Background: Knowledge of the structural features of P450 2C19 that underlie its distinct roles in human drug metabolism is lacking.
Results: The structure of P450 2C19 was determined by x-ray crystallography.
Conclusion: The structure of the enzyme exhibits features that distinguish it from closely related P450s 2C8 and 2C9.
Significance: Structural characterization of P450 2C19 contributes to a better understanding of its role in drug clearance.
Keywords: Cytochrome P450, Drug Metabolism, Membrane Enzymes, Molecular Pharmacology, Protein Structure
Abstract
To identify the structural features underlying the distinct substrate and inhibitor profiles of P450 2C19 relative to the closely related human enzymes, P450s 2C8 and 2C9, the atomic structure (Protein Data Bank code 4GQS) of cytochrome P450 2C19 complexed with the inhibitor (2-methyl-1-benzofuran-3-yl)-(4-hydroxy-3,5-dimethylphenyl)methanone (Protein Data Bank chemical component 0XV) was determined to 2.87 Å resolution by x-ray crystallography. The conformation of the peptide backbone of P450 2C19 is most similar to that of P450 2C8, but the substrate-binding cavity of P450 2C8 is much larger than that of P450 2C19 due to differences in the amino acid residues that form the substrate-binding cavities of the two enzymes. In contrast, the substrate-binding cavity of P450 2C19 is much more similar in size to that of the structure of the P450 2C9 flurbiprofen complex than to that of a modified P450 2C9 or that of P450 2C8. The cavities of the P450 2C19 0XV complex and the P450 2C9 flurbiprofen complex differ, however, because the helix B-C loops of the two enzymes are dissimilar. These conformational differences reflect the effects of adjacent structural elements that interact with the B-C loops and that differ between the two enzymes. The availability of a structure for 2C19 will facilitate computational approaches for predictions of substrate and inhibitor binding to this enzyme.
Introduction
P4503 2C19 is a human cytochrome P450 monooxygenase that contributes significantly to metabolic clearance of several widely prescribed drugs (1, 2). Allelic variation in human populations leads to significant differences among individuals in P450 2C19 expression and metabolic capacity that underlie poor, extensive and ultra-fast phenotypes for drug clearance. These phenotypes are evident for the clearance of S-mephenytoin (3) as well as for a number of antidepressants and serotonin reuptake inhibitors (1, 4, 5). Inter-individual differences in P450 2C19 metabolic capacity can also alter the clearance of omeprazole and structurally related inhibitors of gastric acid secretion leading to differences in clinical outcomes and dosing requirements (6). Omeprazole has also been reported to inactivate P450 2C19, which could contribute to reported drug-drug interactions with clopidogrel (7, 8). Efficient metabolism of clopidogrel by P450 2C19 is important for conversion of this pro-drug to an efficacious inhibitor of platelet aggregation (9–12). Patients carrying loss-of-function alleles for CYP2C19 who were treated for acute myocardial infarction with clopidogrel exhibited higher rates of subsequent cardiovascular events (13). A number of studies support this concern as well as for the adverse effects of loss of function alleles on the efficacy of clopidogrel to suppress stent thrombosis (14). In contrast, ultrafast metabolizers having the CYP2C19*17 allele can experience increased activation of clopidogrel that is associated with an improved therapeutic outcome (15) but also with an increased risk for bleeding (16).
P450 2C19 is one of four P450 2C monooxygenases that are encoded by a cluster of genes that arose from gene duplication and divergence on chromosome 10. Three of these P450s, 2C8, 2C9, and 2C19, are expressed in human liver and significantly contribute in different ways to hepatic capacity to metabolize drugs (1). These differences in the contributions of the three enzymes to drug clearance ultimately reflect structural differences that confer selectivity for substrates and sites of metabolism for shared substrates. Crystal structures of P450s 2C8 (17, 18) and 2C9 (19, 20) reveal extensive amino acid differences for side chains that shape the substrate-binding cavities of the two enzymes as well as conformational differences that underlie differences in their substrate and inhibitor profiles. Moreover, structures of P450 2C9 exhibit significant differences between the substrate-binding site observed for the structure of the P450 2C9 flurbiprofen complex and active sites observed for structures obtained using a modified P450, 2C9m7, with or without S-warfarin bound in the active site (20). P450 2C9m5 has five amino acid substitutions in the helices F, F′, and G′ and intervening connections, K206E, I215V, C216Y, S220P, P221A, I222L, and I223L, that facilitate crystallization (19). P450s 2C19 and 2C9 differ for only 43 out of 490 amino acids, and only two of these differences, V208L and I362L, are predicted to reside inside the active site cavities of the two enzymes based on the structures of P450 2C9. Characterization of chimeras derived from P450s 2C9 and 2C19 (21–25) indicated that most amino acid differences conferring the distinct catalytic properties of one enzyme to the other reside outside the active sites of 2C9 structures. Therefore, crystallization of the P450 2C19 was undertaken to obtain a structure for P450 2C19 to better understand how the unique catalytic properties of P450 2C19 arise.
This study reports the structure of P450 2C19 in complex with inhibitor 0XV (Fig. 1) and compares the structure of 2C19 to structures determined for P450s 2C8 and 2C9. 0XV is structurally and chemically related to the potent P450 2C9 inhibitor benzbromarone ((2-ethyl-1-benzofuran-3-yl)-(4-hydroxy-3,5-dibromophenyl)methanone) (26). The high binding affinity of P450 2C9 for benzbromarone at physiological pH, where the electron withdrawing properties of the bromine substituents lead to ionization of the phenol, is likely to reflect the ability of Arg-108 to complement the charge of the phenolate moiety of benzbromarone as it does for the carboxyl group of flurbiprofen. P450 2C9 oxidizes a large number of organic anions typified by nonsteroidal anti-inflammatory drugs, such as diclofenac, indomethacin, ibuprofen, naproxen, and flurbiprofen (4). In contrast, substitution of a methyl group for each of the two bromine atoms raises the pKa value of the phenol and diminishes the binding affinity of the resulting compound for P450 2C9. Substitution of an iodine atom for each bromine atom and substitution of a methyl group for the ethyl group of benzbromarone produced DMI (2-methyl-1-benzofuran-3-yl)-(4-hydroxy-3,5-diiodophenyl)methanone, a more potent inhibitor. The relatively poor binding affinity of P450 2C19 for these anionic inhibitors led Locuson et al. (26) to examine whether benzbromarone analogues that exhibit higher predicted pKa values for the phenol moiety would function as better inhibitors of P450 2C19 than benzbromarone. Their study identified dimethylbenzarone ((2-ethyl-1-benzofuran-3-yl)-(4-hydroxy-3,5-dibromophenyl)methanone), where the bromines are replaced with methyl groups, to be one of the most potent inhibitors of P450 2C19 with a Ki of 33 nm. The molecule complexed with P450 2C19 in the structure 0XV differs from dimethylbenzarone by replacement of the 2-ethyl substituent on the benzofuran ring with a methyl group and is the dimethyl analogue of DMI. 0XV exhibits a highly similar apparent Ki of 35 ± 5 nm for inhibition of 3-O-methylfluorescein demethylation catalyzed by P450 2C19 as that of dimethylbenzarone. The structure of P450 2C19 0XV complex defines interactions between the enzyme and 0XV and reveals that Arg-108 is located on the outer surface of the helix B-C loop where it is not able to interact with 0XV. This is the first structure of P450 2C19 deposited in the Protein Data Bank (PDB code 4GQS) and provides information regarding structural features that contribute to the unique functional properties of the enzyme.
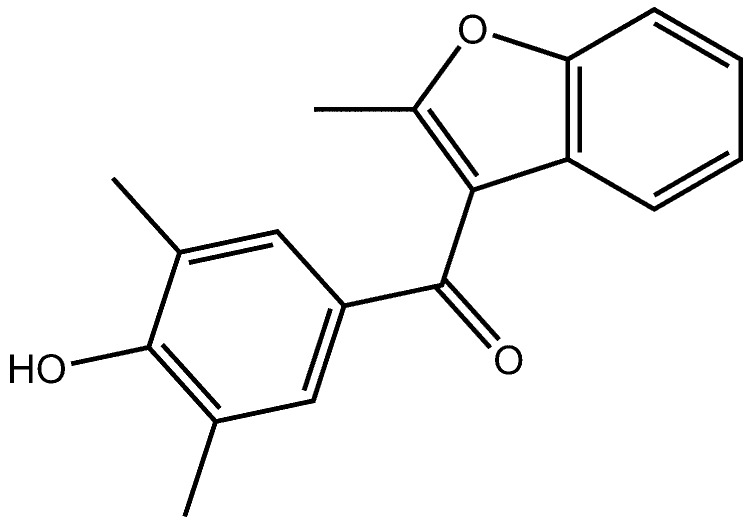
FIGURE 1.
Structure of 0XV.
MATERIALS AND METHODS
Protein Expression, Purification, and Crystallization of P450 2C19
For crystallization, a plasmid was constructed for expression of P450 2C19 in Escherichia coli with modified N and C termini using a strategy described previously for construction of the expression plasmid, P450 2C9dH, which was used for determination of the PDB code 1R9O structure of flurbiprofen complexed with P450 2C9 (20). The P450 2C19dH plasmid was constructed from a cDNA corresponding to the CYP2C19*1B allele (27). This allele corresponds to the major CYP2C19 allele (UniProtKB/Swiss-Prot code P33261). P450 2C19*1A differs from P450 2C19*1B due to a nonsynonymous single nucleotide polymorphism (refSNP rs3758581) that changes codon 331 for valine in the CYP2C19*1B gene to a codon for isoleucine, which is present in NCBI reference sequence for the CYP2C19*1A gene (NG_008384.1). The corresponding proteins do not appear to differ in their catalytic properties (28, 29). A three-fragment ligation reaction was used to insert two fragments, generated by SacI/SphI and SphI/PvuII digestion of pCW2C19*1B plasmid (27), into the pCW2C5LVdH expression vector (30) digested with SacI and PvuII. As a result, the protein is expressed with a modified N-terminal sequence, MAKKT upstream of Ser-23, and substitution of an isoleucine residue and four histidine residues for the native C-terminal valine residue of CYP2C19. The addition of the histidine tag facilitates purification. The modification of the N terminus eliminates a roughly 20-amino acid transmembrane domain and modifies the linker region preceding a proline-rich motif at the beginning of the catalytic domain. E. coli strain DH5α was transformed with pCW2C19dH and pGro7 (Takara) for expression of the proteins. The transformed strain was propagated, and the expression of P450 2C19 was induced as described previously for P450 1A2 (31). The presence of the pGro7 plasmid leads to elevated expression of the chaperones GroEL and GroES, which increases the yield of P450 2C19dH. P450 2C19dH was purified for crystallization in a manner similar to 2C9dH (20) by sequential chromatography on nickel nitrotriloacetate-agarose (Qiagen) and carboxymethyl-Sepharose (Sigma). The concentration of the enzyme was determined from the intensity of the Soret band of the reduced carbon monoxide complex using visible absorption difference spectroscopy (32). The protein was homogeneous as judged by SDS-PAGE, and the preparation exhibited a low spin UV-visible absorption spectrum with a ratio of the absorbance at 418 to that 280 nm of ∼2.
The protein was successfully crystallized by hanging drop vapor diffusion. The protein buffer contained 50 mm potassium phosphate, pH 7.4, 11 mm Nonidet P-40, 20% v/v glycerol, 250 mm NaCl, 1 mm tetrasodium EDTA, and 1 mm dithiothreitol. The protein solution was supplemented with 20 mm 0XV in methanol to a final concentration of 2.5 mm 0XV. The drop consisted of an equal volume of the protein solution containing 300 μm P450 2C19 and a precipitant solution containing 20% w/v polyethylene glycol monomethyl ether 2000, 10% v/v isopropyl alcohol, and 100 mm HEPES, pH 7.5. The hanging drop was equilibrated against the precipitant solution containing 20% v/v glycerol and 0.5 m NaCl.
Data Collection and Structure Solution
Crystals of 2C19dH complexed with 0XV diffracted to greater than 2.9 Å resolution. The diffraction images were integrated and scaled using IMOSFLM and SCALA (33) in the H32 space group 155. Integration and scaling statistics are shown in Table 1. Molecular replacement identified four molecules in the asymmetric unit using PHASER (34) with the structure of 2C9dH flurbiprofen complex, PDB code 1R9O, as the search model. The structural model was refined against data collected from a single crystal at the Stanford Synchrotron Radiation Lightsource, Beamline 11-1 to a resolution of 2.87 Å. The model of the protein structure was built and adjusted using COOT (35) and refined for the four molecules in the asymmetric unit. The software suite, Crystallography and NMR System, version 1.3 (36), was used for refinement of the model by conjugate gradient minimization, simulated annealing, and restrained individual B-factor refinement.
TABLE 1.
Data collection and refinement statistics
1 Values for the highest resolution shell, 3.03 to 2.87 Å, are shown in parentheses.
2 Ramachandran plot is based on analysis using Molprobity (45); 92.2% of residues are in favored regions, and 99.7% of residues are in allowed regions. Five outliers were noted.
3 The polypeptide chain corresponds to residues 29–490 when numbered according to the native protein as well as His-491 and His-492 of chain A. There was insufficient density to model residues 219–224 for chain B, residues 132–141 for chain D, and the side chains for residues 278 and 279 of chain C, and residues 221–226 for chain D.
4 Values are for all waters.
Chain A was defined from residue 29 to 490. Additionally, His-491 and His-492 of the C-terminal expression tag were built for chain A, but the electron density map was not sufficiently definitive to build the histidine expression tag following residue 490 for chains B, C, or D. Similarly, the substitute N-terminal expression tag and linker region preceding amino acid residue 29 could not be built due to conformational disorder. The residue numbering corresponds to that of the native CYP2C19 reference sequence. Noncrystallographic symmetry restraints were used, but they were removed for residues 36–43 and 210–230 because these regions differ between the four molecules in the asymmetric unit as a result of different protein-protein interactions in the crystal lattice. The final model was refined without noncrystallographic symmetry restraints. Residues of 132–141 of chain D in the loop between helices C and D as well as residues 219–224 of chain B between helices F′ and G could not be modeled because of conformational disorder. Additionally, side chains for residues 221–224 of chain D were not modeled, but the trace of the peptide backbone was constructed. Chain A was best defined by computed electron density maps and exhibited the lowest average B values of the four chains (Table 1) with increasing average B values evident for chains C, B, and D, respectively.
RESULTS AND DISCUSSION
Comparison of the Four Molecules in the Asymmetric Unit
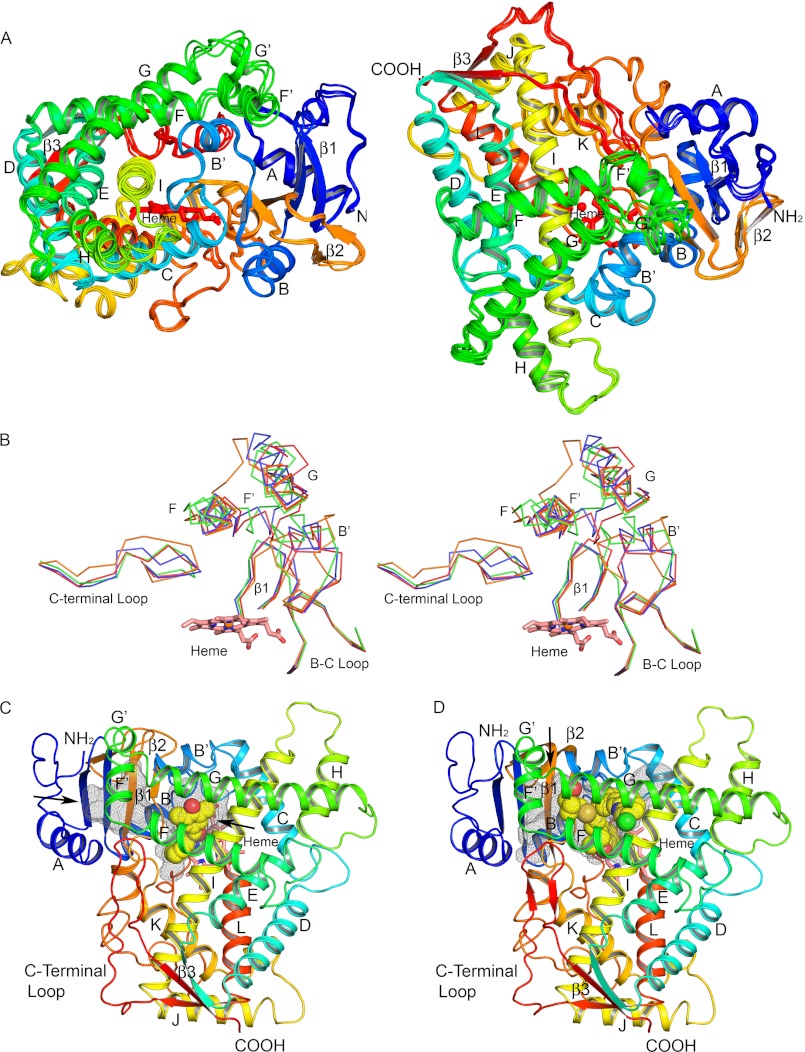
The tertiary structure of the P450 2C19 0XV complex (Fig. 2, A and C) exhibits 12 α-helices designated A–L and three β-sheets designated 1–3 that are typically seen for P450s. Additionally, the helix B-C loop exhibits a B′-helix, and F′- and G′-helices were evident for the loop between helices F and G. These additional helices are generally seen for structures of other family 2 P450s. The P450 2C19 0XV complex crystallized in the H32 space group with four molecules in the asymmetric unit. The root mean square deviations for Cα atoms of chains B, C, and D relative to chain A were generally less than 0.84 Å, but larger differences of 3.5 to 5.5 Å were observed for Cα positions in the helix F′ and G′ region and the adjacent portion of the polypeptide chain preceding helix A (Fig. 2A). This divergence reflects the conformational flexibility of these regions and the effects of interactions with other protein molecules in the crystal lattice that differ for each molecule of P450 2C19 in the asymmetric unit.
FIGURE 2.
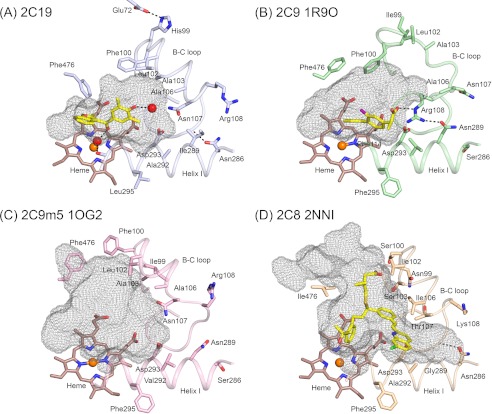
Secondary and tertiary structure of P450 2C19. A, four chains in the asymmetric unit are superimposed, and each chain is colored from blue at the N terminus to red at the C terminus. Helices are designated by letters, and sheets are numbered. The heme prosthetic group is depicted as a stick figure colored red. The chains differ most extensively between helices F′ and G′ and adjacent regions. These conformational differences can be attributed to distinct lattice packing interactions for each chain that can restrain flexible regions to different extents in the crystal. B, traces for Cα atoms of divergent regions evident when the structures of P450s 2C8, PDB code 2NNI chain A (blue trace), 2C9, PDB code 1R9O (orange trace), and 2C9m7, PDB code 1OG2 chain A (green trace), are overlaid on the structure of chain A of the P450 2C19 0XV complex (red trace). Side-by-side images are shown for cross-eyed stereo viewing. C, structure of the P450 2C19 0XV complex has two internal cavities depicted as mesh surfaces. 0XV is rendered as atomic spheres with carbons and oxygens colored yellow, and red occupies the active site cavity above the surface of the heme. A second cavity, which may serve as the substrate access channel, is located under helix F′ and above sheet β-1. D, in contrast, the active site of P450 2C8 (PDB code 2NNI) encompasses both cavities seen in the structure of the P450 2C19 0XV complex. Montelukast, shown as atomic spheres with carbons colored yellow, occupies three branches of the larger active site cavity. Oxygen, nitrogen, chlorine, iron, and sulfur atoms are colored red, blue, green, orange, and gold, respectively. Arrows indicate open solvent channels to the interior of the protein in C and D.
Comparison of the Cα Trace of P450 2C19 with That of P450s 2C8 and 2C9
Interestingly, the Cα trace for P450 2C19 most closely resembles that of P450 2C8, including the helix F-G and helix B-C regions where the 1OG2 and 1R9O structures of P450s 2C9m7 and 2C9 differ extensively from the structure of P450 2C19 and from each other (Fig. 2B). This is remarkable because the amino acid sequence of 2C19 diverges more from 2C8 (25% of residues differ) than from 2C9 (<10% of residues differ). Differences between Cα positions for P450s 2C19 and 2C8 that exceeded 3.0 Å were localized to helix G′ regions of the proteins.
Internal Cavities
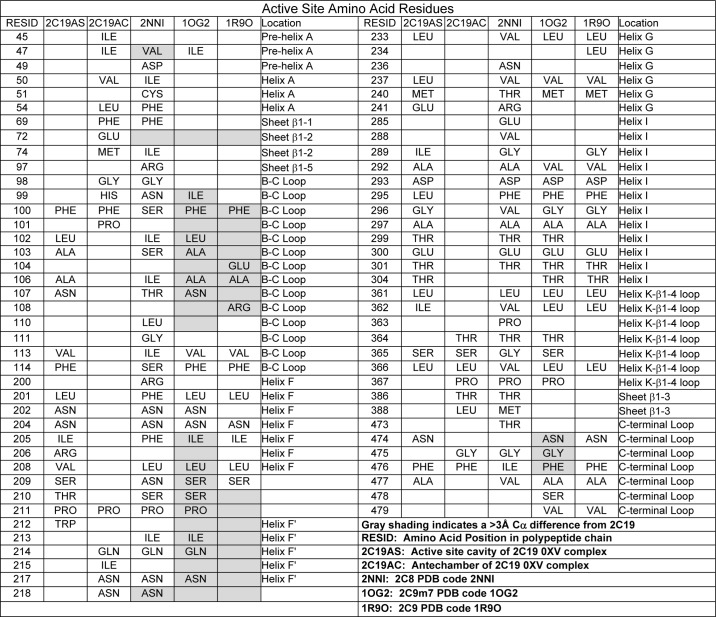
The structure of P450 2C19 exhibits two internal cavities that are depicted in Fig. 2C by mesh surfaces computed using Voidoo with a 1.4-Å probe (37). 0XV resides in the cavity that is positioned above the surface of the heme where the reactive intermediate is formed during catalysis. The second cavity forms an adjacent antechamber located under helix F′ and above β-sheet 1. This is similar to the two cavities observed for the structure (PDB code 3QM4) of the prinomastat complex of P450 2D6 (38). The antechamber may represent a portion of a substrate access channel, and there is an open solvent channel exiting between helix F′ and the N-terminal loop preceding helix A (Fig. 2C, arrow). The antechamber is separated from the active site cavity by the close convergence of the side chains of Phe-100 on the helix B-C loop, Pro-211 in the turn between helix F and helix F′, Leu-366 on the connection between helix K and β-strand 1–4, and Phe-476 at the apex of the turn in the C-terminal loop following helix L. These and other residues forming the boundaries of the two cavities are summarized and compared with residues that line the cavities of P450s 2C8 and 2C9 (Table 2).
TABLE 2.
Active site amino acid residues
In contrast, one large substrate-binding cavity is evident for structures of P450 2C8 as illustrated by the structure of the P450 2C8 montelukast complex (Fig. 2D) and observed for complexes of P450 2C8 with felodipine (PDB code 2NNJ), troglitazone (PDB code 2VNO), or two molecules of 9-cis-retinoic acid (PDB code 2NNH) bound in the active site (18) and with no ligand (PDB code 1PQ2) bound in the active site (17). The reduced sizes of amino acid residues Ser-100, Val-266, and Ile-476 of P450 2C8, which correspond to three of the four residues forming the constriction between the two cavities in 2C19, lead to a single large active site cavity in P450 2C8 without significant differences in Cα positions of the polypeptide backbone. This increase in the volume of the active site is farther augmented by the reduction in the sizes of other residues residing in the cavity. The larger active site cavity of P450 2C8 accommodates montelukast (Mr 586), which is significantly larger than 0XV (Mr 280) and can also bind two molecules of retinoic acid (Mr 300) simultaneously as evident in the PDB code 2NNH structure (18). Additionally, 30 of the 50 residues that line the cavities of the two enzymes differ (Table 2), indicating a significant sequence divergence for the active site residues of the two enzymes that exceeds the overall sequence divergence of 25% exhibited for their complete sequences. This difference of active site residues contributes to functional differences between the two enzymes by altering the number and topology of hydrophobic and polar interactions for substrates as well as the relative sizes of their substrate-binding cavities.
A comparison of the structure of the P450 2C19 0XV complex to structures of apo 2C9m7 (PDB code 1OG2) and the 2C9 flurbiprofen complex (PDB code 1R9O) indicates significant deviations of greater than 3.0 Å for equivalent Cα carbons for the portions of these structures that form the outer surfaces of their substrate-binding cavities (Table 2 and Fig. 2B). These divergent regions include helices F, F′, G′, and G and their connections together with the adjacent turn between the first two strands of β-sheet 1 (Fig. 2B) as well as their helix B-C loops and the turns in their C-terminal loops. Moreover, the P450 2C9m7 1OG2 and 2C9 1R9O structures also differ significantly from each other in these regions. In this regard, the active site cavity is much larger for the 2C9m7 1OG2 structure than that of P450 2C19 0XV complex or the 1R9O structure of the P450 2C9 flurbiprofen complex. This reflects in part a shift in the positions of residues 100 and 476 in the 2C9m5 1OG2 structure (Figs. 3A and 4, B and C), which correspond to residues that form the constriction between the active site and the antechamber in the structure of the P450 2C19 0XV complex. A second structure of P450 2C9m7, PDB code 1OG5, which is very similar to the P450 2C9m7 1OG2 structure, has a molecule of S-warfarin located in the distal portion of the active site cavity (19) that corresponds to the antechamber in the structure of P450 2C19. The flexibility indicated from the comparison of the two structures of P450 2C9 and the presence of the antechamber in the structure of P450 2C19 suggests that residues 100 and 473 might move into the antechamber region of P450 2C19 to open the substrate access channel and to expand the active site cavity of the enzyme to adapt for substrate binding.
FIGURE 3.
A and B, two views of a 2|Fo| − |Fc| σA weighted omit electron density map contoured at 1σ around the model for 0XV and an adjacent water molecule. Amino acid residues contacting 0XV or the water molecule are displayed as stick figures with carbons colored light blue. A portion of helix I is depicted as a cartoon. 0XV and the heme prosthetic group are shown as stick figures, with carbons colored yellow and brown, respectively. Oxygen, nitrogen, and iron atoms are colored red, blue, and orange, respectively. A water molecule represented as a red sphere is hydrogen-bonded (dashed line) to the 0XV hydroxyl moiety. Corresponding residues in the 2C9 1R9O (A and B, green carbons) and 2C9m7 1OG2 structures (A, pink carbons) are shown for residues that differ by amino acid side chain and/or Cα position. C displays an alternative pose for 0XV positioned for oxidation of a methyl group on the phenol moiety. Although this pose is consistent with the formation of one of two alternative products (26), the alternative pose does not fit the density as well as the pose shown in A and B.
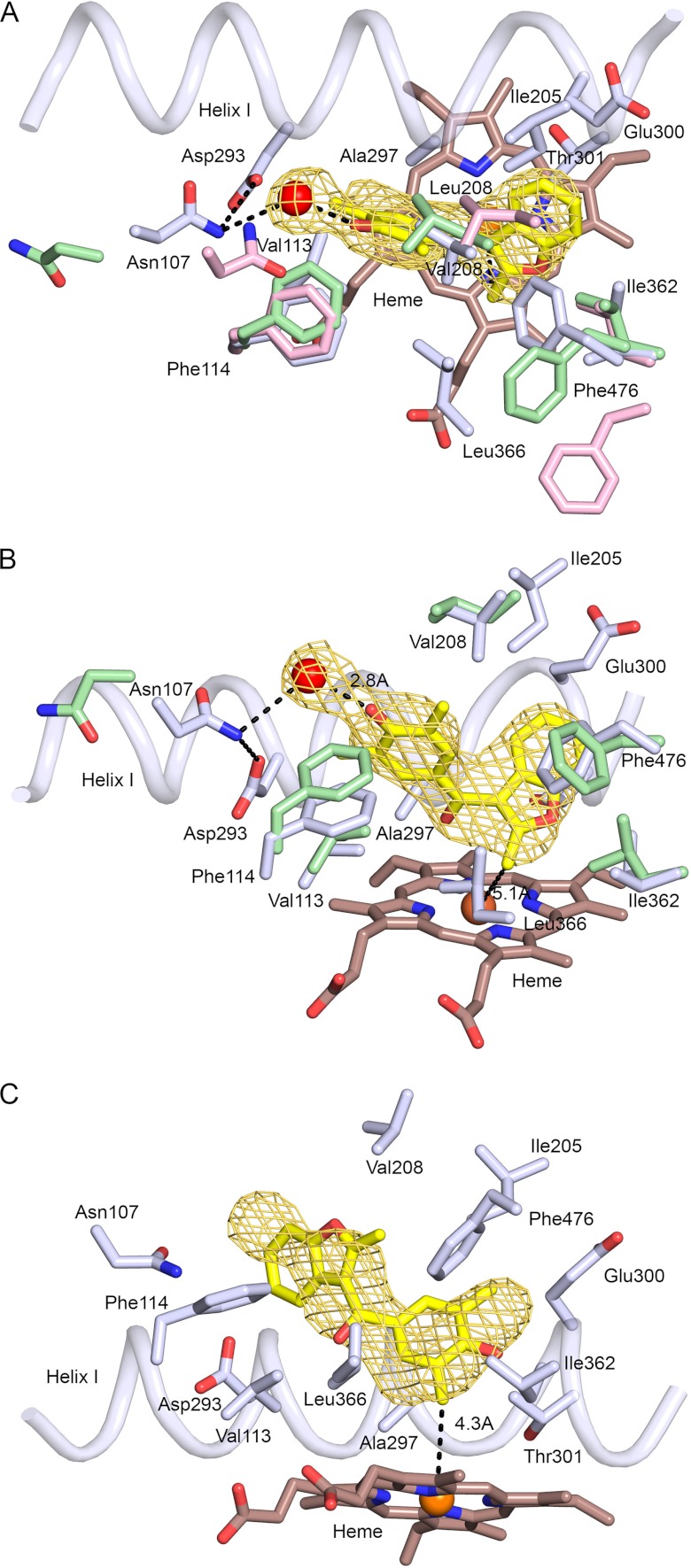
FIGURE 4.
Conformational differences between helix B-C loops significantly alter the active site cavities of the structure of the P450 2C19 0XV complex (A), the structure of the flurbiprofen complex of 2C9, PDB code 1R9O (B), the apo 2C9m7 structure, PDB code 1OG2 (C), and the 2C8 structure, PDB code 2NNI, of the montelukast complex (D). Substrate-binding cavities were calculated with Voidoo and rendered as mesh surfaces. A water molecule represented as a red sphere is hydrogen-bonded (dashed line) to the 0XV hydroxyl moiety, and a second water molecule is bound to the heme iron of chain A, where it forms hydrogen bonds (dashed lines) with the carbonyl of 0XV and the carbonyl of Ala-297 of 2C19. Additional hydrogen bonds discussed in the text are indicated by dashed lines. Amino acid residues that contribute to differential substrate selectivity and/or that directly shape the distal portions of the cavity are displayed as sticks. Color coding is the same as described for Figs. 2 and 3 with the additional use of yellow and purple to color the carbon and fluorine atoms, respectively, of flurbiprofen in the 2C9 1R9O structure.
Interactions between 0XV and P450 2C19 Active Site Residues
0XV occupies a portion of the active site cavity that is closest to the heme iron (Fig. 3, A and B). This portion of the active site is highly similar to that of the 1R9O structure of the P450 2C9 flurbiprofen complex in the identity of the amino acid residues that contact 0XV and their corresponding Cα-positions. Exceptions are shown in Fig. 3, A and B. The positions of several contact residues are altered significantly in the 2C9m7 1OG2 structure as compared with the 2C9 1R9O structure, as shown in Fig. 3A for residues 107, 208, and 476.
The position, orientation, and conformation of the 0XV molecule in the active site cavity are defined by unbiased |Fo|−|Fc| σA-weighted omit maps for the electron density (Fo, observed structure factor; Fc, calculated structure factor) (Fig. 3, A and B). The maps also identified a water molecule that is hydrogen-bonded to the phenol oxygen. The two aromatic groups of 0XV form a “V” shape with a roughly 60° dihedral angle for the planes of the aromatic benzofuran and dimethylphenol moieties. The carbonyl group of 0XV lies in the plane of the phenol group at the apex of the V in close proximity to alanine 297 on helix I and is positioned to accept a hydrogen bond from a water, 2.76 Å, bound to the iron, 2.12 Å, of the heme prosthetic group (Fig. 4A). The water is also hydrogen-bonded to the carbonyl of Ala-297, 2.77 Å. The water bound to the iron was evident in chain A, but it was not modeled in the other chains because the resultant B-factors were too high to establish its presence.
Additionally, the 2-methyl moiety of the benzofuran ring is positioned closest to the heme iron at 5.l Å and would be the most likely site of metabolism based on this binding pose. Although metabolism of 0XV was not examined by Locusun et al. (26), P450 2C19 and 2C9 were shown to catalyze hydroxylation of the benzofuran group of dimethylbenzarone; however, the exact site of metabolism was not identified. Dimethylbenzarone has a 2-ethyl substituent rather than a 2-methyl group on the benzofuran moiety. In addition, P450 2C19 was observed to catalyze the hydroxylation of the phenol moiety of dimethylbenzarone (26). Formation of this alternative product would require that the methyl group of the phenol moiety be positioned close to the reactive oxygen atom of the oxoiron(IV) porphyrin radical cation intermediate in the reaction. 0XV can be modeled in the active site with one of the two methyl groups of the dimethylphenol moiety positioned 4.3 Å from the heme iron (Fig. 3C). Although this pose could account for the observed metabolite, this pose does not fit the omit map very well, and the average real space R value computed for 0XV in the four chains of 0.378 ± 0.029 is more than two standard deviations higher than the average real space R value, 0.186 ± 0.069, for all residues in the protein. In contrast, the pose shown in Fig. 3, A and B, accounted for the observed electron density and exhibited an average real space R value of 0.177 ± 0.037 for the four copies, which is close to the average value for the protein.
The binding pose observed for 0XV in P450 2C19 exhibits extensive contacts with the protein that are likely to contribute to its relatively high binding affinity (Fig. 3, A and B). The benzofuran group of 0XV extends away from the heme into a narrow hydrophobic pocket between helix I and the C-terminal loop. Phe-476 in the turn of the C-terminal loop exhibits an edge to face aromatic interaction with the benzofuran moiety, and the methyl group on the Thr-301 side chain contacts the opposite face. A hydrophobic interaction is evident also between Cβ and Cγ of Glu-300 on helix I and the distal end of the benzofuran ring. Additionally, Ile-205 on helix F contacts one edge of the benzofuran ring, whereas the other edge contacts Ile-362 on the connector between helix K and β-sheet 1. Leu-366 on the same connector contacts the methyl group of the benzofuran moiety. Ile-362 is one of the few amino acids contacting 0XV that differs between 2C19 and 2C9. The longer reach of the corresponding Leu-362 in 2C9 would create a less favorable, close contact with the benzofuran ring. A second residue that contacts 0XV and that differs between the two enzymes is Val-208, which is a leucine residue in 2C9. As shown in Fig. 3A, the position of the Leu-208 in P450 2C9m7 differs in the 1OG2 and 1OG5 structures from that in the 2C9 1R9O structure suggesting that changes in the position of the longer leucine side chain could offset potential effects of this substitution on 0XV binding to P450 2C9. Interestingly, mutation of Leu-208 to valine in P450 2C9 reduces the catalytic efficiency of prochiral oxygenation of phenytoin and reduces the prochiral selectivity to more closely mimic phenytoin oxygenation by P450 2C9 (39). Val-208 contacts one of methyl groups on the 0XV phenolic moiety. The plane of the dimethylphenol moiety is oriented so that the second methyl group of the phenolic moiety contacts Asp-293 on helix I. Val-113 on the helix B′-C loop contacts the adjacent carbon atom of 0XV. The edge of the aromatic ring of the neighboring Phe-114 contacts one face of the phenol ring, but the opposite face of the phenol ring does not exhibit close contacts with the protein.
Structural Differences between P450s 2C19 and 2C9
Although the portion of the active site near the catalytic center of P450 2C19 occupied by 0XV is similar to that of the 1R9O structure of the P450 2C9 flurbiprofen complex with the exceptions noted, the more distal portions of the active site cavity differ between the two enzymes (Fig. 4). The conformation of the helix B-C loop in the 2C9 1R9O and 2C9m7 1OG2 structures differ from each other and from the structure of the B-C loop in the 2C19 0XV complex (Figs. 2B and 4). As a result, the Cα-positions for Val-113, Phe-114, and Asn-107 differ significantly for the three structures (Fig. 3A). In chain C of the asymmetric unit, Asn-107 Nδ donates a hydrogen bond to both the water molecule associated with 0XV and to Asp-293 Oδ, but one or both of these distances are increased to more than 3.5 Å for the other three chains in the asymmetric unit of the P450 2C19 structure. Although the protein backbone of the B-C loop is highly similar for P450 2C8 and P450 2C19, the shorter Thr-107 of P450 2C8 cannot hydrogen-bond to Asp-293 on helix I (Fig. 4D). In the 1OG2 structure of P450 2C9m7, Asn-107 is positioned up and away from Asp-293 with no potential for hydrogen bonding between the two residues (Fig. 4C). In contrast, Asn-107 resides outside of the active site in the 1R9O structure of the P450 2C9 flurbiprofen complex, and Arg-108 resides in the active site where it forms an ionic bond with Asp-293 and hydrogen bonds to Asn-289 and to the carboxyl of the substrate flurbiprofen (Fig. 4B). The presence of Arg-108 in the active site of P450 2C9 is likely to contribute to the high binding affinity observed for benzbromarone and other ionized phenols (40). Mutagenesis studies indicate that substitution of alanine for Arg-108 of P450 2C9 dramatically reduces diclofenac 4-hydroxylase activity (41). Similarly, substitution of a phenylalanine for Arg-108 leads to an almost complete loss of diclofenac and S-warfarin hydroxylation by P450 2C9 while retaining its capacity to catalyze hydroxylation of the nonpolar substrate, pyrene (42).
It is remarkable that B-C loop figures prominently in differential recognition of compounds by P450s 2C9 and 2C19 because the sequence of the loop is identical except for residue 99 where an isoleucine-histidine difference occurs, respectively. The difference in the location of Arg-108 in the two P450 2C9 structures suggests that substrates may play a role in stabilizing the inward facing orientation of Arg-108 observed in the flurbiprofen structure and that the helix B-C loop in P450 2C9 exhibits relatively high mobility compared with P450 2C8 where the effects of substrate binding on the conformation of the helix B-C loop are relatively small (18). The low affinity of P450 2C19 for anionic substrates that are efficiently metabolized by P450 2C9 suggests that Arg-108 does not make a similar transition into the active site of P450 2C19.
As the sequences of the helix B-C loops of P450s 2C9 and 2C19 differ by only one amino acid residue, it is likely that other differences between the two proteins contribute to the more limited mobility of the B-C loop in P450 2C19. Studies with chimeric enzymes have been used to identify amino acid differences between the two enzymes that are determinants of their distinct metabolic properties, and the results of these studies are consistent with this notion. In these studies, residues 286 and 289 were identified as key amino acid differences between P450s 2C9 and 2C19 for selective metabolism of two anionic substrates diclofenac and ibuprofen by P450 2C9 (23) as well as for the high affinity binding of the 2C9 selective inhibitor sulfaphenazole (22, 23). Additionally, the introduction of these two amino acid substitutions into P450 2C19, a serine for Asn-286 and asparagine for Ile-289, conferred high affinity binding for warfarin to P450 2C19, but the two substitutions were not sufficient to confer the enantiomer selectivity and site of metabolism specificity of P450 2C9 to P450 2C19 (22). As shown in Fig. 4B, asparagine 289 interacts directly with Arg-108 in the structure of the flurbiprofen complex of 2C9, and it is likely to contribute to the stability of the inward orientation of the arginine. Additionally, the longer extension of Asn-289 when compared with Ile-289 in P450 2C19 is likely to increase the distance between helix I and the helix B-C loop of P450 2C9. Based on the structure of the 0XV complex of P450 2C19 determined in this study, the substitution of serine for asparagine 286 on helix I would disrupt a hydrogen bonding interaction that is evident between the Asn-286 side chain Oδ and the amide hydrogen of the Gly-111 in the B-C loop of P450 2C19 (Fig. 4A). In the 2C9 1R9O and 2C9m7 1OG2 structures, the amide hydrogen of Gly-111 is oriented in the opposite direction, and the carbonyl oxygen of Phe-110 points toward Ser-286. In both structures, the side chain of Ser-286 is oriented away from the Phe-110 carbonyl in the helix B′-C loop. In the case of the 2C9m7 1OG2 structure, the Phe-110 carbonyl is not sufficiently close for hydrogen bonding to Ser-286. In contrast, steric restrictions prevent rotation of the side chain hydroxyl group of Ser-286 into a position favorable for hydrogen bond donation to the carbonyl of Phe-110. Glycine 111 together with glycines 109 and 98, at the beginning and the end of the B-C loop, are key pivot points for the conformational flexibility exhibited by different structures for the helix B-C loops of 2C9 (20) and of rabbit 2C5 (43, 44). As noted earlier, the conformation of the helix B-C loop in P450 2C8 is very similar to that of 2C19, and it is interesting to note that Asn-286 forms a similar hydrogen bond with the amide hydrogen of Gly-211 and that Gly-289 of 2C8 also permits a close approach of the helix B′-C loop to helix I (Fig. 4D).
Activities of Chimeric Enzymes
Published studies of chimeric enzymes have addressed effects of reciprocal substitutions on 2C9 catalytic activity. The first of these studies demonstrated that introduction of histidine for Ile-99 in the loop between helices B and ′ conferred 50% of the omeprazole 5-hydroxylase activity exhibited by P450 2C19 to 2C9, which normally exhibits very low activity (21). The orientation of Ile-99 differs between the two structures of 2C9 (Fig. 4, B and C), and the larger side chain of histidine might limit the motion implied by this difference in orientation, which is associated with the changes in the orientation of Phe-100 that open and close the P450 2C9 active site and the conformation change in the helix B-C loop allowing Arg-108 to reside in the active site. This torsional freedom in P450 2C9 is evident from different orientations of the side chains of residues 99 and 100 observed for the 2C9 1R9O structure (Fig. 4B) and the 2C9m7 1OG2 structure (Fig. 4C). The difference in the orientation of Phe-100 away from the heme together with the change observed for the position of Phe-476 on the turn in β-sheet 4 greatly enlarges the active site cavity of 1OG2 structure (compare Fig. 4, B and C). Additionally, His-99 hydrogen bonds with Glu-72 of the β-sheet 1 in the structure of P450 2C19, but His-99 derived from P450 2C19 is less likely to hydrogen bond to the corresponding Lys-72 derived from P450 2C9 in the chimeric enzyme.
Additional substitutions of a proline for Ser-220 and threonine for Pro-221 of 2C9 were sufficient to confer rates of omeprazole 5-hydroxylation to 2C9 that were similar to that of 2C19 with the larger of the two effects being attributed to the introduction of Pro-220 > Thr-221. These substitutions effectively change the position of the proline in the turn between helices F′ and G′ to correspond to the position of the proline in P450 2C19 and 2C8. In the structures of the latter two enzymes, helix F′ packs against the N-terminal end of the B′ helix where Oδ of Asn-217 accepts a hydrogen bond from the amide hydrogen of residue 102 in helix B′ in the 2C19 and 2C8 structures, and this interaction may restrain the motions of helix B′. The two substitutions for residues 220 and 221 may work in concert with the His-99 substitution to reduce the dynamic motion of the helix B-C loop in P450 2C9 to more closely mimic P450 2C19. Interestingly, Wada et al. (25) introduced S220P and P221T mutations into P450 2C9 and observed enhanced rates for diclofenac 4′-hydroxylation and tolbutamide p-methylhydroxylation as well as for the two P450 2C19-associated activities, omeprazole 5-hydroxylation and (S)-mephytoin 4-hydroxylation. In this regard, it is interesting to note that a proline substitution for Ser-220 and an isoleucine substitution for Pro-221 were two of seven mutations that were introduced into P450 2C9 to enhance crystallization of the protein for determination of the 2C9m7 1OG2 and 1OG5 structures (19). The conformation of the F′-G′ region of these two structures is highly similar to that of 2C19 and 2C8 but displaced in space by differences in the position of helix F and the helix B-C loop of P450 2C9m7 relative to P450s 2C19 and 2C8 (Fig. 2B). The F′-G′ region exhibits conformational disorder in the 1R9O structure of native P450 2C9 consistent with the notion that the mutations in 2C9m7 produced a more rigid interaction. Interestingly, a significantly larger number of reciprocal substitutions, residues 99, 220, 221, 286, 289, 292, and 295, were necessary to confer S-mephenytoin 4′-hydroxylation equivalent to that of 2C19 to P450 2C9 (21, 24). As discussed earlier, Asn-286 resides outside the cavity and acts to influence the conformation of the helix B′-C loop indirectly through hydrogen bonding interactions with amide hydrogen of Gly-111 (Fig. 4A). Residues 289, 292, and 295 reside on helix I and form boundaries of the active site cavity of P450 2C19 near Asp-293 and Asn-207 (Fig. 4A). Taken together, characterization of the chimeric enzymes suggests that differences in the residues bordering the helix B-C loop confer differences in substrate binding and oxidation through their influence on the conformation and dynamics of the B-C loop that alter, in turn, the shape and chemical properties of the substrate-binding site.
Summary
The first structure of human P450 2C19 deposited in PDB provides insights into substrate and inhibitor binding to the enzyme. The comparison of the structure of P450 2C19 0XV complex with structures of the closely related P450s 2C8 and 2C9 indicates that the tertiary structures of P450s 2C8 and 2C19 are highly similar, although the active site cavities differ greatly due to amino acid differences that directly alter the topography of the active sites as well as the hydrophobic and polar landscapes of the cavities. In contrast, the active site cavity of the 1R9O structure of the P450 2C9 flurbiprofen complex is more similar to that of P450 2C19 in the region where 0XV is bound, but it diverges in the more distal regions that include the helix B-C loop and the turn in the C-terminal loop. The divergence in the tertiary structure of the 1OG2 structure of P450 2C9m7 from that of the P450 2C19 0XV complex is even greater, and the active site of the P450 2C9m7 1OG2 structure is significantly enlarged relative to that of P450 2C19. Nevertheless, the shift of the helix B-C loop away from helix I, which contributes to relocation of Arg-108 into the active site in the 1R9O structure of the flurbiprofen complex, is conserved in the 1OG2 and 1OG5 structures of apo- and warfarin-bound P450 2C9m7 suggesting that this is an inherent property of 2C9 that is not greatly affected by ligand binding or mutations in the helix F-G region. Similarly, this portion of the structure of P450 2C8, which is similar to that of 2C19, is not affected by ligand binding to 2C8 (18). Although some solvent channels are evident in the structure of P450 2C19, it is also likely that a larger channel would open to allow the passage of 0XV in and out of the active site cavity and that closure around structurally distinct substrates and inhibitors would lead to differences for contact residues from the structure of the 0XV complex when other substrates and inhibitors bind to P450 2C19. These comparisons identify the basis for the selectivity of P450s 2C19 and 2C9 for binding 0XV and flurbiprofen, respectively, and reveal how sequence differences within and adjacent to the helix B-C loop affect its conformation and mobility in P450s 2C9, 2C19, and 2C8.
Acknowledgments
We thank Jeffrey Jones (Washington State University) for synthesizing and providing 0XV for this study and Mei Hsu for determination of the Ki value for 0XV inhibition of 3-methoxyfluorescein O-demethylation catalyzed by P450 2C19. The help of the support staff of the Stanford Synchrotron Radiation Lightsource for data collection and Qiping Zhao for excellent technical support is gratefully acknowledged. Portions of this work were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of the SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the United States Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research and by National Institutes of Health Grant P41GM103393 from NIGMS and Grant P41RR001209 from NCRR.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM031001 from NIGMS (to E. F. J.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 4GQS) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- P450 or CYP
- generic term for a cytochrome P450, with individual P450s identified using a number-letter-number designation based on sequence relatedness
- 0XV
- (2-methyl-1-benzofuran-3-yl)-(4-hydroxy-3,5-dimethylphenyl)methanone
- PDB
- Protein Data Bank.
REFERENCES
- 1. Zanger U. M., Turpeinen M., Klein K., Schwab M. (2008) Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 392, 1093–1108 [DOI] [PubMed] [Google Scholar]
- 2. Ingelman-Sundberg M., Sim S. C. (2010) Pharmacogenetic biomarkers as tools for improved drug therapy; emphasis on the cytochrome P450 system. Biochem. Biophys. Res. Commun. 396, 90–94 [DOI] [PubMed] [Google Scholar]
- 3. Goldstein J. A., Faletto M. B., Romkes-Sparks M., Sullivan T., Kitareewan S., Raucy J. L., Lasker J. M., Ghanayem B. I. (1994) Evidence that CYP2C19 is the major (S)-mephenytoin 4′-hydroxylase in humans. Biochemistry 33, 1743–1752 [DOI] [PubMed] [Google Scholar]
- 4. Ingelman-Sundberg M., Sim S. C., Gomez A., Rodriguez-Antona C. (2007) Influence of cytochrome P450 polymorphisms on drug therapies. Pharmacogenetic, pharmacoepigenetic, and clinical aspects. Pharmacol. Ther. 116, 496–526 [DOI] [PubMed] [Google Scholar]
- 5. Spina E., Santoro V., D'Arrigo C. (2008) Clinically relevant pharmacokinetic drug interactions with second generation antidepressants. An update. Clin. Ther. 30, 1206–1227 [DOI] [PubMed] [Google Scholar]
- 6. Shi S., Klotz U. (2008) Proton pump inhibitors. An update of their clinical use and pharmacokinetics. Eur. J. Clin. Pharmacol. 64, 935–951 [DOI] [PubMed] [Google Scholar]
- 7. Boulenc X., Djebli N., Shi J., Perrin L., Brian W., Van Horn R., Hurbin F. (2012) Effects of omeprazole and genetic polymorphism of CYP2C19 on the clopidogrel active metabolite. Drug Metab. Dispos. 40, 187–197 [DOI] [PubMed] [Google Scholar]
- 8. Zvyaga T., Chang S. Y., Chen C., Yang Z., Vuppugalla R., Hurley J., Thorndike D., Wagner A., Chimalakonda A., Rodrigues A. D. (2012) Evaluation of six proton pump inhibitors as inhibitors of various human cytochromes P450. Focus on cytochrome P450 2C19. Drug Metab. Dispos. 40, 1698–1711 [DOI] [PubMed] [Google Scholar]
- 9. Nishiya Y., Hagihara K., Kurihara A., Okudaira N., Farid N. A., Okazaki O., Ikeda T. (2009) Comparison of mechanism-based inhibition of human cytochrome P450 2C19 by ticlopidine, clopidogrel, and prasugrel. Xenobiotica 39, 836–843 [DOI] [PubMed] [Google Scholar]
- 10. Dansette P. M., Libraire J., Bertho G., Mansuy D. (2009) Metabolic oxidative cleavage of thioesters. Evidence for the formation of sulfenic acid intermediates in the bioactivation of the antithrombotic prodrugs ticlopidine and clopidogrel. Chem. Res. Toxicol. 22, 369–373 [DOI] [PubMed] [Google Scholar]
- 11. Kazui M., Nishiya Y., Ishizuka T., Hagihara K., Farid N. A., Okazaki O., Ikeda T., Kurihara A. (2010) Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab. Dispos. 38, 92–99 [DOI] [PubMed] [Google Scholar]
- 12. Dansette P. M., Rosi J., Bertho G., Mansuy D. (2012) Cytochromes P450 catalyze both steps of the major pathway of clopidogrel bioactivation, whereas paraoxonase catalyzes the formation of a minor thiol metabolite isomer. Chem. Res. Toxicol. 25, 348–356 [DOI] [PubMed] [Google Scholar]
- 13. Simon T., Verstuyft C., Mary-Krause M., Quteineh L., Drouet E., Méneveau N., Steg P. G., Ferrières J., Danchin N., Becquemont L., and French Registry of Acute ST-Elevation and Non-ST-Elevation Myocardial Infarction (FAST-MI) Investigators (2009) Genetic determinants of response to clopidogrel and cardiovascular events. N. Engl. J. Med. 360, 363–375 [DOI] [PubMed] [Google Scholar]
- 14. Cuisset T., Morange P. E., Alessi M. C. (2012) Recent advances in the pharmacogenetics of clopidogrel. Hum. Genet. 5, 693–664 [DOI] [PubMed] [Google Scholar]
- 15. Tiroch K. A., Sibbing D., Koch W., Roosen-Runge T., Mehilli J., Schömig A., Kastrati A. (2010) Protective effect of the CYP2C19*17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am. Heart J. 160, 506–512 [DOI] [PubMed] [Google Scholar]
- 16. Sibbing D., Koch W., Gebhard D., Schuster T., Braun S., Stegherr J., Morath T., Schömig A., von Beckerath N., Kastrati A. (2010) Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation 121, 512–518 [DOI] [PubMed] [Google Scholar]
- 17. Schoch G. A., Yano J. K., Wester M. R., Griffin K. J., Stout C. D., Johnson E. F. (2004) Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid-binding site. J. Biol. Chem. 279, 9497–9503 [DOI] [PubMed] [Google Scholar]
- 18. Schoch G. A., Yano J. K., Sansen S., Dansette P. M., Stout C. D., Johnson E. F. (2008) Determinants of cytochrome P450 2C8 substrate binding. Structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid. J. Biol. Chem. 283, 17227–17237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Williams P. A., Cosme J., Ward A., Angove H. C., Matak Vinkoviǽ D., Jhoti H. (2003) Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature 424, 464–468 [DOI] [PubMed] [Google Scholar]
- 20. Wester M. R., Yano J. K., Schoch G. A., Yang C., Griffin K. J., Stout C. D., Johnson E. F. (2004) The structure of human microsomal cytochrome P450 2C9 complexed with flurbiprofen at 2.0 Å resolution. J. Biol. Chem. 279, 35630–35637 [DOI] [PubMed] [Google Scholar]
- 21. Ibeanu G. C., Ghanayem B. I., Linko P., Li L., Pederson L. G., Goldstein J. A. (1996) Identification of residues 99, 220, and 221 of human cytochrome P450 2C19 as key determinants of omeprazole hydroxylase activity. J. Biol. Chem. 271, 12496–12501 [DOI] [PubMed] [Google Scholar]
- 22. Jung F., Griffin K. J., Song W., Richardson T. H., Yang M., Johnson E. F. (1998) Identification of amino acid substitutions that confer a high affinity for sulfaphenazole binding and a high catalytic efficiency for warfarin metabolism to P450 2C19. Biochemistry 37, 16270–16279 [DOI] [PubMed] [Google Scholar]
- 23. Klose T. S., Ibeanu G. C., Ghanayem B. I., Pedersen L. G., Li L., Hall S. D., Goldstein J. A. (1998) Identification of residues 286 and 289 as critical for conferring substrate specificity of human CYP2C9 for diclofenac and ibuprofen. Arch. Biochem. Biophys. 357, 240–248 [DOI] [PubMed] [Google Scholar]
- 24. Tsao C. C., Wester M. R., Ghanayem B., Coulter S. J., Chanas B., Johnson E. F., Goldstein J. A. (2001) Identification of human CYP2C19 residues that confer S-mephenytoin 4′-hydroxylation activity to CYP2C9. Biochemistry 40, 1937–1944 [DOI] [PubMed] [Google Scholar]
- 25. Wada Y., Mitsuda M., Ishihara Y., Watanabe M., Iwasaki M., Asahi S. (2008) Important amino acid residues that confer CYP2C19 selective activity to CYP2C9. J. Biochem. 144, 323–333 [DOI] [PubMed] [Google Scholar]
- 26. Locuson C. W., 2nd, Suzuki H., Rettie A. E., Jones J. P. (2004) Charge and substituent effects on affinity and metabolism of benzbromarone-based CYP2C19 inhibitors. J. Med. Chem. 47, 6768–6776 [DOI] [PubMed] [Google Scholar]
- 27. Richardson T. H., Jung F., Griffin K. J., Wester M., Raucy J. L., Kemper B., Bornheim L. M., Hassett C., Omiecinski C. J., Johnson E. F. (1995) Universal approach to the expression of human and rabbit cytochrome P450s of the 2C subfamily in Escherichia coli. Arch. Biochem. Biophys. 323, 87–96 [DOI] [PubMed] [Google Scholar]
- 28. Blaisdell J., Mohrenweiser H., Jackson J., Ferguson S., Coulter S., Chanas B., Xi T., Ghanayem B., Goldstein J. A. (2002) Identification and functional characterization of new potentially defective alleles of human CYP2C19. Pharmacogenetics 12, 703–711 [DOI] [PubMed] [Google Scholar]
- 29. Wang H., An N., Wang H., Gao Y., Liu D., Bian T., Zhu J., Chen C. (2011) Evaluation of the effects of 20 nonsynonymous single nucleotide polymorphisms of CYP2C19 on S-mephenytoin 4′-hydroxylation and omeprazole 5′-hydroxylation. Drug Metab. Dispos. 39, 830–837 [DOI] [PubMed] [Google Scholar]
- 30. Cosme J., Johnson E. F. (2000) Engineering microsomal cytochrome P450 2C5 to be a soluble, monomeric enzyme. Mutations that alter aggregation, phospholipid dependence of catalysis, and membrane binding. J. Biol. Chem. 275, 2545–2553 [DOI] [PubMed] [Google Scholar]
- 31. Sansen S., Yano J. K., Reynald R. L., Schoch G. A., Griffin K. J., Stout C. D., Johnson E. F. (2007) Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J. Biol. Chem. 282, 14348–14355 [DOI] [PubMed] [Google Scholar]
- 32. Omura T., Sato R. (1964) The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J. Biol. Chem. 239, 2379–2385 [PubMed] [Google Scholar]
- 33. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 35. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography and NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 37. Kleywegt G. J., Jones T. A. (1994) Detection, delineation, measurement, and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 50, 178–185 [DOI] [PubMed] [Google Scholar]
- 38. Wang A., Savas U., Hsu M. H., Stout C. D., Johnson E. F. (2012) Crystal structure of human cytochrome P450 2D6 with prinomastat bound. J. Biol. Chem. 287, 10834–10843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mosher C. M., Tai G., Rettie A. E. (2009) CYP2C9 amino acid residues influencing phenytoin turnover and metabolite regio- and stereochemistry. J. Pharmacol. Exp. Ther. 329, 938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Locuson C. W., 2nd, Wahlstrom J. L., Rock D. A., Rock D. A., Jones J. P. (2003) A new class of CYP2C9 inhibitors. Probing 2C9 specificity with high affinity benzbromarone derivatives. Drug Metab. Dispos. 31, 967–971 [DOI] [PubMed] [Google Scholar]
- 41. Ridderström M., Masimirembwa C., Trump-Kallmeyer S., Ahlefelt M., Otter C., Andersson T. B. (2000) Arginines 97 and 108 in CYP2C9 are important determinants of the catalytic function. Biochem. Biophys. Res. Commun. 270, 983–987 [DOI] [PubMed] [Google Scholar]
- 42. Dickmann L. J., Locuson C. W., Jones J. P., Rettie A. E. (2004) Differential roles of Arg97, Asp293, and Arg108 in enzyme stability and substrate specificity of CYP2C9. Mol. Pharmacol. 65, 842–850 [DOI] [PubMed] [Google Scholar]
- 43. Wester M. R., Johnson E. F., Marques-Soares C., Dansette P. M., Mansuy D., Stout C. D. (2003) The structure of a substrate complex of mammalian cytochrome P450 2C5 at 2.3 Å resolution. Evidence for multiple substrate binding modes. Biochemistry 42, 6370–6379 [DOI] [PubMed] [Google Scholar]
- 44. Wester M. R., Johnson E. F., Marques-Soares C., Dijols S., Dansette P. M., Mansuy D., Stout C. D. (2003) The structure of mammalian cytochrome P450 2C5 complexed with diclofenac at 2.1Å resolution. Evidence for an induced fit model of substrate binding. Biochemistry 42, 9335–9345 [DOI] [PubMed] [Google Scholar]
- 45. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallgr. 66, (Pt. 1) 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]