Background: Galloylated catechins, including (−)-epigallocatechin gallate and (−)-epicatechin gallate, comprise up to 76% of catechins in tea plant; their biosynthesis is unknown.
Results: An enzyme involved in galloylated catechin biosynthesis was purified and identified in tea plant.
Conclusion: Galloylated catechin was biosynthesized via a newly discovered enzyme, epicatechin:1-O-galloyl-β-d-glucose O-galloyltransferase.
Significance: This work improves our understanding of flavan-3-ols biosynthesis.
Keywords: Enzyme Catalysis, Flavonoids, Metabolism, Plant Molecular Biology, Protein Purification, Camellia Sinensis, Enzyme Properties, Enzyme Purification, Galloylated Catechins, Galloyltransferase
Abstract
Catechins (flavan-3-ols), the most important secondary metabolites in the tea plant, have positive effects on human health and are crucial in defense against pathogens of the tea plant. The aim of this study was to elucidate the biosynthetic pathway of galloylated catechins in the tea plant. The results suggested that galloylated catechins were biosynthesized via 1-O-glucose ester-dependent two-step reactions by acyltransferases, which involved two enzymes, UDP-glucose:galloyl-1-O-β-d-glucosyltransferase (UGGT) and a newly discovered enzyme, epicatechin:1-O-galloyl-β-d-glucose O-galloyltransferase (ECGT). In the first reaction, the galloylated acyl donor β-glucogallin was biosynthesized by UGGT from gallic acid and uridine diphosphate glucose. In the second reaction, galloylated catechins were produced by ECGT catalysis from β-glucogallin and 2,3-cis-flavan-3-ol. 2,3-cis-Flavan-3-ol and 1-O-galloyl-β-d-glucose were appropriate substrates of ECGT rather than 2,3-trans-flavan-3-ol and 1,2,3,4,6-pentagalloylglucose. Purification by more than 1641-fold to apparent homogeneity yielded ECGT with an estimated molecular mass of 241 to 121 kDa by gel filtration. Enzyme activity and SDS-PAGE analysis indicated that the native ECGT might be a dimer, trimer, or tetramer of 60- and/or 58-kDa monomers, and these monomers represent a heterodimer consisting of pairs of 36- or 34- of and 28-kDa subunits. MALDI-TOF-TOF MS showed that the protein SCPL1199 was identified. Epigallocatechin and epicatechin exhibited higher substrate affinities than β-glucogallin. ECGT had an optimum temperature of 30 °C and maximal reaction rates between pH 4.0 and 6.0. The enzyme reaction was inhibited dramatically by phenylmethylsulfonyl fluoride, HgCl2, and sodium deoxycholate.
Introduction
Flavonoids, a major class of secondary metabolites in plants, have a number of important physiological roles as endogenous auxin transport regulators (1–3), root development (4, 5), seed germination (6), allelopathy (7), plant-bacterium interaction (8, 9), UV-B protection (10), and plant defense against pathogens and environmental stress (11).
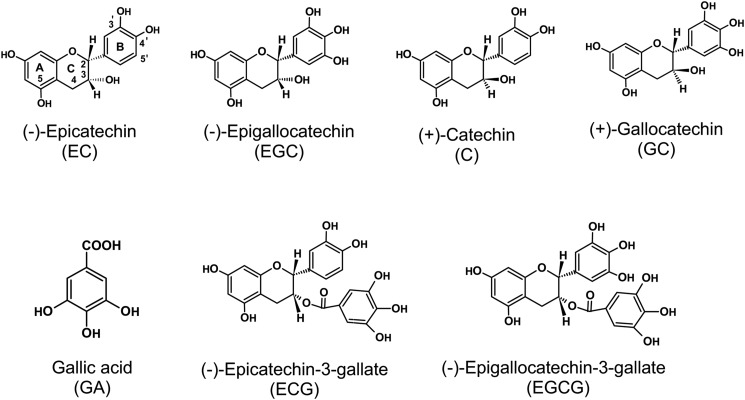
Flavonoids can be grouped into several subgroups including chalcone, flavone, flavonol, flavandiol, anthocyanin, proanthocyanidin (oligomer or polymer of flavan-3-ols and flavan-3,4-diol units) and other specialized forms (12). Flavan-3-ols (catechins), which comprise ∼70–80% of tea polyphenols, are rich in young leaves and shoots of the tea plant (Camellia sinensis (L.) O. Kuntze). Catechins, with a basic 2-phenylchromone structure, are characterized by the di- or tri-hydroxyl group substitution of the B ring, the 2,3-position isomer of the C ring, and presence of a galloyl group at the 3-postion of the C ring (Fig. 1). On the basis of the classical definition proposed of galloyl group structural features, catechins are divided into galloylated and nongalloylated compounds. Galloylated catechins, including (−)-epigallocatechin gallate (EGCG)3 and (−)-epicatechin gallate (ECG), esterified often with gallic acid (GA) in the 3-hydroxyl group of the flavan-3-ol units are major catechin compounds that account for up to 76% of catechins in the tea plant (13, 14).
FIGURE 1.
Basic units of typical catechins.
Catechins, especially EGCG, possess antioxidant activity, antimutagenic, anticarcinogenic, antidiabetic, antibacterial, and anti-inflammatory potential, antihypertensive and anticardiovascular disease effects, solar UV protection, body weight control effects, and therapeutic properties for Parkinson disease (15). The health-promoting effects of galloylated catechins are stronger than those of nongalloylated catechins (16, 17).
Flavonoid biosynthesis has been a major focus of investigation in recent decades (12). As the building blocks of most proanthocyanidins, the 2,3-trans-flavan-3-ol (+)-catechin and 2,3-cis-flavan-3-ol (−)-epicatechin biosynthetic pathways have been investigated intensively at the biochemical and genetic levels (18, 19). The biosynthetic pathway of nongalloylated catechins, which include (+)-catechin (C), (−)-epicatechin (EC), (+)-gallocatechin (GC), and (−)-epigallocatechin (EGC) is well documented. Some key genes and enzymes in the pathway include dihydroflavonol 4-reductase (EC 1.1.1.219), leucoanthocyanidin reductase (EC 1.3.1.77), anthocyanidin synthase (EC 1.3.1.77), and anthocyanidin reductase (EC 1.3.1.77) (20–23). Despite recent progress in improving our understanding of flavan-3-ol synthesis, the mechanism involved in galloylation of catechins remains a mystery (19). In the 1980s, the biosynthesis of galloylated catechins and GA in the tea plant was investigated using radioactive tracer techniques. It was found that GA was presumably esterified with epigallocatechin and epicatechin to form catechin gallates in young tea shoots, and the amount of GA might be a key limiting factor for the formation of EGCG and ECG (24).
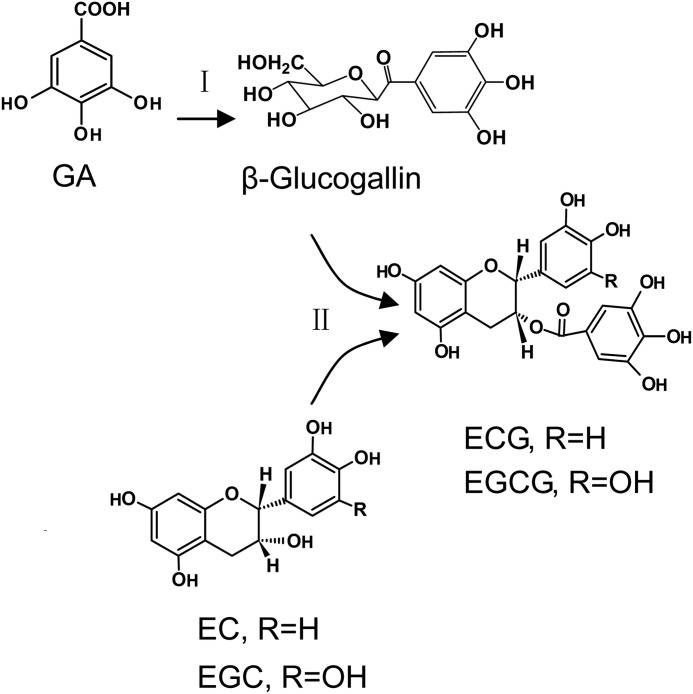
Understanding the galloylation of flavan-3-ols has been hindered by an absence of spontaneous genetic mutants for catechin biosynthesis. Niemetz and Gross (25) have done much research on the biogenetic pathways of hydrolyzable tannins. Their research confirmed that β-glucogallin (1-O-galloyl-β-d-glucose (βG)) exerts a dual role functioning not only as an acyl acceptor but also as an efficient acyl donor. This work indicates βG plays the same role in biosynthesis of galloylated catechin (Fig. 2). Galloylation of glucose with GA to yield βG, the first specific metabolite in the route to hydrolyzable tannins, was catalyzed by enzyme extracts from oak leaves with UDP-glucose serving as an activated substrate (26). This result indicated that, in galloylated catechin biosynthesis, βG rather than GA might be a precursor of galloylated flavan-3-ols.
FIGURE 2.
Reaction diagram of the galloylated catechin biosynthetic pathway. In the first reaction (I), the galloylated acyl donor βG was biosynthesized by UGGT from the substrates GA and UDPG. In the second reaction (II), galloylated catechins ECG or EGCG were produced by ECGT from the substrates βG and nongalloylated catechins EC or EGC.
In this study we sought to identify the enzymatic reactions and purify the key enzyme involved in galloylated catechin biosynthesis. Enzyme assays in vitro were designed to investigate the biosynthesis of galloylated catechins, and the enzymes involved in galloylated catechin biosynthesis were purified and identified. In addition, the enzyme properties were investigated.
EXPERIMENTAL PROCEDURES
Plant Materials
Leaves of the tea plant (C. sinensis (L.) O. Kuntze) were plucked from the experimental tea garden of Anhui Agricultural University during early summer. All of the samples were immediately frozen in liquid nitrogen and stored at −80° before analysis.
Enzyme Extraction and Enzyme Assays
Enzyme extraction was performed in accordance with the method of Zhang et al. (27). All enzymes assays were conducted in phosphate buffer. In the multienzyme incorporative reaction system, the UGGT/ECGT (UDP glucose:galloyl-1-O-β-d-glucosyltransferase/epicatechin:1-O-galloyl-β-d-glucose O-galloyltransferase) assay solution was incubated at 30 °C for 1.5 h in a total volume of 1.5 ml containing 50 mm phosphate buffer (pH 6.0), 0.4 mm EGC or EC, 1.4 mm GA, 2.3 mm UDP glucose (UDPG), 4 mm ascorbic acid, and crude enzyme extract (0.55 mg of total protein).
The UGGT reaction solution was incubated at 30 °C for 1.5 h in a total volume of 1.5 ml containing 50 mm phosphate buffer (pH 6.0), 2.3 mm UDPG, 1.4 mm GA, 4 mm ascorbic acid, and crude enzyme extract (0.55 mg of total protein).
The ECGT assay solution was incubated at 30 °C for 1 h in a total volume of 1.5 ml containing 50 mm phosphate buffer (pH 6.0), 0.4 mm EGC or EC, 0.96 mm βG (Advanced Technology and Industrial Company, Hong Kong, China), 4 mm ascorbic acid, and crude enzyme extract (0.55 mg of total protein).
The above enzyme reactions were terminated by the addition of ethyl acetate. Each of the reaction products was extracted three times with 3 ml of ethyl acetate. The ethyl acetate extract was evaporated and redissolved in 500 μl of methanol and then used directly for analyses of the enzymatic reaction products.
For gel filtration-purified enzyme activity analysis, the ECGT assay solution was incubated at 30 °C for 10 min in a total volume of 100 μl containing 50 mm phosphate buffer (pH 6.0), 0.4 mm EGC or EC, 0.96 mm βG, and 5.6 μg of enzyme. Enzymatic reactions were terminated by the addition of 10 μl of 5 m HCl to the assay solution, and then the solution was used directly for analysis of the enzymatic reaction products.
The protein concentration was determined by the Bradford method (28) using bovine serum albumin as a standard. In the control treatment, crude enzyme extract was heated to 100 °C to inactivate enzyme activities.
Analysis of UGGT and ECGT Enzyme Reaction Products
Extraction of enzyme reaction products was performed according to the method of Liu et al. (29). The solution (20 μl) of reaction products was spotted on a silica GF254 TLC sheet (5 × 20 cm; HeFei BoMei Biotechnology Co., Hefei, China) that was developed in chloroform:methanol:formic acid (28:10:1, v/v) and then sprayed with 1% vanillin-HCl (w/v) reagent. The spots of reaction products in the methanol extract were identified by Rf values, and their visual color compared with those of catechin standards.
The reaction products extract was analyzed by HPLC on a Phenomenex Synergi 4u Fusion-RP80 column (5 μm, 250 × 4.6 mm) with detection at 280 nm. Ultraviolet spectra were recorded with a Waters 2487 UV array detector (Waters Corp., Milford, MA). For HPLC analysis, the solvent system consisted of 1% (v/v) acetic acid (A) and 100% acetonitrile (B). After injection (5 μl), a linear gradient at a flow rate of 1.0 ml/min was set as follows: B from 10 to 13% (v/v) in 20 min was initiated, then B from 13 to 30% (v/v) between 20 and 40 min; B from 30 to 10% (v/v) between 40 and 41 min. Peaks were identified by comparison of the retention times with those of standards.
Analysis of enzyme reaction products by LC-MS followed the method of Miketova et al. (30). Enzymatic products were analyzed by HPLC, and the area of the product peaks were collected and identified by LC-MS. Liquid chromatography electrospray-ionization-MS analyses were performed on a Thermo Finnigan LCQ Advantage instrument using the following conditions: negative ion detection mode, centroiding mode, multiplier at 1600 keV, 1000 atomic mass units/s, source at 4.5 kV, sheath gas at 70 p.s.i., auxiliary gas at 25 p.s.i., capillary temperature at 220 °C, and UV detection at 220 nm.
Preparation and Identification of β-Glucogallin from Tea Plant
For βG analysis, 1 g of fresh leaves was crushed in liquid nitrogen and extracted with 5 ml of methanol by sonication at room temperature for 10 min followed by centrifugation at 4000 × g for 15 min, and the residues were re-extracted twice as above. The supernatants were pooled and evaporated and redissolved in 5 ml of water. The pooled supernatants were then extracted three times with chloroform. The supernatant water phase was purified further with a SEP-PAK C18 cartridge, and after filtration the supernatant was separated by HPLC, and the βG peak was collected and freeze-dried. The powder was used for chemical identification by HPLC, MS, and 1H and 13C NMR spectroscopy.
For HPLC analysis, the solvent system consisted of 1% acetic acid in water (A) and acetonitrile (B). After injection (5 μl), a linear gradient at a flow rate of 1.0 ml/min was established as follows: B from 0 to 10% (v/v) in 10 min was initiated, then B from 10 to 30% between 10 and 30 min. Peaks were identified by comparing the retention times with those of standards.
The LC-MS analysis of βG employed the same method as that used for analysis of enzyme reaction products described above. 1H and 13C NMR spectra were recorded in methanol-d4 on a Bruker Avance 400 MHz spectrometer using TMS as an internal standard. Chemical shifts were expressed in ppm (δ).
ECGT Isolation and Purification
ECGT purification consisted of acetone powder preparation, ammonium sulfate precipitation, hydrophobic interaction chromatography, concanavalin A (ConA) chromatography, and gel filtration. The first two steps were performed at 4 °C, and the last three steps were conducted at room temperature. Gel filtration was performed to estimate the relative molecular mass of the enzyme.
Step 1; Ammonium Sulfate Precipitation
The acetone powder was prepared by homogenization of 50 g of tea leaves in cold acetone (−20°) with a Waring blender. The finely ground precipitate was collected by vacuum filtration. The precipitate was washed several times with acetone until the washings were colorless. This precipitate powder was used as a crude material for ECGT preparation. Precipitate powder (20 g) was homogenized with 400 ml of extraction buffer (50 mm phosphate buffer (pH 7.0), 4 mm β-mercaptoethanol, 1% (w/v) polyvinylpolypyrrolidone (Sigma)) and filtered through cheesecloth. The homogenate was centrifuged at 15,000 × g for 15 min at 4 °C, and the supernatant was fractionated with 20–40% ammonium sulfate.
Step 2; Hydrophobic Interaction Chromatography
The precipitate was dissolved in 20 mm phosphate buffer (pH 7.0) containing 1 m ammonium sulfate and loaded onto a butyl-Sepharose column (20 cm × 2.5-cm inner diameter, Bio-Rad). For hydrophobic interaction chromatography, the solvent system consisted of 20 mm phosphate buffer (pH 7.0) including 1 m ammonium sulfate (A) and phosphate buffer (pH 7.0) (B). After application of the enzyme solution, the column was washed with five volumes of buffer A and subsequently eluted with a stepped gradient of 38, 95, and 100% B at a flow rate 2.5 ml/min.
Step 3; Affinity Chromatography
The active fractions were subjected to ConA-Sepharose 4B chromatography (column 10 cm × 1.6 cm inner diameter; GE Healthcare). For ConA chromatography, the solvent system consisted of 20 mm Tris-HCl (pH 7.0) containing 0.5 m NaCl (A) and 20 mm Tris-HCl (pH 7.0) containing 0.5 m α-d-methylglucoside (B). After applying the enzyme solution, the column was washed with 5 volumes of buffer A and subsequently eluted with 100% B at a flow rate of 1 ml/min.
Step 4; Gel Filtration Chromatography
The active fractions were subjected to gel filtration on a Superdex 200 column (50 cm × 1.6 cm inner diameter; GE Healthcare) and eluted with 20 mm phosphate buffer (pH 7.0) containing 0.15 m NaCl at a flow rate of 0.8 ml/min.
Step 5: SDS-PAGE Assay
SDS-PAGE was performed in accordance with the method of Laemmli (31), after which the proteins were visualized with Coomassie Brilliant Blue using the methods of Oakley (32).
Protein Identification by MALDI-TOF-TOF MS
Protein spots were cut from gels, destained for 20 min in 50 mm NH4HCO3 solution containing 30% acetonitrile, and washed in Milli-Q water until the gels were destained. The spots were incubated in 0.2 m NH4HCO3 for 20 min and then lyophilized. Each spot was digested overnight in 12.5 ng/ml trypsin in 0.1 m NH4HCO3. After trypsin digestion, the peptide mixtures were extracted with 8 μl of extraction solution (50% acetonitrile, 0.5% TFA) at 37° for 1 h. Finally the extracts were dried under the protection of N2. Samples were reconstituted in 3 μl of 50% acetonitrile containing 0.1% TFA before MS analysis. A 1-μl drop of this peptide solution was applied to an Anchorchip target plate. After drying at room temperature, a 0.1-μl droplet of CHCA matrix was applied to the plate at the same position. Samples were analyzed with ultrafleXtreme (Bruker). All acquired spectra of samples were processed using flexControlsoftware (Bruker) in the default mode. Parent mass peaks with a mass range of 500–3500 Da were detected with a minimum S/N filter of 10 for precursor ion selection. The five most abundant MS peaks were selected for MS/MS analysis. The combined MS and MS/MS data from the MALDI-TOF-TOF analysis were submitted to Mascot 2.3.02 for a search against the NCBI C. sinensis protein database (www.ncbi.nlm.nih.gov; 827 sequences), C. sinensis Genome Database “cam.pep” to construct a protein data bank (40,551 sequences, data not shown), and C. sinensis Genome Database “tie.pep” to construct a protein data bank (49,413 sequences, data not shown).The identification was accepted based on results from three biological replicates.
Properties of the ECGT Enzyme
For characterization of ECGT enzyme properties, gel filtration-purified enzyme was used. For determination of the optimum pH for the ECGT, citrate-phosphate buffer (pH 4.0–5.5), phosphate buffer (pH 6.0–7.0), and Tris-HCl buffer (pH 7.5–8.0) were used. The optimum temperature range for ECGT activity was tested from 0 ° to 70 °C at pH 6.0. Other assay conditions were identical to those used in the routine assay.
To test the effect of inhibitors on ECGT activity, the enzyme was incubated with the inhibitors for 5 min at 30 °C before the enzyme assay. Enzymatic activity was measured in the presence of PMSF, ZnCl2, EDTA, and β-mercaptoethanol at a final concentration of 0–50 mm, and sodium deoxycholate and HgCl2 were used at a final concentration of 0–5 mm. Other assay conditions were identical to those used in the routine assay.
For investigation of the effects of temperature and pH on enzyme stability, ECGT activity was tested after enzyme storage at −20, 0, 4, 10, 20, 30, 40, or 50° for 48 h and after storage at pH 4.0 to 9.0 at 4° for 48 h. In addition, the temporal stability of ECGT was determined after storage at 4 °C for 0, 2, 7, 20, or 40 days.
RESULTS
Evidence for Biosynthetic Enzymes of Galloylated Catechins
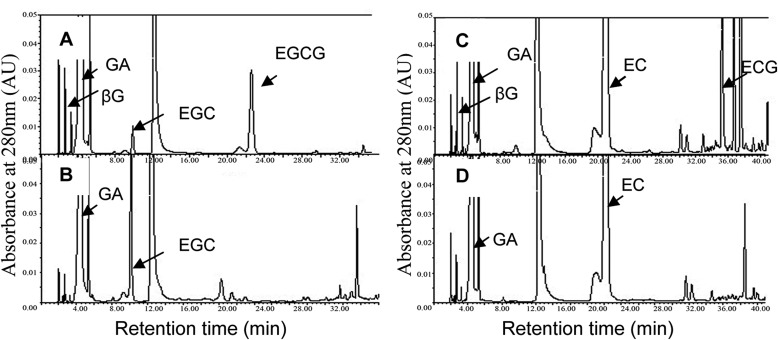
Niemetz and Gross (25) confirmed that βG acts not only as an acyl donor but as an acceptor in the biosynthesis of hydrolyzable tannins. To determine whether catechin galloylation was similar to that of hydrolyzable tannin biosynthesis, a two-step enzyme assay incorporating the substrates GA, EC, or EGC and cosubstrate UDPG was designed, and the enzymatic products were analyzed via TLC and HPLC. The assay showed that UDPG was indispensable in the two-step enzymatic reaction, and a significant amount of βG was detected in the enzymatic products by HPLC analysis (Fig. 3). This result suggested a UDPG-dependent glucosyltransferase existed in the tea plant, and βG was the enzymatic product. In addition, the galloylated catechins EGCG and ECG (Fig. 3), but not (+)-gallocatechin-3-gallate (GCG), were detected by the two-step enzyme assay via TLC and HPLC (data not shown), which indicated that the cis-catechins EGC and EC were appropriate substrates of a galloyltransferase instead of the trans-catechin GC or C. These data further confirmed that EGCG and ECG in the tea plant are biosynthesized via enzymatic galloylation of EGC and EC with βG, whereas GCG in green tea beverages is derived from isomerization of EGCG during green tea production (33).
FIGURE 3.
HPLC analysis of UGGT/ECGT enzyme assay extracts. A and C, the UGGT/ECGT assay solution was incubated at 30 °C for 1.5 h in a total volume of 1.5 ml containing 50 mm phosphate buffer (pH 6.0), 0.4 mm nongalloylated catechins (EGC or EC), 1.4 mm GA, 2.3 mm UDPG, 4 mm ascorbic acid, 1.5 mm salicylic acid, and crude enzyme extract (0.55 mg total of protein). The products βG and galloylated catechins EGCG or ECG were detected clearly in this two-step reaction enzyme assay. Peaks were identified by comparing the retention times with standards. B and D, control treatments of the UGGT/ECGT assay extracts with the crude enzyme extract were heated to 100 °C to inactivate enzyme activities. There were no enzymatic products in control treatments. AU, absorbance units.
To test the above assumptions further, two separate enzyme-reaction assays were performed. The first enzyme assay was designed to detect UDPG-glucosyltransferase activity with the substrates GA and UDPG, and the second assay was to detect galloyltransferase activity with the substrates βG (or 1,2,3,4,6-pentagalloylglucose) and EGC (or EC and GC). The enzymatic products were identified by TLC, HPLC, and LC-MS.
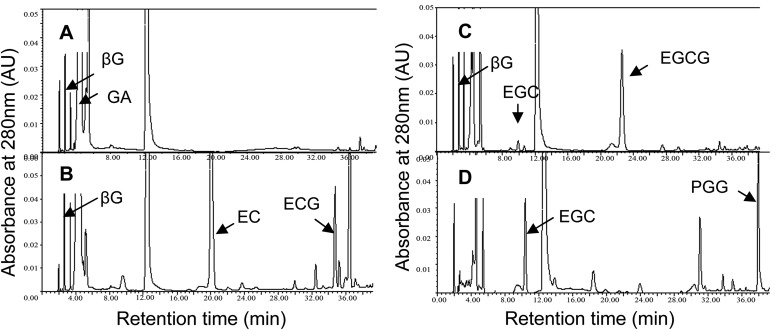
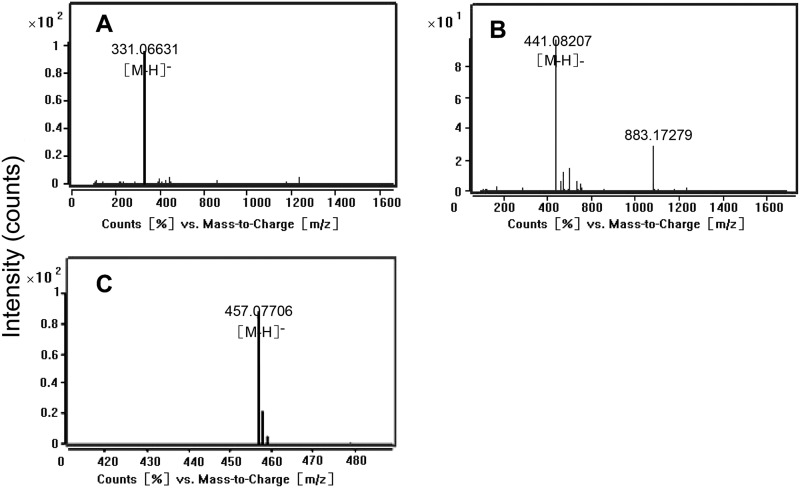
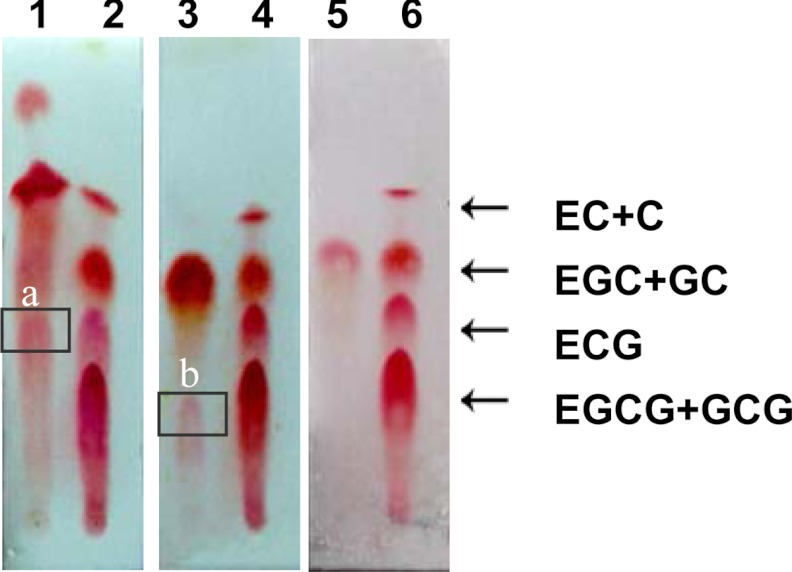
In the UDPG-glucosyltransferase assay, the product βG could not be analyzed effectively by TLC for lack of an appropriate staining reagent. However, HPLC (Fig. 4A) and LC-MS confirmed that the product was βG (Fig. 5A) and indicated the enzyme UGGT existed in the tea plant. In the galloyltransferase assay, TLC analysis of the enzyme reaction products showed two magenta spots with Rf values of 0.43 and 0.28 corresponding to the ECG and EGCG standards that were displayed in TLC sheets by staining with 1% (w/v) vanillin-HCl reagent (Fig. 6). This conclusion was confirmed by LC-MS. The parent ions with m/z 457 and 441 corresponding to EGCG and ECG standards, respectively, were observed (Fig. 5, B and C). EGC, EC, and βG were appropriate substrates of the galloyltransferase instead of GC (Fig. 6) and 1,2,3,4,6-pentagalloylglucose (PGG; Fig. 4D). The deduced galloylated catechin biosynthetic pathway is depicted in Fig. 2.
FIGURE 4.
HPLC analysis of UGGT and ECGT enzyme assay extracts. A, the UGGT assay solution was incubated at 30 °C for 1.5 h in a total volume of 1.5 ml containing 50 mm phosphate buffer (pH 6.0), 2.3 mm UDPG, 1.4 mm GA, 4 mm ascorbic acid, 1.5 mm salicylic acid, and crude enzyme extract (0.55 mg of total protein). The product βG was detected clearly in this assay. B and C, the ECGT assay solution was incubated at 30 °C for 1 h in a total volume of 1.5 ml containing 50 mm phosphate buffer (pH 6.0), 0.4 mm nongalloylated catechins (EGC or EC), 0.96 mm βG, 4 mm ascorbic acid, and crude enzyme extract (0.55 mg of total protein). The galloylated catechins EGCG or ECG were detected clearly in this assay. D, the ECGT assay solution was conducted with substrates 0.96 mm 1,2,3,4, 6-pentagalloylglucose (PGG), 0.4 mm nongalloylated catechins (EGC), and conditions otherwise identical to those of the ECGT assay. No galloylated catechins were produced effectively from the substrates 1,2,3,4,6-pentagalloylglucose and EGC. The product peaks of βG in A, ECG in B, and EGCG in C were collected and identified by LC-MS (see Fig. 5). AU, absorbance units.
FIGURE 5.
Mass spectra of products in the UGGT, ECGT, reaction assays. A, shown are mass spectra of peaks corresponding to βG in the UGGT enzyme reaction assay (see Fig. 4A). The ions of full MS correspond to the βG standard. B and C, MS assay of products peaks in the ECGT enzyme reaction assay (see Fig. 4, B and C) are shown. The ions of 441 and 457 correspond to galloylated catechins ECG and EGCG standards, respectively.
FIGURE 6.
TLC assay of the ECGT reaction extract. Lane 1, ECG was produced with substrates βG and EC. Lane 3, EGCG was produced with substrates βG and EGC. Lane 5, no product was produced with substrates βG and GC. Lanes 2, 4, and 6 are catechin standards. Boxed band a, ECG; boxed band b, EGCG.
Identification of β-Glucogallin in Tea Plant
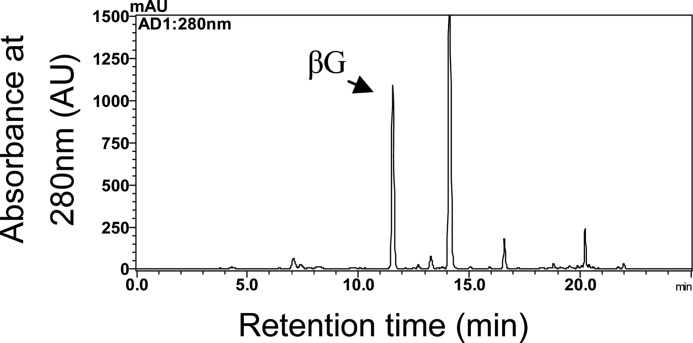
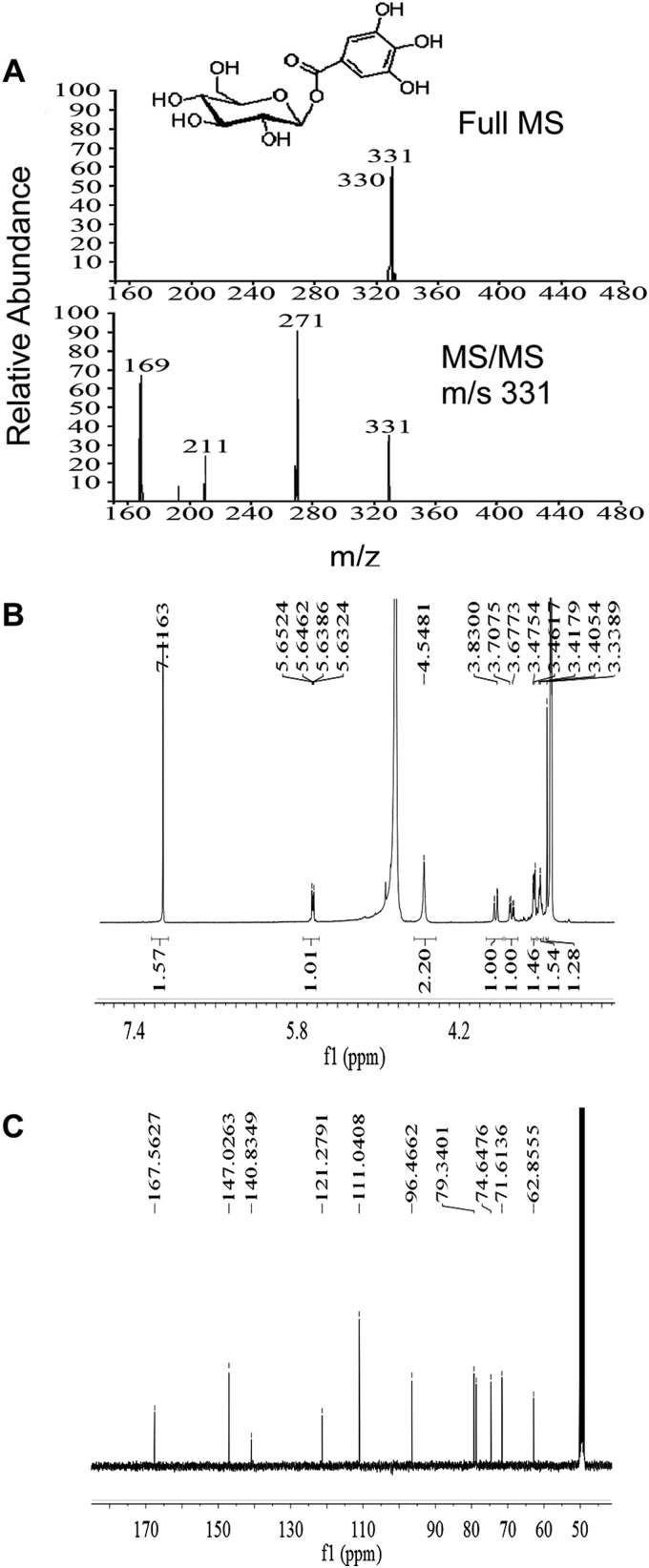
To gain further evidence for the existence of ECGT and UGGT in the tea plant, βG was extracted and identified from the leaves. An improved method for extraction and quantification of βG was established. A SEP-PAK C18 cartridge was used in sample preparation, and the purity of βG in the solvent was enhanced markedly. To prevent interference from noisy peaks, the linear gradient of the solvent system was optimized for βG analysis based on the method used for HPLC analysis of catechins. A single peak with retention time and spectral information consistent with those of the βG standard appeared at 11.75 min in the chromatogram (Fig. 7). The βG peak was collected largely via HPLC and identified by MS and NMR (Fig. 8). The parent ion of the compound was detected at m/z 331, and major fragments were detected at m/z 271 and 169, which were consistent with those of the βG standard. 1H and 13C NMR spectra were recorded in methanol-d4 on a Bruker Avance 400 MHz spectrometer with TMS as an internal standard. Chemical shifts were expressed in ppm (δ). 1H NMR δ: 7.086 (2H, s, H-2′ and H-6′), 5.615 (1H, d, J = 8 Hz, H-1), 3.813 (1H, dd, J = 12.0, 1.6 Hz, H-6a), 3.658 (1H, dd, J = 12.0, 4.8 Hz, H-6b), 3.35 to 3.45 (4H, m, H-2-H-5). 13C NMR δ: 167.06 (C = O), 146.53 (C-5′), 140.34 (C-4′), 120.78 (C1′), 110.54 (C-2′, C-6′), 95.97 (C-1), 78.84 (C-5), 78.23 (C-3), 74.15 (C-2), 71.11 (C-4), 62.36 (C-6). These data were consistent with those of the standard compounds, and thus the compound was identified as βG.
FIGURE 7.
β-Glucogallin assay in tea leaves by HPLC. For HPLC analysis, the solvent system consisted of 1% acetic acid in water (A) and 100% acetonitrile (B). After injection (5 μl), a linear gradient at a flow rate of 1.0 ml min−1 was established as follows: B from 0 to 10% (v/v) in 10 min was initiated, then B from 10 to 30% (v/v) between 10 min to 30 min. Peaks were identified by comparing the retention times with the standard. mAU, absorbance units.
FIGURE 8.
Mass spectrometry and NMR assay of βG in the tea plant. A, shown are mass spectra of the peak corresponding to βG in Fig. 7. B and C, shown are 1H and 13C NMR spectra of the peak corresponding to βG in Fig. 7. 1H and 13C NMR spectra were recorded in methanol-d4 on a Bruker Avance 400 MHz spectrometer with TMS as an internal standard. Chemical shifts were expressed in ppm (δ). The 1H and 13C NMR spectra were consistent with those of the standard. The compound was identified as βG.
Purification of ECGT
Activity of ECGT was monitored throughout the purification. An ∼1.64-fold purification was obtained by ammonium sulfate precipitation. The main active parts existed in the 20–60% ammonium sulfate fraction. About 10-fold specific activity was increased by hydrophobic interaction chromatography separation (Table 1). Monitoring of ECGT activity showed that ECGT enzyme was eluted from the column with a low ion eluent (Fig. 9A), which suggested ECGT was a highly hydrophobic protein. ConA-affinity chromatography was the most effective purification step for ECGT (Fig. 9B). Separation by ConA-affinity chromatography yielded an ∼46-fold increase in purification (Table 1) and indicated that ECGT was a glycoprotein.
TABLE 1.
Purification of ECGT from tea plant
| Purification step | Total protein(mg) | Total activity | Specific activity | Enzyme activity yield | Purification -fold |
|---|---|---|---|---|---|
| pKat | pKat mg−1 | % | |||
| Extraction | 425.20 | 1703.96 | 4.01 | ||
| 20–60% ammonium sulfate fraction | 221.11 | 1450.02 | 6.56 | 85.09 | 1.64 |
| t-Butyl hydrophobic interaction chromatography | 16.77 | 1106.10 | 65.94 | 64.91 | 16.45 |
| Con A-Sepharose | 0.14 | 431.73 | 3084.02 | 25.34 | 769.55 |
| Superdex 200 | 0.05 | 306.76 | 6576.82 | 0.18 | 1641.09 |
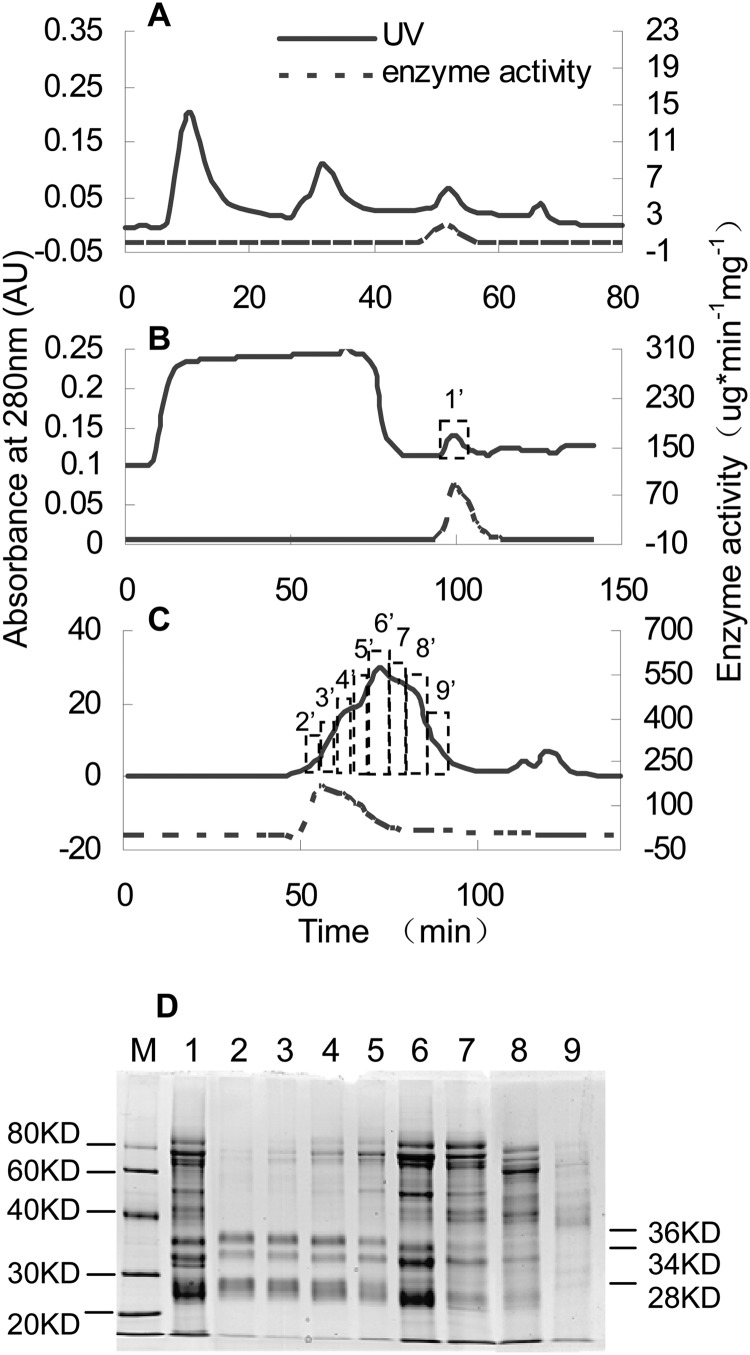
FIGURE 9.
Protein purification of ECGT enzyme. A, t-butyl hydrophobic interaction chromatography of ECGT enzyme is shown. B, affinity chromatography of ECGT enzyme on ConA-Sepharose is shown. C, gel chromatography of ECGT enzyme on Superdex 200 is shown. D, SDS-PAGE analysis of the ECGT purification steps of affinity and gel chromatography is shown. Lane M, protein markers; lane 1, ConA-Sepharose; lane 2–9, separations of peak in Superdex 200. AU, absorbance units.
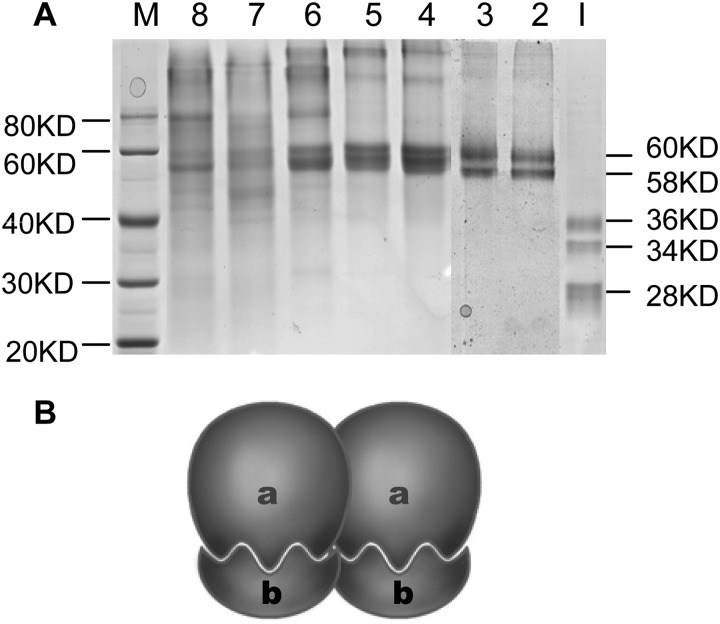
An ∼2-fold increase in specific activity was achieved by separation on a Superdex 200 column (Table 1). Obvious enzyme activities were detected approximately from 52 min (fraction 2′) to 74 min (fraction 6′, Fig. 9C) with estimated molecular masses of 241 to 121 kDa based on a standard curve. To investigate the subunit molecular masses of this enzyme, SDS-PAGE was routinely used for identification. The enzyme activity of lane 2 was about double that of lane 6, whereas there were a large number of superfluous bands in lane 6 from 40 to 80 kDa, so we speculated that three bands of estimated molecular masses of 36, 34, and 28 kDa were the subunits of this enzyme (Fig. 9D). One noteworthy phenomenon was that the SDS-PAGE Coomassie Brilliant Blue-stained bands changed with the degree of protein denaturation. SDS-PAGE analysis of lane 2 showed that only two bands of 60 and 58 kDa remained after the enzyme was denatured in loading buffer containing 1% SDS, and three bands of 36, 34, and 28 kDa were present when the enzyme was denatured in loading buffer containing 5% SDS (Fig. 10A).
FIGURE 10.
SDS-PAGE analysis of the ECGT with different depth denaturation (A) and a predicted model for ECGT oligomer (B). Lane M, protein markers; lanes 2–8, enzyme fractions 2′-8′ in Fig. 9C were denatured in loading buffer with 1% SDS; lane I, enzyme fraction 2′ in Fig. 9C was thoroughly denatured in loading buffer with 5% SDS. The 36- or 34-kDa (a) and 28-kDa (b) subunits constitutes 60- or 58-kDa ECGT monomer.
The above-mentioned enzyme activities and SDS-PAGE analysis indicated that the native ECGT might be a dimer, trimer, or tetramer of 60- and/or 58-kDa monomers. A predicted model for native ECGT oligomer was showed in Fig. 10B. The bands of 60 or 58 kDa were a heterodimer consisting of a pairs of 34 or 36 and 28 kDa subunits, which is similar to a serine carboxypeptidase-like (SCPL) protein encoded by an acyltransferase gene reported by Li et al. (34, 35).
Identification of ECGT Protein by MALDI-TOF-TOF MS
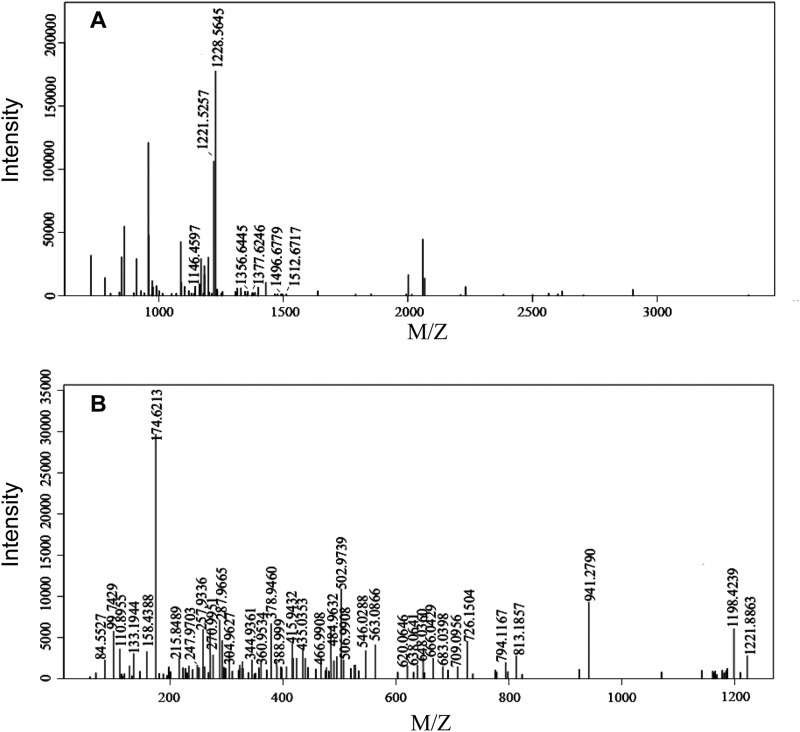
The gel spots of molecular mass 60, 58, 36, 34, and 28 kDa digested by trypsin were analyzed by MALDI-TOF-TOF MS. The same peptides appeared repeatedly in the bands of mass 60, 58, 36, and 34 kDa. Some peptides of the SCPL1199 protein were matched to 60-, 58-, 36-, 34-, and 28-kDa bands (Tables 2 and 3; Fig. 10A). However, some other peptides were not matched to any proteins in the NCBI protein and C. sinensis Genome databases (Fig. 11). The low coverage rate of MS might come from the post-translational processing of the primary translation product. This post-translational processing might be the reason for the difference in molecular masses suggested from the encoding gene and measured from the purified protein in the tea plant.
TABLE 2.
Proteins from SDS-PAGE spots identified by MALDI-TOF-TOF mass spectrometry
| SDS-PAGEa spot | Index | Protein ID | Protein mass | Protein score | Significance score | Isoelectric point | Coverage rate |
|---|---|---|---|---|---|---|---|
| 60 kDa | 1 | SCPL1199 | 55268.57 | 77.4 | 43 | 5.81 | 9.69% |
| 58 kDa | 1 | SCPL1199 | 55268.57 | 62.3 | 43 | 5.81 | 11.34% |
| 36 kDa | 1 | SCPL1199 | 55268.57 | 85.4 | 43 | 5.81 | 11.96% |
| 34 kDa | 1 | SCPL1199 | 55268.57 | 52 | 43 | 5.81 | 9.69% |
| 28 kDa | 1 | SCPL1199 | 55268.57 | 79.9 | 43 | 5.81 | 11.13% |
a SDS-PAGE spots in Fig. 10A.
TABLE 3.
Peptides matched in 60-kDa-spot protein in Fig. 10A. “(M)@1” means oxidative modification at the first amino acid
| Start | End | Observed | Mr (exp.) | Mr (calc.) | Delta | Miss | Modification | Sequence |
|---|---|---|---|---|---|---|---|---|
| 155 | 165 | 1146.46 | 1145.45 | 1145.54 | −0.0829 | 0 | TPGGWPTSDTK | |
| 166 | 175 | 1228.56 | 1227.56 | 1227.59 | −0.0312 | 0 | SAEQSYQFLR | |
| 166 | 176 | 1356.64 | 1355.64 | 1355.68 | −0.0461 | 1 | SAEQSYQFLRK | |
| 459 | 470 | 1377.62 | 1376.62 | 1376.66 | −0.0412 | 1 | GAGHTAPEYYRR | |
| 248 | 260 | 1496.68 | 1495.67 | 1495.72 | −0.0531 | 0 | MALISDEIYENAK | |
| 248 | 260 | 1512.67 | 1511.66 | 1511.72 | −0.0533 | 0 | Oxidation (M)@1 | MALISDEIYENAK |
FIGURE 11.
Mass spectrum analysis of 60-kDa spot protein in Fig. 10A by MALDI-TOF-TOF. A, MS of 60-kDa spot protein in Fig. 10A is shown. B, mass spectra of the peak corresponding to 1228.56 in Fig. 11A is shown.
Properties of the ECGT Enzyme
Gel filtration-purified enzyme was used for determination of its dynamic properties. The activities of ECGT were assayed at EGC or EC concentrations ranging from 0.1 to 0.5 mm in saturated βG, and at βG concentrations ranging from 0.1 to 0.9 mm in saturated EGC. Kinetic parameters were obtained with hyperbolic Michaelis-Menten saturation curves. The Km values for EGC, EC, and βG were 0.26, 0.23, and 0.73 mm, respectively (Table 4). These results indicated that ECGT exhibited higher substrate affinities and catalytic rates for EGC and EC than for βG. No substrate inhibition phenomenon was observed.
TABLE 4.
Michaelis-Menten kinetics (Km, Vmax values, and catalytic efficiency katals) of ECGT
| Substrate specificity of ECGT enzyme from the tea plant | Km | Vmax | kcat |
|---|---|---|---|
| mm | picokatal | min−1 | |
| EC | 0.26 ± 0.04 | 27.6 ± 3.20 | 52.56 ± 5.83 |
| EGC | 0.23 ± 0.03 | 32.48 ± 3.24 | 72.04 ± 2.19 |
| βG | 0.73 ± 0.03 | 59.02 ± 6.28 | 40.18 ± 4.37 |
To determinate the characteristics of ECGT, the effects of pH and temperature on its activity and stability were examined. The optimum temperature for ECGT activity appeared at 30 °C, whereas a high level of the enzyme activity was detected over the broad range of 20–50 °C. The optimum pH values were from 4.0 to 6.0. To study the enzyme stability, ECGT activity was tested after enzyme storage at different temperatures (−20 to 50°) and pH values (4.0–9.0 at 4 °C) for 48 h. The ECGT enzyme retained >95% activity between 10 ° and 30 °C and 80% relative activity between −20 ° and 0 °C, and only 40% relative activity was observed after storage at 50 °C. In acidic buffers with pH 4.0 to 6.0, the activity of ECGT was not affected. The purified enzyme stored at 4 °C was stable for 20 days without significant loss of activity.
To investigate the structural properties and mechanism of protein action, the inhibitory effects of several inhibitors and metal ions were assessed. PMSF, a potent inhibitor which phosphorylates the seryl residues of enzymes, was found inhibiting ECGT activity distinctly when present at 1, 10, and 50 mm. This indicated an active site of serine residue might be important for ECGT catalysis. ECGT activity was also inhibited by β-mercaptoethanol, which implied that ECGT contained disulfide (S-S) bonds in the tertiary and quaternary structure of its protein. Enzymatic activity was not significantly affected by EDTA or Zn2+, which indicated that no ions were present in active sites and that Zn2+ was not necessary for the enzyme metal ion activator. Enzymatic activity was significantly affected by sodium deoxycholate, which might change the configuration of ECGT. ECGT activity was inhibited greatly by 1 mm HgCl2, because the heavy metal ion Hg2+ has a strong affinity for the sulfhydryl (-SH) group. Consequently, this implied that sulfhydryl groups in ECGT were essential for maintaining an active conformation of the enzyme.
DISCUSSION
Galloylation of Catechins in Tea Plant
Flavonoids exhibit a diversity of biological activities and functions and have attracted a great deal of research attention in recent years. Several remarkable advances are reported in reviews (12, 19, 20, 36, 37). Stafford (20, 38, 39) revealed that dihydroflavonol 4-reductase and leucoanthocyanidin reductase catalyzed dihydroflavonols to leucoanthocyanidins, then to 2,3-trans-2R,3S-flavan-3-ol (C or GC) and speculated that 2,3-trans-2R,3S-flavan-3-ol was converted to 2,3-cis-2R,3R-flavan-3-ol (EC or EGC) by an epimerase. However, Xie et al. (21) reported a new flavan-3-ol biosynthesis pathway in which the anthocyanidin reductase, encoded by the BANYLUS gene in Arabidopsis and Medicago truncatula (21), converted anthocyanidins into their corresponding 2, 3-cis-2R, 3R-flavan-3-ols such as EC, epiafzelechin, and EGC in the presence of NADPH. In the present and previous experiments, LAR (leucoanthocyanidin reductase) and ANR (anthocyanidin reductase) genes were demonstrated to exist in the tea plant and may be responsible for the biosynthesis of nongalloylated catechins (40–42).
With the rapid progress in molecular biological techniques, genetic mutants provide a powerful tool to identify functional genes in plants. However, mutants with a high endogenous content of galloylated catechins have not been discovered, and relevant genetic transformation systems in the tea plant have not been established successfully. Differential expression patterns of genes provided an effective method to discover useful functional genes, but in a preliminary experiment the physicochemical factors of sucrose and light, which are used to promote flavonoid biosynthesis, were unable to stimulate biosynthesis of galloylated catechins (data not shown). Furthermore, the accumulation of GA is less affected by light intensity, as reported by Saijo (24). Biochemical studies are presently the best means of studying galloylated catechin biosynthesis.
The biosynthesis of galloylated catechins is based on galloyl transacylation reactions at the 3-position in the C ring of nongalloylated catechins. The transacylation reactions in plants are accomplished via activated donor molecules. Coenzyme A thioesters in the BAHD family, and 1-O-glucose esters also serve as activated donors due to their high free energy of hydrolysis (43). Among 1-O-glucose esters, such as βG in Quercus robur and Rhus typhina (25), isobutyryl-β-glucose in Solanum berthaultii and Solanum lycopersicum (44), and sinapoylglucose in Raphanus sativus (45), glycosylation of UDPG is achieved by UDP-glucose:glucosyltransferases (EC 2.4.1.-). Gallotannins, isobutyryl-β-glucose, sinapoylcholine, and sinapoylmalate were biosynthesized via 1-O-glucose ester-dependent acyltransferase reactions, which involved several acyltransferases such as βG-dependent galloyltransferases (EC 2.3.1.90) (25), 2,3,4-isobutyryl-glucose acyltransferase (34), sinapoylglucose:choline sinapoyltransferase (EC 2.3.1.91) (46), and sinapoyl-glucose:malate sinapoyltransferase (SMT; EC 2.3.1.92) (47), respectively.
The present study is the first to demonstrate that the biosynthesis of galloylated catechins is completed in a similar manner including two reaction steps involving UGGT and a newly discovered enzyme ECGT in the tea plant. β-Glucogallin, rather than GA, serves as the activated donor molecule in transacylation reactions.
Does ECGT Belong to the SCPL Acyltransferase Family?
Serine carboxypeptidases (SCPs; EC 3.4.16.-) are members of the α/β hydrolase family, which catalyze the hydrolysis of the C-terminal peptide bond in proteins or peptides making use of a Ser-Asp-His catalytic triad. However, because the 1990s, many studies have shown that SCPL proteins that share sequence similarity to SCPs function not as peptidases but as acyltransferases and lyases (35, 46, 48), such as acyltransferases in wild tomato (Solanum pennellii) that catalyze the formation of 2,3,4-isobutyryl-glucose (48, 49), sinapoyl-glucose:malate sinapoyltransferase and sinapoylglucose:choline sinapoyltransferase in Arabidopsis thaliana are responsible for sinapate ester biosynthesis (50). In the present analysis, the SCPL1199 protein was identified by MALDI-TOF-TOF MS.
Teutschbein (51) recently reported a unique chlorogenate-dependent caffeoyl transferase, the first example of a GDSL lipase-like protein that lost hydrolytic activity and has acquired a completely new function in plant metabolism as an acyltransferase in the synthesis of hydroxycinnamate esters. However, no GDSL lipase-like protein was matched by MALDI-TOF-TOF MS identification.
Some data show that SCPL acyltransferase proteins have a relative molecular mass 110–120 kDa heterotetramer containing both 34 and 24 kDa bands (34, 35), or a monomeric protein(s) with a relative molecular mass of 52 and 51 kDa for the two isoforms (sinapoyl-glucose:malate sinapoyltransferase I (SMT I) and SMT II, respectively) (52). It is notable that glycosylation resulting from post-translational processing may contribute to the differences between the molecular masses calculated from the nucleotide or amino acid sequences and the molecular mass estimated from the elution position of the purified protein (35).
Five β-glucogallin-dependent galloyltransferases that synthesize gallotannin were highly purified from sumac leaves, and their native enzymes exist as tetramers with molecular masses ranging from 170 to 360 kDa (25). For example, β-glucogallin:pentagalloylglucose galloyltransferase, with a Mr value of 170 kDa, was identified by gel filtration, whereas a single polypeptide band of Mr 42 kDa was detected by SDS-PAGE (53). One special type was β-glucogallin:hexagalloylglucose 3-O-galloyltransferase with an Mr value of 260 kDa, which contained both 36- and 24-kDa bands (54). On the basis of the present results, the native ECGT might be a dimer, trimer, or tetramer composed of 60- and/or 58-kDa monomers. And the 60-kDa monomer was speculated as a heterodimer consisting of a pair of 36- and 28-kDa subunits, whereas 34- and 28-kDa subunits might form a 58-kDa monomer. However, the hydrophobic polypeptides might combine with each other during the isolation and purification processes, thus causing the difficulty of evaluating the native molecular weight ranges.
Few studies have investigated SCPL acyltransferase genes, which are most likely to be involved in the biosynthesis of proanthocyanidins (catechin polymers); examples are the SCPL genes identified in persimmon and grapevine (55, 56). However, the functions of these genes have not been verified, and their correlations with galloylation of catechins are uncertain. In our study, information on the structural properties and mechanism of catalytic action of ECGT proteins in the tea plant was obtained by biochemical analysis. The ECGT enzyme is a hydrophobic glycoprotein, and more than one serine residue rather than metal ions are involved in the active sites of the enzyme. These properties are similar to those of SCPL proteins. Further experiments focused on elucidating the reaction mechanism of ECGT are in progress.
Glycosylation of Gallic Acid
The amount of GA might be a key limiting factor for the formation of EGCG and ECG (24). In the present study we found that the derivative of glycosylation of GA, rather than GA per se, was the precursor of galloylated flavan-3-ols. Most previous research reports of glycosylation of organic acids have focused on benzoate derivatives, such as salicylic acid, hydroxybenzoic acid, and dihydroxybenzoic acid, and few have investigated GA (3,4,5-trihydroxybenzoic acid) (57, 58). Lim et al. (57) expressed 90 genes from Arabidopsis that probably encode glucosyltransferases in Escherichia coli, and their enzymatic catalytic activities toward benzoates were analyzed, of which 14 proteins displayed activity toward 2-hydroxybenzoic acid, 4-hydroxybenzoic acid, and 3,4-dihydroxybenzoic acid, and two enzymes were active toward 2-hydroxybenzoic acid. Khater (58) reported recently that three genes coding for glucosyltransferases displayed capability to nonspecifically catalyze the formation of glucose esters of different phenolic compounds including derivatives of hydroxybenzoic and hydroxycinnamic acids.
Three candidate genes have been screened from the C. sinensis Genome database4 that were clustered into the subgroup of glucosyltransferases that are able to catalyze the formation of glucose esters. Identification of the functions of these candidate genes is in progress.
Acknowledgments
We thank Prof. Huarong Tan for assistance with LC/MS analysis, Prof. Tiejun Ling for technical assistance with 1H and 13C NMR spectroscopy, and Prof. Liwen Niu for protein purification. We thank BGI Tech Solutions Co., Ltd., for mass spectrometric analysis of proteins. We also thank Prof. Shu Wei for helpful comments during the preparation of the manuscript.
This work was supported by the Natural Science Foundation of China (30972401, 31170647, and 31170282), the Natural Science Foundation of Anhui Province (11040606M73), the Collegiate Natural Science Foundation of Anhui Province (KJ2012A110), the Program for Changjiang Scholars and Innovative Research Team in University (IRT1101), and the Major Project of Chinese National Programmes for Fundamental Research and Development (2012CB722903).
Key Laboratory of Tea Biochemistry and Biotechnology, Ministry of Education in China, Anhui Agricultural University, data not published.
- EGCG
- (−)-epigallocatechin gallate
- ECG
- (−)-epicatechin gallate
- ECGT
- epicatechin:1-O-galloyl-β-d-glucose O-galloyltransferase
- GA
- gallic acid
- C
- (+)-catechin
- EC
- (−)-epicatechin
- GC
- (+)-gallocatechin
- EGC
- (−)-epigallocatechin
- βG
- β-glucogallin, 1-O-galloyl-β-d-glucose
- ConA
- concanavalin A
- GCG
- (+)-gallocatechin-3-gallate
- SCPL
- serine carboxypeptidase-like
- UDPG
- UDP glucose.
REFERENCES
- 1. Jacobs M., Rubery P. H. (1988) Naturally occurring auxin transport regulators. Science 241, 346–349 [DOI] [PubMed] [Google Scholar]
- 2. Peer W. A., Bandyopadhyay A., Blakeslee J. J., Makam S. N., Chen R. J., Masson P. H., Murphy A. S. (2004) Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16, 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buer C. S., Muday G. K., Djordjevic M. A. (2007) Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol. 145, 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buer C. S., Muday G. K. (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16, 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buer C. S., Sukumar P., Muday G. K. (2006) Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol. 140, 1384–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Debeaujon I., Léon-Kloosterziel K. M., Koornneef M. (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122, 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bais H. P., Park S. W., Weir T. L., Callaway R. M., Vivanco J. M. (2004) How plants communicate using the underground information superhighway. Trends Plant Sci. 9, 26–32 [DOI] [PubMed] [Google Scholar]
- 8. Redmond J. W., Batley M., Djordjevic M. A., Innes R. W., Kuempe P. L., Rolfe B. G. (1986) Flavones induce expression of nodulation genes in Rhizobium. Nature 323, 632–635 [Google Scholar]
- 9. Wasson A. P., Pellerone F. I., Mathesius U. (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18, 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li J., Ou-Lee T. M., Raba R., Amundson R. G., Last R. L. (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5, 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Treutter D. (2005) Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biology 7, 581–591 [DOI] [PubMed] [Google Scholar]
- 12. Winkel-Shirley B. (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Forrest G. I., Bendall D. S. (1969) The distribution of polyphenols in the tea plant (Camellia sinensis L.). Biochem. J. 113, 741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He Q., Yao K., Jia D., Fan H., Liao X., Shi B. (2009) Determination of total catechins in tea extracts by HPLC and spectrophotometry. Nat. Prod. Res. 23, 93–100 [DOI] [PubMed] [Google Scholar]
- 15. Cabrera C., Artacho R., Giménez R. (2006) Beneficial effects of green tea. A review. J. Am. Coll. Nutr. 25, 79–99 [DOI] [PubMed] [Google Scholar]
- 16. Wolfram S., Wang Y., Thielecke F. (2006) Anti-obesity effects of green tea. From bedside to bench. Mol. Nutr. Food Res. 50, 176–187 [DOI] [PubMed] [Google Scholar]
- 17. Kao Y. H., Chang H. H., Lee M. J., Chen C. L. (2006) Tea, obesity, and diabetes. Mol. Nutr. Food Res. 50, 188–210 [DOI] [PubMed] [Google Scholar]
- 18. Winkel B. S. J. (2006) in The Science of Flavonoids, (Grotewold E., eds) pp. 123–146, Springer Science & Business Media, New York [Google Scholar]
- 19. Dixon R. A., Xie D. Y., Sharma S. B. (2005) Proanthocyanidins. A final frontier in flavonoid research? New Phytol. 165, 9–28 [DOI] [PubMed] [Google Scholar]
- 20. Stafford H. A. (1990) in Flavonoid Metabolism (Stafford H. A., eds) pp. 63–100, CRC, Boca Raton, FL [Google Scholar]
- 21. Xie D. Y., Sharma S. B., Paiva N. L., Ferreira D., Dixon R. A. (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299, 396–399 [DOI] [PubMed] [Google Scholar]
- 22. Xie D. Y., Jackson L. A., Cooper J. D., Ferreira D., Paiva N. L. (2004) Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol. 134, 979–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pang Y., Peel G. J., Wright E., Wang Z., Dixon R. A. (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol. 145, 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saijo R. (1983) Pathway of gallic acid biosynthesis and its esterification with catechins in young tea shoots. Agric. Biol. Chem. 47, 455–460 [Google Scholar]
- 25. Niemetz R., Gross G. G. (2005) Enzymology of gallotannin and ellagitannin biosynthesis. Phytochemistry 66, 2001–2011 [DOI] [PubMed] [Google Scholar]
- 26. Gross G. G. (1983) Partial purification and properties of UDP-glucose:vanillate 1-O-glucosyl transferase from oak leaves. Phytochemistry 22, 2179–2182 [Google Scholar]
- 27. Zhang X., Liu Y., Gao K., Zhao L., Liu L., Wang Y., Sun M., Gao L., Xia T. (2012) Characterization of anthocyanidin reductase from Shuchazao green tea. J. Sci. Food Agric. 92, 1533–1539 [DOI] [PubMed] [Google Scholar]
- 28. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 29. Liu Y., Gao L., Xia T., Zhao L. (2009) Investigation of the site-specific accumulation of catechins in the tea plant (Camellia sinensis (L.) O. Kuntze) via vanillin-HCl staining. J. Agric. Food Chem. 57, 10371–10376 [DOI] [PubMed] [Google Scholar]
- 30. Miketova P., Schram K. H., Whitney J. L., Kerns E. H., Valcic S., Timmermann B. N., Volk K. J. (1998) Mass spectrometry of selected components of biological interest in green tea extracts. J. Nat. Prod. 61, 461–467 [DOI] [PubMed] [Google Scholar]
- 31. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 32. Oakley B. R., Kirsch D. R., Morris N. R. (1980) A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal. Biochem. 105, 361–363 [DOI] [PubMed] [Google Scholar]
- 33. Wang R., Zhou W., Jiang X. (2008) Reaction kinetics of degradation and epimerization of epigallocatechin gallate (EGCG) in aqueous system over a wide temperature range. J. Agric. Food Chem. 56, 2694–2701 [DOI] [PubMed] [Google Scholar]
- 34. Li A. X., Eannetta N., Ghangas G. S., Steffens J. C. (1999) Glucose polyester biosynthesis. Purification and characterization of a glucose acyltransferase. Plant Physiol. 121, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li A. X., Steffens J. C. (2000) An acyltransferase catalyzing the formation of diacylglucose is a serine carboxypeptidase-like protein. Proc. Natl. Acad. Sci. U.S.A. 97, 6902–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winkel B. S. (2004) Metabolic channeling in plants. Annu. Rev. Plant Biol. 55, 85–107 [DOI] [PubMed] [Google Scholar]
- 37. Lepiniec L., Debeaujon I., Routaboul J. M., Baudry A., Pourcel L., Nesi N., Caboche M. (2006) Genetics and biochemistry of seed flavonoids. Annu Rev. Plant Biol. 57, 405–430 [DOI] [PubMed] [Google Scholar]
- 38. Stafford H. A., Lester H. H. (1984) Flavan-3-ol Biosynthesis. The conversion of (+)-dihydroquercetin and flavan-3,4-cis-diol (leucocyanidin) to (+)-catechin by reductases extracted from cell suspension cultures of Douglas fir. Plant Physiol. 76, 184–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stafford H. A., Lester H. H. (1985) Flavan-3-ol biosynthesis. The conversion of (+)-dihydromyricetin to its flavan-3,4-diol (leucodelphinidin) and to (+)-gallocatechin by reductases extracted from tissue cultures of Ginkgo biloba and Pseudotsuga menziesii. Plant Physiol. 78, 791–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Punyasiri P. A., Abeysinghe I. S., Kumar V., Treutter D., Duy D., Gosch C., Martens S., Forkmann G., Fischer T. C. (2004) Flavonoid biosynthesis in the tea plant Camellia sinensis. Properties of enzymes of the prominent epicatechin and catechin pathways. Arch. Biochem. Biophys. 431, 22–30 [DOI] [PubMed] [Google Scholar]
- 41. Eungwanichayapant P. D., Popluechai S. (2009) Accumulation of catechins in tea in relation to accumulation of mRNA from genes involved in catechin biosynthesis. Plant Physiol. Biochem. 47, 94–97 [DOI] [PubMed] [Google Scholar]
- 42. Ashihara H., Deng W. W., Mullen W., Crozier A. (2010) Distribution and biosynthesis of flavan-3-ols in Camellia sinensis seedlings and expression of genes encoding biosynthetic enzymes. Phytochemistry 71, 559–566 [DOI] [PubMed] [Google Scholar]
- 43. Fraser C. M., Thompson M. G., Shirley A. M., Ralph J., Schoenherr J. A., Sinlapadech T., Hall M. C., Chapple C. (2007) Related Arabidopsis serine carboxypeptidase-like sinapoylglucose acyltransferases display distinct but overlapping substrate specificities. Plant Physiol. 144, 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuai J. P., Ghangas G. S., Steffens J. C. (1997) Regulation of triacylglucose fatty acid composition (uridine diphosphate glucose. Fatty acid glucosyltransferases with overlapping chain-length specificity). Plant Physiol. 115, 1581–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Strack D., Knogge W., Dahlbender B. (1983) Enzymatic synthesis of sinapine from 1-O-sinapoyl-β-d-glucose and choline by a cell-free system from developing seeds of red radish (Raphanus sativus L. var. sativus). Z. Naturforsch Teil C. 38, 21–27 [Google Scholar]
- 46. Shirley A. M., McMichael C. M., Chapple C. (2001) The sng2 mutant of Arabidopsis is defective in the gene encoding the serine carboxypeptidase-like protein sinapoylglucose:choline sinapoyltransferase. Plant J 28, 83–94 [DOI] [PubMed] [Google Scholar]
- 47. Stehle F., Brandt W., Milkowski C., Strack D. (2006) Structure determinants and substrate recognition of serine carboxypeptidase-like acyltransferases from plant secondary metabolism. FEBS Lett. 580, 6366–6374 [DOI] [PubMed] [Google Scholar]
- 48. Wajant H., Mundry K. W., Pfizenmaier K. (1994) Molecular cloning of hydroxynitrile lyase from Sorghum bicolor (L.). Homologies to serine carboxypeptidases. Plant Mol. Biol. 26, 735–746 [DOI] [PubMed] [Google Scholar]
- 49. Lehfeldt C., Shirley A. M., Meyer K., Ruegger M. O., Cusumano J. C., Viitanen P. V., Strack D., Chapple C. (2000) Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12, 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fraser C. M., Rider L. W., Chapple C. (2005) An expression and bioinformatics analysis of the Arabidopsis serine carboxypeptidase-like gene family. Plant Physiol. 138, 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Teutschbein J., Gross W., Nimtz M., Milkowski C., Hause B., Strack D. (2010) Identification and localization of a lipase-like acyltransferase in phenylpropanoid metabolism of tomato (Solanum lycopersicum). J. Biol. Chem. 285, 38374–38381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gräwe W., Bachhuber P., Mock H. P., Strack D. (1992) Purification and characterization of sinapoylglucose:malate sinapoyltransferase from Raphanus sativus L. Planta 187, 236–241 [DOI] [PubMed] [Google Scholar]
- 53. Niemetz R., Gross G. G. (1998) Gallotannin biosynthesis. Purification of β-glucogallin:1,2,3,4,6-pentagalloyl-β-d-glucose galloyltransferase from sumac leaves. Phytochemistry 49, 327–332 [Google Scholar]
- 54. Niemetz R., Gross G. G. (2001) Gallotannin biosynthesis. β-Glucogallin:hexagalloyl 3-O-galloyltransferase from Rhus typhina leaves. Phytochemistry 58, 657–661 [DOI] [PubMed] [Google Scholar]
- 55. Ikegami A., Eguchi S., Kitajima A., Inoue K., Yonemori K. (2007) Identification of genes involved in proanthocyanidin biosynthesis of persimmon (Diospyros kaki) fruit. Plant Sci. 172, 1037–1047 [Google Scholar]
- 56. Terrier N., Torregrosa L., Ageorges A., Vialet S., Verriès C., Cheynier V., Romieu C. (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lim E. K., Doucet C. J., Li Y., Elias L., Worrall D., Spencer S. P., Ross J., Bowles D. J. (2002) The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 277, 586–592 [DOI] [PubMed] [Google Scholar]
- 58. Khater F., Fournand D., Vialet S., Meudec E., Cheynier V., Terrier N. (2012) Identification and functional characterization of cDNAs coding for hydroxybenzoate/hydroxycinnamate glucosyltransferases co-expressed with genes related to proanthocyanidin biosynthesis. J. Exp. Bot. 63, 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]