Background: Gdh1 and Gdh3 are glutamate-synthesizing isofunctional NADP-GDH in S. cerevisiae.
Results: Stationary phase-specific GDH3 expression and degradation of Gdh1 were responsible for the Gdh3-dependent glutamate supply and resistance to stress-induced apoptosis in stationary phase.
Conclusion: Gdh3 plays a role distinct from Gdh1 by rendering cells resistant to stress and aging.
Significance: This provides mechanistic insight into apoptosis and protein degradation in response to stress.
Keywords: Apoptosis, Glutamate, Glutamate Dehydrogenase, Glutathione, Oxidative Stress, Apoptosis, GDH1, GDH3, Saccharomyces cerevisiae, Thermal Stress
Abstract
Glutamate metabolism is linked to a number of fundamental metabolic pathways such as amino acid metabolism, the TCA cycle, and glutathione (GSH) synthesis. In the yeast Saccharomyces cerevisiae, glutamate is synthesized from α-ketoglutarate by two NADP+-dependent glutamate dehydrogenases (NADP-GDH) encoded by GDH1 and GDH3. Here, we report the relationship between the function of the NADP-GDH and stress-induced apoptosis. Gdh3-null cells showed accelerated chronological aging and hypersusceptibility to thermal and oxidative stress during stationary phase. Upon exposure to oxidative stress, Gdh3-null strains displayed a rapid loss in viability associated with typical apoptotic hallmarks, i.e. reactive oxygen species accumulation, nuclear fragmentation, DNA breakage, and phosphatidylserine translocation. In addition, Gdh3-null cells, but not Gdh1-null cells, had a higher tendency toward GSH depletion and subsequent reactive oxygen species accumulation than did WT cells. GSH depletion was rescued by exogenous GSH or glutamate. The hypersusceptibility of stationary phase Gdh3-null cells to stress-induced apoptosis was suppressed by deletion of GDH2. Promoter swapping and site-directed mutagenesis of GDH1 and GDH3 indicated that the necessity of GDH3 for the resistance to stress-induced apoptosis and chronological aging is due to the stationary phase-specific expression of GDH3 and concurrent degradation of Gdh1 in which the Lys-426 residue plays an essential role.
Introduction
Glutamate dehydrogenase (GDH)4 (EC 1.4.1.3) catalyzes the reversible oxidative deamination of glutamate to α-ketoglutarate and ammonia using NAD(H) and NADP(H) as cofactors. The enzyme is present in almost all living organisms and plays a pivotal role in important cellular processes, such as the tricarboxylic acid (TCA), ammonia management, and energy metabolism (1). Although the equilibrium of mammalian GDH reaction favors the synthesis of glutamate, this reaction hardly occurs in mammalian cells because it requires high ammonia (Km = 12–25 mm) and α-ketoglutarate (Km = 1.0–2.0 mm) levels not likely to be seen under base-line conditions (1–3). When a high ammonia concentration prevails, however, glutamate synthesis via reductive amination of α-ketoglutarate by GDH may function as a detoxification process (4). The generation of α-ketoglutarate via oxidative deamination of glutamate leads to the production of NAD(P)H, GTP, and ATP through the TCA cycle in mitochondria. Therefore, GTP and ATP are allosteric inhibitors of GDH, whereas ADP is an activator (5).
In all mammals, except for humans and some closely related species, GDH is encoded by a single gene. However, humans and other primates have two distinct genes, GLUD1 and GLUD2, that encode two isoforms of GDH, hGDH1 and hGDH2, respectively (6, 7). GLUD1 is widely expressed in almost all human tissues including liver, brain, pancreas, and kidney, but not muscle. In pancreatic β-cells, immoderate generation of α-ketoglutarate due to an activating mutation of hGDH1 leads to increased insulin exocytosis through ATP overproduction (8–10). The activity of hGDH1 in β-cells is repressed by ADP-ribosylation catalyzed by SIRT4, one of seven homologs of yeast Sir2, which subsequently results in the down-regulation of insulin secretion (11). GLUD2 is expressed predominantly in a limited range of tissues including retina, brain, and testis (7). Despite the high similarity between hGDH1 and hGDH2, as they share all but 16 of their 505 amino acid residues, they show definite differences in their basic catalytic activities and allosteric regulation (3, 5, 12, 13). Thus, the enzymes may contribute differentially to cellular processes. Deregulation of the activity of hGDH2 caused by an S445A substitution in the regulatory domain enhances glutamate oxidation, which results in enhanced nigral cell degeneration (14).
In contrast to mammals in which the reductive amination of α-ketoglutarate by GDH does not occur to an appreciable extent, the yeast Saccharomyces cerevisiae cannot only biosynthesize glutamate but also utilize it via the reactions catalyzed by three distinct GDH isoenzymes. The NAD+-dependent GDH (NAD-GDH; Gdh2) encoded by GDH2 catalyzes reversible oxidative deamination of glutamate to α-ketoglutarate and ammonia (15). Glutamate anabolism via amination of α-ketoglutarate is catalyzed by two different NADP+-dependent GDH (NADP-GDH), Gdh1 and Gdh3, encoded by GDH1 and GDH3, respectively (16). Yeast cells lacking Gdh2 show impaired glutamate utilization and poor growth in minimal glucose media containing glutamate as a nitrogen source (15). Although Gdh2 is responsible for the reversible deamination of glutamate, it is not involved in glutamate biosynthesis during normal growth (15). On the contrary, Gdh1 and Gdh3 catalyze glutamate biosynthesis through the reductive amination of α-ketoglutarate (16). An alternative pathway for glutamate biosynthesis is accomplished by the combined activities of GLN1-encoded glutamine synthetase and the GLT1-encoded glutamate synthase (17, 18). However, both GDH3 and GLT1 are dispensable for yeast growth in minimal glucose medium containing ammonia as a sole nitrogen source, indicating that Gdh1 is the primary enzyme for glutamate biosynthesis (16). Gdh1 uses α-ketoglutarate at a higher rate than does Gdh3 and almost solely contributes to the NADP-GDH activity under fermentative growth conditions with glucose as the sole carbon source. However, during post-diauxic growth, the Gdh1/Gdh3 ratio decreases, and the majority of the total NADP-GDH activity is due to Gdh3, even though GDH1 transcription proceeds during this growth phase (19). This phenomenon is in accordance with a previous observation in that NADP-GDH is degraded during glucose starvation (20).
In the present study, we examined the differential roles of two NADP-GDH, Gdh1 and Gdh3, in sustaining stress resistance. Our results indicate that Gdh3, but not Gdh1, is responsible for tolerance to stress-induced apoptosis in stationary phase cells, as there is stationary phase-specific expression of GDH3 and degradation of Gdh1.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Transformation
S. cerevisiae strains used in this study are listed in Table 1. The YPD medium consisted of 1% yeast extract, 2% peptone, 75 μm adenine sulfate, and 2% glucose. The synthetic complete dextrose (SCD) medium consisted of 0.67% yeast nitrogen base without amino acids (Difco, Detroit, MI), 0.14% yeast synthetic drop-out medium supplement without leucine and uracil (Sigma-Aldrich), and 2% glucose. When necessary, SCD was supplemented with 2 mm uracil and 1 mm amino acids (glutamate and leucine). For solid media, 2% agar (Difco) was added. Transformation of yeast strains was performed using the lithium acetate method (21).
TABLE 1.
S. cerevisiae strains and plasmids used in this study
| Strain and plasmid | Relevant genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | ATCC |
| BY4741-YOR375C | Δgdh1::KanMX4 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | ATCC |
| BY4741-YDL215C | Δgdh2::KanMX4 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | ATCC |
| BY4741-YAL062W | Δgdh3::KanMX4 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | ATCC |
| MCBY004 | Δgdh3::URA3 Δgdh1::KanMX4 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | This study |
| MCBY005 | Δgdh3::URA3 Δgdh2::KanMX4 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | This study |
| MCBY006 | Δgdh1::URA3 Δgdh2::KanMX4 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | This study |
| YCpGdh1 | GDH1 LEU2 | This study |
| YCpGdh3 | GDH3 LEU2 | This study |
| YCpPGDH1-Gdh3 | gdh1P::gdh3ORF LEU2 | This study |
| YCpPGDH3-Gdh1 | gdh3P::gdh1ORF LEU2 | This study |
| YEpGdh2 | GDH2 URA3 | This study |
| YEpPGDH1-LacZ | gdh1P′::LACZ URA3 | This study |
| YEpPGDH3-LacZ | gdh3P′::LACZ URA3 | This study |
| YCpGdh1-FLAG | GDH1::FLAG LEU2 | This study |
| YCpGdh3-FLAG | GDH3::FLAG LEU2 | This study |
| YCpPGDH1-Gdh3-FLAG | gdh1P::gdh3ORF::FLAG LEU2 | This study |
| YCpPGDH3-Gdh1-FLAG | gdh3P::gdh1ORF::FLAG LEU2 | This study |
Plasmids
Plasmids used in this study are listed in Table 1. For construction of YCpGdh1, a 2.4-kb DNA fragment containing the promoter and coding sequence of GDH1 was PCR-amplified from the genomic DNA of an S. cerevisiae WT strain (BY4741) with the primers P1-1 and P1-2 (Table 2). The resulting fragment was cloned into the low copy yeast/Escherichia coli shuttle vector YCp111. In parallel, to construct YCpGdh3, a 2.4-kb fragment containing the complete GDH3 gene was PCR-amplified with the primers P3-1 and P3-2 (Table 2) and cloned into YCP111. To construct YEpGdh2, a 4.3-kb fragment containing the complete GDH2 gene was amplified by PCR with the primers P2-1 and P2-2 (Table 2) and cloned into the high copy yeast/E. coli shuttle vector YEp352.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Oligonucleotide sequence (5′-3′) |
|---|---|
| P1-1 | GGATCCAAGAATGACAGCTTCCCAAGa |
| P1-2 | GGATCCTTAAAATACATCACCTTGa |
| P1-3 | aactatggttcgcttgtcatTTCTTTTTCTTTTTGGTCTCb |
| P1-4 | gaaaaggtaaaaagtaaaaaATGTCAGAGCCAGAATTTCAAc |
| P1-5 | GAGCTCCGAAAACTTCTCTTAATGATGa |
| P1-6 | GAGCTCtcagatcttatcgtcgtcatccttgtaatcAAATACATCACCTTGGTCAAAa,d |
| P1-7 | AAGCTTCGAAACTTCTCTTAATGATG |
| P1-8 | AAGCTTGTTGAAATTCTGGCTCTGACATa |
| P1-9 | GGCATCGTTTACGATTGGCT |
| P1-10 | ttatatgtagctttcgacatTTCTTTTT CTTTTTGGTCTCe |
| P1-11 | gatgcggccagcaaaactaa ATAGTCTAAAAGAAAGAAAAe |
| P1-12 | AGACATGAGAATTGTCAAAG |
| P1-13 | GAATTCATACGGGTTGGCTGCTGGTAa |
| P1-14 | GAATTCGGTTCCATGACTCCATGGAAa |
| P1-419s | CAATGAATGTATCGACTATGCC[underln]GCGAAGTACACTAAGGACGGTAAGf |
| P1-419a | CTTACCGTCCTTAGTGTACTT[underln]CGCGGCATAGTCGATACATTCATTGf |
| P1-420s | AATGTATCGACTATGCCAAG[underln]GCGTACACTAAGGACGGTAAGGf |
| P1-420a | CCTTACCGTCCTTAGTGTA[underln]CGCCTTGGCATAGTCGATACATTf |
| P1-423s | TATCGACTATGCCAAGAAGTACACT[underln]GCGGACGGTAAGGTCf |
| P1-423a | GACCTTACCGTC[underln]CGCAGTGTACTTCTTGGCATAGTCGATAf |
| P1-426a | AGAAGTACACTAAGGACGGT[underln]GCGGTCTTGCCATCTTTGGTf |
| P1-426s | ACCAAAGATGGCAAGAC[underln]CGCACCGTCCTTAGTGTACTTCTf |
| P2-1 | GAGCTCGAGCACTTGACGTTTGGTCCa |
| P2-2 | GAGCTCTCAAGCACTTGCCTCCGCTTa |
| P3-1 | GGATCCTAAAAACCGTCAAGGCATa |
| P3-2 | GGATCCCTAAAAAACGTCTCCCTGGTa |
| P3-3 | gagaccaaaaagaaaaagaaATGACAAGCGAACCAGAGTTg |
| P3-4 | tgaaattctggctctgacatTTTTTACTTTTTACCTTTTCh |
| P3-5 | AAGCTTGCGGTTATATGATCTTCa |
| P3-6 | AAGCTTtcagatcttatcgtcgtcatccttgtaatcAAAAACGTCTCCCTGGTCAAGa,d |
| P3-7 | GGATCCtcagatcttatcgtcgtcatccttgtaatcAAAAACGTCTCCCTGGTCAAGa,d |
| P3-8 | GAGCTCTAAAAACCGTCAAGGCATa |
| P3-9 | AAGCTTCTGGTTCGCTTGTCATa |
| P3-10 | TAAACATACTTGTGGCAGCT |
| P3-11 | ttatatgtagctttcgacatTTTTTACTTTTTACCTTTTCe |
| P3-12 | gatgcggccagcaaaactaaCCGTAAGCGCTATTTTCTTTe |
| P3-13 | AATCACAAGCTCATCGGGCG |
| P3-14 | GAATTCGAGCACTTGCCAAAGTAATTa |
| P3-15 | GAATTCTATGTTCAATGAATTTATTGa |
| P4-1 | ATGTCGAAAGCTACATATAA |
| P4-2 | TTAGTTTTGCTGGCCGCATC |
a Underlined small capitals indicate additional restriction sites.
b Lowercase letters indicate additional Gdh3 N-terminal coding sequence.
c Lowercase letters indicate additional GDH3 promoter region.
d Lowercase letters indicate additional FLAG-coding sequence.
e Lowercase letters indicate additional URA3 sequence.
f Underlined bold capitals indicate mutations (Lys → Ala) incorporated into the primers.
g Lowercase letters indicate additional GDH1 promoter region.
h Lowercase letters indicate additional N-terminal coding sequence for Gdh1.
Two chimeric genes were constructed by a reciprocal exchange of the GDH1 and GDH3 promoter regions. For construction of YCpPGDH1-Gdh3, a 1.0-kb fragment containing the promoter region of GDH1 (gdh1P) was PCR-amplified with the primers P1-1 and P1-3 (Table 2). A 1.4-kb fragment containing the complete coding sequence of Gdh3 (gdh3ORF) was amplified with the primers P3-3 and P3-2 (Table 2). The two PCR products were fused by overlapping PCR, and the resulting 2.4-kb PCR product (gdh1P::gdh3ORF) was cloned into YCp111 to yield YCpPGDH1-Gdh3. Similarly, for construction of YCpPGDH3-Gdh1, a 1.0-kb fragment containing the promoter region of GDH3 (gdh3P) was amplified with the primers P3-1 and P3-4 (Table 2). A 1.4-kb fragment containing the complete coding sequence of Gdh1 (gdh1ORF) was amplified with the primers P1-4 and P1-2 (Table 2). The two PCR products were combined by overlapping PCR, and the resulting 2.4-kb PCR product (gdh3P::gdh1ORF) was cloned into YCp111 to yield YCpPGDH3-Gdh1.
The genes coding for FLAG-tagged derivatives of Gdh1 and Gdh3 were constructed as follows. A 2.3-kb PCR fragment containing the promoter and coding region of GDH1 followed by a FLAG-coding sequence was amplified with the primers P1-5 and P1-6 (Table 2) and cloned into YCp111 to yield YCpGdh1-FLAG. A 2.6-kb PCR fragment containing the promoter and coding region of GDH3 followed by the FLAG-coding sequence was amplified with the primers P3-5 and P3-6 (Table 2) and cloned into YCp111 to yield YCpGdh3-FLAG.
The FLAG-tagged hybrid genes derived from GDH1 and GDH3 by promoter swapping were constructed as follows. A 2.4-kb DNA fragment containing the FLAG-tagged derivative of the gdh1P::gdh3ORF hybrid (gdh1P::gdh3ORF::FLAG) was PCR-amplified from YCpPGDH1-Gdh3 with the primers P1-1 and P3-7 (Table 2). The amplified fragment was cloned into YCp111 to yield YCpPGDH1-Gdh3-FLAG. Similarly, the FLAG-tagged derivative of the gdh3P::gdh1ORF hybrid gene (gdh3P::gdh1ORF::FLAG) was PCR-amplified from YCpPGDH3-Gdh1 with the primers P3-8 and P1-6 (Table 2) and cloned into YCp111 to yield YCpPGDH3-Gdh1-FLAG.
To construct the LacZ reporter plasmids for analysis of the expression of GDH1 and GDH3, the promoter regions and short N-terminal coding sequences (6–8 amino acids) of GDH1 (gdh1P′) and GDH3 (gdh3P′) were fused in-frame to the lacZ gene. A 0.9-kb gdh1P′ fragment was PCR-amplified with the primers P1-7 and P1-8 (Table 2). The resulting PCR product was fused in-frame to a promoter-less lacZ gene to construct the gdh1P′::lacZ fusion in YEp353 (YEpPGDH1-LacZ). In parallel, a 1.2-kb DNA fragment containing the gdh3P′ fragment was PCR-amplified with the primers P3-5 and P3-9 (Table 2). The PCR fragment was then fused in-frame to a promoter-less lacZ gene to form a gdh3P′::lacZ fusion in YEp353 (YEpPGDH3-LacZ).
Site-directed Mutagenesis
Point mutations resulting in single amino acid substitutions in Gdh1 (K419A, K420A, K423A, and K426A) were introduced directly into the YCpGdh1-FLAG vector harboring the GDH1 gene followed by the FLAG-coding sequence to yield YCpGdh1K419A-FLAG, YCpGdh1K420A-FLAG, YCpGdh1K423A-FLAG, and YCpGdh1K426A-FLAG. Site-directed mutagenesis was performed using a QuikChange II XL site-directed mutagenesis kit (Agilent, Santa Clara, CA) according to the manufacturer's instructions. The mutagenic oligonucleotide primers are as listed in Table 2: for K419A mutation, P1-419s and P1-419a; for K420A mutation, P1-420s and P1-420a; for K423A mutation, P1-423s and P1-423a; and for K426A mutation, P1-426s and P1-426s. All mutations were verified by sequencing to confirm that only the intended mutations had been introduced.
Gene Disruption
For construction of the Δgdh1Δgdh3 and Δgdh2Δgdh3 double mutants, a 1.8-kb DNA fragment containing Δgdh3::URA3 fusion construct was amplified by double-joint PCR with the following primers: for the 5′ flanking region of GDH3, P3-10 and P3-11; for the 3′ flanking region of GDH3, P3-12 and P3-13; for the URA3 gene, P4-1 and P4-2; and for the final PCR round, P3-14 and P3-15 (Table 2). The Δgdh1 (BY4741-YOR375C) and Δgdh2 (BY4741-YDL215C) mutants were transformed with the Δgdh3::URA3 fusion construct to yield the Δgdh1Δgdh3 (MCBY004) and Δgdh2Δgdh3 (MCBY005) strains, respectively. Similarly, to construct the Δgdh1Δgdh2 double mutant, a 1.8-kb PCR product containing the Δgdh1::URA3 fusion construct was amplified by double-joint PCR with the following primers: for the 5′ flanking region of GDH1, P1-9 and P1-10; for the 3′ flanking region of GDH1, P1-11 and P1-12; for the URA3 gene, P4-1 and P4-2; and for the final PCR round, P1-13 and P1-14 (Table 2). The Δgdh2 mutant cells were then transformed with the Δgdh3::URA3 fusion construct to yield the Δgdh1Δgdh2 (MCBY006) strain.
Survival Tests
For cell survival experiments, yeast cells were grown in SCD broth at 30 °C for 2 days (unless otherwise indicated) and suspended in PBS to a concentration of 1.0 × 108 cells ml−1. Two-milliliter samples were taken and subjected to heat (50 °C, 30 min) or oxidative stress (1 mm H2O2, 1 h). One hundred-microliter aliquots of each sample were taken and serially diluted with 10-fold steps. For the colony-forming unit (CFU) assay, a 100-μl aliquot of each dilution was spread onto an SCD plate, and colonies were scored after incubation at 30 °C for 3 days.
Test for Apoptotic Markers
TUNEL assays were performed as described by Madeo et al. (22). In brief, cells were fixed with 3.7% formaldehyde, digested with lyticase (Sigma-Aldrich), applied to a polylysine-coated slide, treated with 0.3% H2O2 to block endogenous peroxidases, permeabilized with 0.1% Triton X-100, incubated with 10 μl of TUNEL reaction mixture (Roche Diagnostics) for 60 min at 37 °C, and incubated with 10 μl of Converter POD (Roche Diagnostics) for 10 min. A coverslip was mounted with a drop of Kaiser's glycerol gelatin (Merck, Darmstadt, Germany). Bright-field images of cells were acquired using a BX51 universal research microscope (Olympus Corp., Tokyo) equipped with a DP11 digital camera (Olympus).
To monitor the levels of intracellular reactive oxygen species (ROS), cells were stained with 10 μg ml−1 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) for 2 h with shaking at 30 °C (23). For analysis of nuclear fragmentation, cells were fixed in 3.7% formaldehyde and stained with 0.5 μg ml−1 DAPI (Sigma-Aldrich) (22). Externalization of phosphatidylserine was assayed using FITC-coupled annexin V (BD Biosciences) as described by Madeo et al. (22). Cells were suspended in digestion buffer, and cell walls were digested as described above. The spheroplasts were stained with both FITC-annexin V and 500 μg ml−1 propidium iodide (PI; BD Biosciences).
Fluorescence microscopy was performed under a BX51 universal research microscope (Olympus) equipped with a DP11 digital camera (Olympus) using the appropriate filter sets (FITC filter for DCFH-DA and FITC-annexin V; DAPI filter for DAPI; and rhodamine filter for PI staining). To determine the frequencies of morphological phenotypes (TUNEL, DCFH-DA, DAPI, FITC-annexin V, and PI), at least 300 cells from three independent experiments were evaluated.
NADP-GDH and β-Gal Assay
To prepare whole cell extracts for NADP-GDH assays, yeast cells grown in liquid medium were harvested by centrifugation and washed twice with distilled water and once with extraction buffer (0.1 m potassium phosphate (pH 7.5), 1 mm EDTA, 1 mm DTT, 1 mm PMSF, and 50 μl ml−1 N-α-p-tosyl-l-lysine chloromethyl ketone) (24). Cell pellets were stored at −20 °C. Soluble extracts were prepared by mechanical disruption of cells suspended in extraction buffer by agitation with glass beads in a vortex mixer (five cycles of 1 min of agitation and 1 min of incubation on ice) followed by brief centrifugation to clarify the lysate. NADP-GDH activity was assayed by measuring the oxidation of NADPH according to the method of Doherty (25). One unit of NADP-GDH activity corresponds to 1 μmol of NADP+ produced/min. Protein was measured by the method of Lowry et al. (26) using BSA as a standard.
β-Gal assays were performed using a yeast β-gal assay kit (Thermo Scientific) according to the manufacturer's instructions. β-Gal activity was calculated using the following equation: β-gal activity = (1000 × A420)/(t × V × OD660) in which t = time (in minutes) of incubation and V = volume of cells (ml) used in the assay.
Glutathione and Glutamate Assays
Intracellular levels of the total glutathione pool, glutathione (GSH) and glutathione disulfide (GSSG), were determined by measuring the rate of 2-nitro-5-thiobenzoic acid formation from 5,5′-dithiobis-(2-nitrobenzoic acid) (Sigma-Aldrich) in the glutathione recycling system (27). Five microliters of cell lysate was added to 1 ml of 100 mm phosphate buffer (pH 7.5) containing 0.6 mm 5,5′-dithiobis-(2-nitrobenzoic acid), 5 mm EDTA, 0.2 mm NADPH, and 1 unit ml−1 glutathione reductase (Sigma-Aldrich). The rate of increase in A412 was monitored. To determine the levels of GSSG, NADPH consumption was monitored by measuring the rate of decrease in A340. Intracellular levels of glutamate were determined using a colorimetric glutamate analysis kit (R-Biopharm, Darmstadt, Germany) according to manufacturer's instructions.
Immunoblotting
To prepare cell extracts for immunoblotting, 1 × 109 cells suspended in 50 μl of lysis buffer (0.5% Nonidet P-40, 20 mm HEPES (pH 7.4), 84 mm KCl, 10 mm MgCl2, 0.2 mm EDTA, 0.2 mm EGTA, 1 mm DTT, 5 μg ml−1 aprotinin, 1 μg ml−1 leupeptin, 1 μg ml−1 pepstatin, and 1 mm phenylmethylsulfonyl fluoride) were mechanically disrupted by agitation with glass beads in a vortex mixer. Proteins were separated by gradient SDS-PAGE and electroblotted onto a PVDF membrane (R-Biopharm). Blots were processed as described by Lauber et al. (28).
Gdh1 and Gdh3 proteins were probed with polyclonal anti-Gdh1 and anti-Gdh3 antibodies (Young In Frontier Co., Seoul, Korea) derived from rabbits immunized with the synthetic peptides CIDYAKKYTKDGKV and CIQAAQEYSTEKNTNT, respectively. FLAG-tagged proteins were probed with a rabbit polyclonal anti-FLAG antibody (Sigma-Aldrich). Tubulin, a loading control, was probed with a rabbit anti-tubulin antibody (Sigma-Aldrich). Membranes were then incubated with HRP-conjugated affinity-purified goat anti-rabbit secondary antibody (Sigma-Aldrich) followed by enhanced chemiluminescent staining using ECL reagents (R-Biopharm).
RESULTS
Deletion of GDH3 Causes Increased Sensitivity to Thermal and Oxidative Stress in Stationary Phase Cells and Accelerates Chronological Aging
To address the differential roles of the two NADP-GDH, Gdh1 and Gdh3, we examined the resistance against heat and oxidative stress of NADP-GDH yeast mutants and their derivatives that harbor either ectopically expressed GDH1 or GDH3 genes. Although the survival rates of the WT, Δgdh1, and Δgdh1Δgdh3/YCpGdh3 yeast strains grown in SCD for 2 days were ∼70% after 30 min of exposure to heat stress at 50 °C, less than 10% of the initial population of the Δgdh3 (BY4741-YAL062W), Δgdh1Δgdh3/YCp111, and Δgdh1Δgdh3/YCpGdh1 strains survived following the same treatment (Fig. 1A). In addition, only 10–15% of the initial population of Δgdh3, Δgdh1Δgdh3/YCp111, and Δgdh1Δgdh3/YCpGdh1 cells cultured for 2 days survived after a 1-h exposure to 1 mm H2O2, whereas more than 75% of the WT, Δgdh1, and Δgdh1Δgdh3/YCpGdh3 cells subjected to the same treatment remained alive. These results indicate that deletion of GDH3 causes increased sensitivity to both heat and oxidative stress in stationary phase cells, whereas impairment of GDH1 leads to no significant stress-sensitive phenotype. Additionally, the stress sensitivity of the Gdh3-null mutants was not suppressed by ectopic expression of GDH1.
FIGURE 1.
Deletion of GDH3 results in increased sensitivity of yeast cells to thermal, oxidative, and aging stress. A, survival rates of yeast strains exposed to thermal or oxidative stress. Yeast cells (WT, Δgdh1, Δgdh3, Δgdh1Δgdh3/YCp111, Δgdh1Δgdh3/YCpGdh1, and Δgdh1Δgdh3/YCpGdh3) grown in SCD for 2 days were exposed to thermal (50 °C, 30 min) or oxidative (10 mm H2O2, 1 h) stress. Surviving cells were evaluated by CFU assays. Values are the means ± S.E. of three independent experiments. *, p < 0.001 (two-tailed Student's t test versus WT). B, survival rates of yeast strains during chronological aging. The yeast strains listed in A were grown in SCD for 13 days. Surviving cells were evaluated as described in A. ■, WT; □, Δgdh1; ●, Δgdh3; ○, Δgdh1Δgdh3/YCp111; ♦, Δgdh1Δgdh3/YCpGdh1; ♢, Δgdh1Δgdh3/YCpGdh3.
Intracellular ROS is a mediator of chronological aging in yeast (29, 30). Accordingly, we have shown previously that ROS accumulation caused by depletion of intracellular glutamate in the yeast cells lacking Cit1, which catalyzes the first step of the TCA cycle, leads to accelerated chronological aging (31). To determine whether the deletion of GDH1 or GDH3 affects the survival of cells in chronologically aged yeast cultures, WT and mutant cells were cultured in SCD for 13 days, and the surviving cells were quantified by CFU assay. Although ∼50% of the initial populations of the WT, Δgdh1, and Δgdh1Δgdh3/YCpGdh3 strains remained viable after a 7-day culture, only 2–5% of the Δgdh3, Δgdh1Δgdh3/YCp111, and Δgdh1Δgdh3/YCpGdh1 cells survived after the same culture period (Fig. 1B). In addition, the survival rates of Δgdh3, Δgdh1Δgdh3/YCp111, and Δgdh1Δgdh3/YCpGdh1 cultures reached their minimum levels after 7 days and then increased continuously up to 20–30% of the initial population during a subsequent 6-day cultivation. This result indicates that the Gdh3-null strains experienced a much faster and more severe aging process than did their isogenic WT population. Furthermore, Gdh3-null cells were subjected to accelerated aging-dependent cell death followed by adaptive regrowth upon long-term cultivation. Our data support a previous study that reported that during the chronological aging process of yeast, premature apoptotic death promotes the regrowth of a subpopulation of better adapted mutants (29).
Oxidative Stress-induced Death of Stationary Phase Gdh3-null Cells Is Caused by ROS-mediated Apoptosis
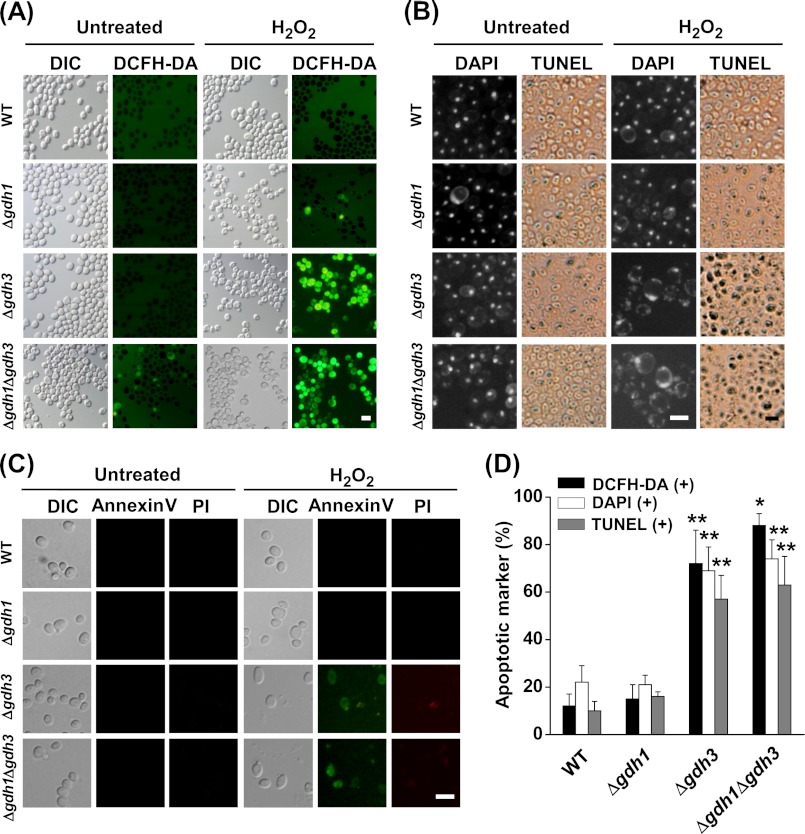
To clarify whether the increased sensitivity of the Gdh3-null cells to heat and oxidative stress is caused by a higher tendency of the mutant populations toward stress-induced apoptosis, we looked for cytological and biochemical features of apoptosis in the mutant cells. We analyzed the cells for the presence of intracellular ROS, which are both necessary and sufficient for inducing apoptosis in yeast (32). As a probe for ROS production, we used DCFH-DA, which can be oxidized to the fluorescent chromophore 2′,7′-dichlorofluorescein primarily by the action of peroxide (H2O2). As seen in the fluorescence micrographs (Fig. 2, A and D), a vast majority of the Δgdh3 (72%) and Δgdh1Δgdh3 cells (88%) grown for 2 days in SCD broth at 30 °C were fluorescent after a 1-h exposure to 10 mm H2O2. On the other hand, the WT and Δgdh1 cells did not show any detectable levels of 2′,7′-dichlorofluorescein fluorescence under these conditions. Therefore, these results indicate that deletion of GDH3 causes oxidative stress-dependent accumulation of ROS in stationary phase cells.
FIGURE 2.
Yeast Gdh3-null cells display apoptotic markers under stress conditions. A, ROS accumulation in yeast cells exposed to oxidative stress. Yeast cells (WT, Δgdh1, Δgdh3, and Δgdh1Δgdh3) grown in SCD for 2 days were exposed to 10 mm H2O2 for 1 h. Cells were stained with DCFH-DA and examined by fluorescence microscopy. Bar, 10 μm. DIC, differential interference contrast. B, nuclear or DNA fragmentation in yeast cells exposed to oxidative stress. Yeast cells prepared and treated as described in A were stained with DAPI or TUNEL reagent and examined by fluorescence microscopy. Bar, 10 μm. C, phosphatidylserine externalization in yeast cells exposed to oxidative stress. Yeast strains prepared and treated as described in A were stained with FITC-annexin V and PI and examined by fluorescence microscopy. Bar, 10 μm. D, percentage of yeast cells exhibiting the typical hallmarks of apoptosis after exposure to oxidative stress (10 mm H2O2, 1 h). The number of cells with accumulated ROS (DCFH-DA(+)), fragmented nuclei (DAPI(+)), and fragmented DNA (TUNEL(+)) were determined from ∼500–700 cells in three independent experiments. Values are the mean ± S.E. *, p < 0.001; **, p < 0.005 (two-tailed Student's t test versus WT).
Because Δgdh1 and Δgdh1Δgdh3 cells exposed to oxidative stress have elevated ROS levels, we assayed for nuclear fragmentation, a well established cytological hallmark of apoptosis. A normal, single-round nucleus was detected by DAPI staining in WT and mutant cells under normal conditions (untreated) (Fig. 2, B and D). On the other hand, ∼66 and 72% of the Δgdh3 and Δgdh1Δgdh3 cells, respectively, displayed irregularly shaped and fragmented nuclei at 1 h after exposure to H2O2. In contrast, only ∼20% of the similarly treated WT and Δgdh1 strains had abnormal nuclei.
We also examined nuclear DNA fragmentation, another feature of apoptosis, using TUNEL analysis in which fluorescent nucleotides were added to the 3′-OH ends of the DNA fragments making the phenomenon visible by fluorescence microscopy. None of the WT or mutant cells had TUNEL-positive nuclei under normal conditions (untreated) (Fig. 2, B and D). However, after 1 h of exposure to H2O2, ∼58 and 62% of Δgdh3 and Δgdh1Δgdh3 cells, respectively, showed intensely stained nuclei indicating DNA strand breakage. On the other hand, the nuclei of both the WT and Δgdh1 cells remained unstained or only slightly stained after the same treatment.
In yeast cells as well as mammalian cells, translocation of phosphatidylserine, which is predominantly located on the inner leaflet of the plasma membrane under normal conditions, to the outer leaflet serves as a sensitive marker for the early stages of apoptosis (33). For detection of phosphatidylserine in the outer layer of the cytoplasmic membrane, spheroplasts formed from the cells subjected to oxidative stress were stained with both FITC-annexin V and PI. The stained cells were then observed by fluorescence microscopy. When exposed to H2O2 for 1 h, a portion of the stationary phase cells from the Δgdh3 and Δgdh1Δgdh3 strains were stained exclusively with FITC-annexin V, indicating that they were undergoing an early stage of apoptosis (Fig. 2C). In addition, some of the oxidatively stressed cells were dually stained with both FITC-annexin V and PI, suggesting the late apoptotic phases or necrosis. On the contrary, only a negligible portion of the similarly treated WT and Δgdh1 cells were stained solely with FITC-annexin V and thus were considered to be apoptotic. These results suggest that stationary phase Gdh3-null cells are much more susceptible to oxidative and thermal stress-induced cell death (bearing the structural attributes of apoptosis) than are the isogenic WT strains.
Stationary Phase Gdh3-null Cells Exhibit GSH and Glutamate Depletion, Which Leads to Increased Susceptibility to Oxidative Stress-induced Apoptosis
GSH synthesized from glutamate, glycine, and cysteine by the action of γ-glutamylcysteine synthase (Gsh1) and GSH synthase (Gsh2) is one of the most prevalent reducing thiol compounds functioning as an antioxidant in nearly every aerobic organism (34). Accordingly, depletion of GSH or its precursor, glutamate, causes oxidative or thermal stress-induced apoptosis in yeast cells (31).
To determine whether the ROS accumulation in stressed Gdh3-null cells is facilitated by depletion of GSH, which is used by many peroxidases to scavenge H2O2 and a multitude of organic hydroperoxides (35), we evaluated the levels of GSH in the mutant cells (Table 3). When Δgdh3 and Δgdh1Δgdh3 cells were grown for 2 days in SCD containing 1 mm glutamate, the levels of GSH were ∼26 and 29% of that in WT cells, respectively. In contrast, Δgdh3 and Δgdh1Δgdh3 mutants exhibited GSH levels similar to those of WT cells when grown in SCD supplemented with a higher concentration of either glutamate or GSH (88 and 92% in the presence of 10 mm glutamate and 76 and 80% in the presence of 10 mm GSH, respectively). Accordingly, the intracellular levels of glutamate were similar to those of GSH. Glutamate concentrations in the cells of Δgdh3 and Δgdh1Δgdh3 mutants were 20% less than that of WT cells when cultured in SCD containing 1 mm glutamate. The depletion of glutamate in Δgdh3 and Δgdh1Δgdh3 mutant cells recovered to at least 75% of the normal level when the medium was supplemented with either 10 mm GSH or 10 mm glutamate.
TABLE 3.
Intracellular levels of GSH and glutamate in S. cerevisiae strains grown in different media
| Strain | GSHa + additions to SCD |
Glutamateb + additions to SCD |
||||
|---|---|---|---|---|---|---|
| 1 mm Glutamate | 10 mm Glutamate | 10 mm GSH | 1 mm Glutamate | 10 mm Glutamate | 10 mm GSH | |
| nmol/mg protein | nmol/mg protein | |||||
| WT | 121.4 ± 8.6 | 131.5 ± 10.6 | 137.3 ± 15.1 | 11.1 ± 3.3 | 19.6 ± 3.8 | 13.2 ± 4.6 |
| Δgdh1 | 118.7 ± 10.5 | 123.7 ± 5.8 | 139.4 ± 7.9 | 9.9 ± 2.2 | 18.9 ± 6.7 | 14.4 ± 3.3 |
| Δgdh2 | 125.9 ± 3.4 | 133.7 ± 11.5 | 134.5 ± 19.3 | 13.4 ± 4.4 | 22.6 ± 5.5 | 15.7 ± 2.1 |
| Δgdh3 | 31.7 ± 10.6 | 120.5 ± 18.3 | 105.1 ± 32.1 | 2.1 ± 0.7 | 17.4 ± 4.2 | 10.1 ± 4.2 |
| Δgdh1Δgdh2 | 120.4 ± 5.8 | 125.7 ± 10.4 | 136.7 ± 4.7 | 11.5 ± 1.9 | 20.5 ± 0.8 | 16.2 ± 2.0 |
| Δgdh1Δgdh3 | 35.1 ± 6.0 | 115.6 ± 10.1 | 110.6 ± 10.6 | 1.9 ± 0.7 | 16.8 ± 5.2 | 10.8 ± 1.1 |
| Δgdh2Δgdh3 | 81.7 ± 19.1 | 124.6 ± 4.1 | 99.8 ± 4.6 | 6.1 ± 2.5 | 18.1 ± 3.8 | 12.5 ± 2.6 |
a Values were obtained from [(GSH + GSSG) − GSSG] and are presented as mean ± S.E. from three independent experiments. The percentage of GSSG to total cellular glutathione pool (GSH + GSSG) was <10% in all strains.
b Values are presented as mean ± S.E. from three independent experiments.
We examined the effect of exogenous GSH and glutamate on stress-induced apoptosis of the yeast strains. In accordance with the restoration of the intracellular GSH and glutamate levels, Δgdh3 and Δgdh1Δgdh3 cells grown in SCD medium supplemented with 10 mm GSH or glutamate showed almost the same level of survival after a 1-h exposure to 10 mm H2O2 as did WT and Δgdh1 cells (Fig. 3A). In addition, when stained with DCFH-DA, Δgdh3 and Δgdh1Δgdh3 cells showed only slightly higher levels of DCF fluorescence compared with WT and Δgdh1 cells under oxidative stress conditions. This indicates significant attenuation of stress-induced ROS accumulation in the presence of 10 mm GSH or glutamate (Fig. 3B). Together, these data suggest that the deletion of GDH3 results in the depletion of intracellular GSH, which, in turn, induces the accumulation of ROS and accompanying apoptotic cell death during stress conditions.
FIGURE 3.
Exogenous supply of GSH or glutamate suppresses the hypersusceptibility of yeast Gdh3-null cells to stress-induced apoptosis mediated by ROS accumulation. A, effect of exogenous GSH and glutamate on survival of yeast cells exposed to oxidative stress. Yeast strains (WT, Δgdh1, Δgdh3, and Δgdh1Δgdh3) grown in SCD with or without 10 mm GSH or glutamate for 2 days were exposed to 10 mm H2O2 for 1 h. Surviving cells were evaluated by CFU assays. Values are the mean ± S.E. of three independent experiments. B, effect of exogenous GSH and glutamate on ROS accumulation in yeast cells exposed to oxidative stress. Yeast cells prepared and treated as described in A were stained with DCFH-DA and examined by fluorescence microscopy. The number of DCFH-DA-positive cells was estimated in fluorescence images and total cells in the corresponding differential interference contrast images. Approximately 500–700 cells were observed in three independent experiments. Values are the mean ± S.E. *, p < 0.005 (two-tailed Student's t test versus untreated).
The Increased Susceptibility of Stationary Phase Gdh3-null Cells to Thermal and Oxidative Stress-induced Apoptosis Is Suppressed by GDH2 Deletion
The NAD-GDH (Gdh2) encoded by GDH2 catalyzes oxidative deamination of glutamate to α-ketoglutarate and ammonia (15), hence the reverse reaction of glutamate synthesis from α-ketoglutarate catalyzed by either Gdh1 or Gdh3. To investigate how Gdh2 function is related to stress-induced apoptosis, we analyzed the effect of GDH2 deletion and the presence of ectopic GDH2 on the hypersusceptibility of Gdh3-null mutants.
The introduction of the Δgdh2 mutation caused increased resistance to both thermal and oxidative stress in Gdh3-null strain (Fig. 4A). The survival rates of Δgdh2Δgdh3 cells grown for 2 days in SCD were ∼55% after a 1-h exposure to oxidative stress (10 mm H2O2) and 47% after a 30-min exposure to thermal stress (50 °C), whereas the survival rates of Δgdh3 cells were ∼15 and 5%, respectively. Correspondingly, Δgdh2Δgdh3 cells exposed to oxidative and thermal stress exhibited significantly lower levels of ROS accumulation (∼44 and 62%, respectively) compared with Δgdh3 cells (∼76 and 92%, respectively) when monitored by fluorescence microscopy using DCFH-DA (Fig. 4B). In addition, the levels of GSH and glutamate in Δgdh2Δgdh3 cells were ∼2.6- and 2.9-fold higher than that of Δgdh3 cells, respectively, when the cells were grown for 2 days in SCD containing a basal level of glutamate (1 mm) (Table 3). On the contrary, all of the yeast strains containing ectopic GDH2 in high copy vectors (WT/YEpGdh2, Δgdh1/YEpGdh2, Δgdh2/YEpGdh2, and Δgdh3/YEpGdh2) showed reduced resistance against both thermal and oxidative stress-induced apoptosis, although to a limited extent, compared with their corresponding host strains (Fig. 4A). Thus, it appears that deletion of GDH2 compensates for the depletion of intracellular glutamate and GSH followed by stress-induced ROS accumulation and apoptotic cell death in stationary phase Gdh3-null cells, whereas ectopic expression of GDH2 enhances the depletion of intracellular glutamate and GSH. These results are in agreement with the fact that Gdh2 catalyzes the oxidative deamination of glutamate to α-ketoglutarate resulting in decreased intracellular glutamate and GSH levels.
FIGURE 4.
Deletion of GDH2 suppresses the hypersusceptibility of yeast Gdh3-null cells to stress-induced apoptosis mediated by ROS accumulation. A, effect of Δgdh2 mutation on survival of yeast cells exposed to thermal or oxidative stress. Yeast strains (WT, Δgdh1, Δgdh2, Δgdh3, Δgdh2Δgdh3, WT/YEpGDH2, Δgdh1/YEpGDH2, Δgdh2/YEpGDH2, and Δgdh3/YEpGDH2) grown in SCD for 2 days were exposed to either thermal (50 °C, 30 min) or oxidative stress (10 mm H2O2, 1 h). Surviving cells were evaluated by CFU assays. Values are the means ± S.E. of three independent experiments. *, p < 0.001 (two-tailed Student's t test versus Δgdh3). B, effect of the Δgdh2 mutation on ROS accumulation in yeast cells exposed to thermal or oxidative stress. Yeast cells prepared and treated as described in A were stained with DCFH-DA and examined by fluorescence microscopy. The number of DCFH-DA-positive cells was estimated in fluorescence images and total cells in corresponding differential interference contrast images. Approximately 500–700 cells were observed in three independent experiments. Values are the mean ± S.E. *, p < 0.001; **, p < 0.05 (two-tailed Student's t test versus Δgdh3).
GDH1, but Not GDH3, Is Responsible for the Resistance against Stress-induced Apoptosis in Logarithmic Phase Cells
To determine the differential roles of GDH1 and GDH3 in sustaining resistance to stress-induced apoptosis, we monitored the ectopic expression of gdh1P′::lacZ and gdh3P′::lacZ hybrid genes in WT/YEpPGDH1-LacZ and WT/YEpPGDH3-LacZ strains, respectively. Considerably higher levels of β-gal activity (≥1500 units) were detected in the WT/YEpPGDH1-LacZ cell extracts, regardless of the growth stage, than in the WT/YEpPGDH3-LacZ cell extracts (Fig. 5). The level of β-gal activity in WT/YEpPGDH3-LacZ cells was low (∼250 units) after a 24-h culture into late logarithmic phase but gradually increased during the following 48-h period up to levels 4-fold higher than during the logarithmic phase (∼1,000 units). Therefore, GDH1 was expressed consistently throughout all of the growth periods at a relatively high level, whereas GDH3 was expressed at negligible levels during the logarithmic phase but increased gradually to a higher level during stationary phase. These results are in agreement with a previous report indicating that GDH3 transcription is repressed by glucose and is induced only under respiratory conditions or during the stationary phase (36), whereas transcription of GDH1 in cells grown with glucose is regulated by the transcriptional activators Gln3, Hap2, and Hap3 (37).
FIGURE 5.
GDH3 transcription occurs exclusively during stationary phase, whereas GDH1 transcription is consistent throughout all growth periods. Yeast strains (WT/YEpPGDH1-LacZ and WT/YEpPGDH3-LacZ) were grown in SCD, and samples were taken after 24, 48, and 72 h of culture. GDH1 and GDH3 transcription levels were estimated by measuring β-gal activity. β-Gal activity was calculated using the following equation: β-gal activity = (1000 × A420)/(t × V × OD660), where t = time (in minutes) of incubation and V = volume of cells (ml) used in the assay. Three independent experiments were performed in triplicate. Values are the mean ± S.E. *, p < 0.001 (two-tailed Student's t test versus YEpPGDH3-LacZ at 24 h).
To determine whether the transcription profiles of GDH1 and GDH3 are consistent with the subsequent gene expression events, we analyzed the enzymatic activities and protein levels of Gdh1 and Gdh3 in WT cells and a variety of mutant strains. The levels of NADP-GDH activity in the exponential phase (24 h) cells of Δgdh3, Δgdh1Δgdh3/YCpGdh1, and WT strains were ∼10-fold higher than in Δgdh1 and Δgdh1Δgdh3/YCpGdh3 cells. Meanwhile, the levels of NADP-GDH activity in the stationary phase (48 h) cells of Δgdh1, Δgdh1Δgdh3/YCpGdh3, and WT strains were at least seven times higher than in Δgdh3 and Δgdh1Δgdh3/YCpGdh1 cells (Table 4). In immunoblotting, Gdh1 protein was detected in the exponential phase (24 h) cells of Δgdh3, Δgdh1Δgdh3/YCpGdh1, and WT strains but not in the stationary phase (48 h) cells of the strains (Fig. 6A). On the contrary, Gdh3 protein was detected in the stationary phase cells of Δgdh1, Δgdh1Δgdh3/YCpGdh3, and WT strains but not in their exponential phase cells. In addition, a substantial level of Gdh1-FLAG signal was detected in Δgdh1Δgdh3/YCpGdh1-FLAG cells during the exponential phase (12 and 24 h), but no signal was identified during the stationary phase (48 and 72 h) (Fig. 6B). However, Δgdh1Δgdh3/YCpGdh3-FLAG cells showed only negligible levels of Gdh3-FLAG signal during the logarithmic phase followed by a marked increase in Gdh3-FLAG signal to the highest level during stationary phase. Together, these results suggest that a large majority of the NADP-GDH activity is Gdh1-dependent until the cells reach late logarithmic phase and then becomes Gdh3-dependent during the stationary phase.
TABLE 4.
Intracellular levels of NADP-GDH and NAD-GDH activities in S. cerevisiae strains
| Strain | NADP-GDH-specific activityα |
NAD-GDH-specific activityα |
||
|---|---|---|---|---|
| 24 hb | 48 hb | 24 hb | 48 hb | |
| WT | 1.224 ± 0.102 | 1.451 ± 0.126 | 0.042 ± 0.002 | 0.051 ± 0.003 |
| Δgdh1 | 0.121 ± 0.006 | 1.316 ± 0.092 | 0.039 ± 0.001 | 0.057 ± 0.003 |
| Δgdh2 | 1.213 ± 0.083 | 1.413 ± 0.117 | <0.002 | <0.002 |
| Δgdh3 | 1.144 ± 0.088 | 0.159 ± 0.054 | 0.048 ± 0.002 | 0.049 ± 0.004 |
| Δgdh1Δgdh2 | 0.120 ± 0.090 | 1.307 ± 0.125 | <0.002 | <0.002 |
| Δgdh1Δgdh3 | <0.005 | <0.005 | 0.045 ± 0.003 | 0.055 ± 0.005 |
| Δgdh2Δgdh3 | 1.219 ± 0.099 | 0.234 ± 0.016 | <0.002 | <0.002 |
| Δgdh1Δgdh3/YCpGdh1 | 1.145 ± 0.108 | 0.187 ± 0.013 | 0.044 ± 0.001 | 0.054 ± 0.002 |
| Δgdh1Δgdh3/YCpGdh3 | 0.122 ± 0.009 | 1.345 ± 0.117 | 0.041 ± 0.003 | 0.052 ± 0.003 |
| Δgdh1Δgdh3/YCpPGDH1-Gdh3 | 1.253 ± 0.081 | 1.559 ± 0.089 | 0.047 ± 0.003 | 0.058 ± 0.004 |
| Δgdh1Δgdh3/YCpPGDH3-Gdh1 | 0.007 ± 0.001 | 0.222 ± 0.014 | 0.046 ± 0.004 | 0.053 ± 0.005 |
α Values given in μmol/min/mg protein and presented as mean ± S.E. from three independent experiments.
b Length of time cells were cultured in SCD.
FIGURE 6.
Gdh3 protein is stable throughout all growth stages, whereas Gdh1 is subjected to stationary phase-specific degradation. A, immunoblot analysis of the Gdh1 and Gdh3 proteins in yeast cells at different growth stages. Yeast strains (WT, Δgdh1, Δgdh3, Δgdh1Δgdh3/YCp111, Δgdh1Δgdh3/Gdh1, Δgdh1Δgdh3/YCpGdh3) were grown in SCD, and samples were taken after 24 and 48 h of culture. Cell extracts were immunoblotted and probed with anti-Gdh1, anti-Gdh3 and anti-tubulin antibodies. B, immunoblot analysis of the FLAG-tagged Gdh1 and Gdh3 proteins in yeast cells carrying the promoter-swapped derivatives of GDH1 and GDH3 at different growth stages. Yeast strains (Δgdh1Δgdh3/YCpGdh1-FLAG, Δgdh1Δgdh3/YCpGdh3-FLAG, Δgdh1Δgdh3/YCpPGDH1-Gdh3-FLAG, and Δgdh1Δgdh3/YCpPGDH3-Gdh1-FLAG) were grown in SCD and samples were taken after 12, 24, 48, and 72 h of culture. Cell extracts were immunoblotted and probed with anti-FLAG and anti-tubulin antibodies.
Gdh1, but Not Gdh3, Is Subjected to Degradation in Stationary Phase Cells in Which the Lys-426 Residue Plays an Essential Role
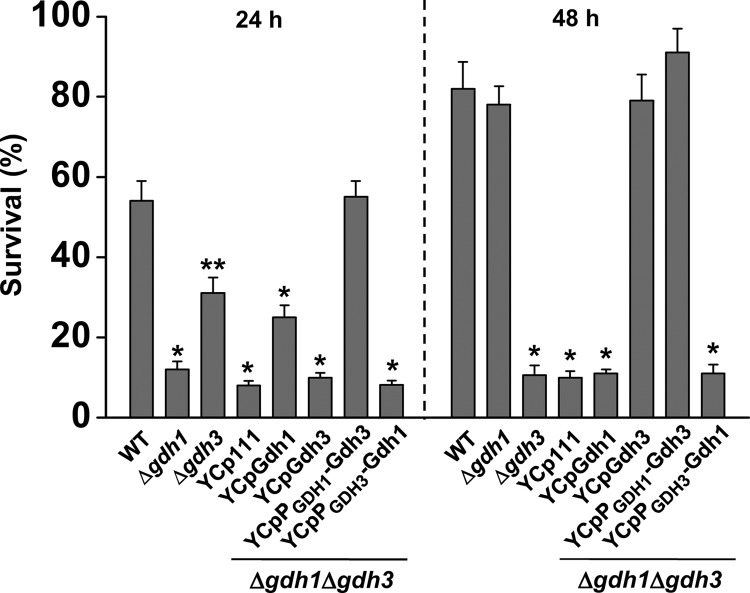
Interestingly, the results of the NADP-GDH activity assays (Table 4) and immunoblotting (Fig. 6) obtained from the stationary phase cells exhibited a significant discrepancy with the results of the β-gal reporter assay (Fig. 5) in that only negligible levels of Gdh1 protein and its corresponding NADP-GDH activity were detected despite relatively high levels of GDH1 transcription. To further investigate the cause for such discrepancy, we monitored the activity and protein levels of NADP-GDH in Δgdh1Δgdh3 cells carrying either of the promoter-swapped derivatives of GDH1 or GDH3. Although the levels of NADP-GDH activity in the YCpPGDH1-Gdh3 transformants were almost the same as those in the WT cells regardless of growth stage, the NADP-GDH activity in cells carrying YCpPGDH3-Gdh1 was negligible (Table 4). Accordingly, no signal for the Gdh1-FLAG protein was detected in Δgdh1Δgdh3/YCpPGDH3-Gdh1-FLAG cells throughout all growth stages, whereas strong signals for the FLAG-tagged Gdh3-FLAG protein was observed in Δgdh1Δgdh3/YCpPGDH1-Gdh3-FLAG cells in all growth stages (Fig. 6B). In agreement with these results, both the late logarithmic (24 h) and stationary (48 h) cells of the Δgdh1Δgdh3/YCpPGDH1-Gdh3 transformant had levels of survival after a 1-h exposure to 10 mm H2O2 similar to those of WT cells, whereas the survival rates of Δgdh1Δgdh3/YCpPGDH3-Gdh1 cells showed only basal levels of resistance against oxidative stress (Fig. 7). This result indicates that ectopic expression of gdh1P::gdh3ORF confers consistent resistance to stress-induced apoptosis in Δgdh1Δgdh3 cells throughout all growth stages, whereas the ectopic expression of gdh3P::gdh1ORF cannot protect the cells from stress-induced damage. Taken together, these results suggest that the Gdh3 protein is stable throughout all growth stages, whereas Gdh1 is subjected to stationary phase-specific degradation.
FIGURE 7.
Ectopic expression of gdh3P::gdh1ORF cannot prevent stress-induced apoptosis in Δgdh1Δgdh3 cells, whereas ectopic expression of gdh1P::gdh3ORF confers stress resistance throughout all growth stages. Yeast strains (WT, Δgdh1, Δgdh3, Δgdh1Δgdh3, Δgdh1Δgdh3/YCpGDH1, Δgdh1Δgdh3/YCpGDH3, Δgdh1Δgdh3/YCpPGDH1-GDH3, and Δgdh1Δgdh3/YCpPGDH3-GDH1) grown in SCD for 24 or 48 h were exposed to oxidative stress (10 mm H2O2, 1 h). Surviving cells were evaluated by CFU assays. Values are the mean ± S.E. of three independent experiments. *, p < 0.001; **, p < 0.005 (two-tailed Student's t test versus WT).
Gdh1 and Gdh3 share an extremely high degree of homology (∼92%) over the entire amino acid sequences except for the Box420Gdh1 and Box420Gdh3 regions located near the C-terminal ends of the proteins (Fig. 8A). The most distinctive feature of Box420Gdh1 is that it has four lysine residues that may be responsible for protein degradation, whereas Box420Gdh3 contains only one lysine residue. Thus, to determine the mechanism of stationary phase-specific Gdh1 degradation, we constructed transformants carrying ectopic hybrid genes encoding the FLAG-tagged Gdh1 derivatives with single alanine (Ala) substitutions for lysine (Lys) in the Box420Gdh1 region (YCpGdh1K419A-FLAG, YCpGdh1K420A-FLAG, YCpGdh1K423A-FLAG, and YCpGdh1K426A-FLAG) and tracked the quantitative change of the fusion proteins by immunoblotting. Although the Gdh1K419A-FLAG, Gdh1K420A-FLAG, and Gdh1K423A-FLAG proteins were detected only in exponential phase cells (12 and 24 h) as WT FLAG-tagged Gdh1 (Gdh1-FLAG), the Gdh1K426A-FLAG protein was detected throughout all of the growth stages (Fig. 8B). Thus, the Lys-426 residue, but not Lys-419, Lys-420, or Lys-423, in the Box420Gdh1 region plays an essential role in stationary phase-specific degradation of Gdh1.
FIGURE 8.
Gdh1, but not Gdh3, is subjected to stationary phase-specific degradation in which the Lys-426 residue in the Box420Gdh1 region plays an essential role. A, amino acid sequences of the C-terminal regions of Gdh1 and Gdh3. The two isoenzymes share an extremely high degree of homology throughout their amino acid sequences except for in the Box420Gdh1 and Box420Gdh3 regions. Point mutations causing single amino acid substitutions in Gdh1 (K419A, K420A, K423A, and K426A) were introduced directly into YCpGdh1-FLAG. B, immunoblot analysis of the FLAG-tagged Gdh1 and its mutant derivatives. Yeast strains (BY4741) carrying YCpGdh1K419A-FLAG, YCpGdh1K420A-FLAG, YCpGdh1K423A-FLAG, or YCpGdh1K426A-FLAG were grown in SCD, and samples were taken after 12, 24, and 48 h of culture. Cell extracts were immunoblotted and probed with anti-FLAG and anti-tubulin antibodies.
DISCUSSION
S. cerevisiae is the first microorganism described in which the NADP-GDH activity is encoded by two genes (16). It has been claimed that coordinated regulation of the two NADP-GDH isoenzymes enables a balanced utilization of α-ketoglutarate for glutamate synthesis during diauxic growth and eventually improves the efficiency of glutamate biosynthesis (19). However, the physiological significance of this apparent redundancy has not yet been fully addressed. In the present study, we attempted to determine the differential role of the two isoforms of NADP-GDH, Gdh1 and Gdh3, in the resistance to stress-induced apoptosis and chronological aging.
The initial clue as to the involvement of Gdh3 in the protection against stress-induced apoptosis was based on the observation that Gdh3-null cells, but not Gdh1- or Gdh3-null cells, were hypersensitive to oxidative and thermal stress compared with WT cells (Fig. 1A). We observed several typical morphological and cytological hallmarks of apoptosis, such as ROS accumulation, nuclear and DNA fragmentation, and phosphatidylserine translocation, in stationary phase Gdh3-null cells following oxidative stress (Fig. 2). A similar phenomenon was observed in Gdh3-null cells exposed to thermal stress (data not shown). In addition, impairment of GDH3 resulted in a higher susceptibility to chronological aging-induced cell death followed by regrowth of a subpopulation that consisted of cells better adapted to prolonged culture conditions (Fig. 1B).
Cells should be equipped with an efficient ROS-scavenging enzymatic system to protect against oxidative damage. Superoxide dismutase catalyzes the destruction of superoxide free radicals to oxygen and H2O2 (38), which is in turn reduced to H2O by catalase or peroxidase. The principal enzyme for H2O2 detoxification is generally considered to be glutathione peroxidase (GPx), which requires GSH as a reducing power (39), rather than catalase, because catalase has a much lower affinity than GPx for H2O2 (40). Thus, GSH is an essential metabolite for stress resistance. When grown in SCD medium, Gdh3-null cells were subject to GSH depletion, which was relieved by an exogenous supply of GSH or glutamate (Table 3). Furthermore, the hypersusceptibility of the stationary cells of Gdh3-null strains to thermal and oxidative stress-induced apoptosis, which is mediated by ROS, was suppressed by exogenous GSH or glutamate (Fig. 3) or by deletion of GDH2 (Fig. 4). Thus, Gdh3 plays a pivotal role in preventing the stress-induced ROS accumulation and subsequent apoptotic events by supplying glutamate, one of the precursors for GSH biosynthesis, in stationary phase cells. It is also suggested that the major form of the ROS accumulated in the stationary phase Gdh3-null cells exposed to oxidative and thermal stress is H2O2, the substrate of GPx. In a human B-lymphoma cell line, decreased GSH alone can act as a potent early activator of apoptotic signaling. Increased ROS production following mitochondrial GSH depletion irreversibly commits cells to apoptosis (41). In addition, both glutamate and glutamine have a significant role not only in the resistance of cells to apoptosis but also in promoting cell proliferation (42). Glutamate can rescue a breast carcinoma cell line from apoptotic cell death (43). Taken together, in both yeast and mammalian cells, glutamate is required for the first committed step in the synthesis of GSH and acts as a suppressor of stress-induced apoptosis. Thus, the present results indicate that yeast strains lacking NADP-GDH could be used as a model system for studying the mechanisms of apoptotic or proliferative defects related to glutamate metabolism in mammals.
Despite the high level of sequence homology between Gdh1 and Gdh3, the transcription patterns of GDH1 and GDH3 are significantly different from each other. Our data from the β-gal reporter assays indicate that GDH3 transcription occurs mainly during the stationary phase, whereas GDH1 is transcribed consistently throughout all growth periods (Fig. 5). Accordingly, GDH3 transcription is strongly repressed by glucose and is highly induced under respiratory conditions and during the stationary phase (36, 37, 44). This implies that Gdh3 may play a crucial role in sustaining oxidative phosphorylation. On the other hand, GDH1 transcription is controlled by transcriptional activators exclusive of either nitrogen (Gln3 and Gcn4) or carbon metabolism (HAP complex) and occurs independently of growth stage and glucose repression (37).
Although consistently high levels of GDH1 transcription were observed regardless of the growth stage (Fig. 5), negligible levels of Gdh1 protein (Fig. 6A) and its NADP-GDH activity (Table 4) were detected during the stationary phase, suggesting that Gdh1 is subjected to stationary phase-specific degradation. The activity and protein levels of the NADP-GDH in Δgdh1Δgdh3 cells carrying ectopic copies of the promoter-swapped derivatives of GDH1 or GDH3 were in agreement with our hypothesis: whereas the NADP-GDH activity of Gdh3 (YCpPGDH1-Gdh3) and the protein level of Gdh3-FLAG (YCpPGDH1-Gdh3-FLAG) were consistently high, the NADP-GDH activity of Gdh1 (YCpPGDH3-Gdh1) and Gdh1-FLAG (YCpPGDH3-Gdh1-FLAG) protein levels were trivial throughout all growth periods (Table 4 and Fig. 6B). In further support, a previous study reported that transfer of S. cerevisiae cultures to medium deficient in a readily utilizable carbon source results in proteolysis of NADP-GDH. However, it has not been determined which of the two NADP-GDH, Gdh1 or Gdh3, is subjected to such degradation (20).
We asked how it is that only Gdh1, but not Gdh3, is subjected to stationary phase-specific degradation despite the extremely high degree of sequence homology between the two enzymes. Thus, we evaluated the importance of degradation of the lysine-rich Box420 region of Gdh1 (Box420Gdh1) that contains a sequence distinct from that of the corresponding Box420 region of Gdh3 (Box420Gdh3) (Fig. 8A). Substituting alanine for the lysine residues in Box420Gdh1 and tracking the stability of the mutant derivatives of Gdh1 revealed that only one lysine residue (Lys-426) of the four lysine residues concentrated in Box420Gdh1 was necessary for the stationary phase-specific degradation of Gdh1 (Fig. 8). A previous study identified Lys-325 and Lys-371 of Gdh1 as ubiquitinated residues through a proteomics approach to enrich, recover, and identify ubiquitin conjugates from S. cerevisiae lysates (45). Although almost all of the ubiquitin-modified lysine residues are exposed at the surface of the molecule and are readily accessible from the outside, Lys-371 is located in a hydrophobic stretch, PPKAA, and is buried inside the protein (46). Thus, this position may be involved in the degradation of misfolded Gdh1 molecules. However, Gdh1 and Gdh3 share a striking homology throughout their amino acid sequences, except for their Box420 regions. The sequences surrounding Lys-325 and Lys-371 in Gdh1 are almost identical to those centered at the corresponding lysine residues, Lys-326 and Lys-372, in Gdh3. Thus neither of the lysine residues is responsible for the stationary phase-specific degradation of Gdh1. The significance of the ubiquitination of Lys-325 and Lys-371 requires further investigation. Future studies will focus on determining whether the degradation of Gdh1 is mediated by the ubiquitin-proteosome pathway in which Lys-426 provides a specific binding site for ubiquitin.
During the stationary phase, yeast cells acquire a variety of features, including a dramatic reduction in the overall rate of growth and protein synthesis, accumulation of the storage carbohydrate glycogen, and increased resistance to a variety of environmental stresses such as oxidative stress and heat shock (47, 48). Specifically, the rate of protein synthesis drops ∼300-fold upon entry into stationary phase (48, 49), which is an essential characteristic for stationary phase survival (50). Thus, it seems that a variety of amino acids become unnecessary for protein synthesis in stationary phase cells. In parallel, cells protect themselves from increasing environmental stress-induced accumulation of ROS during stationary phase. Among the ROS-scavenging systems, the GSH system, which consists of GSH, GPx, and glutathione reductase, is probably the most important intracellular defense mechanism. GPx catalyzes the reduction of H2O2 and oxidizes GSH to GSSG. GSSG is then reduced back to GSH by glutathione reductase. Hence, the ability of the cell to reduce GSSG or synthesize GSH from glutamate is the key to how effectively the cell can eliminate ROS-mediated cell damage (51). In our previous study, we showed that the hypersusceptibility of yeast cells lacking Cit1 to stress-induced apoptosis mediated by ROS does not result from the depletion of reducing power required for glutathione reductase reaction but, instead, is due to an insufficient supply of glutamate, a precursor of GSH biosynthesis (31). Therefore, yeast cells in stationary phase require glutamate for their GSH supply rather than for protein synthesis. The two isofunctional NADP-GDH of S. cerevisiae differ in allosteric properties and rates of α-ketoglutarate utilization. Specifically, Gdh1 exhibits a 3-fold higher rate of α-ketoglutarate utilization than does Gdh3 (19). Thus, Gdh1 is more suitable for functioning as a major glutamate-producing enzyme during the exponential phase in which substantial amounts of amino acids, including glutamate, are necessary for protein synthesis. On the other hand, Gdh3 seems to be more suitable during the stationary phase in which glutamate is mainly required for GSH biosynthesis. Therefore, it may be more beneficial to the cells to substitute Gdh1 with Gdh3 through the stationary phase-specific expression of GDH3 and simultaneous degradation of Gdh1 after exiting from the exponential growth phase.
Acknowledgments
We thank Profs. Jeong-Yoon Kim and Hee-Moon Park for critical reading of the manuscript and valuable advice.
This work was supported by Grant 2009–0077095 from the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology (MEST).
- GDH
- glutamate dehydrogenase
- NADP-GDH
- NADP+-dependent GDH
- CFU
- colony-forming unit
- DCFH-DA
- 2′,7′-dichlorodihydrofluorescein diacetate
- GPx
- glutathione peroxidase
- PI
- propidium iodide
- ROS
- reactive oxygen species
- SCD
- synthetic complete dextrose.
REFERENCES
- 1. Hudson R. C., Daniel R. M. (1993) l-Glutamate dehydrogenases: distribution, properties, and mechanism. Comp. Biochem. Physiol. B 106, 767–792 [DOI] [PubMed] [Google Scholar]
- 2. McKenna M. C., Sonnewald U., Huang X., Stevenson J., Zielke H. R. (1996) Exogenous glutamate concentration regulates the metabolic fate of glutamate in astrocytes. J. Neurochem. 66, 386–393 [DOI] [PubMed] [Google Scholar]
- 3. Plaitakis A., Zaganas I. (2001) Regulation of human glutamate dehydrogenases: implications for glutamate, ammonia, and energy metabolism in brain. J. Neurosci. Res. 66, 899–908 [DOI] [PubMed] [Google Scholar]
- 4. Cooper A. J., Plum F. (1987) Biochemistry and physiology of brain ammonia. Physiol. Rev. 67, 440–519 [DOI] [PubMed] [Google Scholar]
- 5. Plaitakis A., Metaxari M., Shashidharan P. (2000) Nerve tissue-specific (GLUD2) and housekeeping (GLUD1) human glutamate dehydrogenases are regulated by distinct allosteric mechanisms: implications for biologic function. J. Neurochem. 75, 1862–1869 [DOI] [PubMed] [Google Scholar]
- 6. Mavrothalassitis G., Tzimagiorgis G., Mitsialis A., Zannis V., Plaitakis A., Papamatheakis J., Moschonas N. (1988) Isolation and characterization of cDNA clones encoding human liver glutamate dehydrogenase: evidence for a small gene family. Proc. Natl. Acad. Sci. U.S.A. 85, 3494–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shashidharan P., Michaelidis T. M., Robakis N. K., Kresovali A., Papamatheakis J., Plaitakis A. (1994) Novel human glutamate dehydrogenase expressed in neural and testicular tissues and encoded by an X-linked intronless gene. J. Biol. Chem. 269, 16971–16976 [PubMed] [Google Scholar]
- 8. Fahien L. A., MacDonald M. J., Kmiotek E. H., Mertz R. J., Fahien C. M. (1988) Regulation of insulin release by factors that also modify glutamate dehydrogenase. J. Biol. Chem. 263, 13610–13614 [PubMed] [Google Scholar]
- 9. Sener A., Malaisse-Lagae F., Malaisse W. J. (1981) Stimulation of pancreatic islet metabolism and insulin release by a nonmetabolizable amino acid. Proc. Natl. Acad. Sci. U.S.A. 78, 5460–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stanley C. A., Lieu Y. K., Hsu B. Y., Burlina A. B., Greenberg C. R., Hopwood N. J., Perlman K., Rich B. H., Zammarchi E., Poncz M. (1998) Hyperinsulinism and hyperammonemia in infants with regulatory mutations of the glutamate dehydrogenase gene. N. Engl. J. Med. 338, 1352–1357 [DOI] [PubMed] [Google Scholar]
- 11. Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., Wolberger C., Prolla T. A., Weindruch R., Alt F. W., Guarente L. (2006) SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126, 941–954 [DOI] [PubMed] [Google Scholar]
- 12. Shashidharan P., Clarke D. D., Ahmed N., Moschonas N., Plaitakis A. (1997) Nerve tissue-specific human glutamate dehydrogenase that is thermolabile and highly regulated by ADP. J. Neurochem. 68, 1804–1811 [DOI] [PubMed] [Google Scholar]
- 13. Zaganas I., Kanavouras K., Mastorodemos V., Latsoudis H., Spanaki C., Plaitakis A. (2009) The human GLUD2 glutamate dehydrogenase: localization and functional aspects. Neurochem. Int. 55, 52–63 [DOI] [PubMed] [Google Scholar]
- 14. Plaitakis A., Latsoudis H., Spanaki C. (2011) The human GLUD2 glutamate dehydrogenase and its regulation in health and disease. Neurochem. Int. 59, 495–509 [DOI] [PubMed] [Google Scholar]
- 15. Miller S. M., Magasanik B. (1990) Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. J. Bacteriol. 172, 4927–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Avendaño A., Deluna A., Olivera H., Valenzuela L., Gonzalez A. (1997) GDH3 encodes a glutamate dehydrogenase isozyme, a previously unrecognized route for glutamate biosynthesis in Saccharomyces cerevisiae. J. Bacteriol. 179, 5594–5597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benjamin P. M., Wu J. I., Mitchell A. P., Magasanik B. (1989) Three regulatory systems control expression of glutamine synthetase in Saccharomyces cerevisiae at the level of transcription. Mol. Gen. Genet. 217, 370–377 [DOI] [PubMed] [Google Scholar]
- 18. Filetici P., Martegani M. P., Valenzuela L., González A., Ballario P. (1996) Sequence of the GLT1 gene from Saccharomyces cerevisiae reveals the domain structure of yeast glutamate synthase. Yeast 12, 1359–1366 [DOI] [PubMed] [Google Scholar]
- 19. DeLuna A., Avendano A., Riego L., Gonzalez A. (2001) NADP-glutamate dehydrogenase isoenzymes of Saccharomyces cerevisiae. Purification, kinetic properties, and physiological roles. J. Biol. Chem. 276, 43775–43783 [DOI] [PubMed] [Google Scholar]
- 20. Mazón M. J., Hemmings B. A. (1979) Regulation of Saccharomyces cerevisiae nicotinamide adenine dinucleotide phosphate-dependent glutamate dehydrogenase by proteolysis during carbon starvation. J. Bacteriol. 139, 686–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ito H., Fukuda Y., Murata K., Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Madeo F., Fröhlich E., Fröhlich K. U. (1997) A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139, 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu C. Y., Bird A. J., Winge D. R., Eide D. J. (2007) Regulation of the yeast TSA1 peroxiredoxin by ZAP1 is an adaptive response to the oxidative stress of zinc deficiency. J. Biol. Chem. 282, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 24. Cogoni C., Valenzuela L., González-Halphen D., Olivera H., Macino G., Ballario P., González A. (1995) Saccharomyces cerevisiae has a single glutamate synthase gene coding for a plant-like high-molecular-weight polypeptide. J. Bacteriol. 177, 792–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Doherty D. (1970) l-Glutamate dehydrogenase (yeast). Methods Enzymol. 17, 850–856 [Google Scholar]
- 26. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 27. Anderson M. E. (1985) Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol. 113, 548–555 [DOI] [PubMed] [Google Scholar]
- 28. Lauber K., Appel H. A., Schlosser S. F., Gregor M., Schulze-Osthoff K., Wesselborg S. (2001) The adapter protein apoptotic protease-activating factor-1 (Apaf-1) is proteolytically processed during apoptosis. J. Biol. Chem. 276, 29772–29781 [DOI] [PubMed] [Google Scholar]
- 29. Fabrizio P., Battistella L., Vardavas R., Gattazzo C., Liou L. L., Diaspro A., Dossen J. W., Gralla E. B., Longo V. D. (2004) Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J. Cell Biol. 166, 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fabrizio P., Longo V. D. (2003) The chronological life span of Saccharomyces cerevisiae. Aging Cell 2, 73–81 [DOI] [PubMed] [Google Scholar]
- 31. Lee Y. J., Hoe K. L., Maeng P. J. (2007) Yeast cells lacking the CIT1-encoded mitochondrial citrate synthase are hypersusceptible to heat- or aging-induced apoptosis. Mol. Biol. Cell 18, 3556–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Madeo F., Fröhlich E., Ligr M., Grey M., Sigrist S. J., Wolf D. H., Fröhlich K. U. (1999) Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145, 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mayor T., Lipford J. R., Graumann J., Smith G. T., Deshaies R. J. (2005) Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol. Cell. Proteomics 4, 741–751 [DOI] [PubMed] [Google Scholar]
- 34. Grant C. M., MacIver F. H., Dawes I. W. (1996) Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr. Genet. 29, 511–515 [DOI] [PubMed] [Google Scholar]
- 35. Grant C. M. (2001) Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol. 39, 533–541 [DOI] [PubMed] [Google Scholar]
- 36. Avendaño A., Riego L., DeLuna A., Aranda C., Romero G., Ishida C., Vázquez-Acevedo M., Rodarte B., Recillas-Targa F., Valenzuela L., Zonszein S., González A. (2005) Swi/SNF-GCN5-dependent chromatin remodelling determines induced expression of GDH3, one of the paralogous genes responsible for ammonium assimilation and glutamate biosynthesis in Saccharomyces cerevisiae. Mol. Microbiol. 57, 291–305 [DOI] [PubMed] [Google Scholar]
- 37. Riego L., Avendaño A., DeLuna A., Rodríguez E., González A. (2002) GDH1 expression is regulated by GLN3, GCN4, and HAP4 under respiratory growth. Biochem. Biophys. Res. Commun. 293, 79–85 [DOI] [PubMed] [Google Scholar]
- 38. Bermingham-McDonogh O., Gralla E. B., Valentine J. S. (1988) The copper, zinc-superoxide dismutase gene of Saccharomyces cerevisiae: cloning, sequencing, and biological activity. Proc. Natl. Acad. Sci. U.S.A. 85, 4789–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Galiazzo F., Schiesser A., Rotilio G. (1987) Glutathione peroxidase in yeast. Presence of the enzyme and induction by oxidative conditions. Biochem. Biophys. Res. Commun. 147, 1200–1205 [DOI] [PubMed] [Google Scholar]
- 40. Avery A. M., Avery S. V. (2001) Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. J. Biol. Chem. 276, 33730–33735 [DOI] [PubMed] [Google Scholar]
- 41. Armstrong J. S., Steinauer K. K., Hornung B., Irish J. M., Lecane P., Birrell G. W., Peehl D. M., Knox S. J. (2002) Role of glutathione depletion and reactive oxygen species generation in apoptotic signaling in a human B lymphoma cell line. Cell Death Differ. 9, 252–263 [DOI] [PubMed] [Google Scholar]
- 42. Matés J. M., Pérez-Gómez C., Núñez de Castro I., Asenjo M., Márquez J. (2002) Glutamine and its relationship with intracellular redox status, oxidative stress, and cell proliferation/death. Int. J. Biochem. Cell Biol. 34, 439–458 [DOI] [PubMed] [Google Scholar]
- 43. Savolainen K. M., Loikkanen J., Naarala J. (1995) Amplification of glutamate-induced oxidative stress. Toxicol. Lett. 82, 399–405 [DOI] [PubMed] [Google Scholar]
- 44. Cox K. H., Tate J. J., Cooper T. G. (2002) Cytoplasmic compartmentation of Gln3 during nitrogen catabolite repression and the mechanism of its nuclear localization during carbon starvation in Saccharomyces cerevisiae. J. Biol. Chem. 277, 37559–37566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mayor T., Deshaies R. J. (2005) Two-step affinity purification of multiubiquitylated proteins from Saccharomyces cerevisiae. Methods Enzymol. 399, 385–392 [DOI] [PubMed] [Google Scholar]
- 46. Catic A., Collins C., Church G. M., Ploegh H. L. (2004) Preferred in vivo ubiquitination sites. Bioinformatics 20, 3302–3307 [DOI] [PubMed] [Google Scholar]
- 47. Werner-Washburne M., Braun E., Johnston G. C., Singer R. A. (1993) Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57, 383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Werner-Washburne M., Braun E. L., Crawford M. E., Peck V. M. (1996) Stationary phase in Saccharomyces cerevisiae. Mol. Microbiol. 19, 1159–1166 [DOI] [PubMed] [Google Scholar]
- 49. Herman P. K. (2002) Stationary phase in yeast. Curr. Opin. Microbiol. 5, 602–607 [DOI] [PubMed] [Google Scholar]
- 50. Paz I., Choder M. (2001) Eukaryotic translation initiation factor 4E-dependent translation is not essential for survival of starved yeast cells. J. Bacteriol. 183, 4477–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang X. F., Cynader M. S. (2000) Astrocytes provide cysteine to neurons by releasing glutathione. J. Neurochem. 74, 1434–1442 [DOI] [PubMed] [Google Scholar]