Background: De novo protein folding is assisted by essential networks of molecular chaperones.
Results: Overproduction of Hsp33 controls EF-Tu stability thus allowing bacterial growth without trigger factor and DnaK.
Conclusion: Slowing down translation limits protein aggregation and enables bacterial survival in the absence of major chaperones.
Significance: Deciphering networks of chaperones is crucial for understanding how cells respond to severe protein aggregation.
Keywords: Heat Shock Protein, Molecular Chaperone, Molecular Genetics, Protein Aggregation, Protein Folding, Hsp33, Hsp70/DnaK, Protein Quality Control, Toxin-antitoxins, Trigger Factor
Abstract
Intracellular de novo protein folding is assisted by cellular networks of molecular chaperones. In Escherichia coli, cooperation between the chaperones trigger factor (TF) and DnaK is central to this process. Accordingly, the simultaneous deletion of both chaperone-encoding genes leads to severe growth and protein folding defects. Herein, we took advantage of such defective phenotypes to further elucidate the interactions of chaperone networks in vivo. We show that disruption of the TF/DnaK chaperone pathway is efficiently rescued by overexpression of the redox-regulated chaperone Hsp33. Consistent with this observation, the deletion of hslO, the Hsp33 structural gene, is no longer tolerated in the absence of the TF/DnaK pathway. However, in contrast with other chaperones like GroEL or SecB, suppression by Hsp33 was not attributed to its potential overlapping general chaperone function(s). Instead, we show that overexpressed Hsp33 specifically binds to elongation factor-Tu (EF-Tu) and targets it for degradation by the protease Lon. This synergistic action of Hsp33 and Lon was responsible for the rescue of bacterial growth in the absence of TF and DnaK, by presumably restoring the coupling between translation and the downstream folding capacity of the cell. In support of this hypothesis, we show that overexpression of the stress-responsive toxin HipA, which inhibits EF-Tu, also rescues bacterial growth and protein folding in the absence of TF and DnaK. The relevance for such a convergence of networks of chaperones and proteases acting directly on EF-Tu to modulate the intracellular rate of protein synthesis in response to protein aggregation is discussed.
Introduction
In the bacterium Escherichia coli, the folding of newly synthesized proteins is orchestrated by three major chaperone systems, namely trigger factor (TF),2 DnaK (Hsp70), and GroEL (Hsp60). The ribosome-bound TF is the first molecular chaperone to interact with most of the newly synthesized polypeptides (1). TF does this by anchoring itself to the L23 ribosomal protein located at the ribosomal exit port (2), thus enabling its early interaction with nascent polypeptide chains as short as 40 amino acids (3, 4). Following polypeptide chain elongation and release from the ribosome, TF may stay bound to its substrate for a prolonged period, thus facilitating folding or transfer to downstream cytosolic chaperones such as GroEL or DnaK (5). Recently, a quantitative ribosome profiling analysis of nascent chains bound to TF, revealed that in addition to cytosolic proteins, outer membrane proteins are also TF substrates. In addition, deletion of the tig gene, encoding TF, leads to a decrease in the steady state level of outer membrane proteins, suggesting that TF plays an important role in outer membrane proteins processing as well (6–8).
Assisted by its DnaJ and GrpE co-chaperones, the major chaperone DnaK facilitates co- and/or post-translational folding of nearly 15% of the nascent polypeptides (9, 10). A recent analysis of the DnaK protein “interactome” at physiological temperature by Calloni and colleagues (11) revealed that DnaK interacts with more than 600 proteins, including ∼80% of cytosolic proteins, ∼11% of inner membrane proteins, ∼3% of outer membrane proteins, and ∼3% of periplasmic proteins (11). In agreement with such a central role for DnaK in intracellular protein folding, E. coli cells carrying a null mutation in the dnaK gene exhibit multiple phenotypes and readily accumulate extragenic suppressors (12). The third major molecular chaperone involved in de novo protein folding is the chaperonin GroEL assisted by its co-chaperone GroES (Hsp10) (13). It is believed that about 10% of the newly synthesized polypeptides interact post-translationally with GroEL (14). Specifically, more than 250 GroEL substrates have been identified, 85 of which were shown to be obligate GroEL substrates, including 13 essential proteins (15). In agreement with these results, the groEL and groES genes are absolutely essential for E. coli viability, because neither can be deleted under all conditions tested (16).
It has been shown that TF and DnaK act coordinately in the de novo protein folding pathways of many protein substrates (3, 4, 17, 18). However, DnaK is not capable of replacing TF function in the stabilization of presecretory proteins (8). Accordingly, previous genetic studies have shown that the simultaneous deletion of both the tig and dnaK genes causes synthetic bacterial lethality at temperatures above ∼30 °C, depending on the particular genetic background strain (3, 4, 17, 18). Noticeably, more than 1000 proteins, pre-existing and de novo synthesized, were shown to aggregate at 30 °C in this double tig dnaK mutant, indicating that the cooperation between TF and DnaK is indeed crucial for overall cellular protein homeostasis (11).
In agreement with the existence of such a chaperone network, overexpression of GroEL/GroES significantly suppresses both the temperature-sensitive phenotype and the protein folding defect observed in the simultaneous absence of both TF and DnaK (15, 17, 18). In this case, overexpression of GroEL/GroES likely favors interaction with nonobligate GroEL substrates, thus restore cell viability. Interestingly, protein substrates of GroEL and DnaK overlap significantly (15), as DnaK interacts with more than 40% of obligate GroEL substrates (11). This observation is in agreement with a proposed model in which DnaK is working upstream of GroEL to stabilize obligate GroEL substrates in a nonaggregated state for subsequent productive interaction with GroEL. In addition, significantly more GroEL substrates interact with DnaK in vivo upon GroEL depletion and about ∼70% of GroEL substrates aggregate in the double tig dnaK mutant, thus arguing again for a key role of TF and DnaK in targeting substrates to GroEL (11). Interestingly, overexpression of the export chaperone SecB is also capable of replacing DnaK and TF, suggesting that this chaperone can also play an important role in the cellular chaperone network in response to cytoplasmic protein misfolding or aggregation (19). In this respect it is interesting to note that both GroEL and SecB were recently isolated as highly enriched DnaK interacting proteins (“interactors”), further emphasizing the functional cooperation among these chaperones (11).
In this work, we show that the redox regulated and heat shock-induced chaperone Hsp33 can efficiently suppress the bacterial growth and protein folding defects observed in the absence of both TF and DnaK, thus revealing a role for Hsp33 as a major participant of the E. coli chaperone network. However, in contrast with our previous results concerning GroEL and SecB, we now show that Hsp33 acts indirectly, by specifically interacting with the essential elongation factor Tu (EF-Tu) and targeting it for degradation by the protease Lon.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Phages, and Culture Conditions
Genetic experiments were carried out in the E. coli MC4100 (20) or W3110 genetic background strains (21). The MC4100 mutant derivatives Δtig::CmR, ΔdnaKdnaJ::KanR, Δtig::CmR ΔdnaKdnaJ::KanR, dnaJ::Tn10-42, Δtig dnaJ::Tn10-42 (17), Δtig::CmS (7), Δlon::KanR (22), ΔclpP::CmR (23), ΔclpQ::TetR (24), and W3110 Δtig ΔdnaKdnaJ::KanR (17) have been previously described. The ΔhslO::KanR allele was obtained from strain JWK5692 (Keio collection). All mutations described in this study were moved to the appropriate genetic background by bacteriophage P1-mediated transduction. Bacteria were routinely grown in LB medium supplemented when necessary with chloramphenicol (10 μg/ml), kanamycin (50 μg/ml), ampicillin (100 μg/ml), or tetracycline (15 μg/ml).
The construction of the dnaK-protease and dnaK hslO double mutants was performed as follows. The KanR cassettes from MC4100 ΔhslO::KanR and Δlon::KanR were first removed using plasmid pCP20 as described (25). The ΔdnaKdnaJ::KanR mutant allele was then introduced into the hslO and the various protease mutants, thus leading to the construction of strains MC4100 ΔhslO ΔdnaKdnaJ::KanR, Δlon ΔdnaKdnaJ::KanR, ΔclpP::CmR ΔdnaKdnaJ::KanR, and ΔclpQ::TetR ΔdnaKdnaJ::KanR. To construct strain MC4100 ΔhslO Δtig::CmR dnaJ::Tn10-42, the Δtig::CmR mutant allele was first introduced into MC4100 ΔhslO, followed by introduction of dnaJ::Tn10-42(TetR). In this case, selection for transductants was carried out at 30 °C.
The mutant strains used for the Lon-dependent suppression experiments were constructed as follows. The ΔhslO::KanR mutant allele was first introduced into MC4100 Δlon. Next, the ΔdnaK52::CmR mutant allele (12) was introduced into both MC4100 ΔhslO::KanR and MC4100 Δlon ΔhslO::KanR. MC4100 Δtig::CmR Δlon::KanR dnaJ::Tn10-42 was constructed by first moving the Δtig::CmR mutant allele into MC4100 Δlon::KanR. Note that the lon and tig genes are 90% linked by P1 transduction on the E. coli chromosome. The dnaJ::Tn10-42 allele was then introduced into MC4100 Δtig::CmR Δlon::KanR and selection was done at 30 °C. All DNA cloning and transformation experiments were carried out in strain DH5α (Invitrogen).
Plasmid Construction
Plasmids pSE380ΔNcoI, p29SEN (17), and pBAD22 (26) have been previously described. To construct the high-copy number plasmid pSE-Hsp33 (pSE380 ΔNcoI-Hsp33) and the low-copy number plasmid p29-Hsp33 (p29SEN-Hsp33), the 879-bp hslO gene was PCR-amplified using primers Hsp33-for (5′-CGGAATTCTCATGATTATGCCGCAACATGACC-3′) and Hsp33-rev (5′-CGAAGCTTGGATCCTTAATGAACTTGCGGATCTGC-3′) using MG1655 genomic DNA as template. The PCR fragment was digested with EcoRI and HindIII and cloned into either pSE380ΔNcoI or p29SEN previously digested with the same enzymes. A similar cloning procedure was used to construct the pSE-Hsp33(Y12E), pSE-Hsp33(M172S) mutant derivatives, except that in this case, plasmids pET11a-hslO(Y12E) or pET11a-hslO(M172S) DNA were used as a template (27). Plasmid pSE-Hsp33-FLAG (pSE380ΔNcoI-Hsp33-flag tagged) containing Hsp33 with the C-terminal FLAG tag “GSDYKDDDDKSA” was constructed using primers Hsp33-for and 33FT-rev (5′-GCAAGCTTGGATCCTTAGGCGCTTTTATCGTCGTCATCTTTGTAGTCGCTGCCATGAACTTGCGGATCTGC-3′) and pSE-Hsp33 as DNA template. The PCR fragment was cloned as an EcoRI-HindIII-digested PCR fragment in pSE380ΔNcoI digested with the same enzymes. Plasmid pSE-Hsp33(Q151E) was constructed by QuikChange mutagenesis using the appropriate primers. Plasmid pSE-Hsp33(1–235) was constructed using primers Hsp33-for and Hsp33(1–235)-rev (5′-CAAGCTTGGATCCttaCGAGCAGGTGCATTTGAACTC-3′) and pSE-Hsp33 as DNA template. The PCR fragment was cloned as an EcoRI-HindIII-digested fragment into pSE380ΔNcoI digested with the same enzymes.
The p29SEN-derivatives containing either the yrfG hslR hslO or the hslR hslO operon were constructed using yrfG-For (5′-GAGAATTCCATATGCATATCAACATTGCCTG-3′) or hslR-For (5′-GAGAATTCCATATGAAAGAGAAACCTGCTGTTG-3′) forward primers, respectively. In this case, the primer Hsp33-rev was used as a reverse primer.
To construct plasmid p29-EF-Tu, the 1185-bp long tufA gene, encoding EF-Tu, was PCR-amplified using primers tufA-for (5′-GAGAATTCATGTCTAAAGAAAAATTTGAAC-3′) and tufA-rev (5′-GAAAGCTTTTAGCCCAGAACTTTAGCAAC-3′) and MG1655 genomic DNA as template. The PCR fragment was digested with EcoRI and HindIII and cloned into p29SEN digested with the same enzymes.
The toxin gene hipA (1323 bp) was PCR amplified using primers HipA-For (5′-GCGAATCCATGCCTAAACTTGTCACTTG-3′) and HipA-Rev (5′-GCGCATGCTCACTTACTACCGTATTCTC-3′) and MG1655 genomic DNA as template. The hipA gene was then cloned under control of an arabinose-inducible promoter as an EcoRI-SphI fragment into plasmid pBAD22 digested with the same enzymes. The resulting plasmid was named pBAD-HipA. Note that hipA was cloned under the tightly regulated araBADp promoter to minimize the background expression level of HipA in the absence of inducer, thus avoiding killing by the toxin. All constructs generated by PCR were sequence verified.
Bacterial Viability Assay and Genetic Experiments
In vivo complementation of the temperature-sensitive phenotypes of MC4100 Δtig::CmR ΔdnaKdnaJ::KanR, MC4100 Δtig dnaJ::Tn10–42, MC4100 Δtig Δlon dnaJ::Tn10–42, and MC4100 Δtig ΔhslO dnaJ::Tn10–42, was performed as follows. Cultures of fresh transformants were first grown overnight in LB ampicillin/glucose (0.4%) at the permissive temperature (see figure legends), diluted 1/50 into the same medium, further grown to mid-log phase, serially diluted 10-fold, and spotted on LB ampicillin agar plates with or without IPTG (at 5, 50, or 500 μm) and incubated at the indicated temperatures. Complementation of the W3110 ΔdnaKdnaJ::KanR Δtig::CmR temperature-sensitive phenotype by HipA was carried out as described above, except that the l-arabinose inducer (0.1, 0.5, 1, or 2%) was used in this case. To monitor Hsp33 toxicity of the MC4100 and its mutant derivatives, mid-log phase cultures of fresh transformants were grown at the permissive growth temperature (22 or 30 °C; see figure legends) in LB ampicillin/glucose (0.4%), were serially diluted 10-fold and spotted on LB ampicillin agar plates with or without IPTG (100 or 500 μm), and incubated at the indicated temperatures. The co-transduction frequencies between ΔdnaKdnaJ::KanR and the linked thr::TetR alleles were analyzed using bacteriophage P1-mediated transduction as described previously (9), except that MC4100 Δtig and MC4100 Δtig ΔhslO were used as the recipient strains.
Isolation of Protein Aggregates and Cell Fractionation
Cultures of fresh transformants of MC4100 ΔdnaKdnaJ::KanR Δtig::CmR containing plasmids pSE380ΔNcoI, or pSE-Hsp33 wild type or its mutant derivatives were grown at 22 °C, diluted 1/50 and grown to an A600 of 0.3 in LB ampicillin (100 μg/ml), glucose (0.4%). IPTG inducer (0.5 mm) was added and cultures were further incubated for 2 h at 22 °C. Next, the cultures were transferred to a 37 °C water bath, and shaken (180 rpm) for 1 h, and protein aggregates were isolated as described by Tomoyasu and colleagues (28). Protein aggregates were prepared and analyzed by SDS-PAGE using 4–15% Mini-Protean TGX gels (Bio-Rad). For HipA-mediated suppression of protein aggregates, cultures of fresh transformants of W3110 ΔdnaKdnaJ::KanR Δtig::CmS carrying plasmid pBAD22, or pBAD-HipA at 22 °C were diluted 1/50 and grown to an A600 of 0.3 in LB ampicillin (100 μg/ml). l-Arabinose inducer (0.5%) was then added and cell cultures were further incubated for 4 h at 22 °C. Cultures were then transferred to 37 °C in a water bath, shaken (180 rpm) for 1 h, and protein aggregates were prepared as described (28).
Western Blot Analysis
Proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Hybond-P, GE Healthcare) using a semidry transfer system (Trans-Blot SD, Bio-Rad) for 30 min at 10 V. Membranes were blocked for 1 h at RT or overnight at 4 °C with 3% nonfat milk/Tris-buffered saline with Tween 20 (TBS-T; 0.5 m Tris, 1.5 m NaCl, pH 7.4, plus 0.05% Tween 20). Rabbit polyclonal antibody against Hsp33 (1:2,000 dilution; a kind gift of Ursula Jakob, University of Michigan), a mouse monoclonal antibody against EF-Tu (1:10,000 dilution; Hycult Biotech), a rabbit antibody against DnaJ (1:3,000 dilution (29)), and a rabbit antibody against TF (1:8,000 dilution (7)) were used as primary antibodies, and HRP-conjugated rabbit IgG (1:5,000; Sigma) or mouse IgG (1:2,000; Sigma) were used as secondary antibodies. Membranes were developed by chemiluminescence using an ECL Plus (Prime) immunoblotting detection kit following the manufacturer's protocol (GE Healthcare) with a luminescence analyzer (LAS4000, Fuji).
In Vivo Pull-down Assay
E. coli MC4100, MC4100 Δtig CmR, MC4100 ΔdnaKdnaJ::KanR, and MC4100 Δtig::CmR ΔdnaKdnaJ::KanR transformed with pSE-Hsp33FLAG were first selected at 22 °C on LB agar plates supplemented with glucose (0.4%) and ampicillin. Overnight cultures of fresh transformants, grown at 22 °C, were diluted to an A600 of 0.1 and incubated until mid-log phase at 22 °C in LB supplemented with 0.4% glucose and ampicillin. Expression of Hsp33FLAG and Hsp33(Y12E)FLAG was induced by the addition of 0.5 mm IPTG for 30 min at 22 °C and transferred to 30 °C for 1 h and 30 min. Cells were collected by centrifugation (5,000 × g for 5 min at 4 °C) and resuspended in 500 μl of cold PD buffer (150 mm NaCl, 50 mm Tris, pH 7.5, 1 mg/ml lysozyme, and protease inhibitors from Roche Applied Science) supplemented with Benzonase (3 units/ml of culture volume; Invitrogen). Crude cell extracts were obtained following sonication and centrifugation (14,000 × g for 30 min). Pull-down assays were performed with 500-μl aliquots of cell extract and 50 μl of Anti-FLAG M2-agarose resin (Sigma) in TBS buffer (150 mm NaCl, 50 mm Tris, pH 7.5). Suspensions were gently rocked at 4 °C for 2 h, and beads were pelleted by centrifuging for 1 min at 5,000 × g. After washing the beads 3 times with 1 ml of TBS buffer, proteins were eluted in TBS buffer containing 100 μg/ml of FLAG peptide (DYKDDDDK). Samples were separated by SDS-PAGE on 4–20% Mini-Protean TGX gels (Bio-Rad) and stained with Coomassie instant blue (expedeon).
In Vitro Translation and Cross-linking Experiments
Strains MC4100 ΔdnaKdnaJ::KanR and Δtig::CmR ΔdnaKdnaJ::KanR were first transformed with pSE-Hsp33FLAG and grown in LB ampicillin medium at 30 and 25 °C, respectively, were used to obtain Hsp33-FLAG-enriched translation lysates, as previously described for the SecB chaperone (19). RpoB150 mRNA was prepared as previously described (19). In vitro translation in the E. coli cell- and membrane-free S-135 extract was carried out using RpoB150 mRNA as previously described (19, 30). Bifunctional cross-linking was induced by the addition of 1 mm bis(sulfosuccinimidyl) suberate for 10 min at 26 °C and quenched at 4 °C by adding 1/10 volume of quench buffer (1 m glycine, 100 mm NaHCO3, pH 8.5). Ribosome-nascent chain complexes were collected as described (31) and analyzed either directly following SDS-PAGE (15%) separation or after immunoprecipitation (32) with a mouse antibody anti-FLAG (Sigma).
Pulse-Chase and Co-immunoprecipitation Analyses
Overnight cultures of MC4100 ΔhslO ΔdnaKdnaJ::KanR transformed with either pSE380 or pSE-Hsp33 were diluted 1/50 in LB ampicillin, in the presence of 100 μCi/ml of [35S]methionine and grown until the A600 reached 0.4. Unlabeled methionine was then added at a final concentration of 15 mm and IPTG at 500 μm to induce expression of Hsp33. Samples were taken at 0, 60, and 120 min and kept overnight on ice-cold TCA (10%), and analyzed by immunoprecipitation with anti-EF-Tu antibodies as described (33). The gels were scanned using Fuji FLA-3000 phosphorimaging technology.
RESULTS
Hsp33 Overproduction Supports Bacterial Growth and Prevents Protein Aggregation in the Absence of TF and DnaK
The ATP-independent and redox-regulated molecular chaperone Hsp33 (heat-shock protein of 33 kDa), which is specifically activated by oxidative protein unfolding, appears to be a key component of the network of heat-induced stress chaperones and proteases in E. coli (34, 35). Specifically, in the presence of stressors such as hypochlorite or hydrogen peroxide combined with high temperature, sensitive pairs of cysteine residues in Hsp33 are sequentially oxidized, leading to Hsp33 dimerization and activation of its chaperone function. The resulting Hsp33 dimers bind unfolded substrate proteins and protect them from aggregation (34). Upon return to reducing conditions, Hsp33-bound substrates are most likely delivered to DnaK for their subsequent refolding (36).
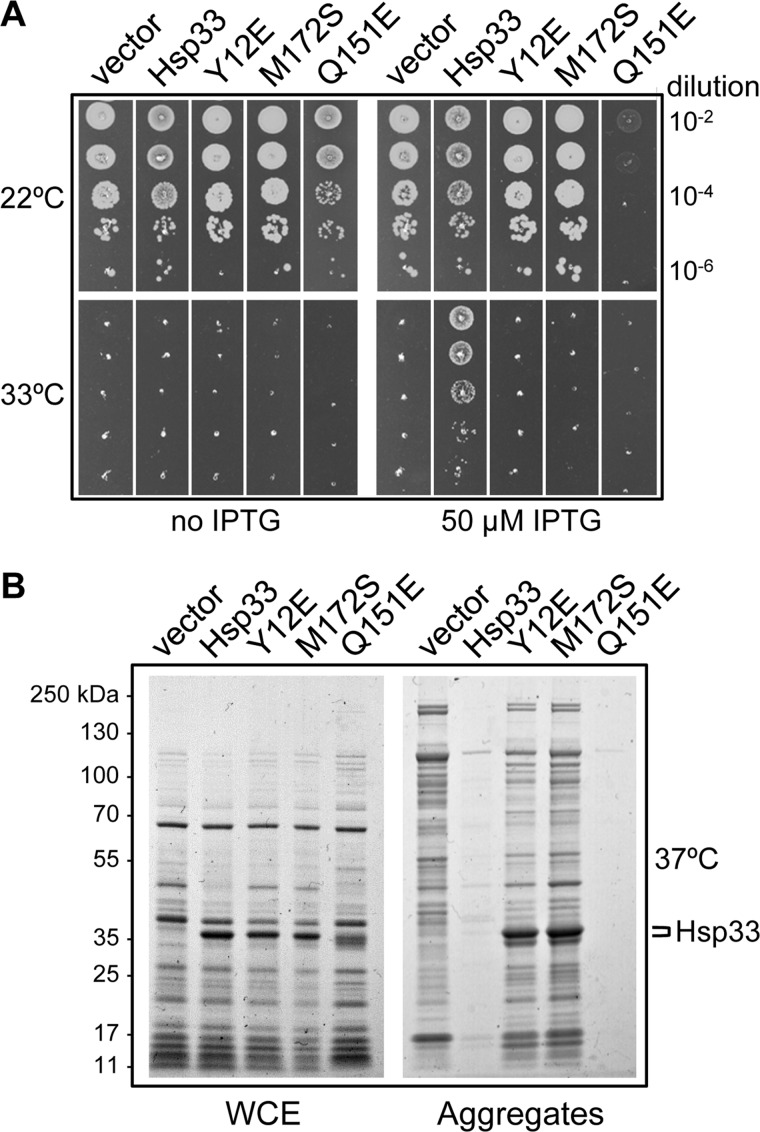
To highlight a possible role of Hsp33 generic chaperone function in the absence of either DnaK or oxidative stress, we asked whether Hsp33, as part of the chaperone network in E. coli, could also restore bacterial growth of the severely compromised Δtig ΔdnaKdnaJ chaperone mutant at higher temperatures (17). This strain lacks the TF/DnaK folding pathway and thus represents a valuable tool for the identification of in vivo chaperone function. The bacterial colony forming experiments presented in Fig. 1A show that indeed, overexpression of Hsp33 (encoded by the hslO gene, the last member of the yrfG-hslR-hslO operon) enables the MC4100 Δtig ΔdnaKdnaJ chaperone mutant to survive at the otherwise nonpermissive temperatures of 33 °C. In addition, we noticed that Hsp33 overexpression interferes somewhat with overall bacterial growth, as judged by smaller colony size formation at the permissive temperatures of growth (Fig. 1A, see below). A Western blot analysis showing Hsp33 expression levels in the presence of IPTG inducer at the permissive temperature of 22 °C is provided in supplemental Fig. S1. Further analysis of the extent of assistance by Hsp33 in the absence of TF and DnaK showed that suppression by Hsp33 is not strain background specific and is not further improved by the expression of the full yrfG-hslR-hslO operon (supplemental Fig. S1).
FIGURE 1.
Hsp33 overexpression rescues bacterial growth and prevents protein aggregation in the absence of DnaK and TF. A, fresh transformants of strain MC4100 Δtig::CmR ΔdnaKdnaJ::KanR containing the plasmid pSE380ΔNcoI parental vector, pSE-Hsp33, pSE-Hsp33(Y12E), pSE-Hsp33(M172S), or pSE-Hsp33(Q151E) were grown at 22 °C, serially diluted 10-fold, and spotted on LB ampicillin agar plates with or without IPTG inducer. Plates were incubated for 1 day at 33 °C or 2 days at 22 °C. B, protein aggregates and their corresponding whole cell extracts (WCE) were isolated from strain MC4100 Δtig::CmR ΔdnaKdnaJ::KanR expressing the pSE-based Hsp33 derivatives, grown at 22 °C, and transferred for 1 h at 37 °C in the presence of IPTG to induce Hsp33 overexpression. The position of the protein bands corresponding to Hsp33 or its mutant derivatives is indicated.
As stated above, de novo protein folding is severely impaired in the absence of both TF and DnaK chaperones. As a consequence, proteins massively accumulate as cytoplasmic aggregates in the Δtig ΔdnaKdnaJ mutant strain. We took advantage of this observation to ask whether Hsp33 overexpression can also prevent the in vivo protein aggregation that occurs in the absence of TF and DnaK, as previously shown for the overproduction of SecB or GroEL (7). Hsp33 was overexpressed in the MC4100 Δtig ΔdnaKdnaJ strain and aggregated proteins were detected following a 1-h incubation at the nonpermissive temperature of 37 °C, as described (37). As anticipated from the bacterial viability results, Hsp33 overexpression efficiently prevented the accumulation of protein aggregates at 37 °C (Fig. 1B).
We next asked whether mutations in the hslO gene, which were previously shown to differentially affect activation of its Hsp33 chaperone function in vitro, could also rescue the defects of the Δtig ΔdnaKdnaJ mutant in the absence of oxidizing conditions. Specifically, we chose the following hslO mutations: Hsp33(Y12E), which is constitutively active in vitro but mainly insoluble in vivo; Hsp33(M172S), activated by high temperature in the absence of oxidizing conditions (27); Hsp33(Q151E), constitutively monomeric and redox-regulated with preference for slowly unfolding protein substrates (38); and Hsp33(1–235), constitutively active as a dimer in vitro (39). The corresponding mutant genes were cloned in a multicopy plasmid under control of an IPTG-inducible promoter and tested for their ability to suppress the growth defects exhibited by the MC4100 Δtig ΔdnaKdnaJ mutant strain. We found that, in sharp contrast to Hsp33 wild-type, overexpression of Hsp33(Y12E), Hsp33(M172S), or Hsp33(1–235) mutant proteins did not rescue bacterial growth at 33 °C (Fig. 1A and supplemental Fig. S2). These results suggest that the integrity of the redox switch mechanism is critical for Hsp33-mediated suppression of the Δtig ΔdnaKdnaJ phenotype. Consistently, neither the Hsp33(Y12E) nor the Hsp33(M172S) mutant was capable of preventing protein aggregation at 37 °C in vivo (Fig. 1B). Instead, these two mutant Hsp33 proteins co-purified with the protein aggregates. Although it is known that Hsp33(Y12E) is mainly insoluble in vivo (27), it is possible that the presence of the Hsp33(M172S) in protein aggregates represents a bona fide chaperone-substrate interaction.
The monomeric and redox-regulated Hsp33(Q151E) mutant displayed a sharply different behavior. It turned out that even at the permissive temperature of growth Hsp33(Q151E) was highly toxic to bacterial growth at all temperatures tested in the MC4100 Δtig ΔdnaKdnaJ mutant background when induced (Fig. 1A). In sharp contrast to its deleterious effect on bacterial growth, monomeric Hsp33(Q151E) fully prevented the accumulation of protein aggregates at the nonpermissive temperature of 37 °C, as well as, or even better than Hsp33 wild type (Fig. 1B). These findings raise the possibility that the monomeric Hsp33 species, which is believed to be transiently populated during the Hsp33 activation cascade process and which shows strong preference for slowly unfolding substrates in vitro (40, 41), may exhibit robust substrate binding properties in the absence of the TF/DnaK pathway.
Hsp33 Function Is Critical in the Absence of Both TF and DnaK
The suppression of both the Δtig ΔdnaKdnaJ temperature-sensitive growth phenotype and the intracellular protein folding defects following Hsp33 overproduction reveal an important functional cooperation among these three major chaperones. To get additional biological clues for such interplay we examined the phenotypes of various mutational combinations among these chaperone genes in the MC4100 strain background. A phenotypic analysis of the double Δtig ΔhslO and ΔdnaKdnaJ ΔhslO mutants revealed only minor growth differences compared with their respective single mutant parents, i.e. the double ΔdnaKdnaJ ΔhslO mutant was more temperature-sensitive for growth at 40 °C than its isogenic single ΔdnaKdnaJ mutant (supplemental Fig. S3).
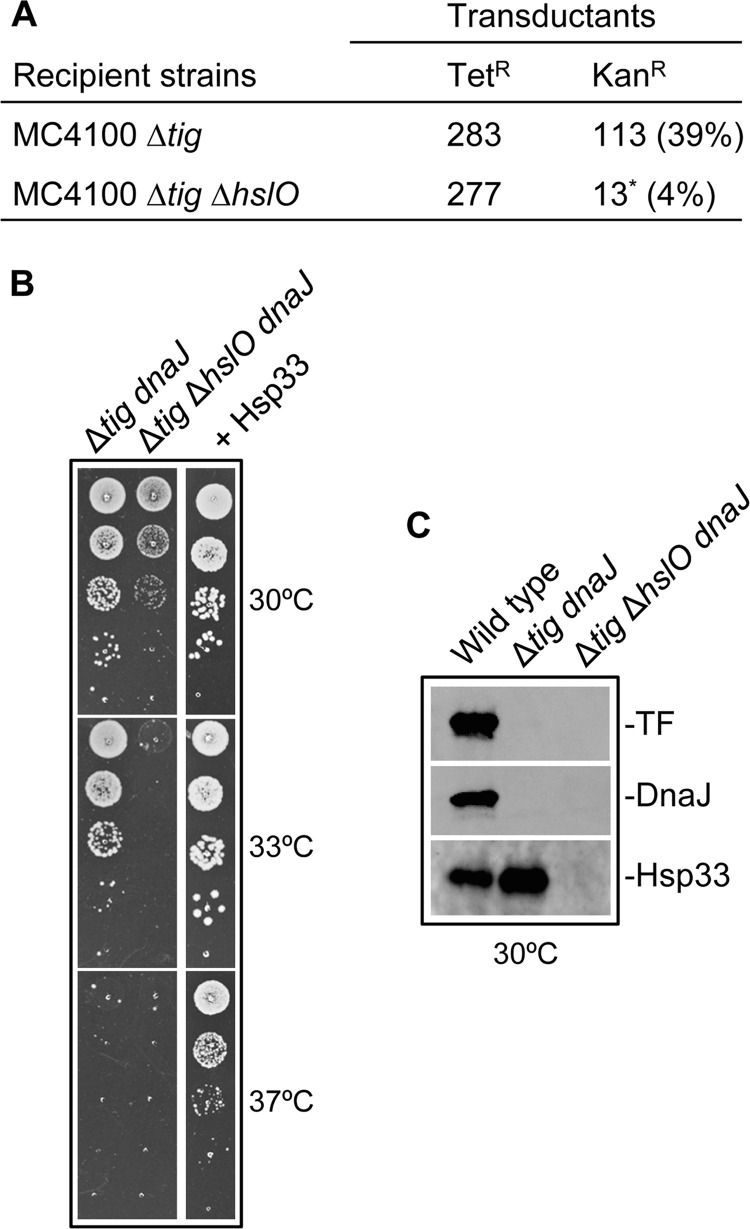
Next, we tested the viability of the triple Δtig ΔdnaKdnaJ ΔhslO mutant compared with its isogenic Δtig ΔdnaKdnaJ mutant parent. To do so, we used the same methodology as we previously described for the construction of the Δtig ΔdnaKdnaJ chaperone mutant (9, 17). Specifically, we first investigated the co-transduction frequency between the ΔdnaKdnaJ::KanR mutant allele and its nearby, ∼40% linked thr::TetR resistance marker using either MC4100 Δtig or MC4100 Δtig ΔhslO as recipient strains (Fig. 2A). As anticipated from our previous work, the ΔdnaKdnaJ mutation was efficiently transduced into the Δtig single mutant with the expected 40% co-transduction frequency. In contrast, the occasional ΔdnaKdnaJ::KanR thr::TetR co-transductants formed extremely small colonies and, in addition, appeared at a 10-fold lower frequency (4% instead compared with the expected ∼40%) when the Δtig ΔhslO strain was used as the recipient. Most likely, the few, slowly growing transductants are due to the continuous accumulation of unknown extragenic suppressors, which enable the occasional survival of the Δtig ΔdnaKdnaJ ΔhslO triple mutant.
FIGURE 2.
Hsp33 is essential for bacterial viability in the absence of DnaK and TF. A, synthetic lethality among the dnaK, tig, and hslO genes was analyzed by P1-mediated transduction experiments. Transductants were first selected on tetracycline plates following a 2-day incubation at 22 °C (the permissive temperature for the tig dnaK double mutant) and subsequently tested for possession of the kanamycin resistance carried by the linked ΔdnaKdnaJ::KanR mutant allele. The co-transduction frequencies of the KanR marker are given as percentage of the total number of TetR transductants. The asterisk indicates the formation of very small colonies. B, synergistic effect of the tig, dnaJ, and hslO mutations on bacterial growth. Mid-log phase cultures of either (i) MC4100 Δtig dnaJ::Tn10-42 and MC4100 Δtig ΔhslO dnaJ::Tn10-42, or (ii) MC4100 dnaJ::Tn10-42 Δtig ΔhslO containing p29SEN-Hsp33 were serially diluted 10-fold and spotted on LB agar plates supplemented with 500 μm IPTG. C, whole cell extracts of MC4100, MC4100 Δtig dnaJ::Tn10-42, and MC4100 Δtig ΔhslO dnaJ::Tn10-42 were separated by SDS-PAGE and analyzed by Western blot using anti-TF, -DnaJ, and -Hsp33 antibodies.
To further demonstrate such a synergy among the various chaperones, we took advantage of the phenotype exhibited by null mutations in the dnaJ gene (the main DnaK co-chaperone), which exhibit a significantly less severe growth phenotype defect due to the presence of the djlA and cbpA genes, known to code for two additional DnaJ-like family members, capable of partially replacing DnaJ function (17, 42). We previously showed that the double tig dnaJ mutant is temperature-sensitive for growth above 34 °C in a strictly DnaK-dependent manner (17). Based on these earlier results, we introduced the dnaJ::Tn10–42 mutation into either the MC4100 Δtig or MC4100 Δtig ΔhslO mutant backgrounds at 30 °C and subsequently compared their bacterial growth phenotypes at various temperatures (Fig. 2B). In agreement with the synthetic lethality observed in the absence of DnaK (Fig. 2A), the triple Δtig ΔhslO dnaJ mutant growth was shown to be significantly more temperature-sensitive than its isogenic Δtig ΔdnaJ double mutant parent. As expected, the colony-forming ability of this triple mutant was fully restored following Hsp33 expression from a plasmid (Fig. 2B). The absence of TF, Hsp33, or DnaJ protein antigens in the corresponding null mutant strains was confirmed by Western blot analysis (Fig. 2C). Taken together, our above data highlight for the first time the essential functional interplay between the TF, DnaK, and Hsp33 chaperones in vivo.
Hsp33 Specifically Interacts with EF-Tu
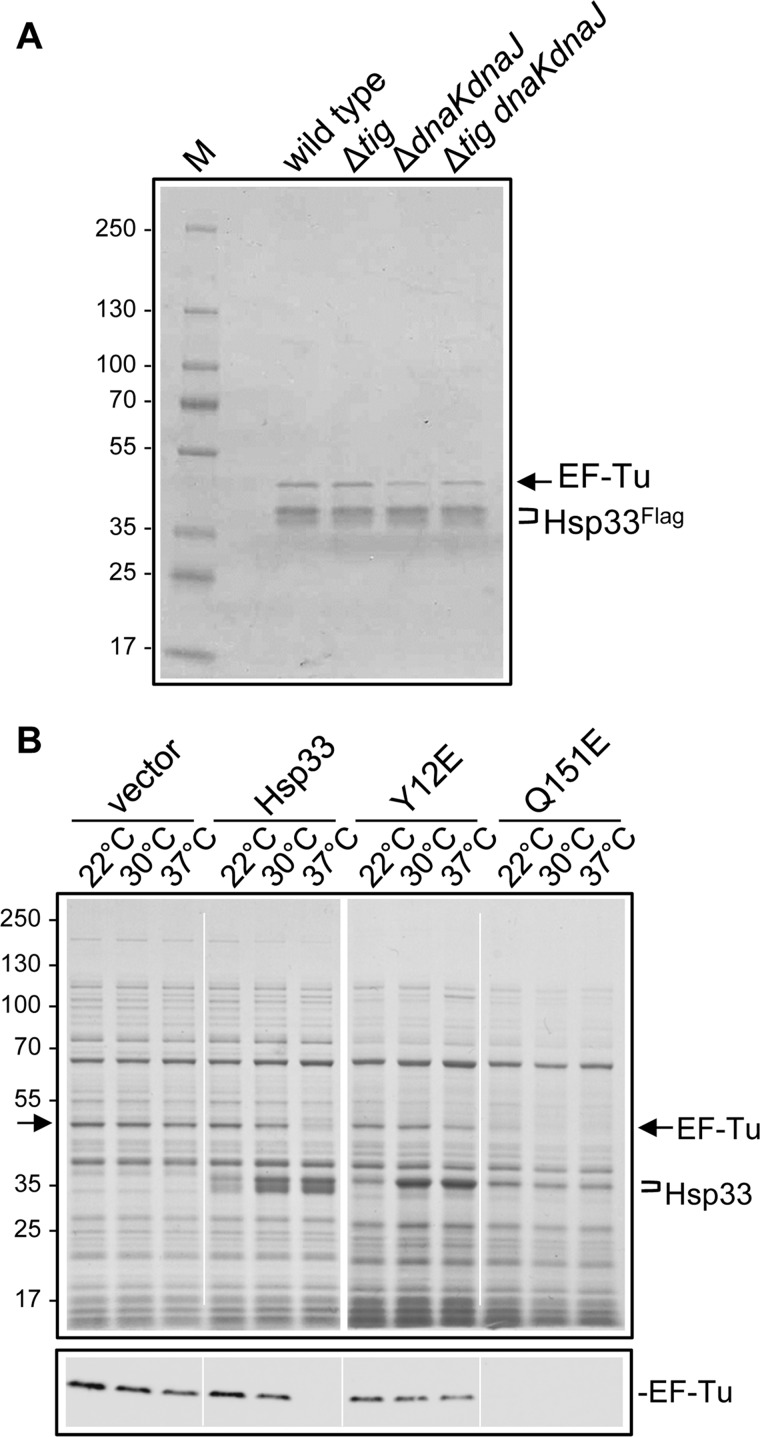
The genetic suppression results suggested that, as observed for SecB and GroEL (17, 19), Hsp33 could bind de novo substrates of TF and DnaK and efficiently prevent their aggregation. However, in contrast to SecB, in vitro cross-linking experiments did not reveal any co-translational interaction with our model substrate RpoB150, previously shown to interact with TF, DnaK, and SecB, and to aggregate in the absence of TF and DnaK (supplemental Fig. S4) (19). We next proceeded to search for Hsp33 interacting proteins in four isogenic strains, namely, MC4100 wild type and its Δtig, ΔdnaKdnaJ, and Δtig ΔdnaKdnaJ mutant derivatives. In these strains, in vivo pull-down experiments were carried out using overexpressed FLAG-tagged Hsp33 as bait (Fig. 3A). An analogous pull-down experiment with a lysate expressing untagged Hsp33 is shown as a control in supplemental Fig. S4. We found that one major Hsp33 interacting partner could be detected in all four genetic backgrounds tested, which was identified by mass spectrometry analysis to be the essential elongation factor EF-Tu, a key regulator of the polypeptide chain elongation cycle during the translation process (Fig. 3A) (43). This finding suggested that the specific interaction between EF-Tu and Hsp33 might be relevant for suppression of the Δtig ΔdnaKdnaJ mutant phenotypes by Hsp33.
FIGURE 3.
Hsp33 specifically interacts with EF-Tu. A, identification of the Hsp33 interacting substrates by pull-down analysis using an Hsp33-FLAG version expressed in MC4100, MC4100 Δtig::CmR, MC4100 ΔdnaKdnaJ::KanR, or MC4100 Δtig::CmR ΔdnaKdnaJ::KanR at 30 °C. The main co-precipitated protein obtained in all four strains was EF-Tu as identified by a mass spectrometric analysis. B, endogenous levels of EF-Tu in MC4100 Δtig::CmR ΔdnaKdnaJ::KanR following overexpression of Hsp33 wild type or its mutant derivatives Hsp33(Y12E) and Hsp33(Q151E). Cultures of MC4100 Δtig::CmR ΔdnaKdnaJ::KanR transformed with the various pSE380-based constructs were grown in LB ampicillin, supplemented with 0.4% glucose at 22 °C to an A600 of 0.3. IPTG was then added at 500 μm for 30 min at 22 °C for preinduction and cultures were transferred to 22, 30, or 37 °C for 1 h and 30 min. Whole cell extracts were prepared, separated by SDS-PAGE, and stained with Coomassie Blue. The characterization of the EF-Tu protein was confirmed by Western blot analysis shown at the bottom of the panel.
Next, we monitored the endogenous EF-Tu levels in response to Hsp33 overexpression in the Δtig ΔdnaKdnaJ mutant. In this case, the Hsp33(Y12E) and Hsp33(Q151E) were used as inactive and hyperactive controls, respectively, as deduced from the results shown in Fig. 1. We found that the endogenous EF-Tu levels were rapidly diminished following Hsp33 overexpression, especially at the nonpermissive temperature of 37 °C (Fig. 3B). A prolonged overexpression of Hsp33 at 22 °C also led to a decrease of EF-Tu (supplemental Fig. S5). In addition, the effect on EF-Tu was exacerbated in the presence of the constitutively monomeric and redox-regulated Hsp33(Q151E) mutant (Fig. 3B), which also efficiently binds EF-Tu as shown in supplemental Fig. S4. This last result suggests that distinct intermediates populating the multistep activation cycle of Hsp33 could possess unique functions in vivo (38, 41, 44). In contrast, and as expected, the Hsp33(Y12E) mutant was completely nonfunctional, behaving like the plasmid vector control (Fig. 3B).
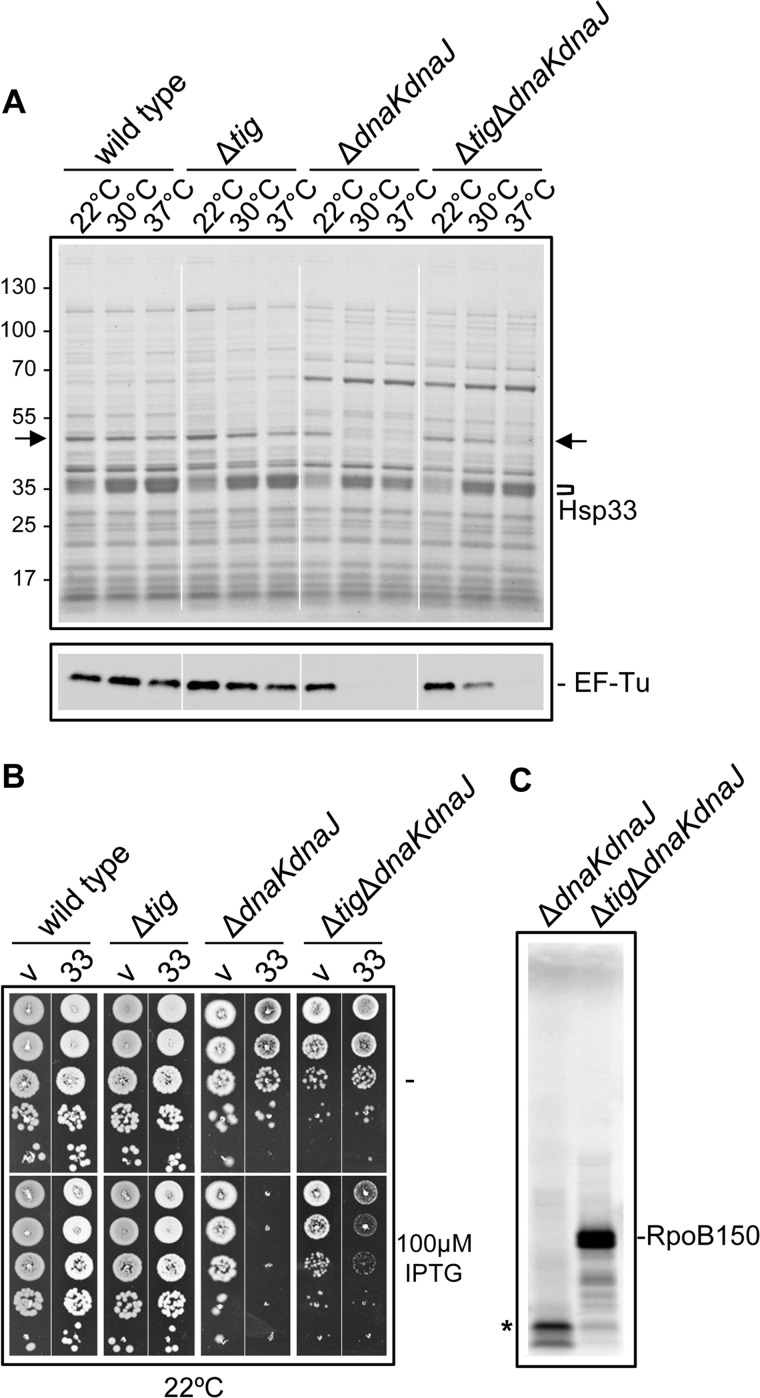
Although at this stage we cannot completely rule out the possibility that Hsp33, as a chaperone, directly prevents protein aggregation in the absence of TF and DnaK, our results strongly suggest that modulation of the rate of polypeptide chain elongation by Hsp33 might be responsible for bacterial survival. Our in vivo pull-down experiments described above never revealed a major difference in the qualitative nature of the Hsp33 interactor pattern. Nevertheless, we reproducibly noticed that the EF-Tu/Hsp33 ratio was higher in the wild type and the Δtig mutant than for the ΔdnaKdnaJ or Δtig ΔdnaKdnaJ isogenic mutant strains (Fig. 3A). Consequently, we tested whether the effect of Hsp33 on EF-Tu was different in the absence of TF, DnaK, or both. To do so, we overexpressed Hsp33 in the four isogenic strains and monitored the EF-Tu levels in whole cell extracts. The results presented in Fig. 4A show that upon Hsp33 overexpression, the EF-Tu levels are not significantly affected in the wild-type strain, besides the slight decrease in EF-Tu observed for the single Δtig mutant at 30 and 37 °C. In contrast, the EF-Tu intracellular levels dropped very rapidly when Hsp33 was overexpressed in the ΔdnaKdnaJ mutant, even at 30 °C. The expression of Hsp33 in the Δtig ΔdnaKdnaJ mutant resulted in an intermediate behavior. Note that the Hsp33 overexpression levels were comparable in all instances (Fig. 4A). The results presented in Fig. 4B suggest that the diminution of the endogenous EF-Tu directly correlates with bacterial growth inhibition at the otherwise permissive temperature of 22 °C, coupled with a marked toxicity of Hsp33 in the ΔdnaKdnaJ mutant.
FIGURE 4.
Hsp33-mediated control of EF-Tu in the absence of DnaK. A, endogenous EF-Tu levels in MC4100, MC4100 Δtig::CmR, MC4100 ΔdnaKdnaJ::KanR, or MC4100 Δtig::CmR ΔdnaKdnaJ::KanR following Hsp33 overexpression were analyzed as described in the legend to Fig. 3B. Arrows indicate the position of EF-Tu in whole cell extracts. B, overexpression of Hsp33 (pSE-Hsp33) in MC4100 and its mutant derivatives. Bacterial cultures grown at 22 °C were serially diluted 10-fold and spotted on LB ampicillin agar plates at 22 °C with or without 100 μm IPTG. The letter v stands for the vector and the number 33 represents for the Hsp33-carrying plasmid. C, in vitro translation of the first 150 amino acids of RpoB using cell- and membrane-free extracts of MC4100 ΔdnaKdnaJ::KanR and MC4100 Δtig::CmR ΔdnaKdnaJ::KanR mutants overexpressing Hsp33-FLAG. After in vitro translation, radiolabeled samples were resuspended in loading buffer and separated by a SDS-15% PAGE. The asterisk indicates the position of the truncated RpoB150 polypeptide (left lane). The position of the full-length RpoB150 is indicated as RpoB150 (right lane).
These findings suggest that in the absence of TF and DnaK, overexpressed Hsp33 may target EF-Tu for degradation by proteases. Moreover, the effect on EF-Tu was exacerbated in the ΔdnaKdnaJ mutant, thus suggesting that TF may also facilitate interaction of EF-Tu with Hsp33, leading to its subsequent degradation. We first examined whether Hsp33 overexpression differentially affects polypeptide chain elongation in vitro. To do so, we prepared translation lysates from both ΔdnaKdnaJ and Δtig ΔdnaKdnaJ mutant strains overexpressing Hsp33, and subsequently compared their ability to support translation of the RpoB150 mRNA in vitro (19). In agreement with our previous work, we found that cell lysates obtained from the Δtig ΔdnaKdnaJ mutant strain indeed efficiently translated RpoB150 mRNA, as judged by the presence of labeled, full-length RpoB150 nascent chains (Fig. 4C) (19). In sharp contrast, lysates prepared from the ΔdnaKdnaJ mutant strain expressing Hsp33 were not capable of translating the RpoB150 mRNA to completion, as judged by the absence of the full-length RpoB150 nascent chain, with concomitant appearance of a putative short truncated RpoB150 translation product (marked with an asterisk, left lane of Fig. 4C). These results are in agreement with the observed dramatic decrease in the levels of endogenous EF-Tu in the absence of DnaK with the simultaneous overproduction of Hsp33 and the resulting strong toxicity on bacterial growth (Fig. 4, A and B).
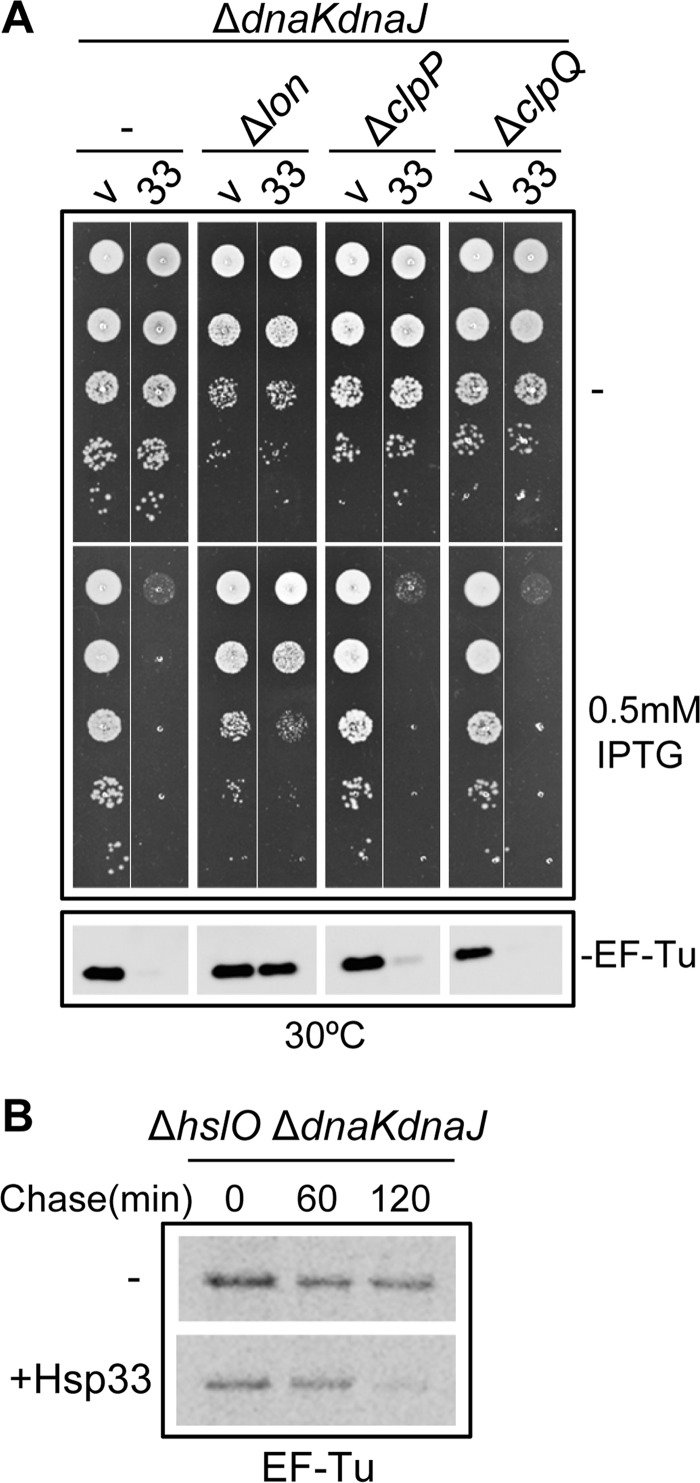
Hsp33 Triggers EF-Tu Degradation by the Stress Protease Lon
Our results suggest that in the absence of DnaK, and to a lesser extent in the absence of both DnaK and TF, the chaperone Hsp33 specifically binds and delivers EF-Tu to yet unknown protease(s). To identify such protease(s), we took advantage of the strong DnaK-dependent toxicity of Hsp33 observed in Fig. 4B. We reasoned that mutation in the specific protease might abolish the Hsp33-dependent decrease of endogenous EF-Tu and the resulting toxicity. Therefore, we combined single deletions of Δlon, ΔclpP, or ΔclpQ major cytoplasmic AAA+ proteases (45) with the ΔdnaKdnaJ mutation at 30 °C, overexpressed Hsp33, and monitored its effect on bacterial growth and EF-Tu. The results presented in Fig. 5A clearly show that in contrast to ΔclpP and ΔclpQ, the Δlon mutation efficiently suppresses Hsp33 toxicity and restores endogenous EF-Tu close to the wild type level. Yet, a low amount of EF-Tu was also recovered in the absence of clpP, suggesting some overlap between Lon and ClpP for this substrate (Fig. 5A, bottom).
FIGURE 5.
Hsp33 stimulates Lon protease-mediated degradation of EF-Tu. A, cultures of MC4100 ΔdnaKdnaJ::KanR, MC4100 Δlon ΔdnaKdnaJ::KanR, MC4100 ΔclpP::CmR ΔdnaKdnaJ::KanR, and MC4100 ΔclpQ::TetR ΔdnaKdnaJ::KanR transformed with pSE380 vector (indicated as v) or pSE-Hsp33 (indicated as 33), were serially diluted 10-fold and spotted on LB ampicillin agar plates with or without 500 μm IPTG inducer at 30 °C. Steady state levels of EF-Tu in the corresponding strains were analyzed by Western blot analysis using liquid cultures following a 2-h incubation in the presence of 500 μm IPTG inducer at 30 °C. B, the kinetics of EF-Tu degradation in vivo in MC4100 ΔhslO ΔdnaKdnaJ::KanR bacteria transformed with either the pSE380 vector or pSE-Hsp33 was analyzed by first labeling with [35S]methionine, followed by a 0, 60-, or 120-min chase with excess, unlabeled methionine and co-immunoprecipitation.
To ensure that the decreased EF-Tu levels were due to EF-Tu degradation and not to an effect on transcription, the stimulation of EF-Tu degradation by overexpressed Hsp33 was further demonstrated using pulse-chase experiments followed by immunoprecipitation of EF-Tu in the ΔdnaKdnaJ strain expressing Hsp33. The results presented in Fig. 5B show that indeed, Hsp33 significantly accelerates EF-Tu degradation in the absence of DnaK, suggesting that Hsp33 maintains EF-Tu in a conformation competent for degradation.
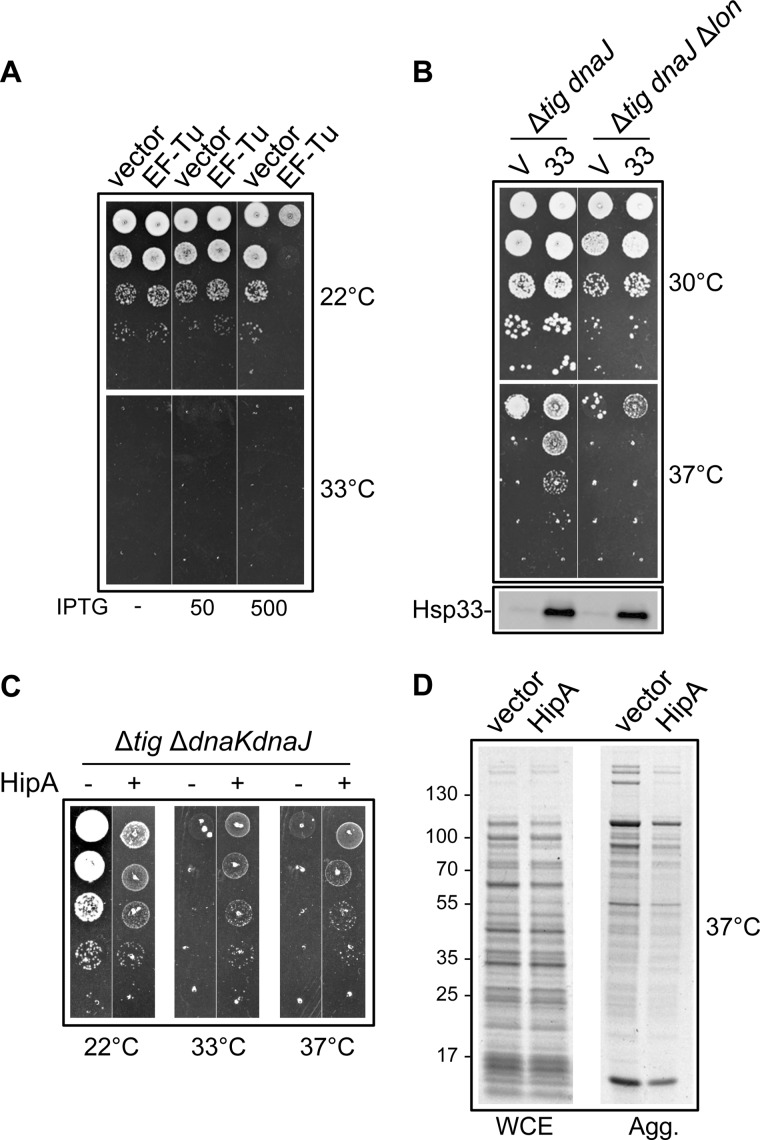
EF-Tu Inhibition Helps Bacterial Growth in the Absence of TF and DnaK
The proposed control of polypeptide chain elongation by a network of chaperones and proteases suggests that the suppression by Hsp33 in the absence of TF and DnaK is linked to EF-Tu degradation, and not to the previously observed in vitro chaperone activity of EF-Tu (46). In agreement with such a hypothesis, EF-Tu overexpression does not rescue the growth defect of the MC4100 Δtig ΔdnaKdnaJ mutant strain (Fig. 6A). As a control, the steady state level of plasmid-encoded EF-Tu is shown in supplemental Fig. S6. We next examined whether Hsp33-mediated degradation of EF-Tu by Lon was responsible for the suppression in the absence of TF and DnaK/DnaJ. We expressed Hsp33 both in the Δtig dnaJ and Δtig Δlon dnaJ mutants, and compared suppression of the temperature-sensitive phenotype by Hsp33. Note that as observed for MC4100 Δtig ΔdnaKdnaJ, overexpression of Hsp33 in the less sensitive MC4100 Δtig dnaJ mutant strain similarly stimulated EF-Tu degradation by Lon at a high growth temperature (supplemental Fig. S6). Remarkably, the results presented in Fig. 6B clearly show that in contrast with the expected suppression observed for the Δtig dnaJ (lon+) double mutant, Hsp33 was not capable of rescuing bacterial growth in the absence of Lon (Fig. 6B). These results strongly suggest that control of the endogenous EF-Tu levels is critical when the TF/DnaK pathway is compromised.
FIGURE 6.
EF-Tu inactivation facilitates growth and protein folding in the absence of TF and DnaK chaperones. A, EF-Tu overexpression in MC4100 Δtig::CmR ΔdnaKdnaJ::KanR. Cultures of MC4100 Δtig::CmR ΔdnaKdnaJ::KanR transformed with p29SEN or p29-EF-Tu were grown at 22 °C, serially diluted 10-fold, and spotted on LB ampicillin agar plates with or without IPTG (μm) and incubated at 22 or 33 °C. B, cultures of MC4100 Δtig::CmR dnaJ::Tn10-42 or MC4100 Δtig::CmR dnaJ::Tn10-42 Δlon::KanR transformed with either p29SEN (indicated as v) or p29-Hsp33 (indicated as 33) were grown in LB ampicillin, supplemented with glucose 0.4% at 30 °C, serially diluted 10-fold, and spotted on LB ampicillin agar plates with or without 50 μm IPTG at 30 or 37 °C. Protein levels were analyzed by Western blot analysis of the extracts following a 2-h induction with 50 μm IPTG at 30 °C. The genetic background of each strain used is shown on top of each panel. C, the HipA toxin efficiently rescues bacterial growth in the absence of TF and DnaK. Strain W3110 Δtig::CmR ΔdnaKdnaJ::KanR pBAD-HipA was grown without (−) or with (+) 0.5% l-arabinose inducer at the indicated temperatures. D, prevention of protein aggregation by HipA toxin expressed from the pBAD-HipA plasmid in strain W3110 Δtig::CmR ΔdnaKdnaJ::KanR. HipA expression was induced with 0.5% l-arabinose for 6 h at 22 °C followed by 1 h and 30 min at 33 °C. Whole cell extracts (WCE) and aggregates were separated by SDS-PAGE and stained with Coomassie Blue.
Next, we took advantage of the HipA toxin of the HipAB toxin-antitoxin module from E. coli, known to bind to and inhibit EF-Tu by phosphorylation at residue Thr-382, thus compromising polypeptide chain elongation (47, 48). We first asked whether HipA overexpression could mimic Hsp33-mediated control of EF-Tu in the absence of both TF and DnaK. To do so, the gene encoding HipA was cloned in a plasmid under control of the tightly regulated araBADp promoter (to avoid growth inhibition by the toxin) and expressed in the sensitive Δtig ΔdnaKdnaJ mutant. As expected for a toxin affecting translation, HipA expression significantly slowed down bacterial growth at the permissive temperature of 22 °C (Fig. 6C). In contrast, at the nonpermissive temperature of 37 °C, a mild overexpression of HipA efficiently rescued bacterial growth and prevented protein aggregation in the absence of both TF and DnaK (Fig. 6, C and D). These results indicate that a partial inactivation of EF-Tu function may be beneficial to bacterial survival in the absence of the major TF and DnaK chaperones and highlights a possible role for the toxin-antitoxin system in controlling the rate of protein synthesis in response to protein aggregation (see “Discussion”).
DISCUSSION
The heat shock protein Hsp33 is a highly specialized redox-regulated molecular chaperone, which is activated by a combination of both oxidative stress and protein unfolding or aggregation (49). In this work, we propose that Hsp33 can also perform key, housekeeping cellular functions in the absence of exogenous stressors as part of the network of stress chaperones and proteases in E. coli. This conclusion was reached by demonstrating that overexpression of Hsp33 can efficiently support bacterial growth and prevent accumulation of protein aggregates in the absence of TF and DnaK. However, in contrast with other chaperones, such as SecB and GroEL (17–19), suppression by Hsp33 does not seem to rely on its general chaperone function. Instead, it appears that Hsp33 efficiently binds to the essential elongation factor EF-Tu and specifically targets it for degradation by the Lon protease. The subsequent decrease in the level of endogenous EF-Tu may result in a decrease of the intracellular rate of nascent polypeptide synthesis, thus minimizing the need for action by downstream chaperones TF and DnaK.
Although we do not know yet about the relevance of such a pathway under physiological conditions where all the chaperones are present, our results point toward the existence of an intricate network of stress chaperones and proteases that result in an inhibition of the rate of translation in response to severe protein aggregation. It is known that under stress conditions that affect protein folding, several major, stress-induced chaperones, including DnaK, are recruited to the accumulating protein aggregates (50). Importantly, such a recruitment of DnaK to protein aggregates results in the rapid stabilization of the heat shock sigma factor σ32, thus leading to a prolonged induction of heat shock proteins, including Hsp33 and Lon. Note that in our case, both the induction of heat shock proteins and the aggregation-prone conditions are, at least in part, artificially recreated by the introduction of the tig dnaKdnaJ mutations (17). Yet, in the absence of these dominant chaperones, we cannot exclude that induction of the stress response or accumulation of extragenic suppressors that help bacterial growth without TF and DnaK could force the bacteria to use pathways that are not utilized under normal conditions (51, 52). In our working model, the increased recruitment of the major molecular chaperones to pre-existing aggregates and newly synthesized proteins rapidly overpowers the cellular chaperone capacity available for de novo protein folding. This, in turn, results in a signal that triggers a transient inhibition of the rate of translation and a subsequent reduction in the intracellular levels of newly synthesized polypeptides that normally necessitate molecular chaperones for their proper folding (53). Such a possible mechanism is supported by previous studies showing that slowing down translation facilitates de novo protein folding in E. coli (54, 55) and in mammalian cells, as shown recently (56).
If such an intricate control of translation rate by the availability of stress chaperones and proteases indeed exists, then the results presented in this work clearly point to elongation factor EF-Tu as one of the major targets of such an inhibition. Interestingly, in addition to the Hsp33·EF-Tu complex demonstrated in this study, an interaction between EF-Tu and Lon has also been found in vivo (57), further supporting the proposed interplay among chaperones and proteases. However, it remains to be determined whether EF-Tu is the sole protein directly targeted for degradation by the Hsp33/Lon pathway and whether other minor interactions that our experimental procedures are missing could also participate.
It is known that EF-Tu adopts several conformations and populates various intermediate stages during its functional cycle in polypeptide chain elongation (38, 41, 44). At this time, the actual conformational state(s) of EF-Tu that is recognized by Hsp33 and Lon remains unknown. Intriguingly, in contrast to our finding that in E. coli Hsp33 stimulates EF-Tu degradation by Lon, it was recently shown in Vibrio cholerae that Hsp33 was instead required to maintain EF-Tu at cellular levels sufficient to allow bacterial growth at high temperature under aerobic conditions (58). In this case, Hsp33 stabilized the highly sensitive V. cholerae EF-Tu protein from oxidative protein degradation in vivo. Interestingly, the E. coli EF-Tu protein was not affected under the same conditions (58). The differential results seen with different bacteria strongly suggest that although the proposed network controlling EF-Tu intracellular levels is conserved, the overall outcome may differ depending on the particular organism and/or stress conditions employed.
The fact that degradation of EF-Tu by Hsp33 and Lon predominantly occurs when DnaK is absent suggests that under normal growth conditions DnaK prevents EF-Tu degradation, either by directly interacting with EF-Tu or indirectly by inhibiting the σ32-dependent synthesis of Lon. In contrast with such a protective effect of DnaK, we found that the presence of TF significantly stimulates EF-Tu degradation, indicating that TF directly or indirectly facilitates the interaction of EF-Tu with Hsp33 and/or its subsequent recognition by Lon. Such a hypothesis is strongly supported (i) by a recent study demonstrating that in contrast with DnaK, the ribosome-bound TF chaperone can unfold substrates and facilitate their degradation in vitro (59), and (ii) by the synthetic lethality observed between the tig, dnaKdnaJ, and hslO mutations (Fig. 2A).
How does Hsp33 contribute to such a network in the absence of oxidative stressors? One possibility is that Hsp33 is only activated by increasing levels of reactive oxygen species stimulated by the severe protein homeostasis collapse induced by the lack of both TF and DnaK major chaperones (11). However, this seems unlikely because (i) Hsp33 efficiently binds EF-Tu in the wild type E. coli strain as well, and (ii) EF-Tu is much more efficiently degraded in the dnaKdnaJ mutant than in the triple tig dnaKdnaJ mutant where proteostasis breakdown is the most severe (9, 10, 17). Therefore, we propose that a significant fraction of endogenous Hsp33 may be active and physiologically relevant even in the absence of oxidative stress. This is supported by the synergistic effects observed among the tig, dnaK (or dnaJ), and hslO mutations. However, the redox and the oligomeric status (monomeric, dimeric, or oligomeric) of cellular Hsp33 that is capable of performing such a function in vivo remains to be determined (38, 41, 44). In addition, it is not yet established whether Hsp33 induces EF-Tu degradation under physiological conditions where all the chaperones are present. As stated above, Hsp33 is activated by oxidative protein unfolding (34, 35), a condition that is known to impede DnaK functions (36). Under such extreme stress conditions, quality control of EF-Tu by activated Hsp33 and Lon could thus become a key to bacterial survival. Further studies are needed to elucidate such possible interplay under severe oxidative conditions.
Analogous to the newly identified cooperation between Hsp33 and Lon in controlling EF-Tu, overexpression of the HipA toxin of the chromosomally encoded type-II HipAB toxin-antitoxin system can also suppress both the growth defect and the accumulation of protein aggregates in the absence of TF and DnaK. Toxin-antitoxin modules are two-component systems composed of a stable toxin that forms an inactive complex with its less stable cognate antitoxin. In response to specific stress conditions the unstable antitoxin is generally degraded by stress proteases and the thus free active toxin subsequently targets important cellular processes such as DNA replication or protein synthesis, resulting in growth inhibition and eventual cell death (60–63). Recent work has shown that E. coli toxin HipA is a protein kinase belonging to the Tor family, which specifically inhibits EF-Tu and blocks translation (47, 64). In agreement with such an activity, overexpression of HipA slows down E. coli growth and increases antibiotic tolerance and persistence (reviewed in Ref. 65). Although it is well established that many stress-responsive toxin-antitoxin systems affect translation in bacteria, the finding that an active toxin can partially bypass major molecular chaperone requirements in vivo is novel. An attractive hypothesis is that severe protein aggregation may trigger the activation of stress-responsive toxin-antitoxin systems to control the level of protein synthesis, thus relieving the overwhelmed downstream network of molecular chaperones until normal conditions resume. In agreement with this hypothesis, four other well characterized antitoxins present in E. coli, namely MazE, RelB, MqsA, and DinJ, whose respective cognate toxins MazF, RelE MqsR, and YafQ are known to affect protein synthesis, have been recently shown to be bona fide DnaK substrates in vivo (11). Clearly, further work is needed to elucidate such a possible role for chromosomally encoded toxin-antitoxin systems in bacteria.
Acknowledgments
We thank Françoise Schwager and Ronald Ullers for their contribution at the early stages of this project, Elsa Perrody for assistance with the pull-down experiments, Ursula Jakob and Claudia Cremers for insightful discussions and for the kind gift of plasmids and antibody, and all members of the Genevaux laboratory for helpful advice and discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R01GM088207 (to C. G.), a grant from the CNRS-ATIP Microbiology program (to P. G), and a joint Region Midi-Pyrénées/CNRS grant (to N. B.).

This article contains supplemental Figs. S1–S6.
- TF
- trigger factor
- EF
- elongation factor
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside.
REFERENCES
- 1. Valent Q. A., Kendall D. A., High S., Kusters R., Oudega B., Luirink J. (1995) Early events in preprotein recognition in Escherichia coli. Interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 14, 5494–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kramer G., Rauch T., Rist W., Vorderwülbecke S., Patzelt H., Schulze-Specking A., Ban N., Deuerling E., Bukau B. (2002) L23 protein functions as a chaperone docking site on the ribosome. Nature 419, 171–174 [DOI] [PubMed] [Google Scholar]
- 3. Lakshmipathy S. K., Tomic S., Kaiser C. M., Chang H. C., Genevaux P., Georgopoulos C., Barral J. M., Johnson A. E., Hartl F. U., Etchells S. A. (2007) Identification of nascent chain interaction sites on trigger factor. J. Biol. Chem. 282, 12186–12193 [DOI] [PubMed] [Google Scholar]
- 4. Merz F., Boehringer D., Schaffitzel C., Preissler S., Hoffmann A., Maier T., Rutkowska A., Lozza J., Ban N., Bukau B., Deuerling E. (2008) Molecular mechanism and structure of trigger factor bound to the translating ribosome. EMBO J. 27, 1622–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaiser C. M., Chang H. C., Agashe V. R., Lakshmipathy S. K., Etchells S. A., Hayer-Hartl M., Hartl F. U., Barral J. M. (2006) Real-time observation of trigger factor function on translating ribosomes. Nature 444, 455–460 [DOI] [PubMed] [Google Scholar]
- 6. Crooke E., Wickner W. (1987) Trigger factor. A soluble protein that folds pro-OmpA into a membrane-assembly-competent form. Proc. Natl. Acad. Sci. U.S.A. 84, 5216–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ullers R. S., Ang D., Schwager F., Georgopoulos C., Genevaux P. (2007) Trigger factor can antagonize both SecB and DnaK/DnaJ chaperone functions in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104, 3101–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oh E., Becker A. H., Sandikci A., Huber D., Chaba R., Gloge F., Nichols R. J., Typas A., Gross C. A., Kramer G., Weissman J. S., Bukau B. (2011) Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teter S. A., Houry W. A., Ang D., Tradler T., Rockabrand D., Fischer G., Blum P., Georgopoulos C., Hartl F. U. (1999) Polypeptide flux through bacterial Hsp70. DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97, 755–765 [DOI] [PubMed] [Google Scholar]
- 10. Deuerling E., Schulze-Specking A., Tomoyasu T., Mogk A., Bukau B. (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400, 693–696 [DOI] [PubMed] [Google Scholar]
- 11. Calloni G., Chen T., Schermann S. M., Chang H. C., Genevaux P., Agostini F., Tartaglia G. G., Hayer-Hartl M., Hartl F. U. (2012) DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 1, 251–264 [DOI] [PubMed] [Google Scholar]
- 12. Bukau B., Walker G. C. (1989) Delta dnaK52 mutants of Escherichia coli have defects in chromosome segregation and plasmid maintenance at normal growth temperatures. J. Bacteriol. 171, 6030–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horwich A. L., Farr G. W., Fenton W. A. (2006) GroEL-GroES-mediated protein folding. Chem. Rev. 106, 1917–1930 [DOI] [PubMed] [Google Scholar]
- 14. Houry W. A., Frishman D., Eckerskorn C., Lottspeich F., Hartl F. U. (1999) Identification of in vivo substrates of the chaperonin GroEL. Nature 402, 147–154 [DOI] [PubMed] [Google Scholar]
- 15. Kerner M. J., Naylor D. J., Ishihama Y., Maier T., Chang H. C., Stines A. P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Hartl F. U. (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122, 209–220 [DOI] [PubMed] [Google Scholar]
- 16. Fayet O., Ziegelhoffer T., Georgopoulos C. (1989) The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J. Bacteriol. 171, 1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Genevaux P., Keppel F., Schwager F., Langendijk-Genevaux P. S., Hartl F. U., Georgopoulos C. (2004) In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep. 5, 195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vorderwülbecke S., Kramer G., Merz F., Kurz T. A., Rauch T., Zachmann-Brand B., Bukau B., Deuerling E. (2004) Low temperature or GroEL/ES overproduction permits growth of Escherichia coli cells lacking trigger factor and DnaK. FEBS Lett. 559, 181–187 [DOI] [PubMed] [Google Scholar]
- 19. Ullers R. S., Luirink J., Harms N., Schwager F., Georgopoulos C., Genevaux P. (2004) SecB is a bona fide generalized chaperone in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101, 7583–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Casadaban M. J. (1976) Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and μ. J. Mol. Biol. 104, 541–555 [DOI] [PubMed] [Google Scholar]
- 21. Bachmann B. J. (1972) Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36, 525–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakr S., Cirinesi A. M., Ullers R. S., Schwager F., Georgopoulos C., Genevaux P. (2010) Lon protease quality control of presecretory proteins in Escherichia coli and its dependence on the SecB and DnaJ (Hsp40) chaperones. J. Biol. Chem. 285, 23506–23514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maurizi M. R., Clark W. P., Katayama Y., Rudikoff S., Pumphrey J., Bowers B., Gottesman S. (1990) Sequence and structure of ClpP, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 265, 12536–12545 [PubMed] [Google Scholar]
- 24. Yamaguchi Y., Tomoyasu T., Takaya A., Morioka M., Yamamoto T. (2003) Effects of disruption of heat shock genes on susceptibility of Escherichia coli to fluoroquinolones. BMC Microbiol. 3, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Datsenko K. A., Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cremers C. M., Reichmann D., Hausmann J., Ilbert M., Jakob U. (2010) Unfolding of metastable linker region is at the core of Hsp33 activation as a redox-regulated chaperone. J. Biol. Chem. 285, 11243–11251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tomoyasu T., Mogk A., Langen H., Goloubinoff P., Bukau B. (2001) Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40, 397–413 [DOI] [PubMed] [Google Scholar]
- 29. Genevaux P., Schwager F., Georgopoulos C., Kelley W. L. (2002) Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ (Hsp40) J-domain. Genetics 162, 1045–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Urbanus M. L., Scotti P. A., Froderberg L., Saaf A., de Gier J. W., Brunner J., Samuelson J. C., Dalbey R. E., Oudega B., Luirink J. (2001) Sec-dependent membrane protein insertion. Sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ullers R. S., Houben E. N., Raine A., ten Hagen-Jongman C. M., Ehrenberg M., Brunner J., Oudega B., Harms N., Luirink J. (2003) Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J. Cell Biol. 161, 679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luirink J., High S., Wood H., Giner A., Tollervey D., Dobberstein B. (1992) Signal-sequence recognition by an Escherichia coli ribonucleoprotein complex. Nature 359, 741–743 [DOI] [PubMed] [Google Scholar]
- 33. Jong W. S., ten Hagen-Jongman C. M., Genevaux P., Brunner J., Oudega B., Luirink J. (2004) Trigger factor interacts with the signal peptide of nascent Tat substrates but does not play a critical role in Tat-mediated export. Eur. J. Biochem. 271, 4779–4787 [DOI] [PubMed] [Google Scholar]
- 34. Jakob U., Muse W., Eser M., Bardwell J. C. (1999) Chaperone activity with a redox switch. Cell 96, 341–352 [DOI] [PubMed] [Google Scholar]
- 35. Chuang S. E., Blattner F. R. (1993) Characterization of twenty-six new heat shock genes of Escherichia coli. J. Bacteriol. 175, 5242–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoffmann J. H., Linke K., Graf P. C., Lilie H., Jakob U. (2004) Identification of a redox-regulated chaperone network. EMBO J. 23, 160–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tomoyasu T., Arsène F., Ogura T., Bukau B. (2001) The C terminus of σ32 is not essential for degradation by FtsH. J. Bacteriol. 183, 5911–5917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chi S. W., Jeong D. G., Woo J. R., Lee H. S., Park B. C., Kim B. Y., Erikson R. L., Ryu S. E., Kim S. J. (2011) Crystal structure of constitutively monomeric E. coli Hsp33 mutant with chaperone activity. FEBS Lett. 585, 664–670 [DOI] [PubMed] [Google Scholar]
- 39. Kim S. J., Jeong D. G., Chi S. W., Lee J. S., Ryu S. E. (2001) Crystal structure of proteolytic fragments of the redox-sensitive Hsp33 with constitutive chaperone activity. Nat. Struct. Biol. 8, 459–466 [DOI] [PubMed] [Google Scholar]
- 40. Jakob U., Eser M., Bardwell J. C. (2000) Redox switch of hsp33 has a novel zinc-binding motif. J. Biol. Chem. 275, 38302–38310 [DOI] [PubMed] [Google Scholar]
- 41. Graf P. C., Martinez-Yamout M., VanHaerents S., Lilie H., Dyson H. J., Jakob U. (2004) Activation of the redox-regulated chaperone Hsp33 by domain unfolding. J. Biol. Chem. 279, 20529–20538 [DOI] [PubMed] [Google Scholar]
- 42. Genevaux P., Georgopoulos C., Kelley W. L. (2007) The Hsp70 chaperone machines of Escherichia coli. A paradigm for the repartition of chaperone functions. Mol. Microbiol. 66, 840–857 [DOI] [PubMed] [Google Scholar]
- 43. Thompson R. C., Dix D. B., Karim A. M. (1986) The reaction of ribosomes with elongation factor Tu·GTP complexes. Aminoacyl-tRNA-independent reactions in the elongation cycle determine the accuracy of protein synthesis. J. Biol. Chem. 261, 4868–4874 [PubMed] [Google Scholar]
- 44. Akhtar M. W., Srinivas V., Raman B., Ramakrishna T., Inobe T., Maki K., Arai M., Kuwajima K., Rao ChM. (2004) Oligomeric Hsp33 with enhanced chaperone activity: gel filtration, cross-linking, and small angle x-ray scattering (SAXS) analysis. J. Biol. Chem. 279, 55760–55769 [DOI] [PubMed] [Google Scholar]
- 45. Sauer R. T., Baker T. A. (2011) AAA+ proteases. ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612 [DOI] [PubMed] [Google Scholar]
- 46. Caldas T. D., El Yaagoubi A., Richarme G. (1998) Chaperone properties of bacterial elongation factor EF-Tu. J. Biol. Chem. 273, 11478–11482 [DOI] [PubMed] [Google Scholar]
- 47. Schumacher M. A., Piro K. M., Xu W., Hansen S., Lewis K., Brennan R. G. (2009) Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323, 396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hansen S., Vulić M., Min J., Yen T. J., Schumacher M. A., Brennan R. G., Lewis K. (2012) Regulation of the Escherichia coli HipBA toxin-antitoxin system by proteolysis. PLoS One 7, e39185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kumsta C., Jakob U. (2009) Redox-regulated chaperones. Biochemistry 48, 4666–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Winkler J., Seybert A., König L., Pruggnaller S., Haselmann U., Sourjik V., Weiss M., Frangakis A. S., Mogk A., Bukau B. (2010) Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 29, 910–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bukau B., Walker G. C. (1990) Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 9, 4027–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kramer G., Rutkowska A., Wegrzyn R. D., Patzelt H., Kurz T. A., Merz F., Rauch T., Vorderwülbecke S., Deuerling E., Bukau B. (2004) Functional dissection of Escherichia coli trigger factor. Unraveling the function of individual domains. J. Bacteriol. 186, 3777–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 54. Agashe V. R., Guha S., Chang H. C., Genevaux P., Hayer-Hartl M., Stemp M., Georgopoulos C., Hartl F. U., Barral J. M. (2004) Function of trigger factor and DnaK in multidomain protein folding. Increase in yield at the expense of folding speed. Cell 117, 199–209 [DOI] [PubMed] [Google Scholar]
- 55. Siller E., DeZwaan D. C., Anderson J. F., Freeman B. C., Barral J. M. (2010) Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J. Mol. Biol. 396, 1310–1318 [DOI] [PubMed] [Google Scholar]
- 56. Meriin A. B., Mense M., Colbert J. D., Liang F., Bihler H., Zaarur N., Rock K. L., Sherman M. Y. (2012) A novel approach to recovery of function of mutant proteins by slowing down translation. J. Biol. Chem. 287, 34264–34272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. (2005) Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 58. Wholey W. Y., Jakob U. (2012) Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibrio cholerae. Mol. Microbiol. 83, 981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hoffmann A., Becker A. H., Zachmann-Brand B., Deuerling E., Bukau B., Kramer G. (2012) Concerted action of the ribosome and the associated chaperone trigger factor confines nascent polypeptide folding. Mol. Cell 48, 63–74 [DOI] [PubMed] [Google Scholar]
- 60. Yamaguchi Y., Park J. H., Inouye M. (2011) Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45, 61–79 [DOI] [PubMed] [Google Scholar]
- 61. Gerdes K., Christensen S. K., Løbner-Olesen A. (2005) Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 62. Van Melderen L., De Bast M. S. (2009) Bacterial toxin-antitoxin systems. More than selfish entities? Plos Genet. 5, e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Moll I., Engelberg-Kulka H. (2012) Selective translation during stress in Escherichia coli. Trends Biochem. Sci. 37, 493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Correia F. F., D'Onofrio A., Rejtar T., Li L., Karger B. L., Makarova K., Koonin E. V., Lewis K. (2006) Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J. Bacteriol. 188, 8360–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lewis K. (2010) Persister cells. Annu. Rev. Microbiol. 64, 357–372 [DOI] [PubMed] [Google Scholar]