Background: Atg9 vesicles are directly involved in autophagosome formation.
Results: Mass spectrometric analysis revealed that Atg9 vesicles contain vesicle-tethering proteins Trs85 and Ypt1. These proteins localize to the autophagosome formation site in an Atg9-dependent manner.
Conclusion: Atg9 vesicles play a role in the recruitment of the vesicle-tethering machinery.
Significance: This study is the first proteomics study of Atg9 vesicles.
Keywords: Autophagy, Mass Spectrometry (MS), Membrane Trafficking, Rab, Vesicles, Atg9 Vesicle, TRAPP
Abstract
Atg9 is a transmembrane protein that is essential for autophagy. In the budding yeast Saccharomyces cerevisiae, it has recently been revealed that Atg9 exists on cytoplasmic small vesicles termed Atg9 vesicles. To identify the components of Atg9 vesicles, we purified the Atg9 vesicles and subjected them to mass spectrometry. We found that their protein composition was distinct from other organellar membranes and that Atg9 and Atg27 in particular are major components of Atg9 vesicles. In addition to these two components, Trs85, a specific subunit of the transport protein particle III (TRAPPIII) complex, and the Rab GTPase Ypt1 were also identified. Trs85 directly interacts with Atg9, and the Trs85-containing TRAPPIII complex facilitates the association of Ypt1 onto Atg9 vesicles. We also showed that Trs85 and Ypt1 are localized to the preautophagosomal structure in an Atg9-dependent manner. Our data suggest that Atg9 vesicles recruit the TRAPPIII complex and Ypt1 to the preautophagosomal structure. The vesicle-tethering machinery consequently acts in the process of autophagosome formation.
Introduction
Autophagy was initially described as a response to nutrient limitation. During starvation, a portion of cytoplasmic components is delivered into lytic compartments (i.e. lysosomes or vacuoles) and degraded (1–3). Because of its involvement in various biological processes including infectious and neurodegenerative diseases, in recent years, autophagy has attracted a great deal of interest (4, 5). Previous studies in yeast have identified over 30 autophagy-related (Atg)2 proteins; knowledge of these proteins has facilitated analysis of molecular mechanisms of autophagy (1). In response to starvation, most of the Atg proteins are localized at a perivacuolar punctate structure called the preautophagosomal structure (PAS) (6). According to the prevailing model, a membrane structure termed the isolation membrane emerges from the PAS, sequesters a portion of cytoplasm, seals, and finally forms a double membrane structure termed the autophagosome.
Atg9, a six-transmembrane protein exposing both the N- and C-terminal domains to the cytoplasm, is essential for autophagosome formation in both yeast and mammals (7–10). Recently, we showed that the majority of Atg9 in the yeast Saccharomyces cerevisiae exists on small cytoplasmic vesicles designated Atg9 vesicles (11). These vesicles, which move throughout the cytoplasm, are single membrane vesicles with a diameter of 30–60 nm. Furthermore, we revealed that Atg9 vesicles assemble to the PAS and become part of autophagosomal membrane, suggesting that Atg9 vesicles are directly involved in autophagosome formation (11). However, it remains unclear how Atg9 vesicles function. In this study, we characterized Atg9 vesicles by immunopurification followed by mass spectrometric analysis, which identified Trs85 and Ypt1 as proteins that reside on Atg9 vesicles. Trs85 is a component of the transport protein particle III (TRAPPIII) complex (12). TRAPP complexes are known to function as a GDP/GTP exchange factor for Rab GTPases, Ypt proteins in yeast, and also as vesicle-tethering complexes required for membrane trafficking between the endoplasmic reticulum and the Golgi apparatus (13). The Rab GTPase Ypt1 also functions in vesicle tethering (14). Although both Trs85 and Ypt1 were reported to be localized to the PAS and involved in autophagy (12, 15, 16), to date it is unclear how these proteins are localized to the PAS and in which step these proteins function during autophagosome formation. Here, we suggest that Atg9 vesicles transport Trs85 and Ypt1 to the PAS, allowing these vesicle-tethering proteins to function in the process of autophagosome formation.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, Plasmids, and Antibodies
The S. cerevisiae strains used in this study were derived from SEY6210 (17) and are listed in supplemental Table S1. Cells were grown at 30 °C in YPD (1% yeast extract, 2% peptone, and 2% glucose) or in SD/CA medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.5% casamino acids, and 2% glucose) supplemented with appropriate amino acids. Autophagy was induced by treatment with 0.2 μg/ml rapamycin (Sigma). Gene deletion, truncation, promoter replacement, and C-terminal protein tagging with GFP, HA, Myc, and TAP were performed as described previously (18). The plasmids for expression of Atg9 and Atg9-6xFLAG and cells expressing Atg9-2xGFP and Atg9-3xBAP were constructed as described previously (11). To construct strains expressing mRFP-Ape1, pPS128 and pPS129 were integrated into the genome after digestion with AflII and AvrII, respectively, as described previously (19). To construct the strain expressing GFP-Ypt1 from the native promoter, a DNA fragment encoding yeGFP-YPT1 was integrated into the YPT1 locus by homologous recombination. In the resulting strain, yeGFP-Ypt1 is transcribed from its own promoter and terminated with the PGK1 terminator. The plasmid for expression of GFP-Atg8 was described previously (6). Antibodies against Vph1, Pgk1, Dpm1, Por1, and Pep12 were purchased from Invitrogen. Antibodies against GFP and HA (3F-10) were purchased from Roche Applied Science. Anti-Myc antibody (9E10) was purchased from Berkeley Antibody Co. Anti-Sed5 antibody was a kind gift from Dr. Koji Yoda (The University of Tokyo, Tokyo, Japan). Polyclonal antibodies against Atg9 and Atg27 were described previously (7, 11). HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories.
Immunoisolation
For immunoisolation of Atg9 vesicles, cells expressing Atg9-6xFLAG from a pRS316 single copy plasmid and Atg9-3xBAP from the ATG9 locus were used. Both Atg9 variants were expressed under the control of the native promoter and terminator. Cells under growing conditions or treated with rapamycin for 1 h were harvested, washed twice, and disrupted in a Multi-beads shocker (Yasui Kikai) with 0.5-mm zirconia beads in HSE buffer (25 mm HEPES-KOH, pH 7.2, 750 mm sorbitol, and 5 mm EDTA) including 0.5 mg/ml BSA and 50 mm NaCl. After centrifugation at 50,000 × g for 20 min at 4 °C, the supernatants were incubated with anti-FLAG antibody-bound Dynabeads® Protein G (Dynal) for 3 h at 4 °C. The beads were collected using a magnetic stand and washed three times with HSE buffer including 0.5 mg/ml BSA and 250 mm NaCl. In the two-step purification, elution was performed using 2 mg/ml 3xFLAG peptide (Sigma), and the eluate was incubated with Dynabeads MyOneTM Streptavidin C1 (Dynal) for 20 min at 4 °C. First, the beads were incubated with 0.5% Triton X-100 for 5 min on ice and then incubated at 65 °C for 10 min with SDS-PAGE sample buffer (50 mm Tris-HCl, pH 6.8, 1.6% SDS, 16 mm DTT, and 4% glycerol). Eluted proteins were subjected to SDS-PAGE and analyzed by immunoblotting or stained with SYPRO® Ruby protein gel stain (Invitrogen) followed by mass spectrometric analysis. For immunoisolation of TRAPP complexes, cells expressing Trs65-TAP or Trs85-TAP were disrupted in a Multi-beads shocker with HSE buffer. After centrifugation at 15,000 × g, the supernatants were solubilized with 0.5% Triton X-100 and incubated with IgG-Dynabeads (rabbit IgG was conjugated to M-270 Epoxy Dynabeads, Dynal) for 2 h at 4 °C. After washing with HSE buffer, bound proteins were eluted with AcTEVTM protease (Invitrogen) or SDS-PAGE sample buffer. For the in vitro binding assay, Trs65- or Trs85-bound beads were mixed with the isolated Atg9 vesicles in HSE buffer and incubated for 2 h at 4 °C. Beads were collected, and the supernatants (unbound sample) and the beads (bound sample) were incubated with SDS-PAGE sample buffer followed by immunoblotting analysis. Proteins were detected using a luminescent image analyzer (LAS-4000 mini, GE Healthcare) and quantified using MultiGauge software (Fujifilm).
Electron Microscopy
Negative staining was performed as follows. The Atg9 vesicles isolated as described above were applied onto a carbon-coated 400-mesh copper grid and stained with 3% uranyl acetate for 1 min at room temperature. The grids were examined using an H-7500 transmission electron microscope (Hitachi).
Identification of Proteins by Mass Spectrometric Analysis
Mass spectrometric analysis of the purified Atg9 vesicles was performed with three different methods. Atg9 vesicle-specific bands and gels at the corresponding positions in negative control samples were cut out. The excised protein bands were in-gel digested with trypsin according to a modified procedure of Ref. 20, and the proteins were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) or liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). In MALDI-TOF-MS analysis, masses of the peptides were determined by Ultraflex TOF/TOF (Bruker Daltonics), and the MS data obtained were searched against a protein sequence database (Swiss-Prot) using the Mascot search program (Matrix Science). LC-MS/MS analyses were performed using a nanoflow multidimensional HPLC system (Paradigm MS4, Michrom Bioresources) coupled to an electrospray ionization ion trap mass spectrometer (LCQ Deca XP, Thermo Fisher Scientific) according to a procedure of Ref. 20 or using a system that consisted of an inlet system (Acquity UPLC, Waters) and a Q-TOF mass spectrometer (SYNAPT G2 HDMS, Waters) by a newly constructed procedure as described below. The in-gel digested samples were injected to an inlet system with C18 column (75-μm diameter; 150-mm length; particle size, 1.7 μm) at a flow rate of 0.5 μl/min. Solutions used for the LC were ultrapure water containing 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B). A gradient program of 1% B (0 min), 40% B (30 min), and 95% B (31 min) was used. Samples separated by LC were introduced into the electrosprayer for MS/MS analyses in order. The mass spectrometer was operated in positive ionization mode, and a capillary voltage of 3.0–3.2 kV and a cone voltage of 30 V were utilized. The inlet LC flow was nebulized using nitrogen gas (700 liters/h). Argon gas was used for collision-induced dissociation. The system was equipped with an integral LockSpray unit with its own reference sprayer that was controlled automatically by the acquisition software to collect a reference scan every 10 s. The reference for LockSpray was Glu-fibrinopeptide B (Sigma) at a concentration of 200 fmol/μl. The reference solution was introduced into the lock mass sprayer at a flow rate of 0.5 μl/min. A single lock mass calibration at m/z 785.8426 in positive ion mode was used during analysis. MS data were acquired over the mass range of 50–1990 Da with a scan time of 1 s and a detector voltage of 1800 V. The MS data obtained were searched against Swiss-Prot through the application of the search program ProteinLynx Global Server (Waters). Proteins that had the appropriate molecular weight and were not detected in negative control samples were selected as candidates, and the three top scoring proteins for each band are listed in supplemental Table S2.
Two-hybrid Assays
The two-hybrid assays were performed using MatchmakerTM Gold (Clontech). The N-terminal region (amino acids 2–318) or C-terminal region (amino acids 747–997) of Atg9 was cloned into the NdeI-BamHI site of plasmid pGADT7. The full length, N-terminal half (amino acids 1–400), or C-terminal half (amino acids 401–698) of Trs85 was cloned into the NdeI-BamHI site of plasmid pGBKT7. AH109 transformants were selected on SD plates lacking leucine and tryptophan and tested for growth on SD plates lacking histidine, leucine, and tryptophan.
Fluorescence Microscopy
Fluorescence microscopy was performed using an inverted fluorescence microscope (IX-71, Olympus) equipped with an electron-multiplying charge-coupled device camera (ImagEM, C9100-13, Hamamatsu Photonics) and 150× total internal reflection fluorescence objective (UAPON 150XOTIRF, Olympus). Images were acquired at 200 ms/frame using AQUACOSMOS software (Hamamatsu Photonics) and processed using either AQUACOSMOS or MetaMorph software (Molecular Devices). The frequencies of the PAS localization of Trs85 and Ypt1 are shown as the percentages of Ape1 puncta colocalized with puncta of these proteins in 100 cells.
Alkaline Phosphatase Assay
Cells harboring pTN3 plasmid were starved for nitrogen for 4 h, harvested, and disrupted with glass beads. Alkaline phosphatase activity was measured as described in Noda et al. (21) using pTN3. The activity measured from wild-type cells was set to 100%, and that of other strains was normalized to wild-type activity.
RESULTS
Purification and Mass Spectrometric Analysis of Atg9 Vesicles
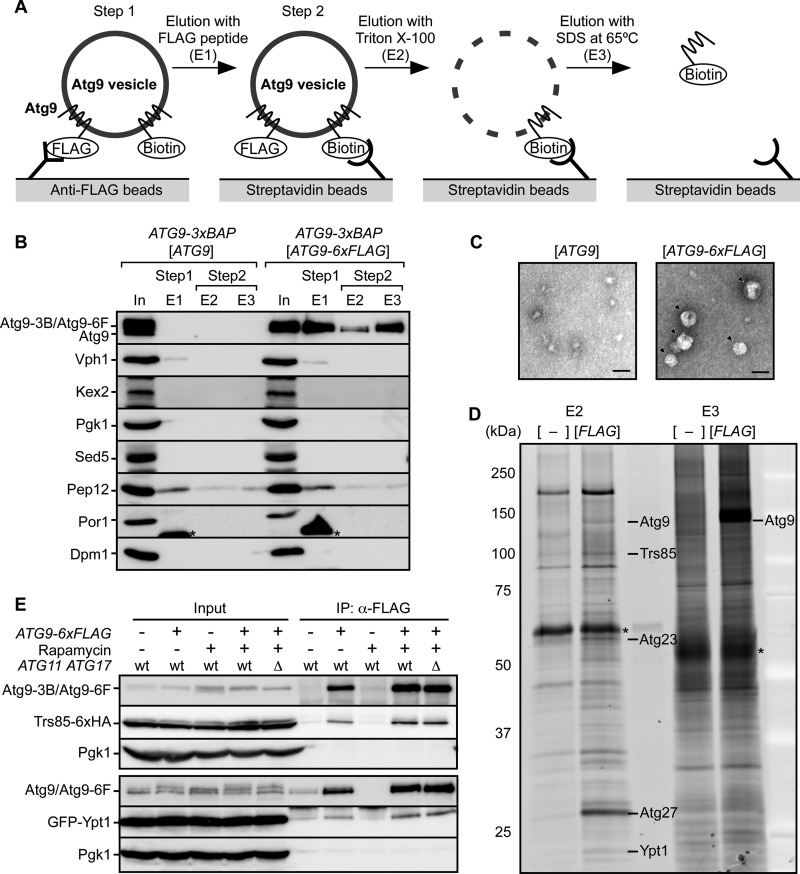
We showed previously that Atg9 exists on cytoplasmic vesicles with a diameter of 30–60 nm (11). To identify proteins associated with the Atg9 vesicles, we carried out immunopurification. For the purification of Atg9 vesicles, we constructed a strain expressing both Atg9-6xFLAG and Atg9-3xBAP (biotinylated tag) (22). Because each Atg9 vesicle contains multiple Atg9 molecules (24–32 molecules) (11), we expected that each vesicle in these cells would contain both FLAG-tagged and BAP-tagged Atg9, and therefore we utilized these tagged proteins for two-step purification. Cells expressing non-tagged Atg9 instead of Atg9-6xFLAG were used as a negative control. The two-step purification was performed as illustrated in Fig. 1A. At the first step, Atg9 vesicles were immunoisolated using anti-FLAG beads and eluted with 3xFLAG peptide (Fig. 1, A and B, Step 1). By this step, Atg9-6xFLAG was isolated specifically in contrast to the negative control sample (E1 fractions in Fig. 1B). Electron microscopy of the E1 fractions revealed vesicular structures with a diameter of 30–50 nm (Fig. 1C). Although Atg9 vesicles are derived from the Golgi apparatus (11, 23), the Golgi marker proteins Kex2 and Sed5 were not detected at all (E1 fractions in Fig. 1B), indicating that the isolated Atg9-containing structures were not a part of the Golgi apparatus. Still, E1 fractions contained Vph1, Pep12, and immunoglobulin as contaminants. At the second step, the Atg9-enriched fraction was further purified using streptavidin-conjugated beads (Fig. 1, A and B, Step 2). After washing the beads, bound proteins were eluted by sequential treatments with Triton X-100 (E2 fractions in Fig. 1B) and SDS-PAGE sample buffer (E3 fractions in Fig. 1B). As a result of the two-step purification, nonspecific binding of other organellar proteins was substantially diminished (Fig. 1B). The proteins in E2 and E3 fractions were visualized using SYPRO Ruby (Fig. 1D). Atg9 vesicle-specific bands (i.e. those not present in the negative control lanes) were cut out and subjected to mass spectrometry by three different procedures (see “Experimental Procedures”). The results of these experiments are shown in supplemental Table S2. Among the candidates with the highest score, Atg23, Atg27, and Ypt1 have been reported to be involved in autophagy (12, 24, 25). The detection of Atg23 and Atg27 was expected because it was reported previously that Atg23 and Atg27 interact with Atg9 (24, 25) and are involved in the biogenesis of Atg9 vesicles (11). Ypt1 has been previously reported to colocalize with Atg9 (12, 26). Interestingly, Trs85, which is known to interact with Ypt1 and be involved in autophagy (12), was also detected (supplemental Table S2). It is therefore possible that Ypt1 and Trs85 cooperatively function on Atg9 vesicles during autophagosome formation. To investigate whether Trs85 and Ypt1 exist on Atg9 vesicles, we subsequently performed co-immunoprecipitation experiments using anti-FLAG beads. We constructed strains expressing Trs85-6xHA or GFP-Ypt1 for detection in immunoblotting analysis and immunoprecipitated Atg9 vesicles from growing cells and cells treated with rapamycin, which mimics starvation conditions and induces autophagy (27). As shown in Fig. 1E, Trs85 and Ypt1 were coprecipitated with Atg9-6xFLAG in both growing and rapamycin-treated cells. We performed an additional experiment using atg11Δ atg17Δ cells in which the PAS is not formed and autophagosome formation is completely blocked. Trs85 and Ypt1 were coprecipitated even in atg11Δ atg17Δ cells (Fig. 1E). From these results, we conclude that Ypt1 and Trs85 are localized on the cytoplasmic Atg9 vesicles irrespective of PAS formation and the induction of autophagy.
FIGURE 1.
Purification and mass spectrometric analysis of Atg9 vesicles. A, an illustration of the two-step purification of Atg9 vesicles. See text for details. B, purification of Atg9 vesicles. Cells expressing Atg9-3xBAP (Atg9-3B) and Atg9-6xFLAG (Atg9-6F) or non-tagged Atg9 under growing conditions were disrupted using beads. Cell lysates (input (In)) were subjected to immunoisolation using anti-FLAG beads followed by elution with 3xFLAG peptide. The eluate (E1) was subjected to a second immunoisolation using streptavidin beads. Bound proteins were eluted by solubilization of membrane structures with Triton X-100 (E2), and the remaining proteins were recovered by denaturation with SDS sample buffer at 65 °C (E3). These fractions were subjected to immunoblotting using antibodies against Atg9, Vph1 (vacuole), Kex2 (late Golgi), Pgk1 (cytoplasm), Sed5 (early Golgi), Pep12 (endosome), Por1 (mitochondria), or Dpm1 (endoplasmic reticulum). Eluted fractions were concentrated 10-fold. Asterisks show bands corresponding to immunoglobulin. C, E1 fractions were subjected to negative staining with uranyl acetate and observed by electron microscopy. Arrowheads indicate isolated vesicles. Scale bars, 50 nm. D, eluted proteins (E2 and E3) were stained with SYPRO Ruby. In comparison with the negative control fractions (−), specific bands in the fractions of FLAG-tagged Atg9 (FLAG) were subjected to mass spectrometric analysis. A representative image is shown, and detected proteins are indicated by name at the corresponding positions. Asterisks show contaminating BSA and immunoglobulin. E, the co-immunoprecipitation assay for Trs85 and Ypt1. Cells expressing Trs85-6xHA or GFP-Ypt1 were harvested under growing or rapamycin-treated conditions. Atg9 vesicles were immunoisolated using anti-FLAG beads. Trs85-6xHA and GFP-Ypt1 were detected using anti-HA and anti-GFP antibodies, respectively. Immunoprecipitation (IP) fractions were concentrated 50-fold.
Trs85 Directly Interacts with Atg9
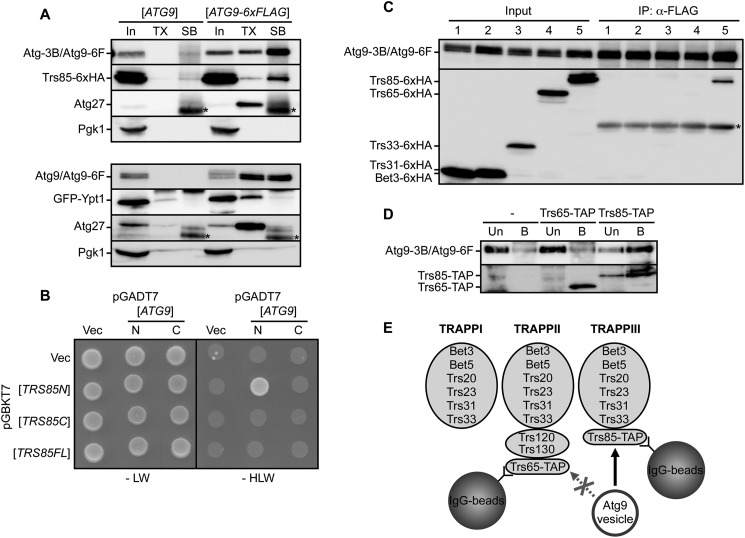
We investigated how Trs85 and Ypt1 are associated with Atg9 vesicles. For this purpose, Atg9 vesicles bound to anti-FLAG beads were treated with 0.5% Triton X-100 on ice (Fig. 2A, TX) and then treated with 1.6% SDS at 65 °C to recover all proteins bound to the beads (Fig. 2A, SB). By treatment with Triton X-100, Atg9-6xFLAG was not efficiently released from the beads (Fig. 2A), indicating that the large majority of Atg9-6xFLAG was directly associated with anti-FLAG beads. Likewise, Trs85 was not efficiently released after treatment with Triton X-100, whereas Ypt1 was released completely as was Atg27. These results imply that Trs85, but not Ypt1, interacts directly with Atg9. Furthermore, yeast two-hybrid assay data suggest an interaction between the N-terminal half of Trs85 and the N-terminal cytoplasmic domain of Atg9 (Fig. 2B and supplemental Fig. S1A), although full-length Trs85 did not show any positive interaction. These results support a direct interaction between Atg9 and Trs85.
FIGURE 2.
Trs85 interacts with Atg9. A, co-immunoprecipitation experiments using anti-FLAG beads and cells expressing Atg9-6xFLAG or non-tagged Atg9. Cells were harvested after rapamycin treatment for 1 h. Immunoisolated Atg9 vesicles were solubilized with Triton X-100 (TX), and remaining proteins were recovered with SDS sample buffer (SB). Proteins were analyzed by immunoblotting. Triton X-100 and SDS sample buffer fractions were concentrated 50-fold. Asterisks show bands corresponding to immunoglobulin. B, two-hybrid assay between Atg9 and Trs85. N-terminal (N) and C-terminal (C) regions of Atg9 were fused with the Gal4 activation domain. The full length and N-terminal and C-terminal halves of Trs85 were fused with the DNA-binding domain. AH109 strains transformed with each vector (Vec) were grown on SD lacking either leucine and tryptophan (−LW) or histidine, leucine, and tryptophan (−HLW). C, cells expressing Atg9-6xFLAG and HA-tagged subunits of TRAPP complexes (lane 1, Bet3; lane 2, Trs31; lane 3, Trs33; lane 4, Trs65; lane 5, Trs85) were treated with rapamycin for 1 h and subjected to a co-immunoprecipitation assay. Immunoprecipitation (IP) fractions were concentrated 50-fold. Asterisks show bands corresponding to immunoglobulin. D, the in vitro binding assay using isolated Atg9 vesicles. Isolated Atg9 vesicles were incubated with Trs65- or Trs85-bound beads. Proteins that were unbound (Un) or bound (B) to the beads were subjected to immunoblotting. Trs65-TAP and Trs85-TAP were detected with HRP-conjugated anti-mouse IgG. As a negative control, cells not expressing TAP-tagged proteins were also subjected to the assay. E, schematic illustration of the in vitro binding assay. Isolated Atg9 vesicles were bound to Trs85 (TRAPPIII) beads but not to Trs65 (TRAPPII) beads. Atg9-3B, Atg9-3xBAP; Atg9-6F, Atg9-6xFLAG.
Trs85 is a subunit of the TRAPPIII complex, one of three distinct TRAPP complexes in yeast (13) (see also Fig. 2E). The TRAPPI complex is composed of six subunits, Bet3, Bet5, Trs20, Trs23, Trs31, and Trs33. These six subunits are common components and are present in both the TRAPPII complex and the TRAPPIII complex. In addition to six common subunits, the TRAPPII complex contains three specific subunits, Trs65, Trs120, and Trs130, whereas the TRAPPIII complex contains one specific subunit, Trs85. Therefore, we conducted a co-immunoprecipitation assay for other subunits of TRAPP complexes. Among the components of TRAPP complexes (Bet3, Trs31, Trs33, Trs65, and Trs85), only Trs85 was coprecipitated with Atg9-6xFLAG, but other subunits were not (Fig. 2C). These results indicate that other TRAPP subunits are not required for the interaction between Trs85 and Atg9, although we cannot exclude the possibility that other subunits were detached from Atg9 vesicles during the immunoprecipitation. For further analysis, we performed an in vitro binding assay between Atg9 vesicles and TRAPP complexes. We prepared Trs85 (TRAPPIII)-bound beads (and Trs65 (TRAPPII)-bound beads as a control) using TAP-tagged Trs85- and IgG-conjugated beads. Then Atg9 vesicles were isolated from yeast cells and incubated with Trs85-bound beads. After the incubation, the Atg9 vesicles were efficiently associated with Trs85-bound beads but not with Trs65-bound beads (Fig. 2D). Although both Trs85-bound beads and Trs65-bound beads contained common subunits of TRAPP complexes (supplemental Fig. S1, B and C), the Atg9 vesicles were associated only with Trs85-bound beads. Therefore, these results suggested that Atg9 vesicles bound directly to Trs85 but not to other TRAPP subunits (Fig. 2E).
The PAS Localization of Trs85 Is Dependent on Atg9
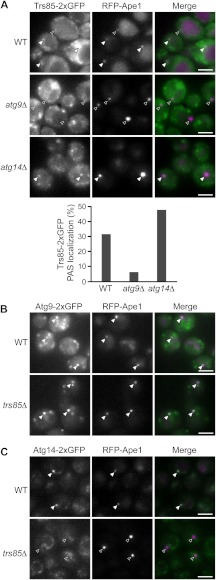
A previous study reported that Trs85 was localized to the PAS (12), but another group could not reproduce this PAS localization (28). Based on our finding that Trs85 is present on the Atg9 vesicles, we assumed that Trs85 would localize to the PAS in an Atg9-dependent manner. To test this idea, we constructed a strain expressing both Trs85-2xGFP and RFP-Ape1, which is the most widely studied cargo of selective autophagy and is frequently used as a PAS marker (1). We examined the localization of these proteins under growing and rapamycin-treated conditions. Fluorescence microscopy revealed that only a minor, but significant portion of Trs85-2xGFP does indeed colocalize with RFP-Ape1 in rapamycin-treated cells (Fig. 3A, top, 31% of Ape1 puncta was colocalized with Trs85). However, the colocalization of Trs85-2xGFP with RFP-Ape1 was significantly diminished in atg9Δ cells (Fig. 3A, middle, 6% colocalized). Under growing conditions, the PAS localization of Trs85 was similarly reduced in atg9Δ cells (supplemental Fig. S2A). This clearly indicates that the PAS localization of Trs85 is dependent on Atg9. Furthermore, the PAS localization of Trs85 was enhanced in atg14Δ cells (Fig. 3A, bottom, 47% and supplemental Fig. S2A, bottom, 35%) in which Atg9 is highly accumulated at the PAS (29). We also observed that Trs85-2xGFP was colocalized with Atg9-2xmCherry at the PAS in atg14Δ cells (supplemental Fig. S3). In addition to Trs85, other subunits of the TRAPPIII complex, Bet3 and Trs33, were colocalized with RFP-Ape1 (supplemental Fig. S2B). From these results, we conclude that Trs85 is localized to the PAS as a component of the Atg9 vesicles and thereby recruits the TRAPPIII complex to the PAS.
FIGURE 3.
The PAS localization of Trs85 is dependent on Atg9. A, the localization of Trs85-2xGFP in wild-type, atg9Δ, and atg14Δ cells. Cells were treated with rapamycin for 1 h. The percentages of the PAS localization of Trs85-2xGFP in 100 cells are shown in the bar graph. B and C, the localization of Atg9-2xGFP (B) and Atg14-2xGFP (C) in wild-type and trs85Δ cells under growing conditions. RFP-Ape1 was used as a PAS marker. Filled arrowheads and open arrowheads indicate dots that were colocalized and not colocalized, respectively. Scale bars, 3 μm.
Trs85 Is Involved in the PAS Localization of Atg14
We next investigated whether the PAS localization of Atg proteins is compromised in trs85Δ cells. We first observed the localization of Atg9-2xGFP in wild-type and trs85Δ cells. Even in trs85Δ cells, Atg9 vesicles were generated normally (supplemental Movies S1 and S2). In addition, the Atg9 vesicles colocalized with RFP-Ape1 in trs85Δ cells under growing and rapamycin-treated conditions with no apparent defect (Fig. 3B and data not shown). These results indicate that Trs85 is not required for the biogenesis of Atg9 vesicles and their PAS localization. As the PAS localization of Atg14, a subunit of phosphatidylinositol 3-kinase (PI3K) complex I (1), is dependent on Atg9 (29), we next asked whether Atg14-2xGFP localizes to the PAS in trs85Δ cells. In trs85Δ cells, Atg14-2xGFP was scarcely localized to the PAS under growing conditions (Fig. 3C). On the contrary, the PAS localization of Trs85 was not blocked in the absence of Atg14 (supplemental Fig. S3). Similar to Atg14-2xGFP, GFP-Atg8 was not localized to the PAS under growing conditions in trs85Δ cells (supplemental Fig. S2C). The failure of PAS localization of GFP-Atg8, which was also reported by Meiling-Wesse et al. (16), may be caused by a defect in the recruitment of PI3K complex to the PAS (29). These results suggest that Trs85 is required for the PAS recruitment of the Atg14-containing PI3K complex and Atg8 under growing conditions. Here, we noted that the PAS localization of Atg14 and Atg8 is not significantly reduced in trs85Δ cells under rapamycin-treated conditions (data not shown; also reported in Ref. 16). Consistently, trs85Δ cells showed a partial defect in autophagosome formation (supplemental Fig. S2D).
Ypt1 Is Localized on Atg9 Vesicles and to the PAS in a Trs85-dependent Manner
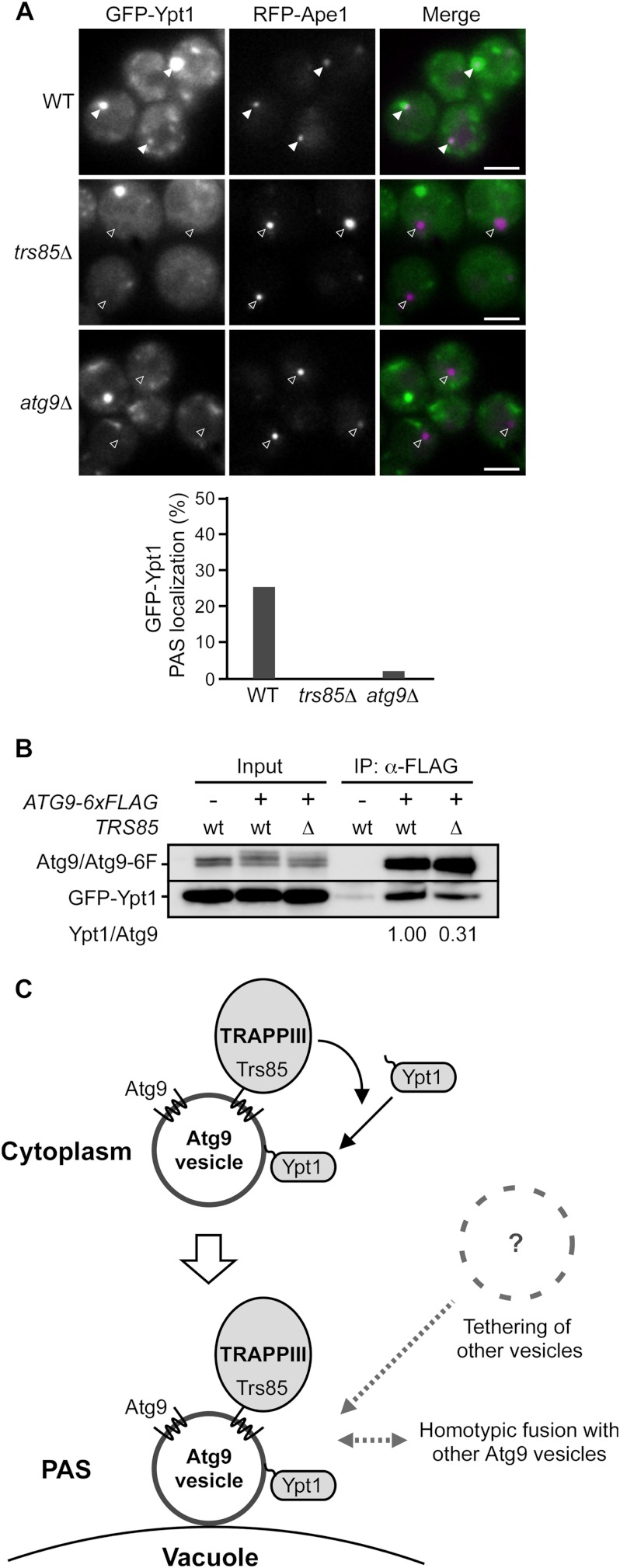
We subsequently examined the localization of GFP-Ypt1. GFP-Ypt1 was observed as several bright dots, one of which was often colocalized with RFP-Ape1 (Fig. 4A, top, 25% colocalized under rapamycin-treated conditions and supplemental Fig. S4, under growing conditions). The PAS localization of GFP-Ypt1 was significantly diminished in trs85Δ cells and atg9Δ cells as expected (Fig. 4A, middle, 0%, and bottom, 2%, respectively, under rapamycin-treated conditions and supplemental Fig. S4, under growing conditions). These results indicate that Ypt1 is localized to the PAS in a Trs85- and Atg9-dependent manner. Given that Ypt1 is located on Atg9 vesicles, these results imply that the localization of Ypt1 onto Atg9 vesicles is dependent on Trs85. We carried out a co-immunoprecipitation assay using wild-type and trs85Δ cells expressing Atg9-6xFLAG. The amount of coprecipitated Ypt1 was decreased in trs85Δ cells relative to wild-type cells (Fig. 4B). From these findings, we conclude that Ypt1 is associated with the Atg9 vesicles with the aid of Trs85 (and the TRAPPIII complex), and thereby Atg9 vesicles recruit Ypt1 to the PAS (Fig. 4C).
FIGURE 4.
Ypt1 is localized on Atg9 vesicles and transported to the PAS in a Trs85-dependent manner. A, the localization of GFP-Ypt1 in wild-type, trs85Δ, and atg9Δ cells under rapamycin-treated conditions. RFP-Ape1 was used as a PAS marker. Filled arrowheads and open arrowheads indicate dots that were colocalized or not colocalized, respectively. The percentages of the PAS localization of GFP-Ypt1 in 100 cells are shown in the bar graph. Scale bars, 3 μm. B, co-immunoprecipitation experiments using wild-type and trs85Δ cells. Cells were treated with rapamycin for 1 h, and the Atg9 vesicles were immunoprecipitated using anti-FLAG beads. Immunoprecipitation (IP) fractions were concentrated 50-fold. The ratios of relative amounts of coprecipitated Ypt1 between wild-type and trs85Δ cells normalized to the amount of precipitated Atg9 are shown. C, schematic diagram of how Ypt1 is transported to the PAS and functions in autophagosome formation. In this model, the Trs85-containing TRAPPIII complex is localized onto Atg9 vesicles via the interaction between Trs85 and Atg9. Prenylated Ypt1 is associated with cytoplasmic Atg9 vesicles with the aid of the TRAPPIII complex. After the PAS localization of Atg9 vesicles, other Atg9 vesicles or other unidentified membrane structures are tethered to the Atg9 vesicle at the PAS by Ypt1 and the TRAPPIII complex.
DISCUSSION
Atg9 is a conserved multispanning transmembrane protein that is essential for autophagosome formation and has been postulated to deliver membrane lipids to the autophagosome. Recently, we reported that Atg9 vesicles become part of the autophagosomal membrane (11). Therefore, we expected that Atg9 vesicles would contain the proteins that mediate vesicle tethering or membrane fusion. In the present study, we report the first proteomics analysis of Atg9 vesicles. As shown in Fig. 1, B and D, Atg9 vesicles contain Atg9 and Atg27 as major protein components but do not contain known organelle marker proteins. These results indicated that Atg9 vesicles are unconventional and specific vesicles responsible for autophagy. Our subsequent mass spectrometric and immunoprecipitation analyses revealed that two vesicle-tethering proteins, Trs85 and Ypt1, are localized on Atg9 vesicles (Fig. 1). Until now, it has been unclear how Trs85 and Ypt1 are localized to the PAS. The interaction between Trs85 and Atg9 suggests that Trs85 is delivered to the PAS by Atg9 vesicles. Consistently, the PAS localization of Trs85 was dependent on Atg9 but not vice versa (Fig. 3, A and B). As Trs85 is the sole specific subunit of the TRAPPIII complex, it is likely that Trs85 directs the TRAPPIII complex to the PAS via its interaction with Atg9 (Fig. 4C). Ypt1 is also localized to the PAS in an Atg9- and Trs85-dependent manner. Ypt proteins (or Rab GTPases) are prenylated and attached to membranes; this membrane association is assumed to be facilitated by GDP/GTP exchange factors such as TRAPP complexes, although the detailed mechanism is controversial (14, 30, 31). It seems likely that the TRAPPIII complex recruits Ypt1 onto Atg9 vesicles, resulting in the PAS localization of Ypt1 (Fig. 4C). Small amounts of Ypt1 are present on Atg9 vesicles even in trs85Δ cells (Fig. 4B); i.e. Ypt1 binds to the Atg9 vesicles to some extent regardless of the aid of the TRAPPIII complex. In proteomics analyses of Atg9 vesicles, we also detected Gyp8 and Yip5 (supplemental Table S2), both of which are Ypt1-interacting proteins (32, 33). These proteins may also be recruited to the PAS as components of Atg9 vesicles and regulate the activity of Ypt1. Our analyses also detected two proteins that are involved in phosphoinositide metabolism, Mss4 and Scs2 (supplemental Table S2). During the preparation of this manuscript, Wang et al. (34) reported that phosphoinositide kinases Pik1 and Mss4 were implicated in autophagy or mitophagy and that impairment of Pik1 affected the trafficking of Atg9. Mss4 and Scs2 might be involved in autophagy through interactions with Atg9 vesicles. Further study is required to elucidate the functions of these proteins in autophagy.
The source(s) of membranes supplied to the autophagosome is one of the most important unresolved issues in autophagy (35); despite substantial advances in our understanding of autophagy, the membrane origin of autophagosomes in yeast remains unknown. We recently showed that Atg9 vesicles are incorporated into the autophagosomal membrane but by themselves do not supply sufficient lipids (11), implying the existence of other lipid sources. Here, we showed that Trs85 and Ypt1 are present on Atg9 vesicles and consequently localize to the PAS. Recently, it was reported that Atg11 is a downstream effector of Ypt1 (26). However, ypt1 mutants and trs85Δ cells showed a more severe defect in autophagy than did atg11Δ cells (12, 26); thus, it is likely that Ypt1 and Trs85 have another function in autophagy. Taking into consideration the known roles of TRAPP complexes and Ypt1, it is reasonable to surmise that the TRAPPIII complex and Ypt1 on Atg9 vesicles tether some kind of membrane structures at the PAS (Fig. 4C), although it remains an open question what membrane structures are tethered to these vesicles. The TRAPPIII complex contains all subunits of the TRAPPI complex, and the TRAPPI complex and Ypt1 act in the tethering of COPII-coated endoplasmic reticulum-derived vesicles (13, 14). Given that endoplasmic reticulum membranes have been considered to be possible sources of autophagosomal membranes in mammalian cells (36), the COPII-coated membrane structure is a possible candidate to be tethered by Ypt1/Trs85 at the PAS. Alternatively, these proteins may tether Atg9 vesicles with each other followed by homotypic fusion. In mammalian cells, it has been reported that homotypic fusion of precursor structures is required for autophagosome formation (37). In either case, the tethering is probably followed by the recruitment of the Atg14-containing PI3K complex to the PAS; this PI3K complex might generate phosphoinositide 3-phosphate on membranes tethered by TRAPPIII and Ypt1. The membrane structures tethered to Atg9 vesicles would be fused during autophagosome formation, and several SNAREs were reported to be involved in autophagy (37, 38). Future investigations are needed to determine which proteins mediate the membrane fusion.
Recently, several groups have reported the existence of the human homolog of Trs85, TRAPPC8 (28, 39–41). Furthermore, TRAPPC8 and Rab1, a mammalian homolog of Ypt1, are involved in autophagy (39, 42, 43). These findings raise the possibility that TRAPPC8 and Rab1 also interact with Atg9-containing membrane structures and function in vesicle tethering at the autophagosome formation sites in mammalian cells. Although several organelles have been reported as putative membrane sources of autophagosomal membranes in mammalian cells (36, 44, 45), the molecular mechanisms of membrane supply remain unclear. Future investigations of Trs85, Ypt1, and their mammalian homologs and identification of the membrane structure tethered by these proteins will provide important clues to the origin of autophagosomal membranes and the mechanism of membrane supply in autophagosome formation.
Acknowledgments
We are grateful to Dr. Akihiko Takasaki and Hisashi Kodama for technical assistance in LC-MS/MS analysis. We thank Dr. Daniel J. Klionsky for providing the mRFP-Ape1 constructs. We also thank the members of the Ohsumi laboratory for materials and helpful discussion and the Bio-Technical Center, Technical Department, Tokyo Institute of Technology for performing MALDI-TOF-MS analysis.
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.

This article contains supplemental Figs. S1–S4, Movies S1 and S2, and Tables S1 and S2.
- Atg
- autophagy-related
- TRAPP
- transport protein particle
- PAS
- preautophagosomal structure
- TAP
- tandem affinity purification
- BAP
- biotin acceptor peptide
- RFP
- red fluorescent protein.
REFERENCES
- 1. Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 2. Inoue Y., Klionsky D. J. (2010) Regulation of macroautophagy in Saccharomyces cerevisiae. Semin. Cell Dev. Biol. 21, 664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mizushima N., Yoshimori T., Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 [DOI] [PubMed] [Google Scholar]
- 4. Levine B., Mizushima N., Virgin H. W. (2011) Autophagy in immunity and inflammation. Nature 469, 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mizushima N., Komatsu M. (2011) Autophagy: renovation of cells and tissues. Cell 147, 728–741 [DOI] [PubMed] [Google Scholar]
- 6. Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Noda T., Kim J., Huang W. P., Baba M., Tokunaga C., Ohsumi Y., Klionsky D. J. (2000) Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148, 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lang T., Reiche S., Straub M., Bredschneider M., Thumm M. (2000) Autophagy and the cvt pathway both depend on AUT9. J. Bacteriol. 182, 2125–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamada T., Carson A. R., Caniggia I., Umebayashi K., Yoshimori T., Nakabayashi K., Scherer S. W. (2005) Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J. Biol. Chem. 280, 18283–18290 [DOI] [PubMed] [Google Scholar]
- 10. Young A. R., Chan E. Y., Hu X. W., Köchl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., Tooze S. A. (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119, 3888–3900 [DOI] [PubMed] [Google Scholar]
- 11. Yamamoto H., Kakuta S., Watanabe T. M., Kitamura A., Sekito T., Kondo-Kakuta C., Ichikawa R., Kinjo M., Ohsumi Y. (2012) Atg9-containing small single-membrane vesicles become part of the autophagosomal membrane. J. Cell Biol. 198, 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lynch-Day M. A., Bhandari D., Menon S., Huang J., Cai H., Bartholomew C. R., Brumell J. H., Ferro-Novick S., Klionsky D. J. (2010) Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 7811–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barrowman J., Bhandari D., Reinisch K., Ferro-Novick S. (2010) TRAPP complexes in membrane traffic: convergence through a common Rab. Nat. Rev. Mol. Cell Biol. 11, 759–763 [DOI] [PubMed] [Google Scholar]
- 14. Hutagalung A. H., Novick P. J. (2011) Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nazarko T. Y., Huang J., Nicaud J. M., Klionsky D. J., Sibirny A. A. (2005) Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy 1, 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meiling-Wesse K., Epple U. D., Krick R., Barth H., Appelles A., Voss C., Eskelinen E. L., Thumm M. (2005) Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 280, 33669–33678 [DOI] [PubMed] [Google Scholar]
- 17. Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 19. Strømhaug P. E., Reggiori F., Guan J., Wang C. W., Klionsky D. J. (2004) Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell 15, 3553–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kurosawa G., Sumitomo M., Akahori Y., Matsuda K., Muramatsu C., Takasaki A., Iba Y., Eguchi K., Tanaka M., Suzuki K., Morita M., Sato N., Sugiura M., Sugioka A., Hayashi N., Kurosawa Y. (2009) Methods for comprehensive identification of membrane proteins recognized by a large number of monoclonal antibodies. J. Immunol. Methods 351, 1–12 [DOI] [PubMed] [Google Scholar]
- 21. Noda T., Matsuura A., Wada Y., Ohsumi Y. (1995) Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210, 126–132 [DOI] [PubMed] [Google Scholar]
- 22. van Werven F. J., Timmers H. T. (2006) The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Res. 34, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D. J., Reggiori F. (2010) An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190, 1005–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tucker K. A., Reggiori F., Dunn W. A., Jr., Klionsky D. J. (2003) Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J. Biol. Chem. 278, 48445–48452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yen W. L., Legakis J. E., Nair U., Klionsky D. J. (2007) Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell 18, 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lipatova Z., Belogortseva N., Zhang X. Q., Kim J., Taussig D., Segev N. (2012) Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc. Natl. Acad. Sci. U.S.A. 109, 6981–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Noda T., Ohsumi Y. (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966 [DOI] [PubMed] [Google Scholar]
- 28. Choi C., Davey M., Schluter C., Pandher P., Fang Y., Foster L. J., Conibear E. (2011) Organization and assembly of the TRAPPII complex. Traffic 12, 715–725 [DOI] [PubMed] [Google Scholar]
- 29. Suzuki K., Kubota Y., Sekito T., Ohsumi Y. (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12, 209–218 [DOI] [PubMed] [Google Scholar]
- 30. Goody R. S., Rak A., Alexandrov K. (2005) The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell. Mol. Life Sci. 62, 1657–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu Y. W., Oesterlin L. K., Tan K. T., Waldmann H., Alexandrov K., Goody R. S. (2010) Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nat. Chem. Biol. 6, 534–540 [DOI] [PubMed] [Google Scholar]
- 32. De Antoni A., Schmitzová J., Trepte H. H., Gallwitz D., Albert S. (2002) Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J. Biol. Chem. 277, 41023–41031 [DOI] [PubMed] [Google Scholar]
- 33. Calero M., Winand N. J., Collins R. N. (2002) Identification of the novel proteins Yip4p and Yip5p as Rab GTPase interacting factors. FEBS Lett. 515, 89–98 [DOI] [PubMed] [Google Scholar]
- 34. Wang K., Yang Z., Liu X., Mao K., Nair U., Klionsky D. J. (2012) Phosphatidylinositol 4-kinases are required for autophagic membrane trafficking. J. Biol. Chem. 287, 37964–37972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Longatti A., Tooze S. A. (2009) Vesicular trafficking and autophagosome formation. Cell Death Differ. 16, 956–965 [DOI] [PubMed] [Google Scholar]
- 36. Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 [DOI] [PubMed] [Google Scholar]
- 37. Moreau K., Ravikumar B., Renna M., Puri C., Rubinsztein D. C. (2011) Autophagosome precursor maturation requires homotypic fusion. Cell 146, 303–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nair U., Jotwani A., Geng J., Gammoh N., Richerson D., Yen W. L., Griffith J., Nag S., Wang K., Moss T., Baba M., McNew J. A., Jiang X., Reggiori F., Melia T. J., Klionsky D. J. (2011) SNARE proteins are required for macroautophagy. Cell 146, 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Behrends C., Sowa M. E., Gygi S. P., Harper J. W. (2010) Network organization of the human autophagy system. Nature 466, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scrivens P. J., Noueihed B., Shahrzad N., Hul S., Brunet S., Sacher M. (2011) C4orf41 and TTC-15 are mammalian TRAPP components with a role at an early stage in ER-to-Golgi trafficking. Mol. Biol. Cell 22, 2083–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zong M., Wu X. G., Chan C. W., Choi M. Y., Chan H. C., Tanner J. A., Yu S. (2011) The adaptor function of TRAPPC2 in mammalian TRAPPs explains TRAPPC2-associated SEDT and TRAPPC9-associated congenital intellectual disability. PLoS One 6, e23350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zoppino F. C., Militello R. D., Slavin I., Alvarez C., Colombo M. I. (2010) Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 11, 1246–1261 [DOI] [PubMed] [Google Scholar]
- 43. Huang J., Birmingham C. L., Shahnazari S., Shiu J., Zheng Y. T., Smith A. C., Campellone K. G., Heo W. D., Gruenheid S., Meyer T., Welch M. D., Ktistakis N. T., Kim P. K., Klionsky D. J., Brumell J. H. (2011) Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy 7, 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hailey D. W., Rambold A. S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P. K., Lippincott-Schwartz J. (2010) Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D. C. (2010) Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12, 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]