Abstract
This article is a survey of my scientific work over 52 years. During my postdoctoral stay in Severo Ochoa's laboratory, I determined the direction of reading of the genetic message, and I discovered two proteins that I showed to be involved in the initiation of protein synthesis. The work I have done in Spain with bacteriophage φ29 for 45 years has been very rewarding. I can say that I was lucky because I did not expect that φ29 would give so many interesting results, but I worked hard, with a lot of dedication and enthusiasm, and I was there when the luck arrived. I would like to emphasize our work on the control of φ29 DNA transcription and, in particular, the finding for the first time of a protein covalently linked to the 5′-ends of φ29 DNA that we later showed to be the primer for the initiation of phage DNA replication. Very relevant was the discovery of the φ29 DNA polymerase, with its properties of extremely high processivity and strand displacement capacity, together with its high fidelity. The φ29 DNA polymerase has become an ideal enzyme for DNA amplification, both rolling-circle and whole-genome linear amplification. I am also very proud of the many brilliant students and collaborators with whom I have worked over the years and who have become excellent scientists. This Reflections article is not intended to be the end of my scientific career. I expect to work for many years to come.
Keywords: DNA, Bacteriophage, DNA Transcription, DNA Replication, DNA Polymerase
Early Years
I was born in November 1938 in Canero, Spain, a small village in the north coast of Asturias, close to Luarca, where my teacher and friend Severo Ochoa, a Nobel Prize winner, was born. When I was one year old, after the Spanish civil war finished, my parents moved to Gijón, a town also located in the north coast of Asturias. There, I attended the school and received the baccalaureate title in 1954. My parents had three children, one boy and two girls, and they had made it very clear that my sister and I would follow a university career, something not very common at that time. Before going to the university, I had to have pre-university studies, and I had to choose whether I wanted to follow a scientific or a humanistic career. I decided to go into science. After I finished my pre-university studies, I had to decide which scientific discipline I wanted to study. I hesitated between chemistry and medicine. Because medicine was not available at the University of Oviedo, close to Gijón, I decided to go to Madrid University (now called Madrid Complutense University) to follow a so-called selective course, common for both careers. This gave me one more year to make a decision. Finally, I decided to study chemistry. I think that was a good decision because I enjoyed the laboratory work, especially the organic chemistry laboratories. We spent many hours for several months doing practical work in the laboratory.
In the summer of 1958, when I had finished my third year of chemistry, I went to Gijón to spend the summer holidays, and I was very lucky to meet Severo Ochoa, which had an important influence on my future. That year, Ochoa went to Gijón, the hometown of his wife, Carmen, and came to our house to visit us. Ochoa and my father were good friends, and they also had a family relationship. I had the opportunity to talk with Ochoa about my projects, and the next day, I attended a conference he gave about his work. Severo Ochoa was a brilliant speaker, and I was fascinated by his talk. I had not yet studied biochemistry, which was taught in the next year, and Ochoa promised to send me a biochemistry book. Indeed, he sent me the book General Biochemistry by Joseph S. Fruton and Sofia Simmonds. I was very happy when I received the book dedicated by Severo Ochoa. When I finished my chemistry studies, I decided to dedicate myself to biochemistry. Ochoa advised me to obtain a Ph.D. degree in Spain with an excellent biochemist, Alberto Sols, who had been trained in the laboratory of Carl and Gerty Cori at the Washington University School of Medicine in St. Louis. Then, I could go to Ochoa's laboratory at the New York University (NYU) School of Medicine for postdoctoral training. Ochoa wrote a reference letter for me to be accepted in Sols' laboratory. Sols could not refuse a request from Severo Ochoa, even if I was a woman, because he had already by that time received the Nobel Prize. During my Ph.D. thesis, I worked on carbohydrate metabolism, mainly glucose-phosphate isomerase from yeast, the enzyme that converts glucose 6-phosphate to fructose 6-phosphate. We found that the enzyme has an anomerase-like activity, producing an intermediate product that seemed to be the open form of glucose 6-phosphate. This was the first finding in my scientific career, something that was very rewarding to me. The results of this work were published in the Journal of Biological Chemistry (1).
At the end of my studies in chemistry, I became engaged to Eladio Viñuela, a brilliant student who also worked in Alberto Sols' laboratory while obtaining his Ph.D. degree (Fig. 1). Eladio's work dealt with yeast phosphofructokinase, and he demonstrated its allosteric properties. In addition, he discovered a new enzyme, liver glucokinase, that converts glucose into glucose 6-phosphate in the liver and disappears in fasted and diabetic rats. I joined Eladio in this work, and we found that the enzyme reappears by refeeding and insulin administration, respectively. This work was also published in the Journal of Biological Chemistry (2, 3). Publishing at that time from Spain in such a prestigious international journal was quite an accomplishment for us.
FIGURE 1.
Margarita Salas (right) and Eladio Viñuela at the Center of Biological Research during their Ph.D. work (1962).
Postdoctoral Years
After I obtained a fellowship from the Juan March Foundation, Eladio and I married in 1963. In August 1964, after finishing our Ph.D. theses, we went to New York to join Severo Ochoa's laboratory in time to attend the International Congress of Biochemistry, where Philip Leder and Marshall Nirenberg presented their work on the use of trinucleotides of specific sequence for the binding of the different aminoacyl-tRNAs. This was the final step in the determination of the genetic code, completing the work carried out in the laboratories of Severo Ochoa, Marshall Nirenberg, and H. Gobind Khorana.
Ochoa decided that Eladio and I should work in different groups. He told us, “At least, you will learn English.” I think that he wanted each of us to develop our work independently. My initial research project was to determine the direction of reading of the genetic message, that is, whether the reading of the mRNA was in the 5′ to 3′ or in the 3′ to 5′ direction. I used a cell-free protein synthesis system that consisted of a high-speed supernatant of Lactobacillus arabinosus that had very low nuclease activity and ribosomes from Escherichia coli that had been washed with 0.5 m NH4Cl and further purified by DEAE-cellulose chromatography. Marvin Smith constructed synthetic polynucleotides that contained the AAC codon at the 3′- or 5′-end. When I used the polynucleotide 5′-AAAAAA … AAAAAC-3′, the amino acids lysine and asparagine were incorporated. Treatment with carboxypeptidase A released asparagine, which indicated that this amino acid was located at the C-terminal end (4). When the polynucleotide 5′-AAAAACAAA … AAA-3′ was used, asparagine was incorporated at the N-terminal end, as indicated by the fact that it was not released by treatment with carboxypeptidase A but with carboxypeptidase B, which hydrolyzes lysines from the C-terminal end (5). I should point out that the AAA triplet at the 5′-end is not translated. The AUG triplet is required to initiate translation at the 5′-end (see below).
I later started a new project, which was the translation of a natural mRNA. I used as messenger the RNA from phage MS2 and a cell-free translation system obtained from E. coli with the ribosomes purified as described above. To my surprise, the system, which was active with poly(A) as messenger, giving rise to the incorporation of lysine, was completely inactive with the MS2 RNA. When I precipitated the ribosomal wash with ammonium sulfate and added this fraction to the purified ribosomes, I recovered the activity with MS2 RNA. I remember that, at that time, Walter (Wally) Gilbert came to NYU to give a seminar, and Ochoa asked me to show him the results. He suggested that the fraction obtained from the ribosomal wash might have something to do with the termination of protein synthesis. This turned out not to be the case. When I used the polynucleotide 5′-AUG(A)24-3′, prepared by Wendell Stanley, Jr., I found that, as in the case of the MS2 RNA, this messenger was not active with the purified ribosomes, but the activity was recovered when I added the ammonium sulfate fraction (6). Because the AUG triplet at the 5′-end of a messenger codes for formyl-methionine, it was likely that the fraction I was adding was involved in the initiation of protein synthesis. I purified two proteins from the ribosomal wash, which I called F1 and F2 (later on called iF1 and iF2), and found that the two proteins were needed for the binding of formyl-methionyl-tRNA to the ribosomes in the presence of the AUG triplet. This result demonstrated that proteins F1 and F2 were required for the initiation of protein synthesis (7). The work on initiation factors for protein synthesis became the future focus in Severo Ochoa's laboratory.
I also collaborated with Jerold (Jerry) Last in determining that UAA was a termination triplet. We used the cell-free system with the purified ribosomes and the initiation factors described above and the synthetic polynucleotides 5′-AUGUUUAAA…AAA-3′ and 5′-AUGUUUUAAAAA…AAA-3′, prepared by Wendell Stanley, Jr. The first polynucleotide gave rise to the synthesis of a polypeptide consisting of formyl-methionyl-phenylalanyl-lysyl…lysine, whereas the second polypeptide produced the peptide formyl-methionyl-phenylalanine, indicating that the UAA triplet is a polypeptide chain termination codon (8).
After working for one year with Charles Weissmann on the replication of MS2 RNA, Eladio proposed to Ochoa a project of his own: characterization of the proteins induced in E. coli after infection with phage MS2. For this project, Eladio developed the technique of polyacrylamide gel electrophoresis (PAGE) in the presence of sodium dodecyl sulfate (SDS), which separates proteins according to their molecular weight (9). In collaboration with Eladio, we showed that the E. coli cell-free system indicated above, using MS2 RNA as messenger, initiated with formyl-methionine the synthesis of two of the three proteins synthesized in vivo after infection of E. coli with phage MS2 (10). These results suggested that formyl-methionine was the initiator of each of the proteins encoded in a polycistronic mRNA.
I have very good memories of my stay in Severo Ochoa's laboratory. The three years I spent there were the best ones of my scientific life. Ochoa taught us (Eladio and me) not only the molecular biology that we would develop and teach in Spain but also his experimental rigor and enthusiasm for research. I would also like to mention some of the scientists from the biochemistry department headed by Ochoa: Bob Warner, Bob Chambers, Charles Weissmann, Albrecht Kleinschmidt, and Dan Lane. We became very good friends with Dan Lane and his wife, Pat.
Back to Spain
After three years in Severo Ochoa's laboratory, Eladio and I decided to return to Spain to try to develop the molecular biology that we learned with Ochoa. We knew it would be difficult to do scientific research in Spain, but we wanted to give a try. Our first decision was the project, which should be different from those we had carried out in Ochoa's laboratory. We also decided to work together because we knew it would be difficult to start a research group in Spain, and it would be easier if we joined our efforts. The previous summer (1966) we had participated in the bacteriophage course at Cold Spring Harbor Laboratory, where we learned how to work with phages. Thus, we decided to choose a phage as a model system to study at the molecular level, including the morphogenesis of the phage particle and the mechanisms of transfer of the genetic information such as replication and transcription. Our choice was a Bacillus subtilis phage called φ29, which Eladio discovered while reading a paper from Dwight Anderson's laboratory in which the morphology of the phage particle and the size of the DNA were described (11). This phage had been initially characterized by Bernard (Bernie) Reilly in John Spizizen's laboratory and had the characteristics we were looking for: it was small in size and had a complex morphology (Fig. 2), and very little was known about it. We sent a grant proposal to the Jane Coffin Childs Memorial Fund for Medical Research and obtained the funding, undoubtedly with the help of Severo Ochoa. Only because of this grant could we start our work in Spain because, at that time (1967), there was no funding for research. After learning that we had been awarded the grant and having obtained phage φ29 from Dwight Anderson, we returned to Madrid in July 1967 to start our scientific adventure.
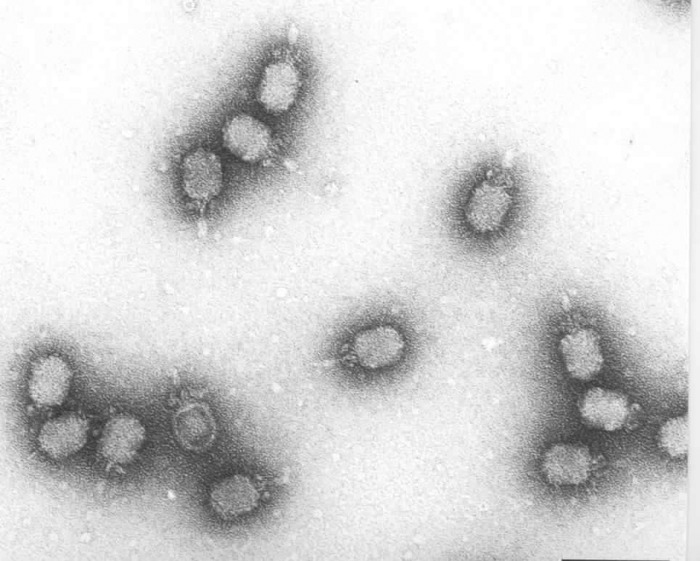
FIGURE 2.
Electron micrograph of bacteriophage φ29.
We started the φ29 work in Madrid in September 1967 at the Center of Biological Research of the Spanish National Research Council (Consejo Superior de Investigaciones Científicas (CSIC)), where we had been appointed as research scientists. The only characteristics known about phage φ29 at that time were the size of the DNA (a molecular mass of ∼12 million Da) and its morphology from an electron micrograph published by Dwight Anderson's laboratory (11). Thus, we had to start with the very basic knowledge of the phage: to perform the genetics to isolate conditional lethal mutants (temperature-sensitive and suppressor-sensitive) and to characterize the structural proteins of the phage as steps previous to the study of the morphogenesis of the phage particle, to isolate the phage DNA, and to study its transcription and replication. Fortunately, a few months after our arrival to Spain, the first predoctoral fellowships were awarded, and we could have our first students. The chairman of the Department of Biochemistry of the Faculty of Chemistry at Madrid Complutense University had invited Eladio and me to teach a course on molecular genetics, which we did. Molecular genetics was thus taught for the first time at a Spanish university. Teaching this course allowed us to select the very best students to carry out the Ph.D. work in our laboratory.
The first student, Enrique Méndez, characterized the structural proteins of the φ29 particle using the SDS-PAGE technique developed by Eladio at NYU. Jesús Avila then joined the group, and he isolated and characterized the B. subtilis RNA polymerase, which was needed for the transcription of φ29 DNA. He showed that the enzyme was composed of several subunits corresponding to E. coli β′, β, σ, and α. The paper describing these results was published in Nature (12), which made us very happy. I was also very excited when I received a letter from James Watson inviting me to attend the Cold Spring Harbor Symposium on Transcription. There, I learned that Richard Losick had obtained results similar to ours.
Another Ph.D. student, Antonio Talavera, later joined the group, and he isolated temperature-sensitive (ts) mutants, which he mapped, and characterized the mutants that were affected in DNA synthesis. Felipe Moreno, a Spanish student who was working in Paris at that time, told me that he wanted to do his Ph.D. research on the genetics of φ29. He isolated suppressor-sensitive (sus) mutants, which he also mapped. In parallel to our work, Bernie Reilly, in Dwight Anderson's laboratory, had also isolated a collection of ts and sus mutants, and we decided to combine the two collections in a linear genetic map, in which 17 genes were characterized (13). Genes 1–6 and 17, coding mainly for replication proteins, were transcribed early after infection by the B. subtilis RNA polymerase, and genes 7–16, coding for structural, morphogenetic, and lytic proteins, were transcribed later and required, in addition to the host RNA polymerase, the early gene 4 product (see below). Using the sus mutants available, several students in the laboratory characterized the morphogenetic route for the construction of the φ29 particle. Our results were very similar to those obtained in Dwight Anderson's laboratory. We also constructed a physical map relative to the genetic map by using marker rescue experiments with the restriction nuclease EcoRI. Because restriction nucleases were not commercially available at that time, José M. Lázaro had to purify the EcoRI enzyme. This was the first time that a restriction nuclease was used in Spain.
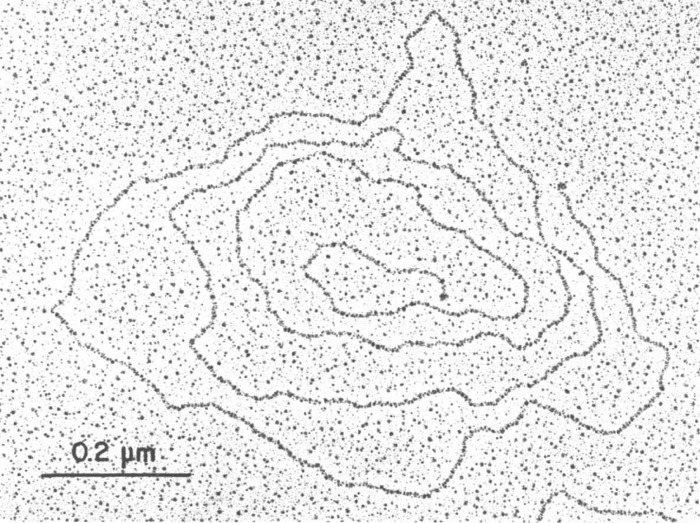
Juan Ortín, another student who joined the group, was involved in the isolation of the phage DNA. To our surprise, the DNA was not obtained in a linear form, as was expected, but as circular DNA and concatemers, as determined by electron microscopy performed by Cesar Vasquez, an Argentinean scientist who had been trained by Albrecht Kleinschmidt at NYU and who joined our laboratory for a few months. We further showed that the circles and concatemers were converted into unit-length linear DNA by treatment with a proteolytic enzyme such as trypsin. This indicated that protein was somehow involved in the formation of the circular and concatemeric DNAs. These results were published in the journal (now defunct) Nature New Biology (14). Interestingly, two years after our publication, Robinson et al. (15) published a similar result with adenovirus DNA. We were very happy to learn that our finding was not restricted to phage φ29 but also occurred in an animal virus. Rekosh et al. (16) later characterized a protein covalently linked at the 5′-ends of adenovirus DNA and proposed a model for the initiation of replication in which a free molecule of the protein would act as a primer by forming a covalent linkage with the 5′-terminal nucleotide dCMP, which would provide the 3′-OH group needed for elongation by the DNA polymerase. We also characterized a protein of 31,000 Da, the product of the viral gene 3, covalently linked to the 5′-ends of φ29 DNA (17), and we later showed that it was involved in the initiation of φ29 DNA replication. This protein was called terminal protein (TP). Fig. 3 is an electron micrograph, taken by José M. Sogo, an excellent electron microscopist, showing a linear φ29 DNA molecule with the TP at the DNA ends.
FIGURE 3.
Electron micrograph of the protein-DNA complex of bacteriophage φ29 (taken by José M. Sogo).
As I mentioned above, the product of the viral gene 4 was required for late transcription because infection with a sus4 mutant was impaired in the transcription of the late genes. Several possibilities existed, among them, that the gene 4 product was a transcriptional activator required, together with the host RNA polymerase, to transcribe the late genes. The next step was to characterize the protein product of gene 4. This was not easy because the protein was not synthesized in amounts high enough to be detected. We were lucky that genetic engineering techniques became available at that time, and we could clone the gene to overproduce and characterize the protein. This was the case not only for the gene 4 product but also for other φ29 proteins. This provided new possibilities for our φ29 work.
Another change took place in the φ29 group in the early 1970s. Eladio decided to start a new project, the study of African swine fever virus, which was a plague in Spain in general and in his homeland, Extremadura, in particular. Eladio also had in mind that by leaving the φ29 work, I would be the only group leader in this project, which we had started together (Fig. 4). This would allow me to show my colleagues that I was able to develop research on my own. At that time, the scientific work of women was very little appreciated. I worked hard, I had very good students, and Eladio helped me continuously. I became a scientist in my own right.
FIGURE 4.
Margarita Salas and Eladio Viñuela (1967).
In 1980, I was fortunate to obtain a National Institutes of Health grant, which was maintained for 24 years. This funding was crucial for the research of my group, complementing the funds I had obtained from Spanish and European sources.
I also would like to mention that every four years from 1980 to 1996 I organized in Salamanca, Spain, the International Workshop on Bacteriophages, which was funded by the European Molecular Biology Organization (EMBO). We had the best phage workers from Europe and the United States. At the first workshop, I showed my foreign colleagues that we were able to do good science in Spain. I think this was the beginning of my international recognition.
Work at the Center of Molecular Biology “Severo Ochoa”
In 1977, we moved to a new center, the Center of Molecular Biology “Severo Ochoa” (Centro de Biología Molecular “Severo Ochoa” (CBMSO)), which was built with the idea of bringing Ochoa back to Spain (Fig. 5). The scientific contribution of Eladio provided the quality of the Center. The work carried out at the CBMSO by my group was mainly the study of the mechanisms of control of φ29 DNA transcription and, in particular, of TP-primed φ29 DNA replication.
FIGURE 5.

Severo Ochoa and Margarita Salas (1986).
Control of φ29 DNA Transcription
The sequences of the four main early promoters, named A1, A2c, A2b, and C2, and that of the late promoter A3 were determined. The early promoters have −10 and −35 hexamers and are recognized by the σA-RNA polymerase, whereas the late promoter lacks the −35 hexamer and requires, in addition to the σA-RNA polymerase, the product of gene 4. We cloned gene 4 in E. coli under the control of the PL promoter of phage λ, and the protein was overproduced and purified. The setup of an in vitro transcription assay allowed us to characterize p4 as a transcriptional activator of the late promoter A3, stabilizing the σA-RNA polymerase as a closed complex. In addition, protein p4 represses promoter A2b by displacing the RNA polymerase from it, as well as promoter A2c through a mechanism that implies the simultaneous binding of p4 and RNA polymerase to the promoter, preventing the escape of the RNA polymerase from it, as shown by María Monsalve and Mario Mencía, two Ph.D. students, and Fernando Rojo, who was trained as a postdoctoral fellow in Ken Timmis' laboratory and who joined my group after coming back to Spain. Interestingly, both promoter A3 activation and promoter A2c repression require interaction between Arg-120 of p4 and the C-terminal domain of the α-subunit of B. subtilis RNA polymerase (18). We also showed that when the −35 hexamer was removed from promoter A2c, protein p4 activated the promoter instead of repressing it, whereas introduction of a −35 hexamer at promoter A3 led to its repression by p4 (19). It was shown later by Monserrat Elías-Arnanz, who joined my group after having been trained in Paul Berg's laboratory as a postdoctoral fellow, that expression of promoters A2b, A2c, and A3 is also regulated by the φ29 protein p6 (20). Protein p6 is a nucleoid-type protein that is involved in the initiation of φ29 DNA replication (see below). In addition, p6 promotes p4-mediated repression of promoter A2b and activation of promoter A3 by enhancing binding of p4 to its recognition site at promoter A3. On the other hand, p4 promotes p6-mediated repression of promoter A2c by favoring the formation of a p6-nucleoprotein complex that interferes with the binding of the RNA polymerase.
The crystal structure of p4 alone and in complex with a 41-bp DNA, including the binding site of promoter A3 and with the target sequence 5′-AACTTTTT-15 bp-AAAATGTT-3′, was determined in collaboration with Miquel Coll's laboratory. Protein p4 has a unique α/β-fold that contains a new DNA recognition motif consisting of two N-terminal β-turn substructures, or N-hooks, located at the tips of an elongated protein homodimer. The two N-hooks, one of each monomer, enter the major groove of the double helix, establishing base-specific contacts. The relevance of the different residues for DNA binding was determined by site-directed mutagenesis. The results indicated that the only base-specific contacts are between Arg-6 of p4 and the G residues at the inverted repeat of the target sequence, with the rest of the contacts being with the phosphate backbone (21). More recently, Ana Camacho, a previous Ph.D. student who later on came back to my laboratory, showed that, in addition to the Arg-6/G-specific contacts, there is DNA sequence-specific recognition through indirect readout of A tracts. Altogether, the results obtained in the study of φ29 DNA transcription indicate that its control is very sophisticated despite the small size of the phage DNA.
Protein-primed φ29 DNA Replication
The ends of φ29 DNA contain an inverted terminal repeat six nucleotides long (5′-AAAGTA-3′), as determined by Cristina Escarmís, who came to my laboratory after postdoctoral training, first with Bob Warner and then with Martin Billeter in Charles Weissmann's laboratory. Cristina sequenced DNA for the first time in Spain. The TP (266 amino acids long) is linked to the DNA ends by a phosphoester bond between the OH group of Ser-232 and 5′-dAMP. Electron microscopy analysis of the replicative intermediates synthesized in φ29-infected B. subtilis showed the presence of two types of replicating molecules, similar to those found in adenovirus-infected cells. Type I molecules are double-stranded DNA with single-stranded tails coming from one or two DNA ends, and type II molecules are partially double-stranded and partially single-stranded. Analysis of these molecules indicated that replication starts at either DNA end, non-simultaneously, and proceeds toward the other end by strand displacement.
When Miguel A. Peñalva, a Ph.D. student, incubated extracts from φ29-infected B. subtilis with [α-32P]dATP and φ29 TP-DNA as template, a 32P-labeled protein with the electrophoretic mobility of the TP was obtained. Treatment of this product with piperidine released 5′-dAMP, indicating the formation of a TP-dAMP covalent complex (22). Genes 2 and 3 were shown, both in vivo and in vitro, to be required for the initiation of φ29 DNA replication. They were initially cloned under the control of the PL promoter of phage λ to overproduce and purify the proteins. The purified protein p2 catalyzed the initiation reaction and, as shown by Luis Blanco, another Ph.D. student, had DNA polymerase activity. In addition, it had 3′–5′ exonuclease activity involved in proofreading. Similar results were obtained in Jun Ito's laboratory. We showed that the purified system consisting of TP, DNA polymerase and TP-DNA as template produced the synthesis of full-length φ29 DNA in a very processive way (23). Thus, the φ29 DNA polymerase is a unique polymerase that is able to use not only the 3′-OH group of a nucleotide but also the OH group of a specific serine residue of the TP. With primed M13 DNA as template, the φ29 DNA polymerase synthesized DNA chains greater than 70 kb (24), indicating that the polymerase has strand displacement capacity. As a result of these two properties, processivity and strand displacement capacity, in addition to its proofreading activity, the φ29 DNA polymerase has been used commercially for isothermal rolling-circle amplification (25) and whole-genome linear amplification (26) (see below).
A Sliding-back Mechanism to Initiate TP-primed DNA Replication
We showed that TP-free φ29 DNA terminal fragments were templates in vitro for the protein-primed initiation and elongation of replication, although their activity was lower than that of TP-DNA. Single-stranded oligonucleotides with sequence corresponding to the 3′ terminus of φ29 DNA also were active templates for TP-primed replication. Thus, Juan Méndez, a Ph.D. student, used as templates single-stranded oligonucleotides with changes in the first, second, or third position relative to the wild-type 3′-terminal sequence. To our surprise, initiation did not take place at the 3′-terminal nucleotide but at the second position from the 3′-end. The DNA ends were recovered by a mechanism that we called sliding back. The TP-dAMP complex formed directed by the second T at the 3′-end slides backward, locating the dAMP in front of the first 3′-terminal nucleotide of the template. Then, the next nucleotide, dAMP, is incorporated into the TP-dAMP initiation complex, again using the second T of the template as director (27). Internal initiation is not a peculiarity of the φ29 system. It also occurs in the φ29-related phages Nf and GA-1, in the Streptococcus pneumoniae phage Cp1, and in the E. coli phage PRD1, as determined in collaboration with Pedro García's and Dennis Bamford's laboratories. Internal initiation also was shown by Peter van der Vliet's laboratory to take place in adenovirus (reviewed in Ref. 28). For the initiation of φ29 DNA replication, the phage DNA polymerase forms a heterodimer with a free molecule of the TP to prime DNA synthesis at each φ29 DNA end. The two proteins in the heterodimer do not dissociate immediately after initiation. There is a so-called transition stage in which the polymerase synthesizes a five-nucleotide-long DNA while still complexed with the primer TP. After some structural change during the incorporation of nucleotides 6–9, the DNA polymerase dissociates from the primer TP when nucleotide 10 is incorporated into the nascent DNA chain (29).
Structure-Function Studies on the φ29 DNA Polymerase and Terminal Protein
At the time we started our work, the three-dimensional structure of the φ29 DNA polymerase was not available, so Luis Blanco, already a postdoctoral fellow in the laboratory and now a group leader at the CBMSO, and Antonio Bernad, a Ph.D. student, made a comparison of the amino acid sequences of prokaryotic and eukaryotic DNA-dependent DNA polymerases that led to the finding of a number of conserved motifs. We proposed that the 3′–5′ exonuclease active site of prokaryotic and eukaryotic DNA polymerases is evolutionarily conserved, formed by three N-terminal amino acid motifs that we called ExoI, ExoII, and ExoIII, invariantly containing four carboxylate groups that bind two metal ions and one tyrosine that is involved in orienting the attacking water molecule (30). Mutants lacking the carboxylic groups of Asp-12, Glu-14, Asp-66, or Asp-169 showed a 100-fold reduction of the exonuclease activity and had lost the strand displacement capacity. Other residues were analyzed by another Ph.D. student, Miguel de Vega, who showed that amino acids such as Asn-62 and Phe-65 were involved in single-stranded DNA binding and had a 10-fold reduction of the exonuclease activity but kept the strand displacement capacity. Site-directed mutagenesis of conserved amino acids in motifs DX2SLYP, KX3NSXYG, TX2GR, YXDTDS, and KXY in the C-terminal domain of φ29 DNA polymerase showed that this domain is involved in both polymerization and protein-primed initiation. In addition, we identified amino acids involved in metal binding and catalysis, DNA binding, TP binding, and dNTP interaction (reviewed in Ref. 31). Several students contributed to this work, among them, María A. Blasco, now director of the National Center of Oncological Research in Madrid.
Later, the crystal structure of φ29 DNA polymerase was determined in collaboration with Thomas (Tom) Steitz's laboratory (32). The structure provided a topological basis for its intrinsic processivity and strand displacement capacity. The main difference between φ29 DNA polymerase and other family B DNA polymerases is the presence of two additional subdomains corresponding to sequence insertions that we had identified as specifically conserved in protein-primed DNA polymerases and called TPR1 and TPR2. When we made a deletion in the TPR2 region, the resulting DNA polymerase mutant showed a decreased DNA-binding capacity and a drastic impairment in its processivity. In addition, the strand displacement capacity of the φ29 DNA polymerase was abolished in the TPR2 deletion mutant (33).
The crystal structure of the φ29 DNA polymerase/TP heterodimer, also obtained in collaboration with Tom Steitz's laboratory (34), showed three domains in the TP: the N-terminal domain, which does not interact with the DNA polymerase; the intermediate domain, which binds the DNA polymerase; and the priming domain, which contains Ser-232 and occupies the same binding cleft in the polymerase as duplex DNA does during elongation. A model was proposed for the transition from initiation to elongation according to which the TP should dissociate from the polymerase after incorporation of approximately six nucleotides, which was in good agreement with existing biochemical data. The specificity of Ser-232 in the TP was determined by a Ph.D. student, Cristina Garmendia (recently the Minister of Science and Innovation of Spain), who showed that a change of this amino acid to threonine completely abolished the primer activity.
The N-terminal domain of the TP was shown by Daniel Muñoz-Espín, then a postdoctoral fellow, to have sequence-independent DNA-binding capacity and to be responsible for the nucleoid association of the TP. In its absence, the efficiency of TP-DNA replication is severely affected. Moreover, the TP recruits the φ29 DNA polymerase to the bacterial nucleoid, and both proteins later are redistributed to enlarged helix-like structures in an MreB cytoskeleton-dependent way (35). Interestingly, we predicted nuclear localization signals within the TPs of bacteriophages from diverse families and hosts, and indeed, of seven TPs tested, those of φ29, Nf, PRD1, Bam35, and Cp1 were found by Modesto Redrejo-Rodríguez, who joined my group after a postdoctoral stay at the Gustave-Roussy Institute in Paris, to localize in the nucleus when expressed in mammalian cells. Analysis of φ29 TP led us to identify a bona fide nuclear localization signal within residues 1–37. Importantly, gene delivery into the eukaryotic nucleus was enhanced by the presence of φ29 TP attached to the 5′-DNA ends (36). These findings show a common feature of TPs from diverse bacteriophages targeting the eukaryotic nucleus and suggest a possible common function by facilitating the horizontal transfer of genes between prokaryotes and eukaryotes.
Viral Proteins p6 and p5, Essential for φ29 DNA Replication
The viral protein p6 is a histone-like protein essential for in vivo φ29 DNA initiation of replication and is highly expressed in φ29-infected cells, amounting up to 700,000 molecules/cell. The purified protein p6 stimulates the in vitro formation of the TP-dAMP initiation complex by facilitating the opening of the DNA ends and decreasing the Km for dATP. Protein p6 binds as a dimer to φ29 DNA, preferentially to the DNA ends, every 24 nucleotides in a cooperative way, forming a nucleoprotein complex. Manuel Serrano, a Ph.D. student who is now a group leader at the National Center of Oncological Research, showed that p6 binding to circular DNA restrains positive supercoiling, supporting a model in which a right-handed superhelix wraps tightly around a multimeric p6 core (37).
Gene 5 of φ29 encodes the single-stranded DNA-binding protein (SSB) p5, which is essential for elongation of replication in vivo and which is very abundant in φ29-infected cells. Crisanto Gutiérrez, who joined the group after a postdoctoral stay in Melvin DePamphilis' laboratory, showed in vitro binding of φ29 SSB to φ29 DNA replicative intermediates by electron microscopy. Binding of SSB to φ29 DNA stimulates dNTP incorporation and increases the elongation rate, mainly when φ29 DNA polymerase mutants impaired in strand displacement are used. This likely occurs because of the helix-destabilizing activity of φ29 SSB.
Phage-Host Interactions in φ29 Development
Although φ29 TP-DNA can be replicated efficiently in vitro with the four proteins already described (TP, DNA polymerase, protein p6, and SSB; see below), there are host proteins that play a role, positive or negative, in φ29 DNA replication. The early gene 56, located at the left side of gene 1, encodes p56, a small protein of 56 amino acids. We found that p56 binds to B. subtilis uracil-DNA glycosylase (UDG), an enzyme involved in the base excision repair pathway. In addition, p56 inhibits the B. subtilis UDG activity. As shown by Gemma Serrano-Heras, a Ph.D. student in the laboratory, inhibition of cellular UDG by protein p56 is a defense mechanism developed by φ29 to prevent the action of the base excision repair pathway if uracil residues arise in their single-stranded replicative intermediates (38). Protein p56 is the first description of a UDG inhibitor encoded by a non-uracil-containing viral DNA. It is likely that other DNAs that replicate by a similar mechanism of strand displacement also encode a UDG inhibitor.
On the other hand, the MreB cytoskeleton plays a crucial role in organizing φ29 DNA replication at the membrane. Thus, phage double-stranded DNA and components of the φ29 replication machinery localize in peripheral helix-like structures in a cytoskeleton-dependent way. We have shown that MreB interacts directly with the φ29 membrane protein p16.7, responsible for attaching the viral DNA at the cell membrane (39).
For φ29 development, we used B. subtilis Spo0A because the phage does not develop efficiently in B. subtilis that is able to sporulate. The Spo0A protein is the master regulator for the initiation of sporulation, and interestingly, we found that it has several binding sites in φ29 DNA. As shown by Wilfried Meijer, a postdoctoral fellow from Holland and later a research scientist of the CSIC, Spo0A suppresses φ29 development by repressing the early promoters A2c, A2b, and C2 and by preventing activation of the late promoter A3. In addition, protein Spo0A inhibits φ29 DNA replication by binding near the φ29 DNA ends, preventing the formation of the protein p6-nucleoprotein complex and thus inhibiting the initiation of φ29 DNA replication. These results explain why we had to use B. subtilis that was unable to sporulate for φ29 development.
From Molecular Biology to Biotechnology
The two intrinsic properties of the φ29 DNA polymerase, processivity and strand displacement capacity, together with its high fidelity due to the combination of high dNMP insertion discrimination and strong proofreading activity, led us to envisage φ29 DNA polymerase as an ideal tool to obtain strand displacement amplification. Using the four purified φ29 proteins described above (TP, DNA polymerase, protein p6, and SSB), Luis Blanco amplified, in vitro, small amounts (0.5 ng) of φ29 TP-DNA (19,285 bp long) by 3 orders of magnitude, obtaining 0.5 μg of DNA after 1 h of incubation at 30 °C. The fidelity of the amplified DNA was demonstrated by transfection experiments in which the infectivity of the amplified DNA, measured as the ability to produce phage particles, was identical to that of the natural φ29 DNA obtained from virions (40).
Recently, Mario Mencía, a former Ph.D. student in the laboratory who came back after a postdoctoral stay in Kevin Struhl's laboratory, obtained TP-primed amplification of heterologous DNA with a minimal replication system based on the phage φ29 DNA replication machinery, producing DNA with TP covalently attached to the 5′-ends. The amplification requires the four φ29 proteins mentioned above (TP, DNA polymerase, protein p6, and SSB). The DNA to be amplified is inserted between two sequences that are the φ29 DNA replication origins, consisting of 191 and 194 bp from the left and right ends of the phage genome, respectively (41). This method provide possibilities regarding amplification of DNA and the generation of hybrid protein-DNA molecules.
On the other hand, the ability of φ29 DNA polymerase to use circular ssDNA as template allowed asymmetric rolling-circle DNA amplification, producing single-stranded concatemeric DNA containing more than ten copies of the initial circular template (24). In a procedure developed by Amersham Biosciences/Molecular Staging, φ29 DNA polymerase is combined with random hexamer primers to achieve isothermal and faithful 104–106-fold amplification of either circular (25) or linear (26) genomes, yielding high-quality amplification products that can be either digested or sequenced directly without further purification. More recently, to enhance the amplification efficiency of φ29 DNA polymerase, Miguel de Vega, a former Ph.D. student and then a research scientist of the CSIC in my group, constructed chimeric DNA polymerases by fusing DNA-binding domains to the C terminus of the φ29 DNA polymerase (42). The addition of (helix-hairpin-helix)2 domains increases DNA binding of the hybrid polymerases without hindering their replication rate. In addition, the chimeric DNA polymerases display an improved and faithful multiply-primed DNA amplification proficiency on both circular plasmids and genomic linear DNA and are unique φ29 DNA polymerase variants with enhanced amplification performance. These chimeric DNA polymerases will contribute to make φ29 DNA polymerase-based amplification technologies one of the most powerful tools for genomics. Our research on φ29 DNA polymerase, carried out for 30 years, has allowed us to exploit the potential of this small viral enzyme, a good example of basic research applied to biotechnology.
Final Comments
I have dedicated 52 years of my life to research, seven of them as a predoctoral and postdoctoral fellow, and 45 of them as an independent investigator working on phage φ29. This work has been very rewarding to me. We have made relevant contributions to unraveling the mechanism of protein-primed φ29 DNA replication as well as the control of φ29 DNA transcription. Although the aim of my work on φ29 has been basic research, I am very happy to say that this basic science led to an important biotechnological application: the use of phage φ29 DNA polymerase for DNA amplification. This is a good example of how basic research can lead to applications that were not foreseen.
My 23 years of teaching molecular genetics at Madrid Complutense University were also very rewarding. I had brilliant students, some of whom came to my laboratory for a Ph.D. degree and are now group leaders doing excellent research.
I would like to stress the fact that the work carried out in my laboratory is the result of the dedication and ideas of the many people who have worked in the φ29 group as pre- and postdoctoral fellows during these exciting 45 years, several of whom I have mentioned in this article. I am very grateful to all of them, especially to those who have helped me in the supervision of the work. Many thanks to José M. Lázaro, who has been working with me since 1972 and who has maintained the laboratory over the years, and to my secretary, Angeles M. Villarraso, who has been with me for the past 16 years and helps me always with great efficiency. I also would like to thank the funding agencies that have supported my work: the Jane Coffin Childs Memorial Fund for Medical Research, which provided the financial support that enabled us to begin our work in Spain; the National Institutes of Health; the European Community; the Spanish Ministry of Education and Science; the Spanish Ministry of Science and Innovation; the Madrid Autonomous Community; and the Fundación Ramón Areces, which supplied an institutional grant to the CBMSO. I thank my teachers: Alberto Sols, for his teaching of enzymology; Severo Ochoa, to whom I owe my decision to do research in biochemistry and who taught us (Eladio and myself) molecular biology so that we could teach and develop it in Spain; and especially Eladio Viñuela, husband, friend, colleague, and always a teacher to me. Eladio, who is no longer with us, has been the most important person in my life, from both the personal and scientific points of view.
Acknowledgment
I dedicate this Reflections article to Eladio.
REFERENCES
- 1. Salas M., Viñuela E., Sols A. (1965) Spontaneous and enzymatically catalyzed anomerization of glucose 6-phosphate and anomeric specificity of related enzymes. J. Biol. Chem. 240, 561–568 [PubMed] [Google Scholar]
- 2. Viñuela E., Salas M., Sols A. (1963) Glucokinase and hexokinase in liver in relation to glycogen synthesis. J. Biol. Chem. 238, 1175–1177 [PubMed] [Google Scholar]
- 3. Salas M., Viñuela E., Sols A. (1963) Insulin-dependent synthesis of liver glucokinase in the rat. J. Biol. Chem. 238, 3535–3538 [PubMed] [Google Scholar]
- 4. Salas M., Smith M. A., Stanley W. M., Jr., Wahba A. J., Ochoa S. (1965) Direction of reading of the genetic message. J. Biol. Chem. 240, 3988–3995 [PubMed] [Google Scholar]
- 5. Smith M. A., Salas M., Stanley W. M., Jr., Wahba A. J., Ochoa S. (1966) Direction of reading of the genetic message. II. Proc. Natl. Acad. Sci. U.S.A. 55, 141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stanley W. M., Jr., Salas M., Wahba A. J., Ochoa S. (1966) Translation of the genetic message. I. Factors involved in the initiation of protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 56, 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salas M., Hille M. B., Last J. A., Wahba A. J., Ochoa S. (1967) Translation of the genetic message. II. Effect of initiation factors on the binding of formyl-methionyl-tRNA to ribosomes. Proc. Natl. Acad. Sci. U.S.A. 57, 387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Last J. A., Stanley W. M., Jr., Salas M., Hille M. B., Wahba A. J., Ochoa S. (1967) Translation of the genetic message. IV. UAA as a chain termination codon. Proc. Natl. Acad. Sci. U.S.A. 57, 1062–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shapiro A. L., Viñuela E., Maizel J. V., Jr. (1967) Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem. Biophys. Res. Commun. 28, 815–820 [DOI] [PubMed] [Google Scholar]
- 10. Viñuela E., Salas M., Ochoa S. (1967) Translation of the genetic message. III. Formylmethionine as initiator of proteins programmed by polycistronic messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 57, 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson D. L., Hickman D. D., Reilly B. E. (1966) Structure of Bacillus subtilis bacteriophage φ29 and the length of φ29 deoxyribonucleic acid. J. Bacteriol. 91, 2081–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Avila J., Hermoso J. M., Viñuela E., Salas M. (1970) Subunit composition of B. subtilis RNA polymerase. Nature 226, 1244–1245 [DOI] [PubMed] [Google Scholar]
- 13. Mellado R. P., Moreno F., Viñuela E., Salas M., Reilly B. E., Anderson D. L. (1976) Genetic analysis of bacteriophage φ29 of Bacillus subtilis: integration and mapping of reference mutants of two collections. J. Virol. 19, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ortín J., Viñuela E., Salas M., Vasquez C. (1971) DNA-protein complex in circular DNA from phage φ29. Nat. New Biol. 234, 275–277 [DOI] [PubMed] [Google Scholar]
- 15. Robinson A. J., Younghusband H. B., Bellett A. J. (1973) A circular DNA-protein from adenoviruses. Virology 56, 54–69 [DOI] [PubMed] [Google Scholar]
- 16. Rekosh D. M., Russell W. C., Bellet A. J., Robinson A. J. (1977) Identification of a protein linked to the ends of adenovirus DNA. Cell 11, 283–295 [DOI] [PubMed] [Google Scholar]
- 17. Salas M., Mellado R. P., Viñuela E. (1978) Characterization of a protein covalently linked to the 5′ termini of the DNA of Bacillus subtilis phage φ29. J. Mol. Biol. 119, 269–291 [DOI] [PubMed] [Google Scholar]
- 18. Rojo F., Mencía M., Monsalve M., Salas M. (1998) Transcription activation and repression by interaction of a regulator with the α subunit of RNA polymerase: the model of phage φ29 protein p4. Prog. Nucleic Acid Res. Mol. Biol. 60, 29–46 [DOI] [PubMed] [Google Scholar]
- 19. Monsalve M., Calles B., Mencía M., Salas M., Rojo F. (1997) Transcription activation or repression by phage φ29 protein p4 depends on the strength of the RNA polymerase-promoter interactions. Mol. Cell 1, 99–107 [DOI] [PubMed] [Google Scholar]
- 20. Elías-Arnanz M., Salas M. (1999) Functional interactions between a phage histone-like protein and a transcriptional factor in regulation of φ29 early-late transcriptional switch. Genes Dev. 13, 2502–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Badía D., Camacho A., Pérez-Lago L., Escandón C., Salas M., Coll M. (2006) The structure of φ29 transcription regulator p4-DNA complex reveals a novel DNA binding motif. Mol. Cell 22, 73–81 [DOI] [PubMed] [Google Scholar]
- 22. Peñalva M. A., Salas M. (1982) Initiation of phage φ29 DNA replication in vitro: formation of a covalent complex between the terminal protein, p3, and 5′-dAMP. Proc. Natl. Acad. Sci. U.S.A. 79, 5522–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blanco L., Salas M. (1985) Replication of φ29 DNA with purified terminal protein and DNA polymerase: synthesis of full-length φ29 DNA. Proc. Natl. Acad. Sci. U.S.A. 82, 6404–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blanco L., Bernad A., Lázaro J. M., Martín G., Garmendia C., Salas M. (1989) Highly efficient DNA synthesis by the phage φ29 DNA polymerase. Symmetrical model of DNA replication. J. Biol. Chem. 264, 8935–8940 [PubMed] [Google Scholar]
- 25. Dean F. B., Nelson J. R., Giesler T. L., Lasken R. S. (2001) Rapid amplification of plasmid and phage DNA using φ29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 11, 1095–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dean F. B., Hosono S., Fang L., Wu X., Faruqi A. F., Bray-Ward P., Sun Z., Zong Q., Du Y., Du J., Driscoll M., Song W., Kingsmore S. F., Egholm M., Lasken R. S. (2002) Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. U.S.A. 99, 5261–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Méndez J., Blanco L., Esteban J. A., Bernad A., Salas M. (1992) Initiation of φ29 DNA replication occurs at the second 3′ nucleotide of the linear template: a sliding-back mechanism for protein-primed DNA replication. Proc. Natl. Acad. Sci. U.S.A. 89, 9579–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salas M., Miller J. T., Leis J., DePamphilis M. L. (1996) in DNA Replication in Eukaryotic Cells (DePamphilis M., ed) pp. 131–176, Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Méndez J., Blanco L., Salas M. (1997) Protein-primed DNA replication: a transition between two modes of priming by a unique DNA polymerase. EMBO J. 16, 2519–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. (1989) A conserved 3′→5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59, 219–228 [DOI] [PubMed] [Google Scholar]
- 31. Blanco L., Salas M. (1996) Relating structure to function in φ29 DNA polymerase. J. Biol. Chem. 271, 8509–8512 [DOI] [PubMed] [Google Scholar]
- 32. Kamtekar S., Berman A. J., Wang J., Lázaro J. M., de Vega M., Blanco L., Salas M., Steitz T. A. (2004) Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage φ29. Mol. Cell 16, 609–618 [DOI] [PubMed] [Google Scholar]
- 33. Rodríguez I., Lázaro J. M., Blanco L., Kamtekar S., Berman A. J., Wang J., Steitz T. A., Salas M., de Vega M. (2005) A specific subdomain in φ29 DNA polymerase confers both processivity and strand displacement capacity. Proc. Natl. Acad. Sci. U.S.A. 102, 6407–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kamtekar S., Berman A. J., Wang J., Lázaro J. M., de Vega M., Blanco L., Salas M., Steitz T. A. (2006) The φ29 DNA polymerase: protein-primed structure suggests a model for the initiation to elongation transition. EMBO J. 25, 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muñoz-Espín D., Holguera I., Ballesteros-Plaza D., Carballido-López R., Salas M. (2010) Viral terminal protein directs early organization of phage DNA replication at the bacterial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 107, 16548–16553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Redrejo-Rodríguez M., Muñoz-Espín D., Holguera I., Mencía M., Salas M. (2012) Functional eukaryotic nuclear localization signals are widespread in terminal proteins of bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 109, 18482–18487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Serrano M., Salas M., Hermoso J. M. (1990) A novel nucleoprotein complex at a replication origin. Science 248, 1012–1016 [DOI] [PubMed] [Google Scholar]
- 38. Serrano-Heras G., Bravo A., Salas M. (2008) Phage φ29 protein p56 prevents viral DNA replication impairment caused by uracil excision activity of uracil-DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 105, 19044–19049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muñoz-Espín D., Daniel R., Kawai Y., Carballido-López R., Castilla-Llorente V., Errington J., Meijer W. J., Salas M. (2009) The actin-like MreB cytoskeleton organizes viral DNA replication in bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 13347–13352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blanco L., Lázaro J. M., de Vega M., Bonnin A., Salas M. (1994) Terminal protein-primed DNA amplification. Proc. Natl. Acad. Sci. U.S.A. 91, 12198–12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mencía M., Gella P., Camacho A., de Vega M., Salas M. (2011) Terminal protein-primed amplification of heterologous DNA with a minimal replication system based on phage φ29. Proc. Natl. Acad. Sci. U.S.A. 108, 18655–18660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Vega M., Lázaro J. M., Mencía M., Blanco L., Salas M. (2010) Improvement of φ29 DNA polymerase amplification performance by fusion of DNA binding motifs. Proc. Natl. Acad. Sci. U.S.A. 107, 16506–16511 [DOI] [PMC free article] [PubMed] [Google Scholar]