 h−1 in comparison with 0.39 ± 0.02 h−1 of wild type C. glutamicum on CGXII (supplemental Fig. 2b). A similar trend was seen on BHI medium, suggesting that AftE is not vital for C. glutamicum but nevertheless is required to ensure optimal growth. Such trends have been observed previously for other glycosyltransferases involved in cell wall synthesis like Emb, MptA, and AftA (32, 39, 40).
Purification and General Features of Lipoglycans
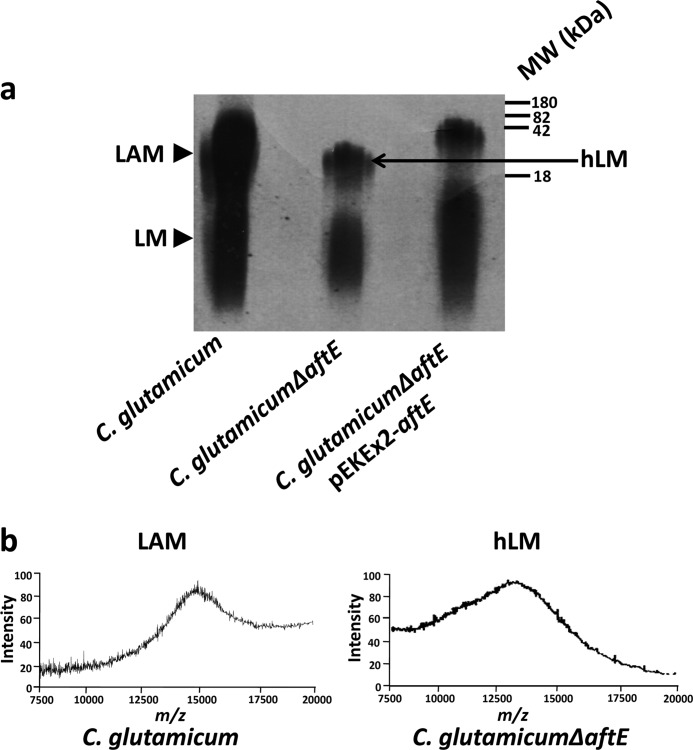
Lipoglycans from C. glutamicum, C. glutamicum ΔaftE, and C. glutamicum ΔaftE pEKEx2-Cg-aftE were extracted from delipidated cells as described previously (31, 35). The crude lipoglycan extract was subjected to hydrophobic interaction chromatography followed by gel permeation chromatography for separation of LM and LAM, which were examined on 15% SDS-PAGE (Fig. 1a and supplemental Fig. 3a). Crude extracts from C. glutamicum showed the presence of LM (a mixture of phosphate-myo-inositol-anchored Cg-LM-A and glucouronic acid-anchored Cg-LM-B with identical mannan backbone (34, 41)) and LAM, whereas lipoglycans from C. glutamicum ΔaftE showed subtle differences in their migration on SDS-PAGE (Fig. 1a). The lipoglycan fraction from C. glutamicum ΔaftE consisted of two species, one with higher molecular weight lipoglycan migrating faster than LAM from wild type and another lower molecular weight lipoglycan migrating similar to LM from wild type (Fig. 1a). Furthermore, MALDI-TOF-MS of the lipoglycans showed that LAM from wild type C. glutamicum exhibited a broad unresolved peak centered at m/z 15,000, indicating a molecular mass of ∼15 kDa with major molecular species consisting of 87 sugar units (based on the molecular mass of Ac1PIM2 with three sugar units, m/z 1398). The MALDI-TOF-MS of higher molecular weight lipoglycan from C. glutamicum ΔaftE showed an unresolved peak that centered around m/z 13,000, suggesting the loss of ∼13 sugar units (Fig. 1b). However, there were no major differences between LM from C. glutamicum and smaller molecular weight lipoglycan from C. glutamicum ΔaftE (data not shown). The presence of a smaller molecular weight lipoglycan, which is similar to LM, and a higher molecular weight lipoglycan, which is shorter than the wild type LAM in C. glutamicum ΔaftE, suggested that AftE is involved in the biosynthesis of LAM and not LM. Complementation of C. glutamicum ΔaftE by transformation with plasmid pEKEx2-Cg-aftE restored the wild type phenotype (Fig. 1a).
FIGURE 1.
Lipoglycan profiles of C. glutamicum, C. glutamicum ΔaftE, and C. glutamicum ΔaftE pEKEx2-aftE. a, lipoglycans were extracted as described under “Experimental Procedures,” analyzed using SDS-PAGE, and visualized by Pro-Q emerald glycoprotein stain (Invitrogen). Lipoglycan profiles are represented with standard molecular mass markers of glycoproteins of 180, 82, 42, and 18 kDa. b, MALDI-TOF-MS spectra of LAM from C. glutamicum and hLM from C. glutamicum ΔaftE. For this, 2,5-dihydrobenzoic acid was used as a matrix, and the spectra were acquired in the linear negative mode with delayed extraction. Lipoglycans were extracted from delipidated cells as described previously (31, 32) and treated with H2O2 before immunological assays to remove any endotoxin contamination (13) and to yield a negative Limulus amebocyte lysate test using E-TOXATETM (Sigma-Aldrich).
Molecular Composition and Linkage Analysis of Lipoglycans
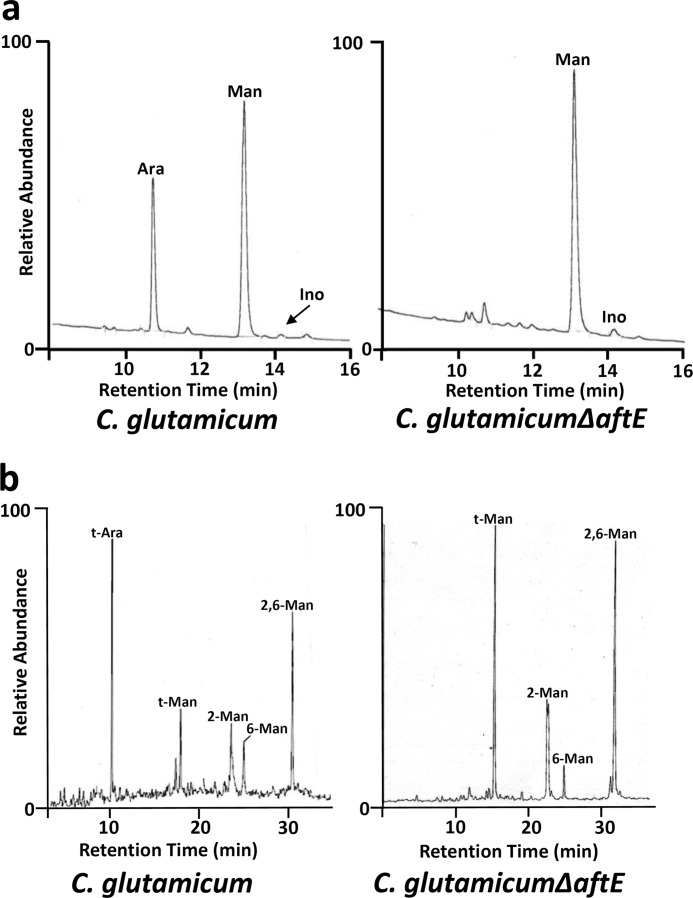
Purified lipoglycans were processed and converted into alditol acetates, and their glycosyl composition was determined by gas chromatography (GC). Based on 1 inositol per molecule of LAM, the molecular composition of LAM from wild type C. glutamicum was determined. It yielded a ratio of 1 inositol, 20 arabinose, and 65 mannose residues, which is in accordance with our previous studies (Fig. 2a) (31, 35). In addition, LM from wild type C. glutamicum consists of 28 sugars with a molecular ratio of 1 inositol (for Cg-LM-A) or 1 glucouronic acid (for Cg-LM-B) over 27 mannose units (32, 41). This observation, which was overlooked in previous studies, suggests that in comparison with LM, LAM has a larger mannan domain made up of ∼65 mannose residues, which might be due to extended activities of MptA, MptC, and MptD (35). Surprisingly, higher molecular weight lipoglycan from C. glutamicum ΔaftE showed a complete absence of the arabinan peak and a ratio of 1 inositol to 74 mannoses, a reduction of 13 sugar units in comparison with LAM from wild type C. glutamicum (Fig. 2a). The complete absence of the arabinan peak should have resulted in the loss of 20 arabinose units instead of 13 units, suggesting that the loss of arabinan units was compensated by some additional mannosylation of LAM intermediate, probably by MptC and MptD. This may be due to a greater availability of branching sites generated due to deletion of aftE. Due to the absence of arabinose and hypermannosylation, this molecule is here referred to as hypermannosylated lipomannan (hLM).
FIGURE 2.
Glycosyl composition and linkage analysis of lipoglycans from C. glutamicum and C. glutamicum ΔaftE. a, for glycosyl compositional analysis, purified lipoglycans were hydrolyzed, reduced, and per-O-acetylated as described under “Experimental Procedures.” Resulting alditol acetates were subjected for GC analysis. b, for determining the sugar linkages present in these lipoglycans species, lipoglycans were per-O-methylated, hydrolyzed, reduced, and per-O-acetylated. The resulting partially per-O-methylated, per-O-acetylated alditol acetates were analyzed using GC-MS. The partially per-O-methylated, per-O-acetylated alditol acetates were identified based on their characteristic alditol cleavage acetal profiles: t-Ara (m/z 118, 161), t-Man (m/z 118, 102, 145, 161, 162, 205), 2-Man (m/z 129, 130, 161, 190), 6-Man (m/z 118, 129, 162, 189, 233), and 2,6-Man (m/z 129, 130, 189, 190).
The glycosyl linkages present in each species of lipoglycans were analyzed by gas chromatography-mass spectrometry (GC-MS) by per-O-methylated alditol acetate derivatives prepared from purified lipoglycans. LAM from wild type C. glutamicum indicated a normal profile of glycosidic linkages corresponding to t-Ara (34%), t-Man (15%), 6-Man (10%), 2-Man (17%), and 2,6-Man (24%). However, mutant hLM from C. glutamicum ΔaftE was found to be completely devoid of t-Ara branching residues and possessed a significant increase in the amount of t-Man (39%), 6-Man (5%), 2-Man (18%), and 2,6-Man (38%) (Fig. 2b). Linkage analysis of LM from C. glutamicum ΔaftE suggested a slightly increased branching degree, ∼87% in comparison with 75% of LM from wild type C. glutamicum (data not shown).
In conclusion, by deleting AftE, we obtained a hypermannosylated intermediate of lipoglycans, devoid of singular arabinan (supplemental Fig. 3b). Next we investigated whether the biological activity of this structure is different from that of LAM from wild type C. glutamicum, particularly in what concerns the modulation of immune responses via TLR2. We focused our study on wild type LAM versus hLM, because these two molecules differ in arabinan branching and degree of mannosylation. Furthermore, the LM from wild type C. glutamicum is dominated by monoacylated glucouronic acid-anchored LM (>90%) and has a poor TLR2 activity and therefore has not been included in this study (data not shown).
Arabinose Branching of LM Impacts T Helper Cell Differentiation
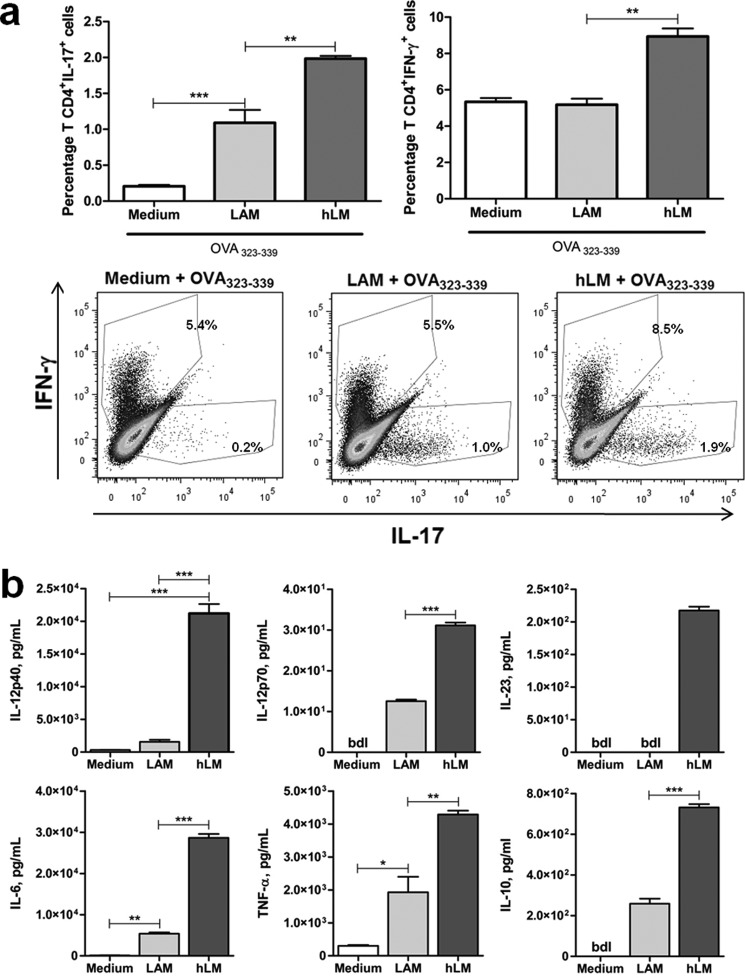
Several factors determine the polarization of Th cell differentiation, including the engagement of specific TLRs (42). The impact of structural variations of LAMs in CD4+ Th cell differentiation has not been addressed before. Taking advantage of the purified wild type LAM and hLM from C. glutamicum and C. glutamicum ΔaftE, respectively, we addressed this issue. For this, primary mouse bone marrow-derived DC were co-cultured with purified CD4+ T cells, from OVA TCR transgenic DO11.10 mice (BALB/c background), in the presence of LAM or hLM and of OVA peptide (OVA(323–339), the antigen recognized by the transgenic TCR in DO11.10 CD4+ T cells). The intracellular cytokine profile of the differentiated CD4+ T cells obtained at the end of the co-culture was evaluated by flow cytometry. Because both Th1 and Th17 have been described in infections with mycobacteria, we focused our analysis on IFN-γ- and IL-17-producing T cells. Both LAM and hLM stimulated CD4+ T cell differentiation into IL-17-producing cells, although in the presence of hLM the percentage of IL-17-producing CD4+ T cells reached significantly higher values (Fig. 3a). Also, hLM, but not LAM, increased the percentage of CD4+ T cells differentiating into Th1 cells producing IFN-γ, although this was a modest effect (Fig. 3a). The differences observed were most likely not due to different proliferation rates of Th cells, because the number of cells recovered at the end of culture was similar to all conditions (supplemental Fig. 4).
FIGURE 3.
hLM is a stronger inducer of Th17 cells than LAM. Naive CD4+ T cells from DO11.10 mice were co-cultured with BALB/c DC in the presence of 2.5 μm peptide OVA(323–339) and/or 10 μg of C. glutamicum LAM or hLM for 10 days. a, graphs and dot plots of FACS data of IL-17 and IFN-γ production by differentiated CD4+ T cells restimulated with PMA and ionomycin in the presence of brefeldin A. Each dot plot and column represents the mean of three wells. b, cytokine production by DC generated from BALB/c mice and stimulated for 24 h with LAM or hLM (10 μg/ml) or left unstimulated (Medium) was evaluated by ELISA. In all cases, results are from one representative of two independent experiments. Each column represents the mean ± S.E. (error bars) of three wells. Statistical significance was calculated with one-way ANOVA post hoc Tukey's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). bld, below detection level.
The differentiation of Th cells is controlled in part by the cytokine environment generated by the antigen-presenting cell. Indeed, the presence of IL-12 (composed of p40 and p35 monomers) is essential for the development of Th1 responses (43), whereas IL-6 together with TGF-β in the microenvironment promotes Th17 differentiation (44). Th17 cells also require as a survival factor IL-23, a heterodimer of p40 and p19 (44). Following our previous findings (Fig. 3a), we sought to understand whether the differential impact of LAM and hLM in Th1 and Th17 cell differentiation was due to the cytokine profile induced by these molecules in DC. To test our hypothesis, we stimulated mouse bone marrow-derived DC (of BALB/c origin) with LAM or hLM and measured the production of IL-12, IL-23, and IL-6 24 h poststimulation. As shown in Fig. 3b, hLM is a stronger inducer of each of these cytokines, which is in line with its effect on Th cell differentiation. Of note, the same pattern was observed for the induction of TNF and IL-10 (Fig. 3b). As expected, unstimulated DC produced only negligible to non-detected cytokine amounts (Fig. 3b). Our findings are in agreement with previously published results, reinforcing the idea that branching of LM and the addition of arabinose residues contribute to a more controlled cellular response in terms of cytokine production (13–15). However, we now additionally implicate variations on lipoglycan structures on the outcome of Th immune responses.
Activation of DC by LAM and hLM Is TLR2-mediated
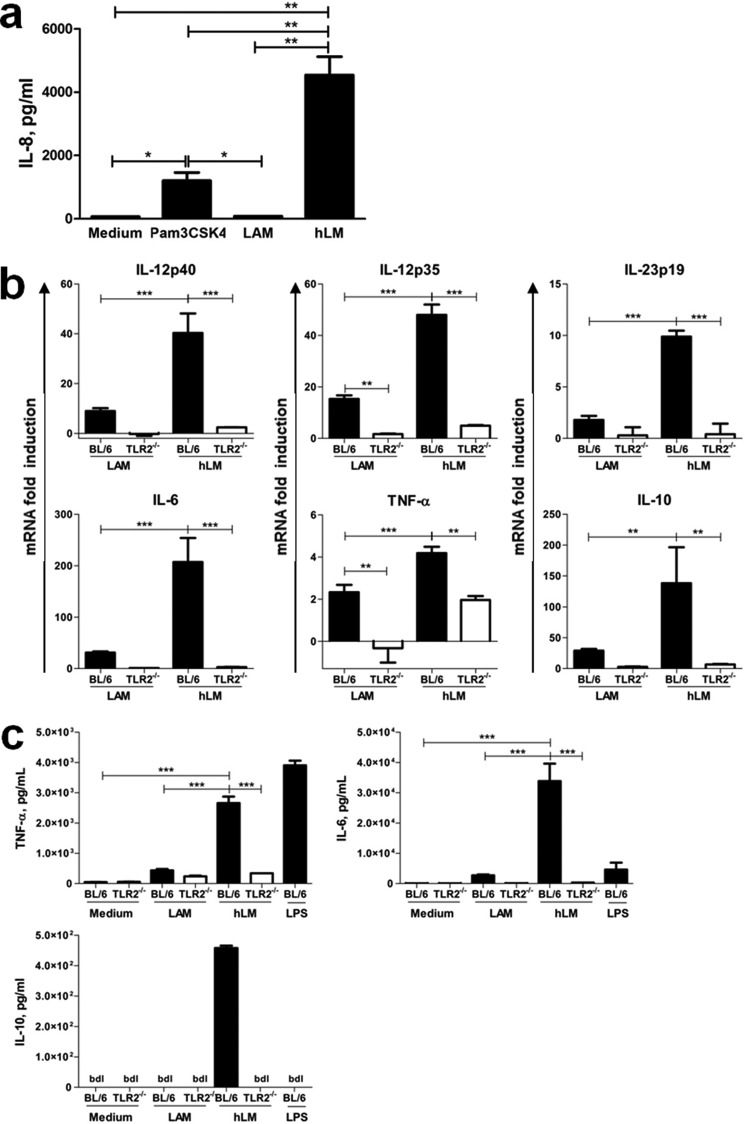
Next we investigated if TLR2 activation was implicated on the differential response of DC to LAM and hLM depended on TLR2 activation. For that, we started by measuring the response of human HEK cells transfected with TLR2 to LAM and hLM. We observed that hLM, as compared with LAM, induced a significantly increased activation of TLR2, as demonstrated by the enhanced TLR2-dependent IL-8 production by the HEK cells (Fig. 4a). Of note, hLM was also more potent in inducing IL-8 than the TLR2 agonist Pam3CSK4 (Fig. 4a). We then stimulated bone marrow-derived DC, generated from WT (BL6) or TLR2-deficient mice, with LAM or hLM and at different time points measured the transcription and production of IL-12p40, p35, p19, and IL-6 as well as of TNF and IL-10. Both transcription and secretion of all cytokines tested were higher upon hLM stimulation of DC than upon LAM stimulation (Fig. 4, b and c). As expected, DC responded to LPS stimulation with the production of several cytokines (Fig. 4c). Furthermore, TLR2 mediated the recognition of both LAM and hLM, because cytokine expression and secretion by stimulated DC was clearly decreased in its absence (Fig. 4, b and c). Of note, PRRs other than TLR2 must also be involved in the recognition of LAM and hLM, because, in the absence of TLR2, cytokine expression was not fully abrogated (Fig. 4, b and c). Interestingly, the increased potency of hLM to activate DC was not only reflected in the expression of proinflammatory cytokines, but also in the expression and production of the anti-inflammatory molecule IL-10 (Fig. 4, b and c). For both IL-6 and TNF, we also determined the response of WT DC to increasing doses of LAM or hLM. For each dose tested, the stimulatory activity of hLM was higher than that of LAM (supplemental Fig. 5). Moreover, a clear difference in the EC50 of the two molecules was observed (285.1 ± 2.0 μg/ml versus 10.7 ± 1.2 μg/ml in the case of TNF and 114.3 ± 1.4 μg/ml versus 15.6 ± 1.1 μg/ml in the case of IL-6, for LAM versus hLM, respectively). These findings suggest that changes on LAMs are discriminated by TLR2, with the stronger induction of TLR2 via hLM impacting the various signaling pathways triggered by this receptor and not only on those leading to proinflammatory cytokines.
FIGURE 4.
Cytokine production induced by LAM or hLM in vitro is TLR2-mediated. a, human HEK293 cells transfected with TLR2 were stimulated with 50 μg/ml LAM or hLM. Non-stimulated cells (Medium) or cells stimulated with 50 ng/ml Pam3CSK4 served as a controls. After 24 h at 37 °C, supernatants were harvested and analyzed for IL-8 using ELISA. b, -fold induction of mRNA expression of IL-12p40, IL-12p35, IL-23p19, IL-6, TNF-α, and IL-10 was evaluated by real-time PCR after 6 h of LAM or hLM stimulation (10 μg/ml) of 1 × 106 BL/6 or TLR2−/− DC, relative to non-stimulated cells. Results are from one representative of two independent experiments. c, production of IL-6, TNF-α, and IL-10 was evaluated by ELISA after 24 h of LAM, hLM (both at 10 μg/ml), or LPS stimulation (25 ng/ml) of 1 × 106 BL/6 and TLR2−/− DC. Unstimulated DC (Medium) were included as a control. In all cases, results are from one representative of two independent experiments. Each column represents the mean ± S.E. of three wells. Statistical significance was calculated with one-way ANOVA post hoc Tukey's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). bld, below detection level.
hLM Is a Stronger Inducer of in Vivo Immune Responses
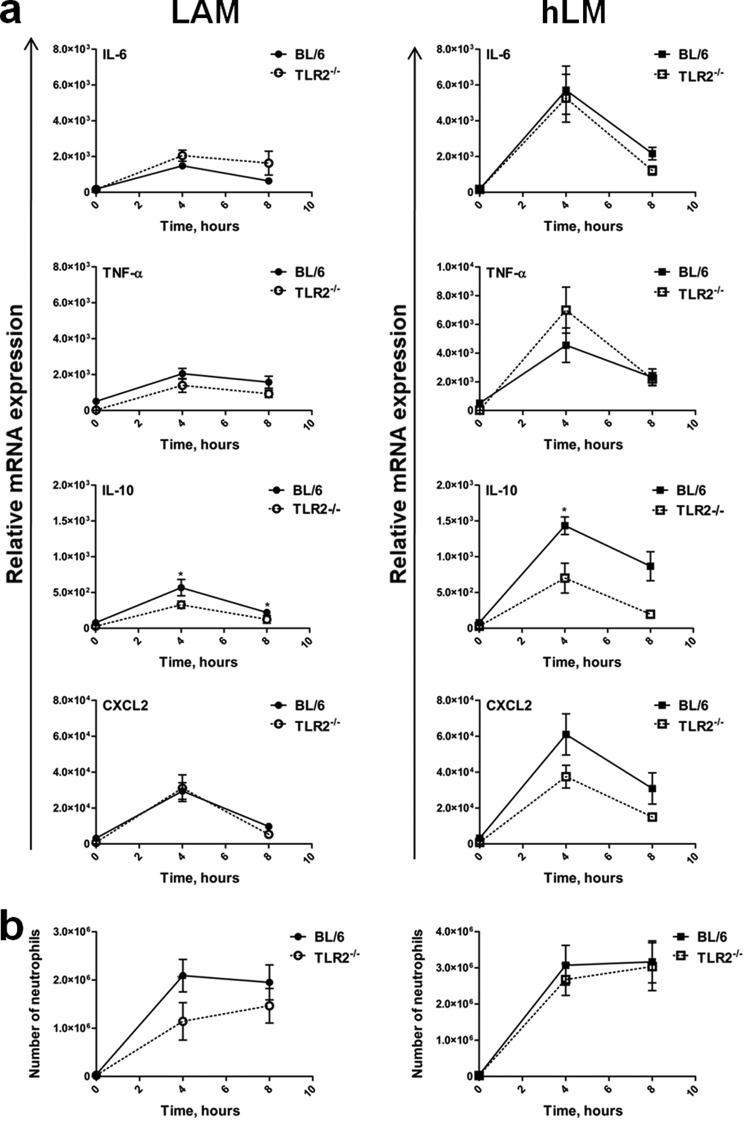
Considering our in vitro data showing that hLM is a stronger inducer of DC responses, with an impact on Th cell differentiation, we next investigated the effect of LAM versus hLM during in vivo immune responses. For this, we injected mice (WT versus TLR2−/−) intraperitoneally with purified LAM or hLM and measured, at different times postinjection, cytokine production at the site of stimulation. Total peritoneal cells were harvested, their RNA was extracted, and the relative mRNA expression of several cytokines was quantified by real-time PCR. In contrast to what was observed in vitro upon DC stimulation with LAM or hLM, in vivo the majority of the molecules tested were induced in a TLR2-independent way. In fact, the absence of TLR2 did not compromise TNF, IL-6, or CXCL2 (the mouse homologue of human IL-8) expression induced by LAM or hLM (Fig. 5a). Lack of TLR2 did, however, contribute to a significant decrease in IL-10 induced in vivo by LAM or hLM (Fig. 5a). Of note, as compared with LAM, hLM induced a higher expression of the tested proinflammatory cytokines and also of the anti-inflammatory cytokine IL-10 and the chemokine CXCL2 (supplemental Fig. 6a). To investigate if the profile of TLR2-independent in vivo responses observed for LAM versus hLM was mainly due to the recruitment of new cells or was a feature of the resident PECs, we collected peritoneal cells from non-stimulated mice, activated them in vitro for 3 or 6 h with LAM or hLM, and analyzed the expression of cytokines in response to this acute stimulation. We found that hLM induced stronger responses than LAM by resident PECs and that, with the exception of IL-10, these responses were TLR2-independent (supplemental Fig. 7).
FIGURE 5.
In vivo cytokine production induced in the peritoneal cavity by LAM and hLM injections requires recognition by PRRs other than TLR2. LAM and hLM (50 μg in 100 μl) were intraperitoneally injected into BL/6 WT or TLR2−/− mice, and PECs were collected 0, 4, and 8 h postinjection. a, mRNA was extracted from 5 × 105 PECs/animal, and IL-6, TNF-α, IL-10, and CXCL2 mRNA expression was evaluated by RT-PCR. The data points represent the mean ± S.E. (error bars) of five animals. b, the number of neutrophils was calculated by taking into account the percentage of neutrophils determined by flow cytometry (Ly6G+GR1+CD11b+) and the total number of cells collected. The data points represent the mean ± S.E. of five animals. Results are from one representative of two independent experiments. Statistical significance was calculated with a two-way ANOVA test with 95% confidence (*, p < 0.05).
Considering that we had detected high IL-8 production by HEK cells stimulated with hLM (Fig. 4a) and also high CXCL2 in our in vivo model upon hLM administration (supplemental Fig. 6a), and because IL-8 is a molecule involved in neutrophil recruitment, we investigated changes in the numbers of neutrophils recruited upon LAM or hLM injection. Although both LAM and hLM induced an increase in the total number of recruited neutrophils throughout time, this increase was significantly more pronounced (∼50% more) in the case of hLM (supplemental Fig. 6b). Furthermore, this increase in neutrophil recruitment was independent of TLR2 (Fig. 5b). Of note, increasing the dose of administered LAM or hLM did not affect the differences observed (i.e. hLM was still a higher inducer of neutrophil recruitment than LAM) (supplemental Fig. 6c). The intraperitoneal injections of either LAM or hLM in BL/6 mice did not influence the total number of cells collected from the peritoneum at any of the time points tested (supplemental Fig. 6d), suggesting that another cellular population was decreased as the influx of neutrophils happened.
Collectively, our data suggest that hLM is a stronger inducer of in vivo and in vitro immune responses than LAM. Although in DC cultures, the effect of both LAM and hLM is mainly mediated by TLR2, additional PRRs to TLR2 must differentially recognize these structures in an in vivo setting.
DISCUSSION
TLRs are among the best studied PRRs, with a recognized role in protecting the host against infection but also in contributing to acute and chronic inflammation, as well as to cancer, if inappropriately activated (45). The need for an appropriate regulation of TLR responses calls for a deep understanding of the structural interaction of TLRs with their ligands. We are particularly interested in understanding the structure-function relationship between TLR2 and lipoglycans, such as LM and LAM. A growing body of evidence strongly suggests that the structure of LM and LAM affects their interaction with TLR2, thus modulating the outcome of the innate immune response (11–15). Understanding the details of these interactions is essential in the context of various infections, including those by M. tuberculosis that represent the second most common infectious cause of mortality and morbidity (46). Importantly, although still controversial, a role for TLR2 has been described during primary and secondary M. tuberculosis infection (18, 19, 22, 23, 25, 47), and polymorphisms in the tlr2 gene have been associated with susceptibility to tuberculosis (48).
Although the structural interactions of TLR2 with lipoprotein ligands have been addressed before by x-ray crystallography (3, 4), those mediating the interaction of TLR2 with lipoglycans are less studied. The detailed study of structural-functional interactions of lipoglycans and TLR2 requires the availability of different structural variants and mimics of lipoglycans. In M. tuberculosis, most of the genetic loci encoding for cell wall biogenesis are essential, which limits the availability of these molecules with variable structures. For this reason, we have in the past successfully used C. glutamicum as a “proof of principle” to understand the biosynthesis of LM and LAM in mycobacteria (8, 32, 34, 35, 49). The biosynthetic pathway of LAM is complex, involving a series of enzymes that coordinately add complexity to the growing lipoglycan (8). Herein, we identified a novel putative glycosyltransferase, AftE, from C. glutamicum, which adds singular arabinose branches on LAM intermediate, resulting in a complete structure of LAM. Biochemical analysis of higher molecular weight lipoglycan from C. glutamicum ΔaftE produced a shorter mutant lipoglycan, hLM, in comparison with LAM from wild type. The glycosyl composition and linkage analysis of hLM suggested complete absence of arabinan or t-Ara. Further biochemical analysis suggested that hLM is a novel lipoglycan with an extended mannose backbone with higher α(1→2)-Man units probably due to the hyperaction of MptC and MptD on branching sites created due to the absence of t-Ara residues in C. glutamicum ΔaftE. Cumulatively, our study sheds light on the synthesis of an additional lipomannan-like lipoglycan, hLM, which may be either an aberrant LM accumulating as a result of lack of arabinosylation or a precursor of LAM before arabinosylation. Presumably, mature LM with mannophosphatidylinositol anchor undergoes further mannosylation in the α(1→6) direction to yield branching sites for α(1→2)-Man and α(1→2)-Ara. There is high probability that one or both of the α(1→6) mannopyranosyltransferases, MptA and MptB, add further α(1→6)-Man units to the mature LM (32, 34), providing further branching sites for arabinose and mannose units for LAM (35).
We therefore generated two different structural variants of LAM, useful for studying the structural-functional relationship between this class of pathogen-associated molecular pattern and TLR2. Our findings show that, independently of the dose tested, hLM significantly induced higher cytokine secretion by DC cultures than LAM. Moreover, hLM was also more efficient than LAM in activating DC, as reflected by its lower values of EC50.
Furthermore, hLM was also a more potent inducer of in vivo responses than LAM, in a model of intraperitoneal injection. Our findings clearly implicate TLR2 as the main PRR involved in in vitro DC recognition of both LAM and hLM, thus suggesting that the structure of lipoglycans, in particular the extent of arabinan branching, influences their interaction with TLR2 and modulates DC responses. These findings are in agreement with previously published studies (13–15). We also provide evidence that, as a result of differential DC activation by LAM and hLM, a Th17-biased T cell response was generated in response to a microenvironment rich in hLM. It will be important in the future to investigate the mechanistic basis for the stronger hLM-induced TLR2-mediated response observed in DC. It is possible that hLM interacts with TLR2 with a higher affinity than LAM or that it induces the recruitment of more intracellular signaling adaptors. It will be interesting to pursue these studies to further understand how TLR2 ligands other than lipoproteins interact with TLR2 and initiate TLR2-mediated immune responses. Interestingly, our data suggest that DC may respond to LAM in a stronger way than HEK cells transfected with TLR2. This finding may provide some hints about the way lipoglycans interact with TLR2. It is tempting to speculate that LAM may preferentially interact with TLR2-TLR1 or TLR2-TLR6 heterodimers (absent in HEK cells expressing TLR2 but existing in DC), whereas hLM may have the ability to trigger cellular responses via TLR2 homodimers.
Our findings show that, similarly to what was observed in vitro, in vivo, lack of arabinose branching correlates with enhanced immune responses. However, in contrast to what was observed for DC cultures, in vivo, other PRRs in addition to TLR2 are certainly involved in the differential recognition of lipoglycans. It is possible that in our in vivo model, cells other than DC are the major sensors for LAM and hLM, thus explaining the difference observed in DC cultures and in the peritoneal cavity. This issue remains to be investigated, with cells expressing DC-SIGN and mannose receptors being good candidates for the recognition of LAM and hLM (50, 51). Further studies will be of interest and important to clarify both of these issues: which cells and which PRRs recognize and differentiate lipoglycan structural variants. Of note, acute stimulation of resident PECs with LAM or hLM induced cytokine expression independently of TLR2, suggesting that at least peritoneal macrophages differ from DC in their way of recognizing and responding to lipoglycans. Neutrophils are also good candidate cells, because we observed an influx of neutrophils to the peritoneal cavity upon LAM or hLM administration. Our data open avenues for the modulation of innate and acquired immune responses by taking advantage of different LAM structures. This is particularly important in the context of infections by M. tuberculosis, where an appropriate balance of Th1 versus Th17 cell responses is critical for bacterial control both in primary infection and vaccination and for pathology prevention (52, 53). It is possible that, by modulating TLR2 signals, a faster and more potent Th17 response is achieved, which has been shown to be fundamental for efficient vaccination (53).
Acknowledgments
We acknowledge scientific discussions with Drs. Anne O'Garra, Luke Alderwick, and Apoorva Bhatt.

This article contains supplemental Table 1 and Figs. 1–7.
- DC
- dendritic cells
- LM
- lipomannan
- hLM
- hypermannosylated lipomannan
- LAM
- lipoarabinomannan
- PRR
- pattern recognition receptor
- Th
- T helper
- TLR
- Toll-like receptor
- PEC
- peritoneal exudate cell
- BL/6
- C57BL/6
- ANOVA
- analysis of variance
- Ara
- arabinose
- Man
- mannose.
REFERENCES
- 1. Medzhitov R. (2007) Recognition of microorganisms and activation of the immune response. Nature 449, 819–826 [DOI] [PubMed] [Google Scholar]
- 2. Kawai T., Akira S. (2010) The role of pattern-recognition receptors in innate immunity. Update on Toll-like receptors. Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 3. Jin M. S., Kim S. E., Heo J. Y., Lee M. E., Kim H. M., Paik S. G., Lee H., Lee J. O. (2007) Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 4. Kang J. Y., Nan X., Jin M. S., Youn S. J., Ryu Y. H., Mah S., Han S. H., Lee H., Paik S. G., Lee J. O. (2009) Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31, 873–884 [DOI] [PubMed] [Google Scholar]
- 5. Krishna S., Ray A., Dubey S. K., Larrouy-Maumus G., Chalut C., Castanier R., Noguera A., Gilleron M., Puzo G., Vercellone A., Nampoothiri K. M., Nigou J. (2011) Lipoglycans contribute to innate immune detection of mycobacteria. PLoS One 6, e28476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pitarque S., Larrouy-Maumus G., Payré B., Jackson M., Puzo G., Nigou J. (2008) The immunomodulatory lipoglycans, lipoarabinomannan and lipomannan, are exposed at the mycobacterial cell surface. Tuberculosis 88, 560–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Briken V., Porcelli S. A., Besra G. S., Kremer L. (2004) Mycobacterial lipoarabinomannan and related lipoglycans. From biogenesis to modulation of the immune response. Mol. Microbiol. 53, 391–403 [DOI] [PubMed] [Google Scholar]
- 8. Mishra A. K., Driessen N. N., Appelmelk B. J., Besra G. S. (2011) Lipoarabinomannan and related glycoconjugates. Structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol. Rev. 35, 1126–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nigou J., Gilleron M., Puzo G. (2003) Lipoarabinomannans. From structure to biosynthesis. Biochimie 85, 153–166 [DOI] [PubMed] [Google Scholar]
- 10. Torrelles J. B., Sieling P. A., Arcos J., Knaup R., Bartling C., Rajaram M. V., Stenger S., Modlin R. L., Schlesinger L. S. (2011) Structural differences in lipomannans from pathogenic and nonpathogenic mycobacteria that impact CD1b-restricted T cell responses. J. Biol. Chem. 286, 35438–35446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Quesniaux V. J., Nicolle D. M., Torres D., Kremer L., Guérardel Y., Nigou J., Puzo G., Erard F., Ryffel B. (2004) Toll-like receptor 2 (TLR2)-dependent-positive and TLR2-independent-negative regulation of proinflammatory cytokines by mycobacterial lipomannans. J. Immunol. 172, 4425–4434 [DOI] [PubMed] [Google Scholar]
- 12. Gilleron M., Nigou J., Nicolle D., Quesniaux V., Puzo G. (2006) The acylation state of mycobacterial lipomannans modulates innate immunity response through Toll-like receptor 2. Chem. Biol. 13, 39–47 [DOI] [PubMed] [Google Scholar]
- 13. Birch H. L., Alderwick L. J., Appelmelk B. J., Maaskant J., Bhatt A., Singh A., Nigou J., Eggeling L., Geurtsen J., Besra G. S. (2010) A truncated lipoglycan from mycobacteria with altered immunological properties. Proc. Natl. Acad. Sci. U.S.A. 107, 2634–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nigou J., Vasselon T., Ray A., Constant P., Gilleron M., Besra G. S., Sutcliffe I., Tiraby G., Puzo G. (2008) Mannan chain length controls lipoglycans signaling via and binding to TLR2. J. Immunol. 180, 6696–6702 [DOI] [PubMed] [Google Scholar]
- 15. Vignal C., Guérardel Y., Kremer L., Masson M., Legrand D., Mazurier J., Elass E. (2003) Lipomannans, but not lipoarabinomannans, purified from Mycobacterium chelonae and Mycobacterium kansasii induce TNF-α and IL-8 secretion by a CD14-Toll-like receptor 2-dependent mechanism. J. Immunol. 171, 2014–2023 [DOI] [PubMed] [Google Scholar]
- 16. Torrelles J. B., Knaup R., Kolareth A., Slepushkina T., Kaufman T. M., Kang P., Hill P. J., Brennan P. J., Chatterjee D., Belisle J. T., Musser J. M., Schlesinger L. S. (2008) Identification of Mycobacterium tuberculosis clinical isolates with altered phagocytosis by human macrophages due to a truncated lipoarabinomannan. J. Biol. Chem. 283, 31417–31428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oliveira-Nascimento L., Massari P., Wetzler L. M. (2012) The role of TLR2 in infection and immunity. Front. Immunol. 3, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bafica A., Scanga C. A., Feng C. G., Leifer C., Cheever A., Sher A. (2005) TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J. Exp. Med. 202, 1715–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drennan M. B., Nicolle D., Quesniaux V. J., Jacobs M., Allie N., Mpagi J., Frémond C., Wagner H., Kirschning C., Ryffel B. (2004) Toll-like receptor 2-deficient mice succumb to Mycobacterium tuberculosis infection. Am J. Pathol. 164, 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jang S., Uematsu S., Akira S., Salgame P. (2004) IL-6 and IL-10 induction from dendritic cells in response to Mycobacterium tuberculosis is predominantly dependent on TLR2-mediated recognition. J. Immunol. 173, 3392–3397 [DOI] [PubMed] [Google Scholar]
- 21. Underhill D. M., Ozinsky A., Smith K. D., Aderem A. (1999) Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc. Natl. Acad. Sci. U.S.A. 96, 14459–14463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reiling N., Hölscher C., Fehrenbach A., Kröger S., Kirschning C. J., Goyert S., Ehlers S. (2002) Cutting edge. Toll-like receptor (TLR)2- and TLR4-mediated pathogen recognition in resistance to airborne infection with Mycobacterium tuberculosis. J. Immunol. 169, 3480–3484 [DOI] [PubMed] [Google Scholar]
- 23. Sugawara I., Yamada H., Li C., Mizuno S., Takeuchi O., Akira S. (2003) Mycobacterial infection in TLR2 and TLR6 knockout mice. Microbiol. Immunol. 47, 327–336 [DOI] [PubMed] [Google Scholar]
- 24. Hölscher C., Reiling N., Schaible U. E., Hölscher A., Bathmann C., Korbel D., Lenz I., Sonntag T., Kröger S., Akira S., Mossmann H., Kirschning C. J., Wagner H., Freudenberg M., Ehlers S. (2008) Containment of aerogenic Mycobacterium tuberculosis infection in mice does not require MyD88 adaptor function for TLR2, -4, and -9. Eur. J. Immunol. 38, 680–694 [DOI] [PubMed] [Google Scholar]
- 25. Teixeira-Coelho M., Cruz A., Carmona J., Sousa C., Ramos-Pereira D., Saraiva A. L., Veldhoen M., Pedrosa J., Castro A. G., Saraiva M. (2011) TLR2 deficiency by compromising p19 (IL-23) expression limits Th 17 cell responses to Mycobacterium tuberculosis. Int. Immunol. 23, 89–96 [DOI] [PubMed] [Google Scholar]
- 26. Kleinnijenhuis J., Oosting M., Joosten L. A., Netea M. G., Van Crevel R. (2011) Innate immune recognition of Mycobacterium tuberculosis. Clin. Dev. Immunol. 2011, 405310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Caws M., Thwaites G., Dunstan S., Hawn T. R., Lan N. T., Thuong N. T., Stepniewska K., Huyen M. N., Bang N. D., Loc T. H., Gagneux S., van Soolingen D., Kremer K., van der Sande M., Small P., Anh P. T., Chinh N. T., Quy H. T., Duyen N. T., Tho D. Q., Hieu N. T., Torok E., Hien T. T., Dung N. H., Nhu N. T., Duy P. M., van Vinh Chau N., Farrar J. (2008) The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 4, e1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alderwick L. J., Birch H. L., Mishra A. K., Eggeling L., Besra G. S. (2007) Structure, function and biosynthesis of the Mycobacterium tuberculosis cell wall. Arabinogalactan and lipoarabinomannan assembly with a view to discovering new drug targets. Biochem. Soc. Trans. 35, 1325–1328 [DOI] [PubMed] [Google Scholar]
- 29. Jankute M., Grover S., Rana A. K., Besra G. S. (2012) Arabinogalactan and lipoarabinomannan biosynthesis. Structure, biogenesis and their potential as drug targets. Fut. Microbiol. 7, 129–147 [DOI] [PubMed] [Google Scholar]
- 30. Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19. Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73 [DOI] [PubMed] [Google Scholar]
- 31. Tatituri R. V., Illarionov P. A., Dover L. G., Nigou J., Gilleron M., Hitchen P., Krumbach K., Morris H. R., Spencer N., Dell A., Eggeling L., Besra G. S. (2007) Inactivation of Corynebacterium glutamicum NCgl0452 and the role of MgtA in the biosynthesis of a novel mannosylated glycolipid involved in lipomannan biosynthesis. J. Biol. Chem. 282, 4561–4572 [DOI] [PubMed] [Google Scholar]
- 32. Mishra A. K., Alderwick L. J., Rittmann D., Tatituri R. V., Nigou J., Gilleron M., Eggeling L., Besra G. S. (2007) Identification of an α(1→6) mannopyranosyltransferase (MptA), involved in Corynebacterium glutamicum lipomanann biosynthesis, and identification of its orthologue in Mycobacterium tuberculosis. Mol. Microbiol. 65, 1503–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Flo T. H., Ryan L., Latz E., Takeuchi O., Monks B. G., Lien E., Halaas Ø., Akira S., Skjåk-Braek G., Golenbock D. T., Espevik T. (2002) Involvement of Toll-like receptor (TLR) 2 and TLR4 in cell activation by mannuronic acid polymers. J. Biol. Chem. 277, 35489–35495 [DOI] [PubMed] [Google Scholar]
- 34. Mishra A. K., Alderwick L. J., Rittmann D., Wang C., Bhatt A., Jacobs W. R., Jr., Takayama K., Eggeling L., Besra G. S. (2008) Identification of a novel α(1→6) mannopyranosyltransferase MptB from Corynebacterium glutamicum by deletion of a conserved gene, NCgl1505, affords a lipomannan- and lipoarabinomannan-deficient mutant. Mol. Microbiol. 68, 1595–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mishra A. K., Krumbach K., Rittmann D., Appelmelk B., Pathak V., Pathak A. K., Nigou J., Geurtsen J., Eggeling L., Besra G. S. (2011) Lipoarabinomannan biosynthesis in Corynebacterineae. The interplay of two α(1→2)-mannopyranosyltransferases MptC and MptD in mannan branching. Mol. Microbiol. 80, 1241–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaur D., Obregón-Henao A., Pham H., Chatterjee D., Brennan P. J., Jackson M. (2008) Lipoarabinomannan of Mycobacterium. Mannose capping by a multifunctional terminal mannosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 105, 17973–17977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hofmann K., Stoffel W. (1993) TMBASE. A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374, 166 [Google Scholar]
- 38. Seidel M., Alderwick L. J., Sahm H., Besra G. S., Eggeling L. (2007) Topology and mutational analysis of the single Emb arabinofuranosyltransferase of Corynebacterium glutamicum as a model of Emb proteins of Mycobacterium tuberculosis. Glycobiology 17, 210–219 [DOI] [PubMed] [Google Scholar]
- 39. Alderwick L. J., Dover L. G., Seidel M., Gande R., Sahm H., Eggeling L., Besra G. S. (2006) Arabinan-deficient mutants of Corynebacterium glutamicum and the consequent flux in decaprenylmonophosphoryl-d-arabinose metabolism. Glycobiology 16, 1073–1081 [DOI] [PubMed] [Google Scholar]
- 40. Alderwick L. J., Seidel M., Sahm H., Besra G. S., Eggeling L. (2006) Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 281, 15653–15661 [DOI] [PubMed] [Google Scholar]
- 41. Tatituri R. V., Alderwick L. J., Mishra A. K., Nigou J., Gilleron M., Krumbach K., Hitchen P., Giordano A., Morris H. R., Dell A., Eggeling L., Besra G. S. (2007) Structural characterization of a partially arabinosylated lipoarabinomannan variant isolated from a Corynebacterium glutamicum ubiA mutant. Microbiology 153, 2621–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mazzoni A., Segal D. M. (2004) Controlling the Toll road to dendritic cell polarization. J. Leukoc. Biol. 75, 721–730 [DOI] [PubMed] [Google Scholar]
- 43. O'Garra A., Robinson D. (2004) Development and function of T helper 1 cells. Adv. Immunol. 83, 133–162 [DOI] [PubMed] [Google Scholar]
- 44. Stockinger B., Veldhoen M. (2007) Differentiation and function of Th17 T cells. Curr. Opin. Immunol. 19, 281–286 [DOI] [PubMed] [Google Scholar]
- 45. Rakoff-Nahoum S., Medzhitov R. (2009) Toll-like receptors and cancer. Nat. Rev. Cancer 9, 57–63 [DOI] [PubMed] [Google Scholar]
- 46. Kirschner D. E., Young D., Flynn J. L. (2010) Tuberculosis. Global approaches to a global disease. Curr. Opin. Biotechnol. 21, 524–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chatterjee S., Dwivedi V. P., Singh Y., Siddiqui I., Sharma P., Van Kaer L., Chattopadhyay D., Das G. (2011) Early secreted antigen ESAT-6 of Mycobacterium tuberculosis promotes protective T helper 17 cell responses in a Toll-like receptor-2-dependent manner. PLoS Pathog. 7, e1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Berrington W. R., Hawn T. R. (2007) Mycobacterium tuberculosis, macrophages, and the innate immune response. Does common variation matter? Immunol. Rev. 219, 167–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mishra A. K., Batt S., Krumbach K., Eggeling L., Besra G. S. (2009) Characterization of the Corynebacterium glutamicum ΔpimB′ ΔmgtA double deletion mutant and the role of Mycobacterium tuberculosis orthologues Rv2188c and Rv0557 in glycolipid biosynthesis. J. Bacteriol. 191, 4465–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Geurtsen J., Chedammi S., Mesters J., Cot M., Driessen N. N., Sambou T., Kakutani R., Ummels R., Maaskant J., Takata H., Baba O., Terashima T., Bovin N., Vandenbroucke-Grauls C. M., Nigou J., Puzo G., Lemassu A., Daffé M., Appelmelk B. J. (2009) Identification of mycobacterial α-glucan as a novel ligand for DC-SIGN. Involvement of mycobacterial capsular polysaccharides in host immune modulation. J. Immunol. 183, 5221–5231 [DOI] [PubMed] [Google Scholar]
- 51. Doz E., Rose S., Nigou J., Gilleron M., Puzo G., Erard F., Ryffel B., Quesniaux V. F. (2007) Acylation determines the Toll-like receptor (TLR)-dependent positive versus TLR2-, mannose receptor-, and SIGNR1-independent negative regulation of pro-inflammatory cytokines by mycobacterial lipomannan. J. Biol. Chem. 282, 26014–26025 [DOI] [PubMed] [Google Scholar]
- 52. Cruz A., Fraga A. G., Fountain J. J., Rangel-Moreno J., Torrado E., Saraiva M., Pereira D. R., Randall T. D., Pedrosa J., Cooper A. M., Castro A. G. (2010) Pathological role of interleukin 17 in mice subjected to repeated BCG vaccination after infection with Mycobacterium tuberculosis. J. Exp. Med. 207, 1609–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Khader S. A., Bell G. K., Pearl J. E., Fountain J. J., Rangel-Moreno J., Cilley G. E., Shen F., Eaton S. M., Gaffen S. L., Swain S. L., Locksley R. M., Haynes L., Randall T. D., Cooper A. M. (2007) IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat. Immunol. 8, 369–377 [DOI] [PubMed] [Google Scholar]