Background: TGF-β induces EMT to regulate tumor invasion and metastasis.
Results: TGF-β induces KDM6B and promotes EMT by activating SNAI1 via removing H3K27me3 marks.
Conclusion: KDM6B is required for TGF-β-induced EMT and breast cancer cell invasion.
Significance: Our findings highlight a novel epigenetic mechanism regulating EMT and cancer cell invasion and have potential implications in targeting metastatic breast cancer.
Keywords: Breast Cancer, E-cadherin, EMT, Epithelial Mesenchymal Transition, Metastasis
Abstract
Epithelial-mesenchymal transition (EMT) is a critical event that occurs in embryonic development, tissue repair control, organ fibrosis, and carcinoma invasion and metastasis. Although significant progress has been made in understanding the molecular regulation of EMT, little is known about how chromatin is modified in EMT. Chromatin modifications through histone acetylation and methylation determine the precise control of gene expression. Recently, histone demethylases were found to play important roles in gene expression through demethylating mono-, di-, or trimethylated lysines. KDM6B (also known as JMJD3) is a histone demethylase that might activate gene expression by removing repressive histone H3 lysine 27 trimethylation marks from chromatin. Here we report that KDM6B played a permissive role in TGF-β-induced EMT in mammary epithelial cells by stimulating SNAI1 expression. KDM6B was induced by TGF-β, and the knockdown of KDM6B inhibited EMT induced by TGF-β. Conversely, overexpression of KDM6B induced the expression of mesenchymal genes and promoted EMT. Chromatin immunoprecipitation (ChIP) assays revealed that KDM6B promoted SNAI1 expression by removing histone H3 lysine trimethylation marks. Consistently, our analysis of the Oncomine database found that KDM6B expression was significantly increased in invasive breast carcinoma compared with normal breast tissues. The knockdown of KDM6B significantly inhibited breast cancer cell invasion. Collectively, our study uncovers a novel epigenetic mechanism regulating EMT and tumor cell invasion, and has important implication in targeting cancer metastasis.
Introduction
Epithelial-mesenchymal transition (EMT)2 is a process by which polarized, immotile epithelial cells acquire the motile mesenchymal phenotype. This important process determines several critical stages involved in embryonic development and also governs tissue repair control mechanisms, organ fibrosis, and cancer progression and invasion through multiple mechanisms (1). In addition to rendering the cells with migratory and invasive properties, EMT also contributes to cancer cell stemness and inhibits cellular apoptosis and senescence (2). Various signaling pathways and molecules have been found to control EMT (3). Among the potential inducers of EMT, TGF-β is a unique pleiotropic cytokine that plays a dual role by acting as a tumor suppressor in the initial stages of tumor growth (inhibiting cell proliferation and inducing apoptosis) and a tumor promoter (by inducing EMT) in advanced stages of tumor growth (4).
In canonical TGF-β signaling, binding of TGF-β ligands to type II TGF-β receptor enables this polypeptide to transphosphorylate and activate type I TGF-β receptor, which in turn binds, phosphorylates, and activates the receptor-associated Smad family transcription factors, Smad2 and Smad3 (5–7). Activated Smad2/3 rapidly form high order complexes with the common Smad, Smad4, which enables the resulting heterotrimeric complexes to translocate to the nucleus to regulate transcription in a gene- and cell-specific manner (5–7). Although it is known that activation of intercellular signaling by different stimuli employs distinct downstream pathways to regulate gene expression, recent breakthroughs highlighting the integration of intercellular signaling with epigenetic mechanisms to regulate gene expression have received greater attention (8, 9). Apart from DNA methylation, epigenetic mechanisms are largely operated by histone modifications, which enable the gene promoter to be accessible or inaccessible to transcription factors, depending on the nature and site of modifications (10, 11). One such important histone modification is histone H3 lysine trimethylation (H3K27me3) on gene promoters mediated by polycomb proteins, which silences gene transcription (12–15). KDM6B is a member of the Fe(II)- and α-ketoglutarate-dependent demethylases reported to activate gene expression by removing H3K27me3 marks on gene promoters (16–22). It regulates the expression of key regulators in development, differentiation, and oncogenic stress-induced cell senescence in cancer (22–24). Despite the fact that signaling pathways driving EMT have been identified, how these signaling networks cooperate with epigenetic mechanisms to regulate the expression of EMT-related transcription factors has not been well investigated. Recently, two different studies highlighted the requirement of KDM6B protein for nodal (TGF-β-related extracellular ligand) Smad2/3 signaling-mediated counteraction of polycomb-associated gene repression in embryonic stem cells, indicating that Smad proteins coordinate with chromatin dynamics at gene promoters to release gene repression (9, 25).
Because TGF-β-Smad is a critical inducer of the developmental and pathological EMT program, we hypothesized that KDM6B might play a role in TGF-β-mediated EMT. We found that KDM6B expression was induced by TGF-β in mammary epithelial cells and was required for the induction of EMT. Mechanistically, we identified that KDM6B controlled EMT by regulating SNAI1, a master transcription factor in EMT. In support of our findings, Oncomine data analysis revealed that KDM6B was highly expressed in invasive breast cancer compared with normal breast tissue. Consistently, the knockdown of KDM6B drastically reduced breast cancer cell invasion by inhibiting SNAI1 expression. Our results unravel a new epigenetic mechanism controlling EMT and cancer cell invasion.
EXPERIMENTAL PROCEDURES
Cell Lines, Antibodies, and Reagents
Normal mouse mammary epithelial cells NMuMG, Madin-Darby canine kidney (MDCK) epithelial cells, and human mammary epithelial cells (MCF-10A) were obtained from ATCC. Immortalized human mammary epithelial (hMLE) cells were provided as a generous gift by Dr. Robert Weinberg at the Whitehead Institute for Biomedical Research (Cambridge, MA). NMuMG, MDCK, and MDA-MB-231 cells were cultured in DMEM containing 10% heat-inactivated fetal bovine serum (FBS) and antibiotics (streptomycin and penicillin) at 37 °C in a 5% CO2, 95% air atmosphere. MCF-10A and hMLE cells were maintained in mammary epithelial growth medium supplemented with essential growth factors (Lonza). For siRNA transfections, 1 × 106 MDA-MB-231 or NMuMG cells were seeded into 6-well plates for 12 h and then transfected using Lipofectamine RNAiMAX reagents according to the manufacturer's protocol (Invitrogen). Two days (48 h) after the transfection, the cells were harvested for various assays. To stably knock down KDM6B, lentiviruses expressing mouse and human KDM6B shRNA were packaged and generated in 293T cells as described previously (26). The cells were seeded in the 6-well plates overnight and then infected with lentiviral particles. 24 h after infection, the cells were selected with puromycin (1 μg/ml) for at least 1 week. Knockdown efficiency was determined by real-time reverse transcription polymerase chain reaction (RT-PCR). Retroviral vector expressing KDM6B was kindly provided by Dr. Paul Khavari (27). KDM6B and Snai1 siRNAs were procured from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). shRNAs targeting KDM6B were prepared using the lentiviral expression vector pLKO.1. Targeting sequences used for mouse Kdm6b shRNA was 5′-CCTGTATATGTCTCTTGTTTA-3′, and human KDM6B shRNAs were 5′-GCGGCTCGTGTATGTACAT-3′ (KDM6Bsh1) and 5′-CTGTTCGTGACAAGTGAGA-3′ (KDM6Bsh2). Anti-E-cadherin, anti-N-cadherin, and anti-fibronectin monoclonal antibodies were purchased from Transduction Laboratories; chromatin immunoprecipitation (ChIP) grade anti-trimethyl-histone H3 (Lys-27) polyclonal antibodies were purchased from Millipore; anti-KDM6B/JMJD3 antibody was from Abgent Inc.; anti-Smad3 antibody was from Abcam; anti-FLAG and anti-tubulin monoclonal antibodies were from Sigma; and anti-vimentin (VIM) monoclonal antibodies were from Cell Signaling Technologies.
Total RNA Extraction and Real-time RT-PCR
Total RNA from cells was purified using TRIzol reagent (Invitrogen) according to the manufacturer's protocol, and cDNA was synthesized with oligo(dT) primers using SuperScript III (Invitrogen). Real-time RT-PCR analysis was carried out with iQ SYBR Green supermix (Bio-Rad) on an iCycler iQ real-time PCR detection system (Bio-Rad). β-actin (mouse) or GAPDH (human and canine) were used as an internal control to calculate the relative expression. Sequences of the primer pairs used were as follows: mouse Kdm6b (5′-CCCCCATTTCAGCTGACTAA-3′, 5′-CTGGACCAAGGGGTGTGTT-3′); mouse Snai1 (5′-CCACTGCAACCGTGCTTTT-3′, 5′-GTGCTTGTGGAGCAAGGACAT-3′); mouse Snai2 (5′-CTCACCTCGGGAGCATACAGC-3′, 5′-TGAAGTGTCAGAGGAAGGCGGG-3′); mouse Twist1 (5′-CGGGTCATGGCTAACGTG-3′, 5′-CAGCTTGCCATCTTGGAGTC-3′); mouse SIP-1 (5′-CACCCAGCTCGAGAGGCATA-3′, 5′-CACTCCGTGCACTTGAACTTG-3′); mouse β-actin (5′-GGGGTGTTGAAGGTCTCAAA-3′, 5′-AGAAAATCTGGCACCCC-3′); human KDM6B (5′-CCTCGAAATCCCATCACAGT-3′, 5′-GTGCCTGTCAGATCCCAGTT-3′); human SNAI1 (5′-TCCCGGGCAATTTAACAATG-3′, 5′-TGGGAGACACATCGGTCAGA-3′); human GAPDH (5′-ATCATCCCTGCCTCTACTGG-3′, 5′-GTCAGGTCCACCACTGACAC-3′); canine SNAI1 (5′-CCCAAGCCCAGCCGATGAG-3′, 5′-CTTGGCCACGGAGAGCCC-3′); canine SNAI2 (5′-CGTTTTCCAGACCCTGGTTA-3′, 5′-TGACCTGTCTGCAAATGCTC-3′); and canine GAPDH (5′-CATCACTGCCACCCAGAAG-3′, 5′-CAGTGAGCTTCCCGTTCAG-3′).
Western Blot Analysis
Cells cultured in 10-cm dishes were washed with PBS and were then collected using the scraper. The cells were lysed using 250 μl of lysis buffer (radioimmuno precipitation assay buffer) for 30 min on ice. After centrifugation at 10,600 × g at 4 °C, the supernatants were collected and stored at −80 °C. The protein concentration of each sample was measured colorimetrically using Bio-Rad reagents, and 25–100 μg of total proteins were resolved on SDS-PAGE. The gel was transferred onto PVDF membrane for 50 min at 15 V using the Bio-Rad semidry transfer system. Western blot analysis was performed as described previously (26).
ChIP Assay
ChIP assays were performed using a ChIP assay kit according to the manufacturer's protocol (Upstate Biotechnology). Cells (2 × 106) were preincubated with a dimethyl 3,3′-dithiobispropionimidate-HCl (Pierce) solution (5 mmol) for 30 min on ice and then treated with formaldehyde. The ChIP-enriched DNA samples were quantified by real-time PCR, and the data are expressed as a percentage of input. The primer pair used for amplifying the mouse Snai1 promoter was as follows: 5′-CGGAGTTGACTACCGACCTT-3′ and 5′-GACCTAGGTAGTCGGGGTCAC-3′.
Immunofluorescence Staining
NMuMG and MDCK cells, grown on Mat-Tek glass bottom culture dishes (MatTek Corp., Ashland, MA) were fixed with 4% paraformaldehyde for 10 min at room temperature. Then the cells were washed in PBS containing 0.05% Tween 20 (PBS-T) three times for 5 min each. Nonspecific reactions were blocked with serum-free protein block (DAKO North America Inc.) and then incubated with the respective primary antibody (anti-N-cadherin, -fibronectin, or -E-cadherin) at 4 °C overnight. After washing, the cells were incubated with goat anti-mouse red IgG (Molecular Probes) for 60 min at room temperature and then counterstained by DAPI after digestion of RNA by RNase. The images were captured using a fluorescence Olympus IX51 microscope.
Oncomine Data Analysis
KDM6B mRNA expression in breast cancers from two independent studies in the Oncomine database (28, 29) was analyzed as described earlier (26). Details of standardized normalization techniques and statistical calculations are described on the Oncomine Web site. Initially, the raw microarray data were analyzed by a standard method using either the robust multichip average for Affymetrix data or the Loess normalization for cDNA arrays. Subsequently, Z score normalization was applied to scale the data and allow comparison of multiple independent studies. This included log2 transformation, setting the array median to 0 and S.D. to 1.
Matrigel Invasion Assays
BioCoat Matrigel invasion chambers (24-well) from BD Biosciences were used for cell invasion assays. Cells (1–2 × 105) suspended in 0.5 ml of serum-free medium were applied on the upper chamber of the Matrigel invasion chamber. In the lower chamber, 0.75 ml of medium containing 0.1% (for MDA-MB-231 and MDCK cells) or 0.5% (for NMuMG cells) FBS was added as a chemoattractant. The chambers were incubated for 48 h in the CO2 incubator. Matrigel was removed from the membrane, and the invaded cells were stained with the HEMA-3 kit (Fisher) and counted.
RESULTS
KDM6B Is Required for TGF-β-induced EMT
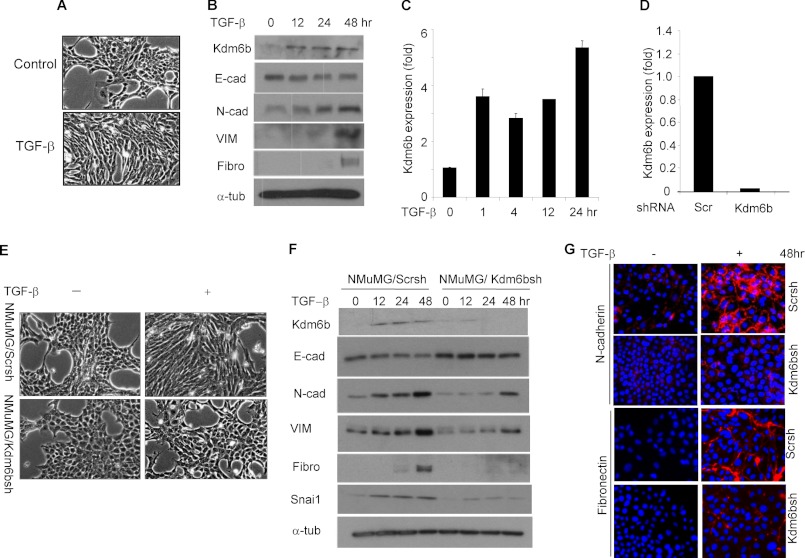
With the impetus that TGF-β-SMAD signaling is a potential inducer of EMT and Nodal-Smad signaling recruits KDM6B to counteract polycomb-mediated repression of developmentally important genes (9), we intended to assess whether KDM6B was associated with TGF-β-induced EMT. Because NMuMG cells are known to undergo EMT in response to TGF-β treatment, we utilized these cells to assess the role of KDM6B in TGF-β-induced EMT. As anticipated, TGF-β treatment induced spindle-shaped morphology in NMuMG cells (Fig. 1A). Western blot showed that TGF-β modestly inhibited the expression of E-cadherin, with concomitant up-regulation of mesenchymal markers, including N-cadherin, VIM, and fibronectin (Fig. 1B), confirming the induction of EMT by TGF-β. Interestingly, TGF-β induced the expression of Kdm6b in NMuMG cells as early as 1 h, and the induction was sustained until 24 h as assessed by real-time RT-PCR (Fig. 1C), implying the possible role for Kdm6b in TGF-β-induced EMT. Western blotting also confirmed the induction of Kdm6b expression in TGF-β-treated NMuMG cells (Fig. 1B).
FIGURE 1.
KDM6B is required for TGF-β-induced EMT. A, NMuMG cells were treated with TGF-β (10 ng/ml) for 24 h. B, Western blotting of EMT markers in the total protein extracted from control and TGF-β-treated NMuMG cells. C, real-time RT-PCR showing the induction of Kdm6b expression in response to TGF-β treatment in NMuMG cells. Values are mean ± S.D. (error bars) of triplicate samples from a representative experiment. D, NMuMG cells were infected with lentiviruses expressing Kdm6b shRNA or scramble shRNA. Kdm6b mRNA was determined by real-time RT-PCR. Values are mean of triplicate samples from a representative experiment. E, NMuMG/Scrsh and NMuMG/Kdm6bsh cells were treated with TGF-β for 24 h. F, NMuMG/Scrsh and NMuMG/Kdm6bsh cells were treated with TGF-β for different time points as indicated. G, NMuMG/Scrsh and NMuMG/Kdm6bsh cells were treated with TGF-β for 48 h, and then immunofluorescence staining was performed.
To determine whether Kdm6b was required for TGF-β-induced EMT, we utilized a lentivirus-based shRNA system to specifically knock down Kdm6b in NMuMG cells. As shown in Fig. 1D, real-time RT-PCR found that Kdm6b mRNA levels were reduced by more than 90% in NMuMG cells expressing Kdm6b shRNA (NMuMG/Kdm6bsh) compared with NMuMG cells expressing scrambled shRNA (NMuMG/Scrsh). Whereas NMuMG/Scrsh cells showed clear spindle-shaped morphological transition after TGF-β treatment, there was no appreciable morphological change in TGF-β-treated NMuMG/Kdm6bsh cells (Fig. 1E). Consistently, Western blot and immunofluorescence staining also showed that the induction of mesenchymal markers in NMuMG/Kdm6bsh cells by TGF-β was significantly inhibited compared with NMuMG/Scrsh cells (Fig. 1, F and G). These results suggest that Kdm6b is required for TGF-β-induced EMT.
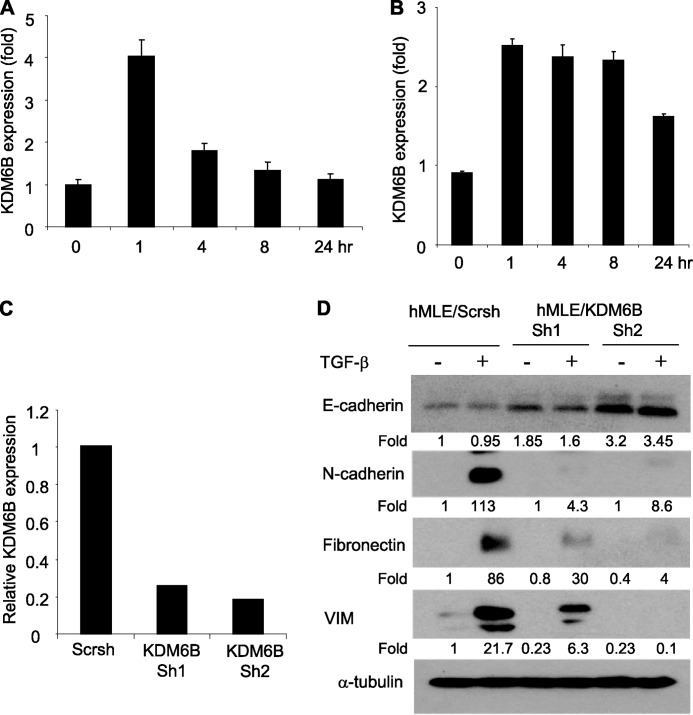
Next, we asked whether KDM6B promoted EMT in human mammary epithelial cell lines. Similar to NMuMG cells, TGF-β treatment also rapidly induced KDM6B expression in hMLE and MCF-10A cells (Fig. 2, A and B). We stably knocked down KDM6B in hMLE cells using two different shRNAs (hMLE/KDM6Bsh1 and -sh2) targeting different sequences on KDM6B mRNA (Fig. 2C). Interestingly, we observed that the basal level of E-cadherin was higher in hMLE/KDM6Bsh1 and hMLE/KDM6Bsh2 cells than in hMLE/Scrsh cells. Moreover, TGF-β-induced expression of mesenchymal markers was significantly reduced in hMLE/KDM6Bsh1 and hMLE/KDM6Bsh2 cells compared with hMLE/Scrsh cells (Fig. 2D). Similarly, we found that TGF-β-induced EMT was also inhibited in hMLE/KDM6Bsh2 cells (data not shown).
FIGURE 2.
KDM6B is required for TGF-β-induced EMT in hMLE cells. A, hMLE cells were treated with TGF-β for different times, and KDM6B expression was quantified by real-time RT-PCR. Values are mean ± S.D. (error bars) of triplicate samples from a representative experiment. B, MCF-10A cells were treated with TGF-β for the indicated times, and KDM6B mRNA was quantified by real-time RT-PCR. C, hMLE cells were infected with lentiviruses expressing scramble shRNA and KDM6B shRNA, and KDM6B mRNA was examined by real-time RT-PCR. D, hMLE/Scrsh and hMLE/KDM6Bsh1 and hMLE/KDM6Bsh2 cells were treated with TGF-β for 8 days. Total cell lysates were prepared and examined by Western blot analysis.
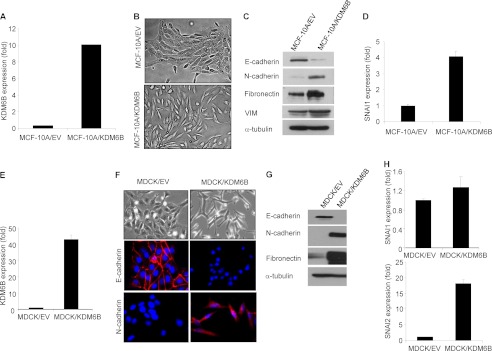
Because we found that TGF-β also induced KDM6B in MCF-10A cells, we knocked down KDM6B in MCF-10A cells. However, KDM6B knockdown did not significantly affect TGF-β-induced Snai1 expression or EMT in MCF-10A cells (data not shown). To explore whether KDM6B promoted the EMT in MCF-10A cells, we overexpressed KDM6B in MCF-10A cells (Fig. 3A). Interestingly, overexpression of KDM6B induced EMT-like phenotype in MCF-10A cells (Fig. 3B). Overexpression of KDM6B strongly inhibited E-cadherin expression and induced the expression of mesenchymal markers (Fig. 3C). Additionally, we found that overexpression of KDM6B also induced spindle-shaped morphology in MDCK cells (Fig. 3, E and F) and inhibited the expression of E-cadherin with concomitant up-regulation of N-cadherin and fibronectin (Fig. 3, F and G).
FIGURE 3.
The overexpression of KDM6B promotes EMT. A, MCF-10A cells were infected with retroviruses expressing KDM6B or empty vector. KDM6B mRNA was analyzed by real-time RT-PCR. B, the overexpression of KDM6B in MCF-10A cells induced an EMT-like phenotype. C, the total cell lysates were prepared from MCF-10A/EV and MCF-10A/KDM6B cells and subjected to Western blotting analysis. D, SNAI1 mRNA was examined by real-time RT-PCR. E, MDCK cells were infected with retroviruses expressing KDM6B or empty vector, and KDM6B mRNA was analyzed by real-time RT-PCR. F, the overexpression of KDM6B in MDCK cells induced spindle-shaped morphology and immunofluorescence staining showed the loss of E-cadherin expression with induction of N-cadherin expression. G, the whole cell extracts were isolated from MDCK/EV and MDCK/KDM6B cells and examined by Western blot. H, the expression of SNAI1 and SNAI2 was examined by real-time RT-PCR. Error bars, S.D.
KDM6B Promotes EMT by Inducing SNAI1
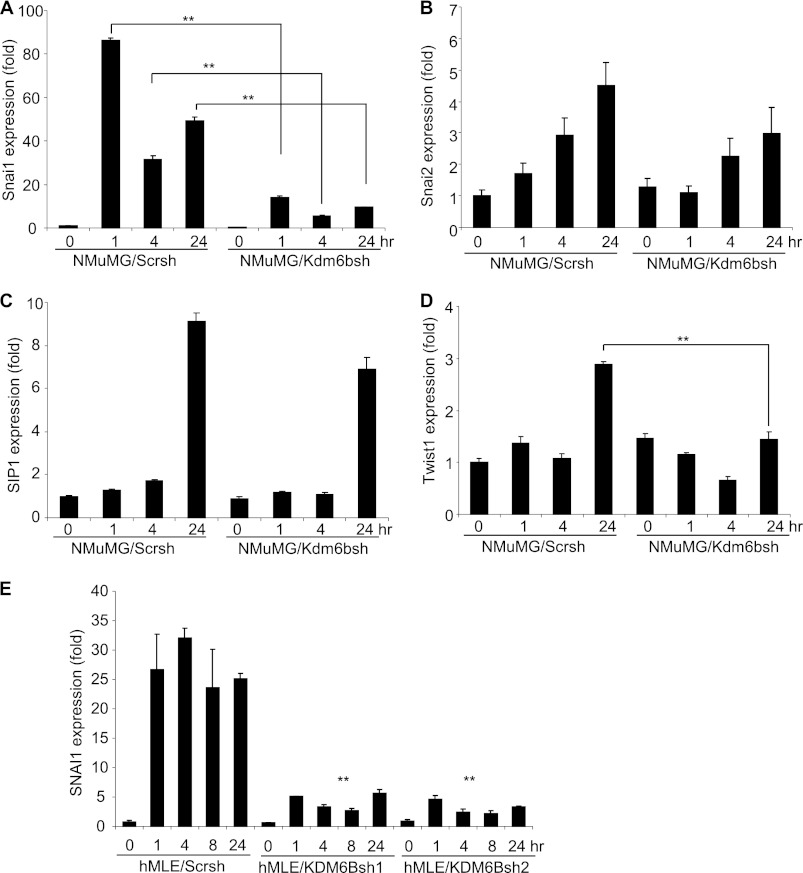
EMT is a complex process because growth factors, cytokines, and oncogenes employ distinct mechanisms to induce and/or to maintain EMT (3, 30). Because the transcription factors, including SNAI1, SNAI2, SIP-1, and TWIST1, are implicated as master transcription factors that mediate EMT (3), we were interested in evaluating whether Kdm6b regulated their expression in NMuMG cells. Real-time RT-PCR profiling revealed that TGF-β potently and rapidly induced Snai1 expression in NMuMG/Scrsh cells, and the induction of Snai1 was significantly suppressed in NMuMG/Kdm6bsh cells, suggesting that Snai1 expression is dependent on Kdm6b (Fig. 4A). Although TGF-β also induced the expression of Snai2 and Sip1, the knockdown of Kdm6b slightly affected their expression (Fig. 4, B and C). Interestingly, although Twist1 was induced by TGF-β at a late point, the knockdown of Kdm6b also inhibited Twist1 expression (Fig. 4D). Our Western blot also confirmed that Kdm6b knockdown also reduced the expression of SNAI1 in NMuMG cells (Fig. 1F). Moreover, we found that TGF-β also potently induced SNAI1 expression, and the knockdown of KDM6B inhibited SNAI1 expression in hMLE cells (Fig. 4E).
FIGURE 4.
The knockdown of Kdm6b inhibits Snai1 expression. A, NMuMG/Scrsh and NMuMG/Kdm6bsh cells were treated with TGF-β, and Snai1 mRNA was quantified by real-time RT-PCR. **, p < 0.01. B, Snai2 mRNA was quantified by real-time RT-PCR. C, Sip1 mRNA was quantified by real-time RT-PCR. D, Twist1 mRNA was quantified by real-time RT-PCR. **, p < 0.01. E, hMLE/Scrsh, hMLE/KDM6Bsh1, and hMLE/KDM6Bsh2 cells were treated with TGF-β, and SNAI1 mRNA was measured by real-time RT-PCR. **, p < 0.01. Error bars, S.D.
To determine whether Kdm6b was sufficient to promote EMT by inducing Snai1, we stably overexpressed Kdm6b in NMuMG cells (NMuMG/Kdm6b) using retrovirus-mediated transduction. Although Kdm6b overexpression alone did not inhibit the expression of E-cadherin, it potently increased the expression of N-cadherin and VIM (Fig. 5, A–C). Kdm6b overexpression significantly enhanced TGF-β-induced Snai1 mRNA (Fig. 5D). Moreover, overexpression of KDM6B alone induced SNAI1 expression in MCF-10A cells (Fig. 3D). Interestingly, although overexpression of KDM6B could not induce SNAI1 in MDCK cells, it potently induced SNAI2 expression (Fig. 3H).
FIGURE 5.
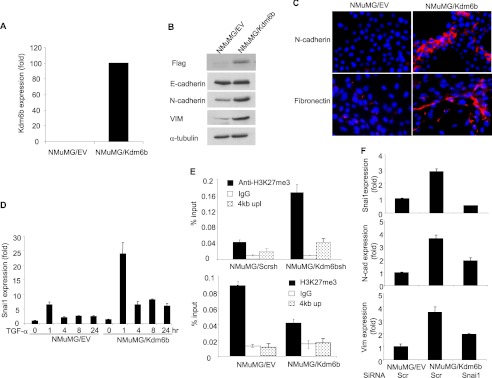
KDM6B promotes SNAI1 expression by removing H3K27me3. A, NMuMG cells were infected with retroviruses expressing Kdm6b or empty vector, and Kdm6b mRNA was quantified by real-time RT-PCR. B, the whole cell extracts were prepared from NMuMG/EV and NMuMG/Kdm6b cells and examined by Western blot. C, immunofluorescence staining of NMuMG/EV and NMuMG/Kdm6b cells for N-cadherin and fibronectin expression. D, NMuMG/EV and NMuMG/Kdm6b cells were treated with TGF-β for different time points as indicated. Snai1 mRNA was quantified by real-time RT-PCR. Values are mean ± S.D. (error bars) of triplicate samples from a representative experiment. E, the knockdown of Kdm6b increased H3K27me3 levels on the Snai1 promoter and overexpression of Kdm6b decreased H3K27me3 levels on the Snai1 promoter, as determined by ChIP assays. The non-target region located on 4 kb upstream of the Snai1 transcriptional start sites was used as a control. F, the knockdown of Snai1 inhibited mesenchymal markers in NMuMG cells induced by Kdm6b. NMuMG/Kdm6b cells were transfected with scramble or Snai1 siRNA. Snai1, N-cadherin, and vimentin expressions were quantified by real-time RT-PCR. Values are the mean of triplicate samples from a representative experiment.
Based on the premise that KDM6B eliminates gene repression by removing the H3K27me3 marks on chromatin, we further investigated the extent of H3K27me3 modification at the SNAI1 promoter by KDM6B using ChIP assays. Previous studies showed that KDM6B can bind to transcription start sites to remove the repressive methylation mark (31), and Smad2/3 can activate Snai1 transcription in response to TGF-β in NMuMG cells (32). Thus, we focused our ChIP assays on the Snai1 promoter. Despite repeated trials, we had technical difficulty in detecting Kdm6b on the Snai1 promoter in NMuMG cells using commercially available anti-Kdm6b antibodies. Alternatively, we examined whether the knockdown of Kdm6b affected the levels of H3K27me3 on the Snai1 promoter. ChIP assays revealed that the levels of H3K27me3 on the Snai1 promoter were significantly increased in NMuMG/Kdm6bsh cells compared with NMuMG/Scrsh cells (Fig. 5E). As a control, the knockdown of KDM6B did not affect H3K27me3 on 4 kb upstream of the Snai1 transcriptional start site. To further confirm our results, we also examined whether overexpression of Kdm6b decreased the levels of H3K27me3 on the Snai1 promoter. Consistently, ChIP assays showed that the levels of H3K27me3 were significantly reduced in NMuMG/Kdm6b cells compared with NMuMG/EV cells (Fig. 5E). Collectively, our results suggest that KDM6B promotes the EMT by inducing SNAI1. Next, we were interested in examining whether the inhibition of Snai1 is sufficient to inhibit the expression of EMT markers induced by Kdm6b. We knocked down Snai1 in NMuMG/Kdm6b cells using siRNA. The depletion of Snai1 significantly inhibited the expression of N-cadherin and VIM induced by Kdm6b, indicating that Snai1 plays a role in Kdm6b-promoted EMT (Fig. 5F).
KDM6B Promotes Breast Cancer Cell Invasion
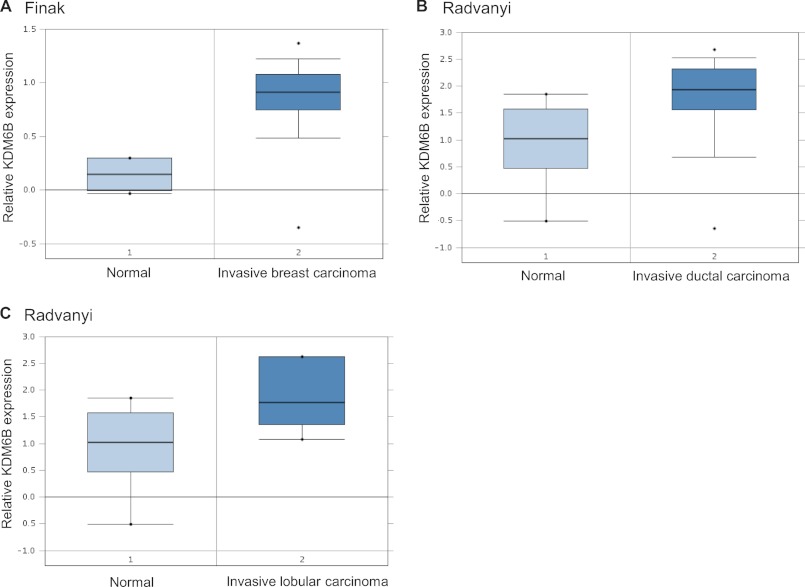
Having established a pro-EMT function for KDM6B, we then sought to determine the biological significance of this mechanism in human cancer because EMT is linked to cancer cell invasion and metastasis (1–4). We screened the expression pattern of KDM6B mRNA in breast cancer using the Oncomine database. In support of our findings, KDM6B was found to be highly expressed in invasive breast carcinoma tissues compared with normal breast tissues in two independent studies (28, 29) (Fig. 6, A–C).
FIGURE 6.
KDM6B is overexpressed in invasive breast carcinoma. A, KDM6B mRNA was higher in invasive breast carcinoma than in normal breast tissues, based on studies reported by Finak et al. (28). *, p < 0.001. B and C, KDM6B mRNA was higher in invasive ductal and lobular breast carcinoma than in normal breast tissues, based on studies by Radvanyi et al. (29). *, p < 0.005.
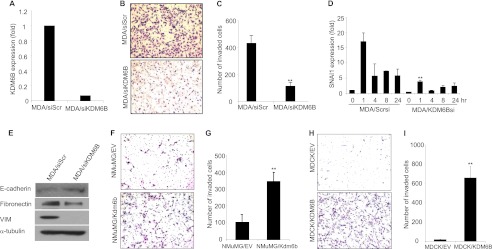
To directly examine whether KDM6B promoted tumor cell invasion, we knocked down KDM6B in the highly invasive breast cancer cell line MDA-MB-231 (Fig. 7A). Matrigel invasion assays revealed that the knockdown of KDM6B significantly inhibited the invasiveness of MDA-MB-231 cells through the Matrigel-coated membrane (Fig. 7, B and C). Consistently, as shown in Fig. 7D, the knockdown of KDM6B significantly reduced SNAI1 expression under basal and TGF-β-treated conditions. In addition, Western blot showed that the knockdown of KDM6B inhibited the expression of VIM and fibronectin in MDA-MB-231 cells and modestly increased E-cadherin expression (Fig. 7E). Finally, we found that the overexpression of Kdm6b promoted the invasion in NMuMG cells (Fig. 7, F and G) and MDCK cells (Fig. 7, H and I) through the Matrigel-coated membrane.
FIGURE 7.
The knockdown of KDM6B reduces SNAI1 expression and inhibits MDA-MB-231 cell invasion. A, MDA-MB-231 cells were transfected with KDM6B siRNA (MDA/siKDM6B) or scramble siRNA (MDA/siScr), and KDM6B mRNA was quantified by real-time RT-PCR. Values are the mean of triplicate samples from a representative experiment. B, photograph of invaded cells after staining with the HEMA-3 kit. C, Quantitative measurement of invaded cells. Each bar represents mean ± S.D. (error bars) of triplicate experiments. **, p < 0.01. D, both MDA/siScr and MDA/siKDM6B cells were treated with TGF-β. SNAI1 mRNA expression was assessed by real-time RT PCR. Values are mean ± S.D. of triplicate samples from a representative experiment. **, p < 0.01. E, the whole cell lysates were extracted from MDA/siScr and MDA/siKDM6B cells and examined by Western blot. F, photograph of invaded NMuMG/EV and NMuMG/Kdm6bcells after staining with the HEMA-3 kit. G, quantitative measurement of invaded cells. Each bar represents the mean ± S.D. of triplicate experiments. **, p < 0.01. H and I, KDM6B overexpression induced invasiveness of MDCK cells. Each bar represents the mean ± S.D. of triplicate experiments. **, p < 0.01.
DISCUSSION
Cancer metastasis, which accounts for major fractions of cancer-related death, is a multistep process associated with EMT. An important feature of EMT is disintegration and disassembly of epithelial cell-cell adhesion complexes that resulted in transitioning of epithelial cells to mesenchymal-like cells, which enables the tumor cells to invade adjacent tissues and blood vessels to metastasize and form secondary tumors at distant sites (4). Recent studies also highlighted that EMT generates tumor cells with cancer stem cell properties and thus favor the tumor recurrence and therapeutic resistance (33). It is well established that SNAI1 and SNAI2 induce EMT in embryonic development, carcinoma progression, and other important biological processes (1–4, 30). Apart from playing a crucial role in EMT, SNAI1 also regulates cell growth by controlling the expression of cyclin D2 and p21Cip in a cell type- and cell context-dependent manner (34, 35). Additionally, SNAI1 promotes cell survival and renders the cells with apoptotic resistance to pro-apoptotic stimuli (34, 36). Despite the fact that the intracellular signaling pathways that regulate the expression of SNAI1 family members are well characterized, our studies showed for the first time that chromatin modification by KDM6B controls SNAI1 expression, thereby promoting EMT.
The finding that KDM6B promotes EMT and cancer cell invasion argues against the tumor suppressor role that it plays in oncogenic signaling-induced cell senescence by controlling the expression of INK4A/ARF (23, 24). Several studies reported that KDM6B acts as a tumor suppressor and is down-regulated in human cancers (23, 24). However, there also were some studies suggesting that KDM6B is highly expressed in human cancers and associated with tumor progression (37, 38). In this regard, our studies provide direct evidence for KDM6B as a promoter of EMT and cancer cell invasion. Conversely, a recent report by Pereira et al. (39) described that KDM6B negatively regulates Snai1 expression and EMT in colon cancer by mediating vitamin D signaling. However, they did not show how KDM6B regulates Snai1 expression. Moreover, they studied vitamin D signaling in colon cancer, whereas our study focused on TGF-β induced EMT in breast cancer. Therefore, the discrepancies between these two studies might be due to differential roles played by KDM6B in different cancer types and/or different signaling mechanisms. However, in support of our findings, Xiang et al. (37) reported that there was a positive correlation between KDM6B expression and prostate cancer progression. In addition, Anderton et al. (38) reported that the aberrant expression of KDM6B was associated with the pathogenesis of Hodgkin's lymphoma. These conflicting results, claiming the opposite functions for KDM6B in cancer, recall the dual functions of TGF-β in cancer development and progression. Although TGF-β employs the same Smad2/3 signaling for its tumor-suppressive and oncogenic functions in cancer, the molecular switch from tumor suppressor to tumor promoter is probably dependent on the binding partner of the Smad transcription complex. Because the recent study uncovered a key mechanism by which KDM6B interacted with and recruited Smad2/3 to the gene promoter in response to nodal signaling to induce the expression of development-associated genes (9) and our co-immunoprecipitation experiment also supports the existence of KDM6B-Smad complex, we speculate that KDM6B might be recruited along with the Smad transcriptional complex in response to TGF-β signaling to regulate tumor development or progression. However, the confirmation of our speculation would require genetic studies using in vivo tumorigenic models, where KDM6B expression and/or function is disrupted in a stage-specific manner in TGF-β-mediated tumor progression.
Contrary to our findings, several earlier studies reported that a histone methyltransferase, EZH2, which mediates H3K27me3, is highly expressed in metastatic breast cancers (40–43). Although it is unclear why the proteins with opposite enzymatic activities are overexpressed in breast cancer, it is possible that breast cancer may require a balanced expression of EZH2 and KDM6B to tightly regulate H3K27me3 status to control the cell plasticity. Alternatively, EZH2 and KDM6B may have different molecular targets in metastatic breast cancers. Intriguingly, many studies have indicated that metastatic lesions and their primary tumors of various carcinomas, including breast cancer, share a similar epithelial feature (44, 45). These observations suggest that the occurrence of mesenchymal-epithelial transition (MET) in the metastatic sites as part of the process of metastatic tumor formation (46, 47). Therefore, it seems that EMT is induced in a spatial and temporal manner during tumor progression and is critical for rendering the tumor cells with the invasive and metastatic phenotype (48), whereas MET (the reverse of EMT) occurs at the metastatic foci and is critical for metastatic tumor formation. Therefore, it is logical to speculate that KDM6B expression might be up-regulated to remove the repressive H3K27me3 marks on the SNAI1 promoter during the EMT. Subsequently, EZH2 expression might be induced to activate MET by enriching the H3K27me3 marks on the SNAI1 promoter. KDM6B may be necessary to initiate tumor metastasis, and EZH2 is required for the late stage by promoting metastatic tumor growth. Hence, it is reasonable that both proteins might be up-regulated in breast cancer tissues because most of the solid tumors are highly heterogeneous in nature because they harbor cells with both epithelial and mesenchymal properties.
Snai1 was rapidly induced by TGF-β in NMuMG cells. Our gain-and-loss function studies suggest that Kdm6b directly regulates Snai1 expression by removing H3K27me3 marks. Interestingly, the knockdown of Kdm6b also potently inhibited Twist1 expression. Because Twist1 was not rapidly induced by TGF-β, Kdm6b could also regulate Twist1 directly or indirectly. Of interest, unlike mammary epithelial cells, overexpression of KDM6B promoted EMT in MDCK cells by inducing SNAI2 but not SNAI1. It suggests that KDM6B can also act as a potential regulator of other EMT-inducing factors in a cell type-dependent manner. Taken together, given the role of EMT in cancer metastasis and cancer cell stemness, our findings reveal a novel epigenetic mechanism regulating EMT and have important implications in devising therapeutic strategies to inhibit breast cancer metastasis.
Acknowledgments
We thank Dr. Robert A. Weinberg for hMLE cells and Dr. Paul A. Khavari for KDM6B plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants CA132134, R3713848, and DE15964.
- EMT
- epithelial-mesenchymal transition
- MET
- mesenchymal-epithelial transition
- H3K27me3
- histone H3 lysine trimethylation
- MDCK
- Madin-Darby canine kidney
- VIM
- vimentin
- hMLE
- human mammary epithelial.
REFERENCES
- 1. Thiery J. P., Acloque H., Huang R. Y., Nieto M. A. (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 2. Singh A., Settleman J. (2010) EMT, cancer stem cells and drug resistance. An emerging axis of evil in the war on cancer. Oncogene 29, 4741–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thiery J. P., Sleeman J. P. (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 4. Wendt M. K., Tian M., Schiemann W. P. (2012) Deconstructing the mechanisms and consequences of TGF-β-induced EMT during cancer progression. Cell Tissue Res. 347, 85–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Feng X. H., Derynck R. (2005) Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21, 659–693 [DOI] [PubMed] [Google Scholar]
- 6. Massagué J., Gomis R. R. (2006) The logic of TGF β signaling. FEBS Lett. 580, 2811–2820 [DOI] [PubMed] [Google Scholar]
- 7. Shi Y., Massagué J. (2003) Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 8. Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495 [DOI] [PubMed] [Google Scholar]
- 9. Dahle Ø., Kumar A., Kuehn M. R. (2010) Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci. Signal. 3, ra48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jenuwein T., Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 11. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 12. Lund A. H., van Lohuizen M. (2004) Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 16, 239–246 [DOI] [PubMed] [Google Scholar]
- 13. Plath K., Fang J., Mlynarczyk-Evans S. K., Cao R., Worringer K. A., Wang H., de la Cruz C. C., Otte A. P., Panning B., Zhang Y. (2003) Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135 [DOI] [PubMed] [Google Scholar]
- 14. Zhao J., Sun B. K., Erwin J. A., Song J. J., Lee J. T. (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schuettengruber B., Cavalli G. (2009) Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136, 3531–3542 [DOI] [PubMed] [Google Scholar]
- 16. Agger K., Cloos P. A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A. E., Helin K. (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734 [DOI] [PubMed] [Google Scholar]
- 17. De Santa F., Totaro M. G., Prosperini E., Notarbartolo S., Testa G., Natoli G. (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094 [DOI] [PubMed] [Google Scholar]
- 18. Lan F., Bayliss P. E., Rinn J. L., Whetstine J. R., Wang J. K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E., Roberts T. M., Chang H. Y., Shi Y. (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449, 689–694 [DOI] [PubMed] [Google Scholar]
- 19. Swigut T., Wysocka J. (2007) H3K27 demethylases, at long last. Cell 131, 29–32 [DOI] [PubMed] [Google Scholar]
- 20. Hong S., Cho Y. W., Yu L. R., Yu H., Veenstra T. D., Ge K. (2007) Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. U.S.A. 104, 18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller S. A., Mohn S. E., Weinmann A. S. (2010) Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 40, 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hübner M. R., Spector D. L. (2010) Role of H3K27 demethylases Jmjd3 and UTX in transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 75, 43–49 [DOI] [PubMed] [Google Scholar]
- 23. Agger K., Cloos P. A., Rudkjaer L., Williams K., Andersen G., Christensen J., Helin K. (2009) The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 23, 1171–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barradas M., Anderton E., Acosta J. C., Li S., Banito A., Rodriguez-Niedenführ M., Maertens G., Banck M., Zhou M. M., Walsh M. J., Peters G., Gil J. (2009) Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 23, 1177–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim S. W., Yoon S. J., Chuong E., Oyolu C., Wills A. E., Gupta R., Baker J. (2011) Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev. Biol. 357, 492–504 [DOI] [PubMed] [Google Scholar]
- 26. Ramadoss S., Li J., Ding X., Al Hezaimi K., Wang C. Y. (2011) Transducin β-like protein 1 recruits nuclear factor κB to the target gene promoter for transcriptional activation. Mol. Cell Biol. 31, 924–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sen G. L., Webster D. E., Barragan D. I., Chang H. Y., Khavari P. A. (2008) Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 22, 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Finak G., Bertos N., Pepin F., Sadekova S., Souleimanova M., Zhao H., Chen H., Omeroglu G., Meterissian S., Omeroglu A., Hallett M., Park M. (2008) Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 14, 518–527 [DOI] [PubMed] [Google Scholar]
- 29. Radvanyi L., Singh-Sandhu D., Gallichan S., Lovitt C., Pedyczak A., Mallo G., Gish K., Kwok K., Hanna W., Zubovits J., Armes J., Venter D., Hakimi J., Shortreed J., Donovan M., Parrington M., Dunn P., Oomen R., Tartaglia J., Berinstein N. L. (2005) The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 102, 11005–11010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sivakumar R., Koga H., Selvendiran K., Maeyama M., Ueno T., Sata M. (2009) Autocrine loop for IGF-I receptor signaling in SLUG-mediated epithelial-mesenchymal transition. Int. J. Oncol. 34, 329–338 [PubMed] [Google Scholar]
- 31. De Santa F., Narang V., Yap Z. H., Tusi B. K., Burgold T., Austenaa L., Bucci G., Caganova M., Notarbartolo S., Casola S., Testa G., Sung W. K., Wei C. L., Natoli G. (2009) Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 28, 3341–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith A. P., Verrecchia A., Fagà G., Doni M., Perna D., Martinato F., Guccione E., Amati B. (2009) A positive role for Myc in TGF-β-induced Snai1 transcription and epithelial-to-mesenchymal transition. Oncogene 28, 422–430 [DOI] [PubMed] [Google Scholar]
- 33. May C. D., Sphyris N., Evans K. W., Werden S. J., Guo W., Mani S. A. (2011) Epithelial-mesenchymal transition and cancer stem cells. A dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 13, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vega S., Morales A. V., Ocaña O. H., Valdés F., Fabregat I., Nieto M. A. (2004) Snai1 blocks the cell cycle and confers resistance to cell death. Genes Dev. 18, 1131–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olmeda D., Jordá M., Peinado H., Fabra A., Cano A. (2007) Snai1 silencing effectively suppresses tumour growth and invasiveness. Oncogene 26, 1862–1874 [DOI] [PubMed] [Google Scholar]
- 36. Kajita M., McClinic K. N., Wade P. A. (2004) Aberrant expression of the transcription factors snai1 and slug alters the response to genotoxic stress. Mol. Cell Biol. 24, 7559–7566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiang Y., Zhu Z., Han G., Lin H., Xu L., Chen C. D. (2007) JMJD3 is a histone H3K27 demethylase. Cell Res. 17, 850–857 [DOI] [PubMed] [Google Scholar]
- 38. Anderton J. A., Bose S., Vockerodt M., Vrzalikova K., Wei W., Kuo M., Helin K., Christensen J., Rowe M., Murray P. G., Woodman C. B. (2011) The H3K27me3 demethylase, KDM6B, is induced by Epstein-Barr virus and over-expressed in Hodgkin's lymphoma. Oncogene 30, 2037–2043 [DOI] [PubMed] [Google Scholar]
- 39. Pereira F., Barbáchano A., Silva J., Bonilla F., Campbell M. J., Muñoz A., Larriba M. J. (2011) KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Hum. Mol. Genet. 20, 4655–4665 [DOI] [PubMed] [Google Scholar]
- 40. Kunju L. P., Cookingham C., Toy K. A., Chen W., Sabel M. S., Kleer C. G. (2011) EZH2 and ALDH-1 mark breast epithelium at risk for breast cancer development. Mod. Pathol. 24, 786–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gong Y., Huo L., Liu P., Sneige N., Sun X., Ueno N. T., Lucci A., Buchholz T. A., Valero V., Cristofanilli M. (2011) Polycomb group protein EZH2 is frequently expressed in inflammatory breast cancer and is predictive of worse clinical outcome. Cancer 117, 5476–5484 [DOI] [PubMed] [Google Scholar]
- 42. Kleer C. G., Cao Q., Varambally S., Shen R., Ota I., Tomlins S. A., Ghosh D., Sewalt R. G., Otte A. P., Hayes D. F., Sabel M. S., Livant D., Weiss S. J., Rubin M. A., Chinnaiyan A. M. (2003) EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 100, 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ding L., Erdmann C., Chinnaiyan A. M., Merajver S. D., Kleer C. G. (2006) Identification of EZH2 as a molecular marker for a precancerous state in morphologically normal breast tissues. Cancer Res. 66, 4095–4099 [DOI] [PubMed] [Google Scholar]
- 44. Kowalski P. J., Rubin M. A., Kleer C. G. (2003) E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res. 5, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chao Y. L., Shepard C. R., Wells A. (2010) Breast carcinoma cells re-express E-cadherin during mesenchymal to epithelial reverting transition. Mol. Cancer 9, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chaffer C. L., Thompson E. W., Williams E. D. (2007) Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs 185, 7–19 [DOI] [PubMed] [Google Scholar]
- 47. Hugo H., Ackland M. L., Blick T., Lawrence M. G., Clements J. A., Williams E. D., Thompson E. W. (2007) Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J. Cell Physiol. 213, 374–383 [DOI] [PubMed] [Google Scholar]
- 48. Thiery J. P. (2002) Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442–454 [DOI] [PubMed] [Google Scholar]