Background: Truly phosphospecific antibodies are difficult to generate and are poorly understood.
Results: Avian single chain Fv library selections yielded fully phosphospecific anti-phospho-tau antibodies, enabling the generation of a 1.9 Å co-crystal structure.
Conclusion: Phosphospecific antibodies were readily generated and can exhibit unique epitope recognition mechanisms.
Significance: High-affinity antibody phosphoepitope recognition has been defined, at high resolution, for the first time.
Keywords: Alzheimer Disease, Disulfide, Phosphorylation, Structural Biology, Tau, Affinity, Chicken, IgG, Specificity
Abstract
Highly specific antibodies to phosphoepitopes are valuable tools to study phosphorylation in disease states, but their discovery is largely empirical, and the molecular mechanisms mediating phosphospecific binding are poorly understood. Here, we report the generation and characterization of extremely specific recombinant chicken antibodies to three phosphoepitopes on the Alzheimer disease-associated protein tau. Each antibody shows full specificity for a single phosphopeptide. The chimeric IgG pT231/pS235_1 exhibits a KD of 0.35 nm in 1:1 binding to its cognate phosphopeptide. This IgG is murine ortholog-cross-reactive, specifically recognizing the pathological form of tau in brain samples from Alzheimer patients and a mouse model of tauopathy. To better understand the underlying binding mechanisms allowing such remarkable specificity, we determined the structure of pT231/pS235_1 Fab in complex with its cognate phosphopeptide at 1.9 Å resolution. The Fab fragment exhibits novel complementarity determining region (CDR) structures with a “bowl-like” conformation in CDR-H2 that tightly and specifically interacts with the phospho-Thr-231 phosphate group, as well as a long, disulfide-constrained CDR-H3 that mediates peptide recognition. This binding mechanism differs distinctly from either peptide- or hapten-specific antibodies described to date. Surface plasmon resonance analyses showed that pT231/pS235_1 binds a truly compound epitope, as neither phosphorylated Ser-235 nor free peptide shows any measurable binding affinity.
Introduction
Many proteins undergo post-translational modifications that regulate their function under normal and pathological conditions (1). Although protein phosphorylation is one of the most extensively recognized and described events during normal and disease-related cell signaling, few significant advances have been made in the generation or molecular characterization of phosphospecific antibody reagents since initial reports, some 30 years ago (2, 3).
In a hyperphosphorylated state, the microtubule-associated protein tau makes up the protein constituent of the paired helical filaments of non-fibrillar tangles in the brains of Alzheimer disease (AD)3 patients and has been the focus of intensive research over the past 2 decades (4, 5). In humans, tau has six isoforms, and all are present in tangles in their hyperphosphorylated state (5–9). The phosphorylation of tau occurs under both normal and pathological conditions and is believed to negatively regulate the ability of tau to promote microtubule assembly (10). Pathological tau has been implicated in generating protein aggregates in the cell body and eventually killing the neuron (6, 11–13).
In the brains of AD patients, tau is found to be phosphorylated at levels 3–4-fold higher than tau from normal brain (14–16). To date, ∼40 tau phosphorylation sites have been identified in association with AD (17). As a result of the association between tau hyperphosphorylation and pathological phenotypes (18), tau has been proposed as both a diagnostic biomarker and a target for therapeutic intervention in AD. Monoclonal antibodies (mAbs) specific for different tau phosphorylation states therefore have the potential to provide valuable research reagents to unravel normal versus pathological tau function and to provide potential diagnostic or even therapeutic tools for AD.
In this study, we chose three phospho-tau epitopes, based on their association with AD pathology (17), as antigens for chicken immunization and subsequent recombinant antibody generation. Chickens are a historically reliable immune host due to their robust immune response against highly conserved mammalian proteins (19), the feasibility of co-immunizing single animals with multiple immunogens (20, 21), and their proven ability to generate highly specific antibodies against both peptides (22) and haptens (23) via display technologies (24).
The chicken V gene repertoire is markedly different from the systems employed by humans, mice, and primates, which all use sequences that are highly diverse in both sequence and structure (25, 26). In chickens, only single functional V genes exist for the light and heavy chains, containing unique VL-JL (λ isotype only), VH-JH (VH3 family), and D segments (27–29). To make such a restricted V gene germ line repertoire highly functional, the chicken has evolved a complex V gene diversification mechanism known as “gene conversion” (29, 30). The gene conversion process in chickens is analogous to that utilized in rabbits (31), where each template V gene is diversified by recombination with segments from numerous upstream pseudogenes.
Surprisingly, chickens also only utilize 15 functional D segments, all of which are highly homologous (28). In contrast to humans and mice, chicken D segments obligately contain cysteine, with the consensus sequence GS(A/G)YC(G/C)(S/W)XA(Y/E) (where X is non-conserved) (28). This limited initial VH CDR3 repertoire is hyperdiversified by the use of D-D junctions, somatic mutation, and the insertion of new sequences via gene conversion. These D-like sequences are donated by pseudogenes and may replace the entire D segment or only a small section, leading to the creation of “mosaic CDRs: (28, 29). The frequent use of cysteine in VH CDR3 of chickens coupled with double D segment insertions leads to >50% of all B-cell clones in the chicken repertoire containing intra-CDR disulfide bonds (32). The high frequency of these probable disulfides suggests that they play an important functional role in CDR structural diversification. The nature of how these bonds are used in chicken antibodies has, however, never hitherto been observed via a crystal structure.
Using phage display technology coupled with a simple deselection method to remove clones with unwanted reactivity to the non-phosphopeptides, we successfully identified chicken single chain Fv (scFv) clones that bind with remarkable specificity to the desired phosphoepitopes. Importantly, the antibodies could be rapidly converted to chimeric human IgGs and were shown to be reactive with both human and murine orthologs of tau, successfully recognizing the phosphoepitopes in the brains of both a rodent AD model and human AD patients. This simple method provides a robust and broadly applicable platform to generate multiple phosphospecific antibodies from a single, small, target-focused, immune phage display library.
Although the recognition of phosphopeptides by protein kinases and phosphatases has been extensively studied (33), there are only a handful of papers describing anti-phosphopeptide antibodies that were selected via display technologies (34, 35). Furthermore, none of the phosphopeptide-antibody complexes currently described in the literature have been structurally characterized beyond in silico modeling and CDR mutagenesis (36–38). To aid our understanding of the molecular mechanisms that mediate phosphospecific epitope recognition, we determined the co-crystal structure of a highly specific, high-affinity, anti-phospho-tau Fab fragment in complex with the pathology-associated phosphoepitope pT231/pS235. The Fab fragment recognizes the six amino acids N-terminal to the phosphorylation site via a long, disulfide-constrained CDR-H3 and forms a “bowl-like” conformation in CDR-H2 to interact directly with the first of two phosphorylated residues (phospho-Thr (pThr)-231). This residue is tightly bound through a network of hydrogen bond interactions, providing a structural basis for the ultra-specific recognition of the phosphoepitope.
To our knowledge, this high-resolution structure represents the first avian antibody and, indeed, the first antibody in complex with its cognate phosphoepitope to be described in the literature. The establishment in this study of a simple methodology to generate ultra-specific and high-affinity phosphoepitope recognition antibodies should make such high-quality reagents accessible to many research groups. When coupled with high-resolution structural analysis of antigen-binding modes, this will facilitate new insights into the recognition of post-translational modifications by novel proteins. It may also inform fundamental comparative immunogenetics and antibody structure-function relationships. In the future, these studies could ameliorate the need for animal immunization by providing sufficient data to create designer, fully synthetic, phospho-biased antibody repertoires, as recently described for libraries biased toward the recognition of haptens (39), proteins (40), and peptides (41).
EXPERIMENTAL PROCEDURES
Peptides, Immunization, and scFv Library Construction
All peptides (supplemental Table 1) were synthesized by Open Biosystems. A cysteine was added to the C terminus of each peptide to allow biotinylation and keyhole limpet hemocyanin conjugation. Immunization of three chickens (in accordance with approved animal review board procedures; Charles River Laboratories) with phosphopeptides conjugated to keyhole limpet hemocyanin (pooled) and scFv phagemid library construction were performed as described previously (42).
Phage Selections and Screening of Peptides
Selections were carried out using a solution-phase protocol with biotinylated peptides and streptavidin bead capture (43) in the presence of a 10-fold molar excess of cognate non-phosphopeptides and scrambled phosphopeptides as competitors (non-biotinylated). Selected bacterial clones were screened via direct binding ELISA as described previously (42, 44).
Immunostaining of AD Brain
Formalin-fixed, paraffin-embedded tissue sections (5 μm thick) from Braak stage VI human AD brain (Sun Health Research Institute) were analyzed by immunohistochemistry. Slides were dehydrated, and heat-induced antigen (epitope) retrieval was performed using a wet-heat treatment at 122 °C in Reveal buffer solution (pH 6) (Biocare Medical). After blocking with 3% H2O2 and 10% (v/v) goat serum (Jackson ImmunoResearch Laboratories), sections were incubated with chimeric anti-phospho-tau IgG (1 μg/ml). Detection was performed with 0.1 μg/ml rabbit anti-human IgG (Jackson ImmunoResearch Laboratories) and an EnVision System+ kit (Dako). Labeled slides were counterstained with Mayer's hematoxylin (Dako).
Preparation and Western Blot Analysis of Mouse Brain Lysates
Frozen hemi-brains without cerebellum were rinsed in PBS, homogenized using a Dounce homogenizer in lysis buffer (Roche immunoprecipitation kit), and processed according to the manufacturer's protocol. Western blotting of brain homogenates (28 μg of total protein/well) was performed using 4–12% NuPAGE gels, nitrocellulose membrane, and an iBlot system (Invitrogen). After blocking (LI-COR block), the blot was incubated with anti-phospho-tau antibodies and anti-GAPDH antibody (LifeSpan Biosciences) as a loading control. Rabbit anti-human IgG was used as a secondary antibody (Jackson ImmunoResearch Laboratories), followed by fluorescently labeled detection antibodies (Alexa Fluor 680 (Invitrogen) and IRDye800 (Rockland)). The blot was visualized using and Odyssey system (LI-COR).
IgG Expression and Fab Preparation
IgGs were produced via transient transfection of CHO-S cells using CHO-MAX transfection reagent and FreeStyle CHO expression medium (Invitrogen). Antibody was then purified from conditioned medium via a 5-ml HiTrap rProtein A FF column (GE Healthcare). A Fab fragment was prepared and purified using a Pierce Fab kit (Thermo Scientific).
Binding Kinetics Analyses
Surface plasmon resonance analysis was performed using a BIAcore T200 biosensor, Series S CM5 chips, an amine coupling kit, 10 mm sodium acetate immobilization buffer at pH 5.0, 1× Hepes Buffered Saline/EDTA/P20 surfactant running buffer (pH 7.4), and 3 m MgCl2 (GE Healthcare). A targeted immobilization program was used to immobilize 4000–8000 response units of anti-human IgG (Fc) (GE Healthcare) at pH 5 on flow cells 1 and 2, followed by the capture of 500 response units of each respective anti-phospho-tau IgG on flow cell 2. For calculation of kinetic constants using global fit analysis, the phospho-tau peptides were diluted in 1× Hepes Buffered Saline/EDTA/P20 surfactant running buffer (2-fold dilution series). Each concentration was injected over both flow cells 1 and 2 for 3 min at a flow rate of 50 μl/min (to minimize the potential for mass transfer or rebinding events) and allowed to dissociate for 10 min, followed by a 5-s regeneration pulse with 5 m MgCl2. Reference-subtracted sensorgrams (flow cell 2 minus 1) for each concentration were analyzed using the kinetics 1:1 binding program within BIAcore T200 evaluation software (version 1.0).
Crystallization and Data Collection
pT231/pS235_1 Fab was formulated at 14.9 mg/ml in 20 mm Tris and 100 mm NaCl (pH 7.0), and 10 mm phosphopeptide (224KKVAVVR(pT231)PPK(pS235)PSSAKC241) was added and mixed before crystallization trials. The Fab/peptide molar ratio was 1:1.2. Crystals of Fab/peptide were grown at 18 ± 1 °C using the sitting-drop vapor diffusion method. Each drop contained 0.15 μl of Fab/peptide mixture and 0.15 μl of reservoir solution containing 200 mm NH4H2PO4 and 40% methyl pentanediol. The crystals appeared after ∼2 months at a final size of 0.1 × 0.4 × 0.05 mm3. X-ray diffraction data were collected remotely at beamline 17-ID of the Industrial Macromolecular Crystallography Association Collaborative Access Team at the Advanced Photon Source, Argonne National Laboratory. Data processing was carried out with the HKL2000 program (45) and AutoPROC (46). Final data statistics were generated using AutoPROC. Crystal data and processing statistics are summarized in supplemental Table 2.
Structure Solution and Refinement
The anti-phospho-tau Fab-peptide complex was solved by molecular replacement using the Phaser program (47), with an ensemble of Protein Data Bank codes 3GJE, 3BN9, and 3KYM for the heavy chain and codes 3MA9, 3G6D, and 3H42 for the light chain as the search models. The structure was initially refined using PHENIX (48) and finalized with BUSTER (49). Bulk solvent correction was used. The 2Fc and Fo − Fc electron density maps were calculated for the inspection and improvement of the structure during refinement. The peptide was built at a later stage of the refinement. Solvent molecules, as peaks greater than or equal to 3σ on the Fo − Fc electron density map with reasonable hydrogen bonding networks, were included as water molecules. Graphic work was carried out using Coot (50). The structure was verified with annealed omit maps and assessed using PROCHECK (51). Illustrations were prepared with PyMOL (DeLano Scientific LLC).
RESULTS
Generation of Chicken Antibodies Specific to Phosphorylated Tau
Antibodies were generated against three separate phosphoepitopes on tau associated with AD pathology (17): pT231/pS235, pT212/pS214, and pS396/pS404 (supplemental Table 1). In particular, conformational changes around Thr-231 have been shown to directly affect the microtubule-binding ability of tau (52). Chicken immunizations were carried out with a mixture of phosphopeptides, each separately conjugated to keyhole limpet hemocyanin before combination. From each immunized chicken, spleen and bone marrow RNAs were isolated, and a pooled scFv phagemid library of 3.8 × 108 cfu was constructed. This library was rescued, and a series of solution-phase selections were carried out using biotinylated versions of each of the phospho-tau peptides in the presence of a 10-fold excess of each of their respective non-biotinylated scrambled phosphopeptides and non-phosphopeptides.
Screening of selected clones was carried out by ELISA for binding of phage scFv to plate-immobilized phospho- and non-phospho-tau peptides. For each of the three peptides, binders specific to phosphopeptide were isolated. A total of 12 unique binders for pT231/pS235, five unique binders for pS396/pS404, and three unique binders for pT212/pS214 were identified. The alignment of the VH and VL sequences for the two most dominant binders for each phosphoepitope is shown in supplemental Fig. S1. Each of these clones was found to have the high framework uniformity that is typical of chicken antibodies (32). They are highly divergent in their VH CDR3 sequences, however, with the exception of pT231/pS235_1 and pT231/pS235_2, which have identical heavy chain sequences but exhibit promiscuity in light chain usage. Clones pT212/pS214_1, pT212/pS214_2, and pS396/pS404_1 are type 2 chicken VH, whereas clones pT231/pS235_1, pT231/pS235_2, and pS396/pS404_2 all contain a pair of non-canonical cysteine residues in CDR-H3 and are representatives of the major structural type 1 chicken VH (32).
Specificity of Reformatted Chimeric IgGs for Phosphopeptides
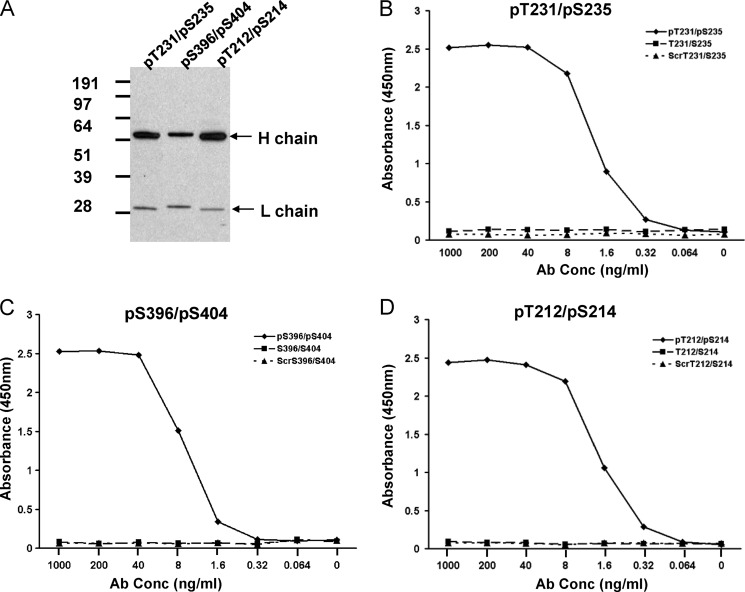
One dominant scFv binder for each of the three phosphopeptides was converted into chimeric human IgG1 containing the VH and VL sequences from the chicken scFv clones grafted onto human IgG1/λ constant domains. As shown in Fig. 1A, the converted IgG molecules express full-length heavy and light chain by Western analysis. Importantly, these chimeric antibodies preserved the binding specificity for each phosphopeptide when tested in a direct ligand binding ELISA. Each of the IgGs showed avid, dose-dependent binding to phosphopeptide but no detectable binding to either non-phosphopeptides or control scrambled peptides (Fig. 1, B–D).
FIGURE 1.
Expression and binding activity of chimeric IgG molecules converted from scFv. A, Western blot analysis of chimeric IgG molecules converted from scFv clones pT231/pS235_1, pS396/pS404_1, and pT212/pS214_1, respectively. The heavy and light chains for each IgG are indicated by arrows. B–D, ELISA analysis of each purified IgG molecule for binding to the phosphopeptide, non-phosphopeptide, and scrambled (Scr) peptide.
To establish the affinities of our three IgGs for their respective phosphopeptide epitopes, binding kinetics analyses were performed using Biacore technology. To ensure 1:1 interaction kinetics, the IgGs were captured on anti-human IgG CM5 chips, and binding responses were measured with peptide in the mobile phase. These analyses showed that the 1:1 affinities ranged from ∼2 μm for pS396/pS404_1 to 98 nm for pT212/pS214_1 and 0.35 nm for pT231/pS235_1 (Table 1).
TABLE 1.
Surface plasmon resonance kinetic analyses of anti-phospho-tau IgGs for phosphopeptides
Kinetics analyses were performed as described under “Experimental Procedures.” NB, no binding signal observed.
| Clone | ka | Kd | KD |
|---|---|---|---|
| m−1 s−1 | s−1 | nm | |
| pS396/pS404_1 | 1.5 × 105 | 3.1 × 10−1 | 2045.0 |
| pT212/pS214_1 | 8.3 × 104 | 8.0 × 10−3 | 97.9 |
| pT231/pS235_1 | 5.3 × 106 | 1.8 × 10−3 | 0.35 |
| pT231 | 5.1 × 105 | 5.8 × 10−4 | 1.1 |
| pT231Δ | 1.7 × 106 | 1.8 × 10−3 | 1.0 |
| pS235 | NB | NB | NB |
As confirmation of the specificity data observed in Fig. 1, Biacore analyses were performed in which the non-phospho and scrambled versions of all peptides used in this study were tested for binding to our highest affinity clone, pT231/pS235_1. A classical binding profile was observed for the pT231/pS235 peptide at 20 nm (supplemental Fig. S2A), but no binding signal was observed for any other peptide at 500 nm (supplemental Fig. S2B). As pT231/pS235_1 shows a binding affinity of 0.35 nm for the pT231/pS235 peptide, we therefore estimate that the binding affinity of pT231/pS235_1 for the non-phosphopeptide is <0.5 μm. This represents (at least) a 1000-fold differential in binding affinity between the phospho and non-phospho versions of the peptide. This finding confirmed that we had identified clones of exemplary phosphospecificity that are incapable of binding to non-cognate phosphoepitopes or non-phosphoepitopes under either avid solid-state (Fig. 1) or 1:1 binding (supplemental Fig. S2) conditions.
On the basis of the co-crystal structure of pT231/pS235_1 Fab and pT231/pS235 peptide (described below), we hypothesized that binding affinity and specificity relied heavily on the phosphorylation at Thr-231. To examine the necessity for pThr-231 and pSer-235 phosphorylation in the binding of pT231/pS235_1, a series of modified peptides were synthesized and subjected to binding affinity analyses as outlined above. Peptide pT231 lacked the phosphorylation at Ser-235, whereas pT231Δ had the full C-terminal sequence after Lys-234 removed. Both peptides retained binding at a slightly reduced affinity in comparison with the pT231/pS235 peptide (Table 1 and supplemental Fig. S3, A–C). Peptide pS235 lacked the phosphorylation at Thr-231 and showed no measurable binding signal, proving the necessity of pThr-231 for the binding affinity of the antibody (Table 1 and supplemental Fig. S3D).
Recognition of Pathological Tau in the Brain of a Human AD Patient and a Transgenic Mouse Model of Tauopathy
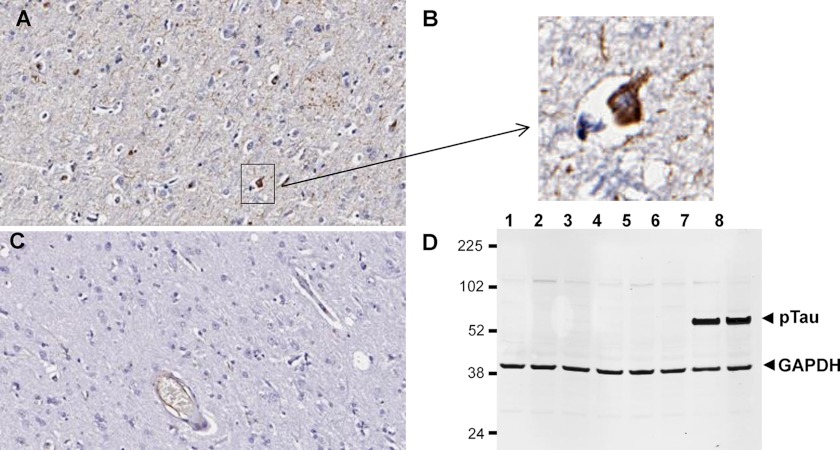
To test whether pT231/pS235_1 can recognize the phospho-tau epitopes in full-length tau, we tested the IgG in immunohistochemistry studies using human AD brain samples. As shown in Fig. 2, the antibody exhibited specific staining patterns in the AD brain samples (Fig. 2A) but not in normal patient brain samples (Fig. 2C). Specific staining of neurofibrillary tangles was also observed (Fig. 2B). In Western blot analysis, the antibody recognized phospho-tau in the brains of 6-month-old tau(P301L) Tg4510 transgenic mice but not in young (3-month-old) transgenic mice or wild-type control mice at either 3 or 6 months of age (Fig. 2D). The results are consistent with the report that these transgenic mice carrying the P301L mutation develop tangles constituting hyperphosphorylated tau at 4–6 months of age (53).
FIGURE 2.
Binding of pathological phospho-tau by antibody pT231/pS235_1 in human AD and Tg4510 transgenic mouse brains. A–C, immunohistochemical staining of human AD brain (A and B) and healthy brain (C). B, magnification of the boxed area in A. D, Western blot analysis of Tg4510 transgenic and wild-type brain lysates with pT231/pS235_1 IgG (plus control anti-GAPDH antibody). Each lane represents a different animal, and the lysate samples are 3-month-old wild-type mice (lanes 1 and 2), 3-month-old transgenic mice (lanes 3 and 4), 6-month-old wild-type mice (lanes 5 and 6), and 6-month-old transgenic mice (lanes 7 and 8).
X-ray Crystallographic Analysis of How Antibody pT231/pS235_1 Binds Its Epitope with High Affinity
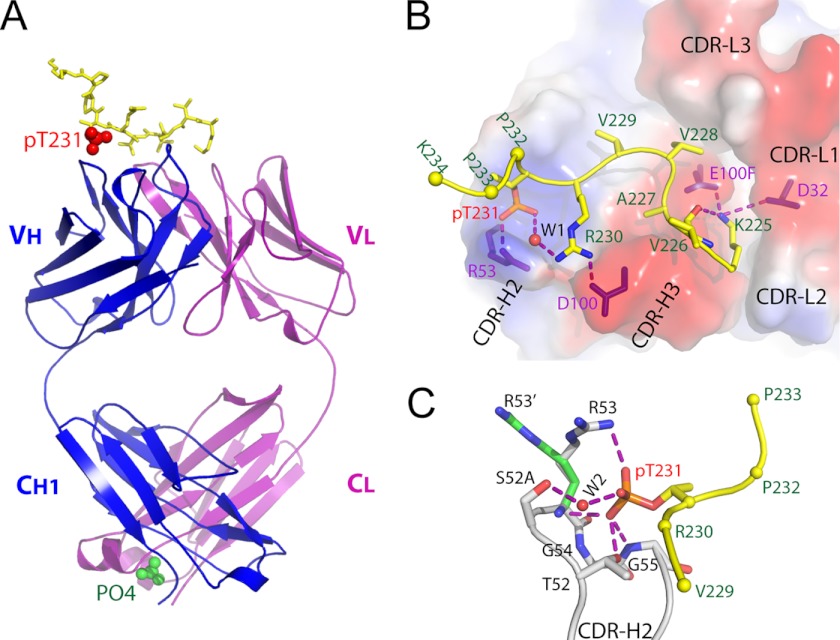
To investigate the structural basis for the high-affinity recognition of the pT231/pS235_1 epitope, we generated purified Fab fragment and used this protein in x-ray crystallography. The sequence of the peptide used for co-crystallization was the same as that of the full-length immunogen, 224KKVAVVR(pT231)PPK(pS235)PSSAKC241, with two phosphorylation sites at Thr-231 and Ser-235. The resulting analyses led to the generation of a co-crystal structure in which the Fab fragment and peptide were resolved at 1.9 Å (Protein Data Bank code 4GLR). In the Fab-peptide co-crystal structure, 10 amino acids (225KVAVVR(pT)PPK234) are visible, of which six (225KVAVVR(pT231)) interact directly with the Fab fragment. The remaining eight residues of the peptide are disordered. The phosphoepitope adopts a specific conformation on top of the CDRs, with two sharp turns at Val-228 and pThr-231 (Fig. 3, A and B, and supplemental Fig. S4). This conformation seems to be maintained by an intramolecular hydrogen bond between the side chain nitrogen of Lys-225 and the carbonyl oxygen of Val-226 and by a water-mediated hydrogen bonding network between a phosphate oxygen and a side chain nitrogen of Arg-230 (Fig. 3B). It is not clear whether this conformation reflects the phosphoepitope's natural state within full-length tau, but our immunohistochemistry data show that the antibody does recognize the phosphorylated epitope in the intact molecule (Fig. 2).
FIGURE 3.
Structure of anti-phospho-tau Fab (pT231/pS235_1) in complex with phosphoepitope pT231/pS235. A, schematic view showing the complex. The Fab heavy chain (VH + CH1) and light chain (VL + CL) are shown in blue and purple, respectively. The phosphoepitope is shown as yellow sticks on top with phosphorylated Thr-231 (pT231) highlighted as red spheres. The solvent PO43− ion is labeled and shown as green spheres. B, transparent electrostatic surface view of the CDRs showing the strong electrostatic interactions between CDR residues and the phosphoepitope. Positively charged areas are shown in blue, and negatively charged areas are shown in red. The backbone of the bound phosphoepitope is shown as a schematic in yellow; the side chains are shown as a stick model in atomic colors (carbon, yellow; nitrogen, blue; oxygen, red; and phosphorus, orange). The side chains of Pro-232, Pro-233, and Lys-234 are omitted for clarity, with Cα shown as spheres. The residues within CDRs that form strong electrostatic interactions with the phosphoepitope are shown as purple sticks. A water molecule is labeled W1. Hydrogen bonds are shown as dashed purple lines. C, interaction details between CDR-H2 and pThr-231. Two conformations of the H/Arg-53 side chain are shown as R53 and R53′, respectively. Hydrogen bonds are shown as dashed purple lines. A water molecule is labeled W2.
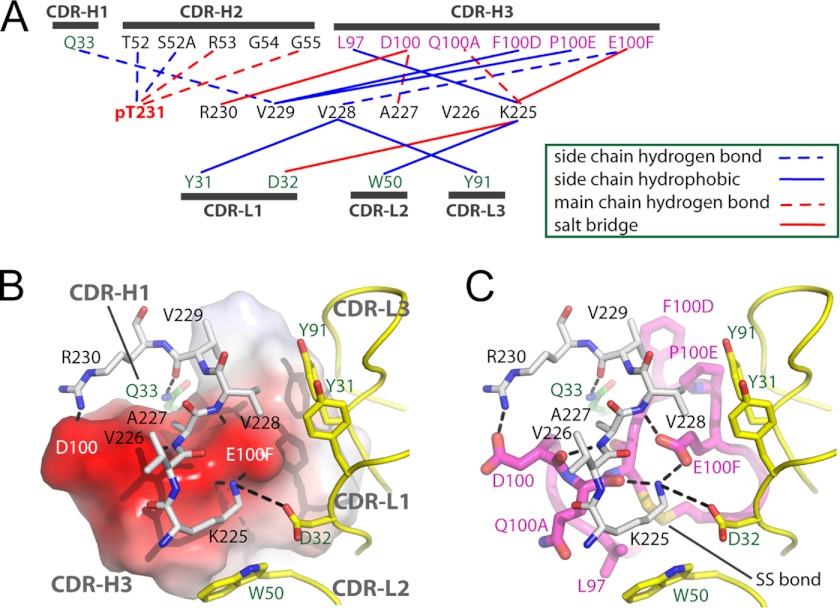
To our knowledge, our structure represents the first anti-phospho antibody in complex with its cognate phosphoepitope to be described in the literature. The mechanism of epitope recognition involves eight hydrogen bonds, three salt bridges, and six hydrophobic interactions (Fig. 4A). The total buried surface area between phosphoepitope and antibody is ∼660 Å2 as calculated by PISA (54). The phosphoepitope is dominated by CDR-H2 and CDR-H3 from the heavy chain, with secondary support from CDR-H1 and the light chain. CDR-H3 provides a platform for binding of the non-phosphorylated 225KVAVVK230 sequence, whereas the light chain CDRs form a hydrophobic wall to support binding of Val-228 and Lys-225 (Figs. 3B and 4B). A total of 16 residues make contact with antigen (Fig. 4A), which is in agreement with the average number of contacts found in anti-peptide antibodies (∼17) (55). In contrast, CDR-H3 makes six contacts, more than the average four. CDR-L2 also makes contact with antigen, which is unusual among antibodies to haptens or peptides, where the paratope is usually too small (55).
FIGURE 4.
Fab-phosphoepitope interaction details. A, schematic representation of all contacts between pT231/pS235_1 Fab and the phosphoepitope pT231/pS235 in the Fab-phosphoepitope co-crystal structure. B and C, interaction details between 225KVAVVR230 and the Fab fragment, with the same orientation in both panels. B, CDR-H3 is shown as an electrostatic surface model, with positively charged areas in blue and negatively charged areas in red. C, CDR-H3 is shown as a schematic in magenta and as sticks in atomic colors (carbon, magenta; nitrogen, blue; and oxygen, red). The CDRs from the light chain are shown as a schematic in yellow, with side chains shown as sticks (carbon, yellow; and oxygen, red). Peptide 225KVAVVR230 is shown as gray sticks (carbon, gray; nitrogen, blue; and oxygen, red). Gln-33 from CDR-H1 is shown as green sticks (carbon, green). Hydrogen bonds are shown as dashed black lines. The disulfide bond within CDR-H3 is shown and labeled SS bond.
The critical phosphorylation site (pThr-231) is exclusively recognized by CDR-H2 (Fig. 3C), which forms a positively charged pocket to accommodate the phosphate. The phosphate group forms hydrogen bonds with the backbone nitrogens of H/Arg-53 (where H = Ab heavy chain, and L = Ab light chain, with Kabat numbering) and H/Gly-55 and the side chain of H/Thr-52 and forms a water-mediated hydrogen bonding network with H/Ser-52A (Kabat numbering scheme (56)). The side chain of H/Arg-53 is well positioned to form a charge-charge interaction with the phosphate, but the guanidinium moiety is not visible in the electron density, suggesting that this interaction is not stable. The fact that four of the eight hydrogen bonds, as well as the water-mediated interaction between the Fab fragment and phosphoepitope, are involved in phosphate recognition (Fig. 4A) might explain why the antibody does not bind to non-phosphorylated peptides with the same sequence (supplemental Fig. S2). Interestingly, a phosphate-binding motif in which the backbone nitrogens of consecutive glycines coordinate the bound phosphate has been reported in a number of enzymes (57). The phosphate recognition motif from CDR-H2 of pT231/pS235_1 also includes two consecutive glycines (52TSRGG55) (Fig. 4A), demonstrating a surprising convergence between an evolutionarily conserved enzyme sequence motif and a sequence with similar function in an immunoglobulin selected by phage display.
In addition to the hydrogen bonding network between CDR-H2 and pThr-231, three salt bridges appear to provide further anchor points for the phosphoepitope binding, forming two charge complementary areas around positively charged side chains of Lys-225 and Arg-230 (Fig. 4B). Two salt bridges (H/Glu-100F–Lys-225 and H/Asp-100–Arg-230) involve CDR-H3, and one salt bridge (L/Asp-32–Lys-225) engages CDR-L1. Only one interaction is found between CDR-H1 and the phosphoepitope (Fig. 4, A and B).
Unique Structural Attributes of Chicken Immunoglobulins Observed in the CDRs of pT231/pS235_1
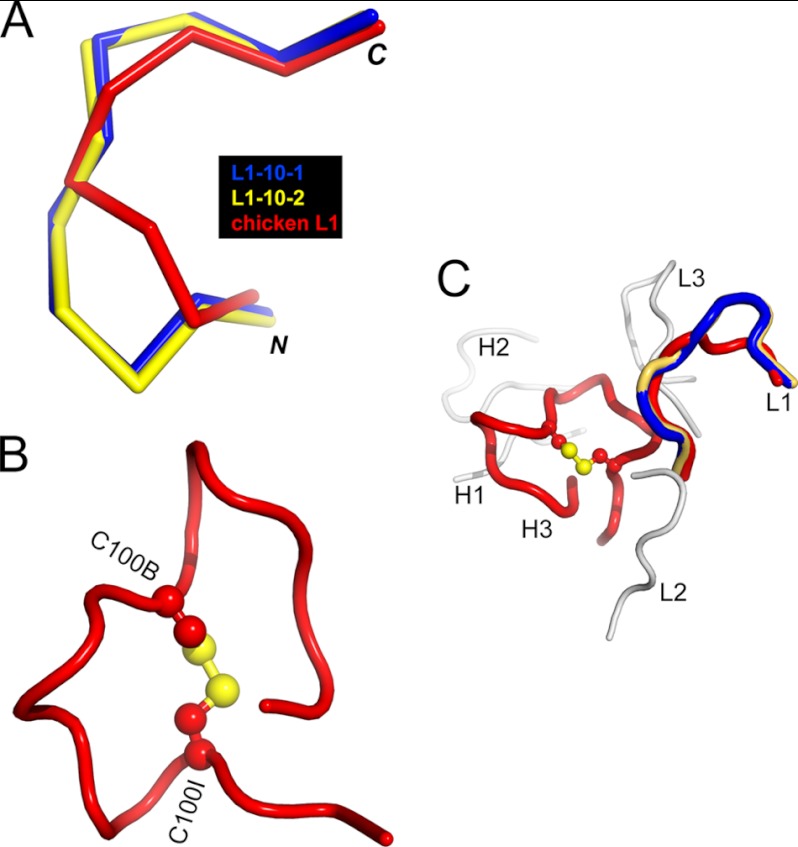
pT231/pS235_1 Fab in complex with its cognate phosphoepitope represents the first chicken antibody structure to be deposited in the Protein Data Bank. We therefore systematically compared the CDR sequences and conformations of this clone with the canonical CDRs from humans and rodents as classified by North et al. (58). CDR-H3 was not included in the analysis because it is known to have diverse lengths and structures (58). We found that chicken CDR-L2, CDR-L3, CDR-H1, and CDR-H2 all adhere to canonical conformations seen in mammalian structures and could be classified into clusters L2–8, L3–11, H1–13, and H2–10, respectively (58). In contrast, chicken CDR-L1 has a conformation that is distinct from all canonical CDR-L1 clusters described in humans and rodents to date (Fig. 5A).
FIGURE 5.
Chicken-specific conformations of CDR-L1 and CDR-H3 from anti-phospho-tau Fab (pT231/pS235_1). A, ribbon view showing the superposition of chicken CDR-L1 (red) with canonical mammalian CDR-L1–10-1 (blue; Protein Data Bank code 1YQV) and CDR-L1–10-2 (yellow; code 1AY1), respectively. CDR-L1–10-1 and CDR-L1–10-2 represent two clusters of mammalian CDR-L1 conformations and show the best structural similarity to chicken CDR-L1. B, schematic view showing the conformation of chicken CDR-H3 stabilized by the intramolecular disulfide bond shown as a ball and stick model in atomic colors (carbon, red; and sulfur, yellow). Two cysteines are labeled C100B and C100I, respectively. C, schematic view showing chicken CDRs with the same coloring scheme as in A and B. CDR-L2, CDR-L3, CDR-H1, and CDR-H2 are shown in gray.
Disulfide bonds are often seen in chicken CDR-H3 loops and are proposed to play important roles in creating chicken paratopes of great structural complexity in the context of a single framework repertoire (32). A large insertion within CDR-H3 (18 amino acids in length) forms a surface that makes numerous interactions with the non-phosphorylated part of the epitope (Fig. 4B). The disulfide bond between Cys-100B and Cys-100I may help to maintain the compact conformation of this large loop (Fig. 5B). The combined CDR-H3, CDR-H2, and CDR-L1 structures therefore create a unique binding site topology not previously described in human or murine antibodies (Fig. 5C).
DISCUSSION
Highly specific mAbs that bind to distinct phosphorylation sites are desirable as research tools to study the physiological and pathological functions related to post-translational modification. Despite this pressing need, the methods used in their generation have progressed little in the last 30 years, and many researchers rely on either commercial or personally generated polyclonal or monoclonal antibodies, which frequently exhibit poor specificity (59). We sought an alternative immune host and antibody selection methodology to generate high-affinity antibodies that are truly specific for individual phosphoepitopes on the model protein tau.
Although phosphospecific antibodies have been obtained successfully from classical naive antibody fragment libraries (35), it is often the case that functional application of such antibodies to therapeutic or diagnostic settings requires a process of in vitro affinity maturation, which can be technically challenging, time consuming, and costly (59, 60). Additionally, if they are generated against full-length phosphorylated protein, then the epitopes recognized by the antibodies cannot be dictated and must be elucidated post hoc (35).
In this study, we have demonstrated that chickens can be used as a model system to enable the rapid generation of a panel of mAbs targeting various phosphoepitopes with exact specificity. Chickens are increasingly used to generate high-affinity antibodies that cross-react across multiple orthologs of a specific mammalian immunogen (20, 21, 61). Although it has been demonstrated that chicken mAbs can be raised against peptide and hapten epitopes (22–24), our report is the first to document the generation of chicken mAbs that specifically recognize phosphoepitopes. Immunizing the same chickens with a mixture of phosphopeptides led to the successful generation of mAbs specific to each of the peptides, demonstrating that this is a feasible technological approach to generate panels of highly specific mAbs against various phosphoepitopes from a single small library. Importantly, despite being raised against linear synthetic peptides, at least one of these antibodies can specifically recognize the pathological epitope of phospho-tau in AD brains analyzed by both immunohistochemistry and Western blotting.
Of the three mAbs tested that bind to the phosphopeptides, one was found to bind both orthologs of phospho-tau in human AD brain and Tg4510 mouse brain, proving the broad potential diagnostic and experimental utility of these recombinant avian antibodies. During our phage selection and screening processes, we also obtained a small number of clones that bind to the non-phosphopeptides. Modification of the phage selection conditions and competition with non-phosphopeptide and scrambled peptides were critical for the isolation of phosphospecific binders. Our combined immunization and selection approach delivered highly phosphospecific clones of up to picomolar affinity despite the tendency of the immune system to predominantly generate a non-phosphospecific response. Our biosensor studies corroborated the binding mechanism suggested by the co-crystal structure, showing the phosphorylation event at Thr-231 to be the critical element in the interaction between the paratope of pT231/pS235_1 and the phosphoepitope. When this residue was dephosphorylated, all affinity for the peptide was lost.
The analysis of co-crystal structures between antibodies and their antigens, combined with natural repertoire characterization, has greatly informed our understanding of antibody structure-function relationships (55, 62). These analyses have led to the recognition that the structural topography and chemical content of the paratope can be distinctly different between antibodies, depending on the size and nature of the antigen (63). Such structural insights have been critical to the adoption of the “designer compound libraries” (64) approach in antibody drug discovery, leading to the creation of synthetic antibody libraries that are tailored to recognize therapeutically or diagnostically relevant proteins (40), haptens (39), or peptides (41). The simplified generation of high-affinity recombinant antibodies to phosphoepitopes, as described here, will improve our ability to provide further structural insights and may potentiate phosphoprotein-targeted designer libraries in the future.
The identification of the high-affinity antibody pT231/pS235_1 facilitated the generation of the Fab-phosphopeptide crystal structure. As truly phosphospecific mAb generation is challenging (34), it may be rare to find such clones with mid-picomolar 1:1 binding affinity that subsequently aid the generation of high-quality crystals. The high-resolution pT231/pS235_1 co-crystal structure showed that the mechanism of interaction used by the antibody does not fit strictly into any one of the classical protein-, peptide-, or hapten-binding modes. In general terms, anti-protein antibodies tend to use broad flat interfaces with antigen (40), whereas anti-hapten antibodies tend to have more limited interaction through a smaller paratope that is buried deep in the VH-VL interface (39, 65). Meanwhile, peptide-binding antibodies are thought to use a “grooved” paratope that is in between the size of the surfaces observed in anti-protein and anti-hapten clones (55). Antibody pT231/pS235_1 contains an unusual bowl-like recess in its CDR-H2 surface, into which the phosphate group of pThr-231 inserts. This stabilizes the specific interaction with the phosphopeptide (but not the unmodified peptide) through a network of hydrogen bond interactions. Antibody pT231/pS235_1 also utilizes a long but highly structured CDR-H3 loop to make the majority of its definitive anti-peptide contacts. Given the issue of entropic penalty during peptide binding (60), it seems likely that the constrained structure of CDR-H3 may be an additional important factor in the high-affinity epitope interaction observed for pT231/pS235_1. Indeed, recent studies of human antibody VH (66), shark VNAR (67), and camelid VHH (68) domains with non-canonical cysteines in their CDRs all support the critical role of disulfide bonding in the maintenance of binding affinity.
The crystal structure of pT231/pS235_1 Fab therefore provides new insight into the CDR structural diversification mechanisms used by chickens. A recent bioinformatic study has shown that the chicken VH domain repertoire is highly homologous to that of humans and mice in both the frameworks and CDR1 and CDR2 (32). In contrast, chickens use a much longer VH CDR3 repertoire and frequently stabilize these long binding loops by introducing non-canonical disulfide bonds (32). In the structure presented here, a descriptive example of that mechanism is highlighted, along with a Vλ CDR1 structure, which may be common in chicken antibodies but is distinctly different from any hitherto described structure in mammals (58, 69).
In summary, this study has validated the use of immunized chickens in a rapid and multiplexed method to generate phosphoepitope-specific mAbs. We have also defined unique structural attributes of the highest affinity antibody generated. Future improvements in the methods utilized here will help increase the success rate of generating phosphoprotein-specific mAbs and allow us to increase the understanding of the nature of phosphoepitope recognition by highly specific antibodies.
Acknowledgments
We thank John Dunlop for support and encouragement on this project. Use of Industrial Macromolecular Crystallography Association Collaborative Access Team beamline 17-ID (or 17-BM) at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357.

This article contains supplemental Figs. S1–S4 and Tables 1 and 2.
The atomic coordinates and structure factors (code 4GLR) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- AD
- Alzheimer disease
- mAb
- monoclonal antibody
- CDR
- complementarity-determining region
- scFv
- single chain Fv
- pThr
- phospho-Thr.
REFERENCES
- 1. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 2. Nairn A. C., Detre J. A., Casnellie J. E., Greengard P. (1982) Serum antibodies that distinguish between the phospho- and dephospho-forms of a phosphoprotein. Nature 299, 734–736 [DOI] [PubMed] [Google Scholar]
- 3. Ross A. H., Baltimore D., Eisen H. N. (1981) Phosphotyrosine-containing proteins isolated by affinity chromatography with antibodies to a synthetic hapten. Nature 294, 654–656 [DOI] [PubMed] [Google Scholar]
- 4. Grundke-Iqbal I., Iqbal K., Quinlan M., Tung Y. C., Zaidi M. S., Wisniewski H. M. (1986) Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J. Biol. Chem. 261, 6084–6089 [PubMed] [Google Scholar]
- 5. Grundke-Iqbal I., Iqbal K., Tung Y. C., Quinlan M., Wisniewski H. M., Binder L. I. (1986) Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. U.S.A. 83, 4913–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alonso A. D., Zaidi T., Novak M., Barra H. S., Grundke-Iqbal I., Iqbal K. (2001) Interaction of tau isoforms with Alzheimer's disease abnormally hyperphosphorylated tau and in vitro phosphorylation into the disease-like protein. J. Biol. Chem. 276, 37967–37973 [DOI] [PubMed] [Google Scholar]
- 7. Brion J. P., Hanger D. P., Bruce M. T., Couck A. M., Flament-Durand J., Anderton B. H. (1991) Tau in Alzheimer neurofibrillary tangles. N- and C-terminal regions are differentially associated with paired helical filaments and the location of a putative abnormal phosphorylation site. Biochem. J. 273, 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goedert M., Spillantini M. G., Cairns N. J., Crowther R. A. (1992) Tau proteins of Alzheimer paired helical filaments: abnormal phosphorylation of all six brain isoforms. Neuron 8, 159–168 [DOI] [PubMed] [Google Scholar]
- 9. Kosik K. S., Joachim C. L., Selkoe D. J. (1986) Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 83, 4044–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindwall G., Cole R. D. (1984) Phosphorylation affects the ability of tau protein to promote microtubule assembly. J. Biol. Chem. 259, 5301–5305 [PubMed] [Google Scholar]
- 11. Abraha A., Ghoshal N., Gamblin T. C., Cryns V., Berry R. W., Kuret J., Binder L. I. (2000) C-terminal inhibition of tau assembly in vitro and in Alzheimer's disease. J. Cell Sci. 113, 3737–3745 [DOI] [PubMed] [Google Scholar]
- 12. Haase C., Stieler J. T., Arendt T., Holzer M. (2004) Pseudophosphorylation of tau protein alters its ability for self-aggregation. J. Neurochem. 88, 1509–1520 [DOI] [PubMed] [Google Scholar]
- 13. Liu F., Li B., Tung E. J., Grundke-Iqbal I., Iqbal K., Gong C. X. (2007) Site-specific effects of tau phosphorylation on its microtubule assembly activity and self-aggregation. Eur. J. Neurosci. 26, 3429–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kenessey A., Yen S. H. (1993) The extent of phosphorylation of fetal tau is comparable to that of PHF-tau from Alzheimer paired helical filaments. Brain Res. 629, 40–46 [DOI] [PubMed] [Google Scholar]
- 15. Köpke E., Tung Y. C., Shaikh S., Alonso A. C., Iqbal K., Grundke-Iqbal I. (1993) Microtubule-associated protein tau. Abnormal phosphorylation of a non-paired helical filament pool in Alzheimer disease. J. Biol. Chem. 268, 24374–24384 [PubMed] [Google Scholar]
- 16. Ksiezak-Reding H., Liu W. K., Yen S. H. (1992) Phosphate analysis and dephosphorylation of modified tau associated with paired helical filaments. Brain Res. 597, 209–219 [DOI] [PubMed] [Google Scholar]
- 17. Hanger D. P., Seereeram A., Noble W. (2009) Mediators of tau phosphorylation in the pathogenesis of Alzheimer's disease. Expert Rev. Neurother. 9, 1647–1666 [DOI] [PubMed] [Google Scholar]
- 18. Hampel H., Blennow K., Shaw L. M., Hoessler Y. C., Zetterberg H., Trojanowski J. Q. (2010) Total and phosphorylated tau protein as biological markers of Alzheimer's disease. Exp. Gerontol. 45, 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamanaka H. I., Inoue T., Ikeda-Tanaka O. (1996) Chicken monoclonal antibody isolated by a phage display system. J. Immunol. 157, 1156–1162 [PubMed] [Google Scholar]
- 20. Finlay W. J., deVore N. C., Dobrovolskaia E. N., Gam A., Goodyear C. S., Slater J. E. (2005) Exploiting the avian immunoglobulin system to simplify the generation of recombinant antibodies to allergenic proteins. Clin. Exp. Allergy 35, 1040–1048 [DOI] [PubMed] [Google Scholar]
- 21. Hof D., Hoeke M. O., Raats J. M. (2008) Multiple-antigen immunization of chickens facilitates the generation of recombinant antibodies to autoantigens. Clin. Exp. Immunol. 151, 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishibori N., Horiuchi H., Furusawa S., Matsuda H. (2006) Humanization of chicken monoclonal antibody using phage-display system. Mol. Immunol. 43, 634–642 [DOI] [PubMed] [Google Scholar]
- 23. Finlay W. J., Shaw I., Reilly J. P., Kane M. (2006) Generation of high-affinity chicken single-chain Fv antibody fragments for measurement of the Pseudonitzschia pungens toxin domoic acid. Appl. Environ. Microbiol. 72, 3343–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andris-Widhopf J., Rader C., Steinberger P., Fuller R., Barbas C. F., 3rd. (2000) Methods for the generation of chicken monoclonal antibody fragments by phage display. J. Immunol. Methods 242, 159–181 [DOI] [PubMed] [Google Scholar]
- 25. Schroeder H. W., Jr. (2006) Similarity and divergence in the development and expression of the mouse and human antibody repertoires. Dev. Comp. Immunol. 30, 119–135 [DOI] [PubMed] [Google Scholar]
- 26. Schroeder H. W., Jr., Hillson J. L., Perlmutter R. M. (1990) Structure and evolution of mammalian VH families. Int. Immunol. 2, 41–50 [DOI] [PubMed] [Google Scholar]
- 27. Parvari R., Ziv E., Lantner F., Heller D., Schechter I. (1990) Somatic diversification of chicken immunoglobulin light chains by point mutations. Proc. Natl. Acad. Sci. U.S.A. 87, 3072–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reynaud C. A., Anquez V., Weill J. C. (1991) The chicken D locus and its contribution to the immunoglobulin heavy chain repertoire. Eur. J. Immunol. 21, 2661–2670 [DOI] [PubMed] [Google Scholar]
- 29. Reynaud C. A., Dahan A., Anquez V., Weill J. C. (1989) Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell 59, 171–183 [DOI] [PubMed] [Google Scholar]
- 30. Reynaud C. A., Anquez V., Grimal H., Weill J. C. (1987) A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell 48, 379–388 [DOI] [PubMed] [Google Scholar]
- 31. Weill J. C., Reynaud C. A. (1992) Early B-cell development in chickens, sheep and rabbits. Curr. Opin. Immunol. 4, 177–180 [DOI] [PubMed] [Google Scholar]
- 32. Wu L., Oficjalska K., Lambert M., Fennell B. J., Darmanin-Sheehan A., Ní Shúilleabháin D., Autin B., Cummins E., Tchistiakova L., Bloom L., Paulsen J., Gill D., Cunningham O., Finlay W. J. (2012) Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids. J. Immunol. 188, 322–333 [DOI] [PubMed] [Google Scholar]
- 33. Tan C. S. (2011) Sequence, structure, and network evolution of protein phosphorylation. Sci. Signal. 4, mr6. [DOI] [PubMed] [Google Scholar]
- 34. Brumbaugh K., Johnson W., Liao W. C., Lin M. S., Houchins J. P., Cooper J., Stoesz S., Campos-Gonzalez R. (2011) Overview of the generation, validation, and application of phosphosite-specific antibodies. Methods Mol. Biol. 717, 3–43 [DOI] [PubMed] [Google Scholar]
- 35. Vielemeyer O., Yuan H., Moutel S., Saint-Fort R., Tang D., Nizak C., Goud B., Wang Y., Perez F. (2009) Direct selection of monoclonal phosphospecific antibodies without prior phosphoamino acid mapping. J. Biol. Chem. 284, 20791–20795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruff-Jamison S., Campos-González R., Glenney J. R., Jr. (1991) Heavy and light chain variable region sequences and antibody properties of anti-phosphotyrosine antibodies reveal both common and distinct features. J. Biol. Chem. 266, 6607–6613 [PubMed] [Google Scholar]
- 37. Ruff-Jamison S., Glenney J. R., Jr. (1993) Molecular modeling and site-directed mutagenesis of an anti-phosphotyrosine antibody predicts the combining site and allows the detection of higher affinity interactions. Protein Eng. 6, 661–668 [DOI] [PubMed] [Google Scholar]
- 38. Ruff-Jamison S., Glenney J. R., Jr. (1993) Requirement for both H and L chain V regions, VH and VK joining amino acids, and the unique H chain D region for the high affinity binding of an anti-phosphotyrosine antibody. J. Immunol. 150, 3389–3396 [PubMed] [Google Scholar]
- 39. Persson H., Lantto J., Ohlin M. (2006) A focused antibody library for improved hapten recognition. J. Mol. Biol. 357, 607–620 [DOI] [PubMed] [Google Scholar]
- 40. Almagro J. C., Quintero-Hernández V., Ortiz-León M., Velandia A., Smith S. L., Becerril B. (2006) Design and validation of a synthetic VH repertoire with tailored diversity for protein recognition. J. Mol. Recognit. 19, 413–422 [DOI] [PubMed] [Google Scholar]
- 41. Cobaugh C. W., Almagro J. C., Pogson M., Iverson B., Georgiou G. (2008) Synthetic antibody libraries focused towards peptide ligands. J. Mol. Biol. 378, 622–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Finlay W. J., Bloom L., Cunningham O. (2011) Optimized generation of high-affinity, high-specificity single-chain Fv antibodies from multiantigen immunized chickens. Methods Mol. Biol. 681, 383–401 [DOI] [PubMed] [Google Scholar]
- 43. Finlay W. J., Cunningham O., Lambert M. A., Darmanin-Sheehan A., Liu X., Fennell B. J., Mahon C. M., Cummins E., Wade J. M., O'Sullivan C. M., Tan X. Y., Piche N., Pittman D. D., Paulsen J., Tchistiakova L., Kodangattil S., Gill D., Hufton S. E. (2009) Affinity maturation of a humanized rat antibody for anti-RAGE therapy: comprehensive mutagenesis reveals a high level of mutational plasticity both inside and outside the complementarity-determining regions. J. Mol. Biol. 388, 541–558 [DOI] [PubMed] [Google Scholar]
- 44. Cummins E., Luxenberg D. P., McAleese F., Widom A., Fennell B. J., Darmanin-Sheehan A., Whitters M. J., Bloom L., Gill D., Cunningham O. (2008) A simple high-throughput purification method for hit identification in protein screening. J. Immunol. Methods 339, 38–46 [DOI] [PubMed] [Google Scholar]
- 45. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 46. Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., Bricogne G. (2011) Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 49. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. (2011) BUSTER Version 2.11.1, Global Phasing Ltd., Cambridge, United Kingdom [Google Scholar]
- 50. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 51. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 52. Lu P. J., Wulf G., Zhou X. Z., Davies P., Lu K. P. (1999) The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature 399, 784–788 [DOI] [PubMed] [Google Scholar]
- 53. Santacruz K., Lewis J., Spires T., Paulson J., Kotilinek L., Ingelsson M., Guimaraes A., DeTure M., Ramsden M., McGowan E., Forster C., Yue M., Orne J., Janus C., Mariash A., Kuskowski M., Hyman B., Hutton M., Ashe K. H. (2005) Tau suppression in a neurodegenerative mouse model improves memory function. Science 309, 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 55. Almagro J. C. (2004) Identification of differences in the specificity-determining residues of antibodies that recognize antigens of different size: implications for the rational design of antibody repertoires. J. Mol. Recognit. 17, 132–143 [DOI] [PubMed] [Google Scholar]
- 56. Kabat E. A., Wu T. T., Reid-Miller M., Perry H. M., Gottesman K. S., Foeller C. (1991) Sequences of Proteins of Immunological Interest, 5th Ed., United States Department of Health and Human Services, Public Health Service, National Institutes of Health, Washington, D.C [Google Scholar]
- 57. Andreeva A., Prlić A., Hubbard T. J., Murzin A. G. (2007) SISYPHUS–structural alignments for proteins with non-trivial relationships. Nucleic Acids Res. 35, D253–D259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. North B., Lehmann A., Dunbrack R. L., Jr. (2011) A new clustering of antibody CDR loop conformations. J. Mol. Biol. 406, 228–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Finlay W. J., Bloom L., Cunningham O. (2011) Phage display: a powerful technology for the generation of high specificity affinity reagents from alternative immune sources. Methods Mol. Biol. 681, 87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zahnd C., Spinelli S., Luginbühl B., Amstutz P., Cambillau C., Plückthun A. (2004) Directed in vitro evolution and crystallographic analysis of a peptide-binding single chain antibody fragment (scFv) with low picomolar affinity. J. Biol. Chem. 279, 18870–18877 [DOI] [PubMed] [Google Scholar]
- 61. Iwamoto S., Nishimichi N., Tateishi Y., Sato Y., Horiuchi H., Furusawa S., Sawamura T., Matsuda H. (2009) Generation and characterization of chicken monoclonal antibodies against human LOX-1. MAbs 1, 357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zemlin M., Klinger M., Link J., Zemlin C., Bauer K., Engler J. A., Schroeder H. W., Jr., Kirkham P. M. (2003) Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J. Mol. Biol. 334, 733–749 [DOI] [PubMed] [Google Scholar]
- 63. MacCallum R. M., Martin A. C., Thornton J. M. (1996) Antibody-antigen interactions: contact analysis and binding site topography. J. Mol. Biol. 262, 732–745 [DOI] [PubMed] [Google Scholar]
- 64. Valler M. J., Green D. (2000) Diversity screening versus focussed screening in drug discovery. Drug Discov. Today 5, 286–293 [DOI] [PubMed] [Google Scholar]
- 65. Tars K., Kotelovica S., Lipowsky G., Bauer M., Beerli R. R., Bachmann M. F., Maurer P. (2012) Different binding modes of free and carrier-protein-coupled nicotine in a human monoclonal antibody. J. Mol. Biol. 415, 118–127 [DOI] [PubMed] [Google Scholar]
- 66. Almagro J. C., Raghunathan G., Beil E., Janecki D. J., Chen Q., Dinh T., LaCombe A., Connor J., Ware M., Kim P. H., Swanson R. V., Fransson J. (2012) Characterization of a high-affinity human antibody with a disulfide bridge in the third complementarity-determining region of the heavy chain. J. Mol. Recognit. 25, 125–135 [DOI] [PubMed] [Google Scholar]
- 67. Fennell B. J., Darmanin-Sheehan A., Hufton S. E., Calabro V., Wu L., Müller M. R., Cao W., Gill D., Cunningham O., Finlay W. J. (2010) Dissection of the IgNAR V domain: molecular scanning and orthologue database mining define novel IgNAR hallmarks and affinity maturation mechanisms. J. Mol. Biol. 400, 155–170 [DOI] [PubMed] [Google Scholar]
- 68. Govaert J., Pellis M., Deschacht N., Vincke C., Conrath K., Muyldermans S., Saerens D. (2012) Dual beneficial effect of interloop disulfide bond for single domain antibody fragments. J. Biol. Chem. 287, 1970–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chailyan A., Marcatili P., Cirillo D., Tramontano A. (2011) Structural repertoire of immunoglobulin λ light chains. Proteins 79, 1513–1524 [DOI] [PubMed] [Google Scholar]