Background: SpChiD, a family 18 glycosyl hydrolase, transglycosylates chitooligosaccharides.
Results: Transglycosylation of SpChiD was improved in terms of the quantity of TG products produced and the extended duration of TG.
Conclusion: The new findings unravel possibilities of modulating chitinases for improved transglycosylation.
Significance: Variants of SpChiD can be used to develop a bio-process for large scale production of longer chain chitooligosaccharides.
Keywords: Chitin, Chitinase, Oligosaccharide, PCR, Polysaccharide, Chitooligosaccharides, Mutagenesis, N-Acetylglucosamine, Serratia proteamaculans, Transglycosylation
Abstract
We describe the improvement of transglycosylation (TG) by chitinase D from Serratia proteamaculans (SpChiD). The SpChiD produced a smaller quantity of TG products for up to 90 min with 2 mm chitotetraose as the substrate and subsequently produced only hydrolytic products. Of the five residues targeted at the catalytic center, E159D resulted in substantial loss of both hydrolytic and TG activities. Y160A resulted in a product profile similar to SpChiD and a rapid turnover of substrate with slightly increased TG activity. The rest of the three mutants, M226A, Y228A, and R284A, displayed improved TG and decreased hydrolytic ability. Four of the five amino acid substitutions, F64W, F125A, G119S, and S116G, at the catalytic groove increased TG activity, whereas W120A completely lost the TG activity with a concomitant increase in hydrolysis. Mutation of Trp-247 at the solvent-accessible region significantly reduced the hydrolytic activity with increased TG activity. The mutants M226A, Y228A, F125A, S116G, F64W, G119S, R284A, and W247A accumulated approximately double the concentration of TG products like chitopentaose and chitohexaose, compared with SpChiD. The double mutant E159D/F64W regained the activity with accumulation of 6.0% chitopentaose at 6 h, similar to SpChiD at 30 min. Loss of chitobiase activity was unique to Y228A. Substitution of amino acids at the catalytic center and/or groove substantially improved the TG activity of SpChiD, both in terms of the quantity of TG products produced and the extended duration of TG activity.
Introduction
Chitin is the second most abundant natural polysaccharide consisting of β(1→ 4)-linked N-acetyl-d-glucosamine (GlcNAc) units in a linear form. Chitin is insoluble in water and mainly exists in two crystalline (α- and β-) forms. The α-chitin consists of sheets of tightly packed alternating parallel and antiparallel chains (1) and is abundant in the exoskeleton of arthropods, insects, fungi, and yeast cell walls. The chains are arranged in parallel orientation in β-chitin (2), which occurs less frequently in nature and is often extracted from squid pens. The insolubility of chitin is a major limitation in eliciting biological activities.
The polymeric chitin and its breakdown products like chitooligosaccharides (CHOS)5 are of increasing interest due to their potential applications in agriculture and medicine. The CHOS are water-soluble, nontoxic, and biocompatible. Biological properties of CHOS include antimicrobial and antitumor activities, immunoenhancing effects on animals (3), and disease protective responses in plants (4–6). Fully acetylated CHOS with degree of polymerization (DP) from 4 to 10 induced peroxidase activity in wheat, although the fully deacetylated CHOS of DP5 and DP7 induced neither peroxidase nor phenylalanine ammonia lyase activities (4). Oligomers of chitin, but not chitosan, were active elicitors of defense-related lignification in wounded (7) and intact wheat (Triticum aestivum L.) leaves (8) and in suspension-cultured wheat cells (9). Chitin octamer (DP8), by acting as a bivalent ligand, induced dimerization of chitin-elicitor receptor kinase 1 of Arabidopsis thaliana (AtCERK1) that was inhibited by shorter chitin oligomers (10). Dimerization of AtCERK1 was critical to activate immune responses.
In view of the wider biological applications for the CHOS, chemical methods for hydrolysis of chitin or chitosan were frequently used for industrial scale production (11). Conventional chemical approaches for synthesis proved inadequate to produce substantial quantities of CHOS because of the complexities involved in the selective protection and subsequent manipulations of the various monosaccharide donors and acceptors (12). The chemical methods employ organic solvents in larger quantities followed by cumbersome purification methods. Therefore, the enzymatic approach for CHOS production would be an attractive alternative. Chitinases (EC 3.2.1.14) are one of the glycosyl hydrolases (GH), the best preferred biological tools for chitin degradation and production of CHOS with biological activity.
A few GHs show transglycosylation (TG), with which new glycosidic bonds are introduced between donor and acceptor sugar molecules. The regio- and stereo-selective synthesis of glycosidic bond can be achieved through retaining GHs showing TG, which follows double displacement reaction (13). The glycosyl-enzyme intermediate formed during the catalysis is a key step, deciding the direction of reaction toward hydrolysis or TG. Hydrolysis occurs when a water molecule attacks the glycosyl-enzyme intermediate leading to the release of a hydrolyzed sugar. Alternatively, TG occurs when a carbohydrate molecule outcompetes with the water molecule (14).
The TG by chitinases could be improved for an efficient production of CHOS with higher DP by mutagenesis. Mutation of Trp-167 at the −3 subsite in chitinase A of Serratia marcescens (15) and a chemical modification of hen egg white lysozyme at Asp-101 and Trp-62 at −4 and −2 subsites, respectively, enhanced the TG activity (16). The variants of Asp-140/311 and Asp-142/313, analogous residues in ChiB and ChiA from S. marcescens, respectively, showed improved TG activity (14). Similarly, the aromatic side chains of Phe-166 and Trp-197 in class V chitinase from Cycas revoluta were important for the TG acceptor binding than for the substrate binding (17). The mutational effects on TG activity of GH18 family chitinases were fewer and also restricted to the changes in conserved catalytic residues or aromatic residues. Chitinase from Bacillus sp. and family GH85, β-N-acetyl-d-glucosaminidase from Streptococcus pneumoniae-catalyzed TG, provided a potential useful tool for the chemo-enzymatic synthesis of glycoproteins (18, 19).
We have characterized a family 18 TG chitinase D from Serratia proteamaculans (SpChiD) that had a molecular mass of 44.4 kDa, producing TG products like DP7, DP10, DP11–12, and DP11–DP13 from DP3, DP4, DP5, and DP6 substrates, respectively (20). The SpChiD displayed TG activity only for 90 min with 2 mm chitotetraose (DP4) as substrate; later only hydrolytic products with lower DP were produced. The TG products detectable during the initial stages of reaction become substrates for hydrolytic activity of SpChiD in a prolonged incubation. The availability of a TG GH18 chitinase like SpChiD provoked us to further improve the TG activity. We report here that the alteration of amino acid residues at the catalytic center, catalytic groove, and solvent-accessible region substantially improves the TG activity both in terms of increasing the quantity of TG products and in extending the duration of TG activity.
EXPERIMENTAL PROCEDURES
Fold Recognition and Homology Modeling
Fold recognition by FUGUE (21) was done as described previously by Sashidhar et al. (22). The amino acid sequence of SpChiD (NCBI RefSeq, YP_001478954) was obtained from NCBI database (www.ncbi.nlm.nih.gov). A BLAST search of the amino acid sequence against Protein Data Bank (PDB) was performed to identify the three-dimensional structures that could share high sequence homology. The three-dimensional coordinates of the basic structure were obtained from the PDB. We built the three-dimensional structure of the SpChiD protein using the MODELLER (23) implemented in Discovery Studio 2.5 (Accelrys Inc.). The structural validation of the generated three-dimensional models was performed using Verify_3D (24, 25) and Ramachandran plot (26).
Selection of Amino Acid Residues for Point Mutations
The amino acid residues that might influence hydrolytic and TG activities of Sp ChiD were selected based on the sequence alignment and the crystal structures of the homologous proteins (1E15, 1ITX, 1E9L, 1HKM, 1EDQ, and 1D2K). We considered the hydrophobic aromatic residues of chitinases that are mainly involved in catalytic interactions with chitin. Using the three-dimensional model, a few amino acid residues were selected for point mutations based on their probable involvement in hydrolytic and TG activities of SpChiD.
Bacterial Strains, Plasmids, Culture Conditions, Biochemicals, and Enzymes
The plasmid pET-22b(+) and Escherichia coli BL21 (DE3) (Novagen, Madison, WI) were used for heterologous expression. E. coli was grown in LB broth (1% peptone, 0.5% yeast extract, 1% NaCl) at 37 °C. Ampicillin, 100 μg/ml, was added to the LB broth as required. Oligonucleotide primers were purchased from Eurofins India (Bangalore, India). Restriction enzymes, T4 DNA ligase, and Pfu DNA polymerase were obtained from MBI Fermentas (Ontario, Canada). Isopropyl β-d-thiogalactoside, ampicillin, and all other chemicals were purchased from Calbiochem or Merck or Hi-media Labs (Mumbai, India). Ni-NTA His bind resin was procured from Novagen (Madison) for protein purification. Different DP CHOS were obtained from Seikagaku Corp. (Tokyo, Japan), through Cape Cod (East Falmouth).
Generation of SpChiD Mutants
SpChiD mutants were generated as described by Ke and Madison (27) with pET-22b-SpChiD as template (18). Mutagenic primers were designed, and amplification was carried out in two different steps using Pfu DNA polymerase. In the first step of PCR, annealing temperature was 52 °C and polymerization time was 90 s. The product from the 1st step PCR was used as a megaprimer for the 2nd step PCR with an annealing temperature of 60 °C and polymerization time of 150 s. Details of the primers used for the generation of SpChiD mutants were listed (supplemental Table S1). Agarose gel electrophoresis was performed, and the amplicon of desired size was gel-eluted using the gel extraction kit from Qiagen (Duesseldorf, Germany). Both the gel-eluted amplicons and expression vector pET-22b(+) were subjected to double digestion with NcoI and XhoI at 37 °C for 6 h, followed by PCR clean up using the Qiagen gel clean up kit. Double-digested amplicons were ligated to NcoI and XhoI sites of the vector pET-22b (+), at 16 °C for 16 h.
Protein Expression, Isolation, and Purification
Highly efficient competent cells of E. coli BL21 (DE3) were transformed with the respective SpChiD mutants, and positive clones were selected on LB-ampicillin plates. Expression of SpChiD and the mutant proteins was done as described by Neeraja et al. (28). Harvested culture pellet was processed for isolation of periplasmic fraction as described in the pET manual (Novagen), concentrated with buffer exchange using Amicon filters (10-kDa cutoff, Millipore, Billerica, MA), and purified as described in Qiagen manual (Duesseldorf, Germany) using Ni-NTA affinity chromatography. The recombinant protein was eluted using buffers containing 50, 150, and 250 mm imidazole. The purified fractions were electrophoresed on 12% SDS-PAGE and visualized using Coomassie Brilliant Blue. The purified protein in 20 mm sodium acetate, pH 5.6, was used for characterization.
Activity Determination and HPLC Analysis for TG
Zymogram analysis for activity determination was done according to Purushotham et al. (29). SpChiD mutants positive for zymogram analysis were considered for HPLC analysis to check the mutational effects on the activity of SpChiD. Two mm DP4 was incubated with 350 nm of the purified SpChiD or its variants. Alternatively, 35 nm SpChiD was incubated with 50 or 100 μm of DP4 to determine the substrate binding with SpChiD. Reaction was performed in 20 mm sodium acetate buffer, pH 5.6, at 40 °C. Fractions were collected up to 6 h at regular intervals, and the reaction was stopped using an equal volume of 70% acetonitrile. The reaction mixture (20 μl) from each fraction was injected into HPLC (Shimadzu, Tokyo, Japan) using a Hamilton syringe (Hamilton Bonaduz, Switzerland). The products were analyzed by using SHODEX Amino-P50 4E column (4.6 inner diameter × 250 mm, Showa Denko K.K.) through isocratic elution with acetonitrile/H2O at 70:30 (29). A flow rate of 0.7 ml/min was maintained, and eluted CHOS were monitored at 210 nm. Quantification of CHOS was done by comparing peak areas of the products with peak areas obtained from known concentration of the CHOS. The mixture containing equal weight of CHOS ranging from DP1 to DP6 was used for standard graph preparation. A linear correlation between peak area and concentration of CHOS in standard samples was established for quantification of reaction products up to DP6. Standard calibration curves for CHOS moieties were constructed separately. These data points yielded a linear curve for each standard sugar with the R2 values of 0.997–1.0 allowing molar concentration of CHOS to be determined with confidence. The decrease in DP4 concentration was considered for specific activity measurements through a linear regression analysis using OriginPro 8 software.
RESULTS
Homology Modeling and Selection of the Mutant Sites
The NCBI BLAST search of the SpChiD at PDB identified the crystal structure of putative chitinase II from Klebsiella pneumoniae (PDB code 3QOK). The SpChiD had 91.1% of sequence similarity with the template crystal structure (supplemental Fig. S1). The modeled structure of the SpChiD (Fig. 1) was validated using PROCHECK (99.4% of the residues are in the allowed region) and Verify_3D (94.9% of residues had an average three- to one-dimensional score of >0.2). Thus, the compatibility of SpChiD three-dimensional model structure with its one-dimensional amino acid sequence was confirmed. The coordinates of chitotetraose were built based on the crystal structure of PDB code 1NH6.
FIGURE 1.
Three-dimensional model of SpChiD. Interactions of chitin tetramer with the catalytic residues (Asp-155, Asp-157, and Glu-159) at the active site are highlighted. The side chains of catalytic residues are shown as sticks, and subsites are indicated. Pictures used for representation were made with PyMOL.
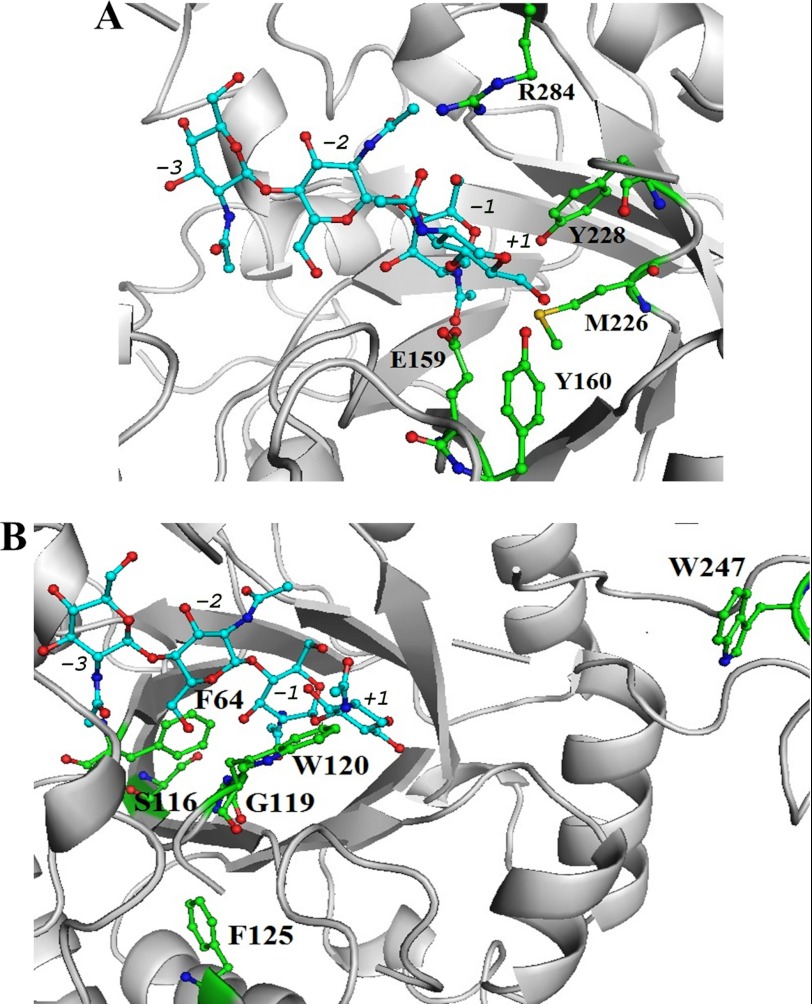
To improve the TG activity of SpChiD, the amino acid residues in the catalytic center, catalytic groove, and in the surface-exposed region were selected for mutagenesis. In the catalytic center, the residues Met-226, Tyr-228, Arg-284, and Tyr-160 were closer to the catalytic triad DXDXE (Fig. 2A). We have mutated Glu-159 from the catalytic triad, Phe-125 close to the catalytic triad, and a few residues present in the catalytic groove, including Ser-116, Gly-119, Trp-120, and Phe-64, where the substrate molecules show major interactions (Fig. 2B).
FIGURE 2.
Close-up view of three-dimensional model of SpChiD representing the residues targeted for mutation. A, catalytic center (Met-226, Tyr-228, Arg-284, Glu-159, and Tyr-160). B, catalytic groove (Phe-64, Phe-125, Gly-119, Ser-116, and Trp-120) and one residue at the solvent-accessible region Trp-247. The side chains of residues mutated in this study are shown as sticks and subsites are indicated. Pictures used for representation were made with PyMOL.
Furthermore, Trp-247 on the surface of the protein, which is expected to play a role in threading of chitin molecules to the catalytic center, was also selected for point mutation.
Amplification and Cloning of SpChiD Mutants
All the SpChiD mutants were amplified using gene-specific mutagenic primers with pET-22b-SpChiD as template. The amplicons of 1.2 kb were double-digested and cloned into NcoI and XhoI sites of pET-22b(+) expression vector. The clones were confirmed through double digestion. The inserted mutation was confirmed by automated DNA sequencing.
Expression, Purification, and Dot Blot Assay for SpChiD and the Mutants
E. coli BL21 (DE3) cells harboring the plasmids of SpChiD and the mutants were used for protein expression. Induced cells were harvested and processed for isolation of periplasmic fraction. Soluble proteins of SpChiD and its variants in the periplasmic fractions were passed through Ni-NTA affinity matrix to obtain pure proteins. SDS-PAGE analysis confirmed the purity of collected protein fractions, and the mutant protein molecular mass was also around 44.4 kDa (supplemental Fig. S2A). The dark blue spots in the in gel assay confirmed the activity of SpChiD and the mutant proteins that had single/double amino acid substitutions (supplemental Fig. S2B).
Hydrolytic and TG Activities of SpChiD
The results with SpChiD showed a very sharp decrease in the initial concentration of DP4 substrate starting from 5 to 45 min (1.26% decreased to 0.2%) and was completely depleted by the end of 120 min (supplemental Fig. S3A). The TG activity of SpChiD was detectable up to 90 min, and the production of TG products (DP5 and DP6) was relatively more up to 30 min only (DP5 from 1.3 to 6.3% and DP6 from 1.9 to 4%). Later, the TG activity decreased and completely extinguished after 90 min (DP5 and DP6 were 1.0 and 0.3%, respectively) (supplemental Fig. S3A). SpChiD produced chitobiose (DP2) as the major end product (56.4%) at the end of 120 min, along with the other two degradation products N-acetylglucosamine (DP1) and chitotriose (DP3). The DP1 product (42.4%) was much higher than DP3 product (0.8%) at 120 min. Reaction of low enzyme (35 nm) with 50 or 100 μm DP4 substrate resulted in similar product profiles that yielded both DP5 and DP6 as TG products (supplemental Fig. S4), suggesting that −2 to +2 and −1 to +3 are possible for productive binding.
Effect of Mutations on Hydrolytic and TG Activities of SpChiD Residues at the Catalytic Center
Glu-159 to Asp Conversion
E159D mutation resulted in significant loss of both hydrolytic and TG activities as indicated by the presence of 92.5% of DP4 substrate at the end of 6 h (supplemental Figs. S3B and S5B). Only a trace amount of hydrolytic products DP1, DP2, and DP3 were detected even after 6 h. Among the TG products, only DP5 and DP6 were detected (no TG products >DP6), but DP5 was detectable at 150 min and DP6 at 60 min. The proportion of quantifiable oligomers, DP1 to DP6, generated by E159D mutant was 0.6, 2.3, 2.5, 92.5, 1.0, and 1.1% at the end of 6 h.
Tyr-160, Met-226, Tyr-228, and Arg-284 to Ala Conversion
Y160A was unique from all other mutants generated, which retained TG activity even when hydrolytic activity increased. The concentration of DP4 substrate and DP3 product followed the same path from 60 min of reaction incubation with almost equal fractions of DP4 and DP3 (0.6 and 0.4%) (supplemental Figs. S3C and S5C). However, up to 45 min, a gradual reduction in DP4 (7.5%) substrate and a simultaneous increase in DP3 (21.0%) product was observed. Reaction mixtures at 150 min showed DP1 (49.7%) and DP2 (49.6%) as the major end products. Y160A was the only mutant that produced nearly equal concentrations of DP1 and DP2 products. Y160A also displayed TG activity up to 45 min. At 5 min, the concentration of DP5 and DP6 products was 8.8 and 4.6% that decreased to 3.0 and 0.9% by 45 min, respectively.
The product profile of the mutants M226A and Y228A was similar except that the latter lost chitobiase activity. A gradual decrease in the initial DP4 substrate through time was detected (supplemental Figs. S3, D and E, and S5, D and E) for M226A and Y228A. The relative proportion of the products produced by the mutant M226A was as follows DP1, 6.4%; DP2, 13.5%; DP3, 37.7%; DP5, 12.0%, and DP6, 5.8%, by 6 h. No DP1 product was detectable with Y228A, as the chitobiase activity of SpChiD was abolished. The fractions collected at 6 h showed that DP2 (14.4%), DP3 (40.7%), DP5 (13.6%), and DP6 (6.8%) were produced by Y228A. A rapid increase of DP5 and DP6 products up to 45 min was followed by a state of equilibrium, which continued until the end of 6 h. Both M226A and Y228A produced more of DP5 than DP6 product.
R284A substitution resulted in a gradual decrease of DP4 substrate, generating more of DP5 and DP6 products, although DP2 (40.8%) and DP3 (26.6%) were the major end products (supplemental Figs. S3F and S5F) at 6 h. The synthesis of DP5 and DP6 products started right from the beginning, but the concentration varied through time. Fractions collected at 30 min showed DP6 (9.0%) > DP5 (6.4%) whereas the fraction at 150 min had DP5 (9.8%) > DP6 (6.2%). Although there was an alteration in the proportion of TG products, both DP5 (8.2%) and DP6 (4.0%) were detectable until the end of 6 h.
Residues at the Catalytic Groove
Phe-64 to Trp Conversion and the Double Mutant E159D/F64W
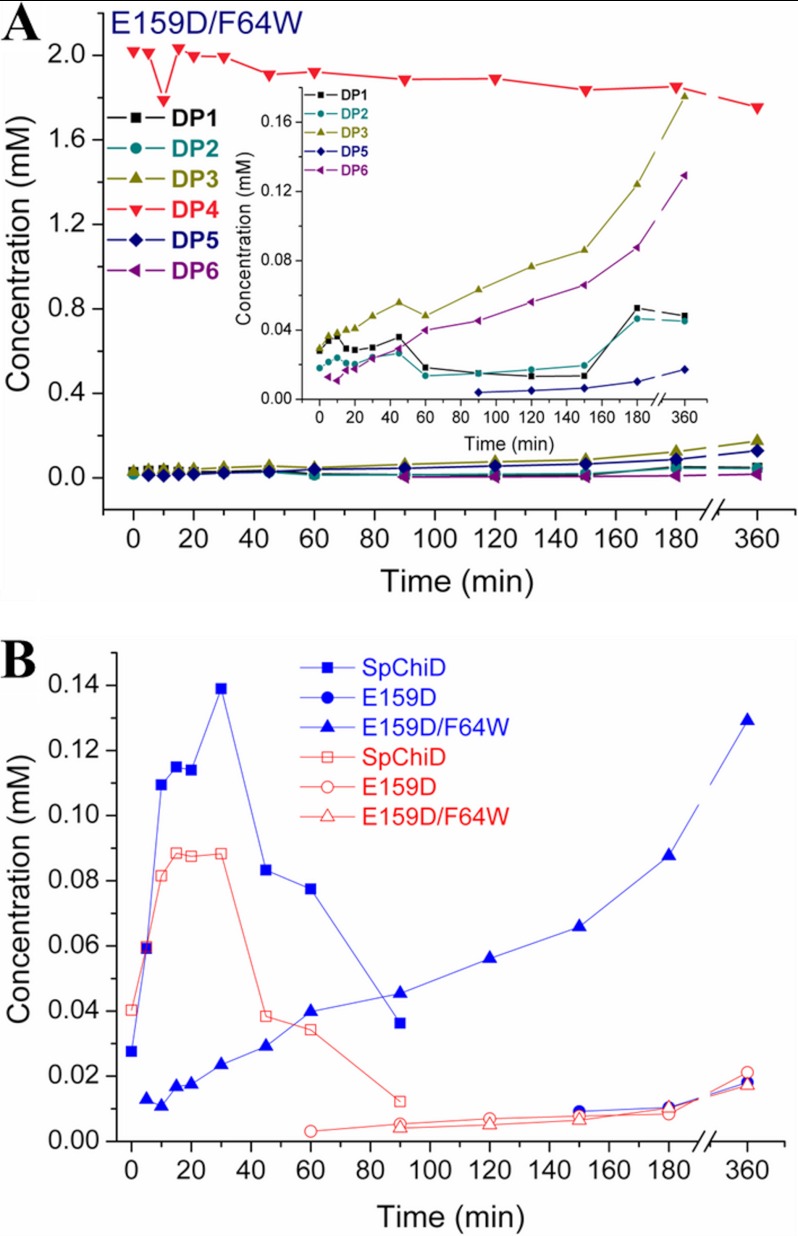
A decrease in the initial concentration of DP4 substrate was detected from 0 to 5 min (62.9 to 48.5%) with F64W (supplemental Figs. S3G and S6A). F64W produced more of the TG products compared with SpChiD, with no decrease in the hydrolytic ability up to 30 min. There was an increase of quantifiable TG products with 12.9% of DP5 and 6.6% of DP6 after 60 min. Although there was a gradual decrease of TG products, more DP5 was detectable than DP6 at 6 h. Hydrolytic products DP3 and DP2 remained as the major end products. When F64W was coupled with E159D, the double mutant E159D/F64W regained both hydrolytic and TG activities (Fig. 3A). Fractions collected at the end of 6 h revealed accumulation of 2.2, 2.1, 8.0, 80.1, 5.9, and 0.8% of oligomers from DP1 to DP6 products, respectively.
FIGURE 3.
Product profile of the double mutant E159D/F64W. Profiles were generated by SpChiD or its variants with 2 mm DP4 substrate in 20 mm sodium acetate buffer, pH 5.6, at 40 °C. A, double mutant (E159D/F64W). Inset represents the differences between hydrolytic (DP1–DP3) and TG (DP5 and DP6) products generated during the course of reaction. B, comparison of quantifiable TG products DP5 (blue solid symbols) and DP6 (red open symbols) by SpChiD, E159D, and E159D/F64W.
Trp-120 to Ala Exchange
The mutation W120A led to a rapid increase in the hydrolysis of DP4 substrate, leaving only 0.5% at 30 min (supplemental Figs. S3H and S6B). The DP3 product also decreased to 2.4% at 30 min, whereas the concentration of DP1 and DP2 products was comparable with those produced by SpChiD. DP2 (56%) was the major end product. Substantial loss of TG activity, with a feeble activity up to 5 min (1.0% of DP5 and 0.5% of DP6), suggested that Trp-120 is an important residue contributing to TG activity of SpChiD.
Phe-125, Gly-119, and Ser-116 Exchange to Ala, Ser, and Gly, Respectively
The product profiles of F125A, G119S, and S116G showed decreased hydrolytic ability by the three mutants. F125A and G119S produced more or less equal quantities of DP2 with 24.0 and 22.2% and DP3 with 21.0 and 18.7%, respectively, as the major end products at 6 h (supplemental Figs. S3, I and J, and S6, C and D). These two mutants also produced more DP5 and DP6 products, but the proportion was less in the early stage and increased gradually through time compared with SpChiD. The proportion of DP5 and DP6 products produced by F125A at 0 min was 1.3 and 1.5%, respectively, which increased to 10.4% of DP5 and 7.8% of DP6 by the end of 6 h. The product profile of G119S was also similar, and 1.5% of DP5 and 0.9% of DP6 products were detectable at 0 min and increased up to 9.5% of DP5 and 8.4% of DP6 by the end of 6 h. S116G produced both DP5 and DP6 TG products right from 0 min with 2.2 and 4.1%, respectively (supplemental Figs. S3K and S6E). The concentration of TG products increased gradually up to 180 min with accumulation of 11.7% of DP5 and 8.8% of DP6 products. Nearly similar quantities of TG products were detectable up to 6 h with accumulation of 11.7 and 7.2% of DP5 and DP6, respectively.
Residue at the Solvent-accessible Region
Trp-247 to Ala
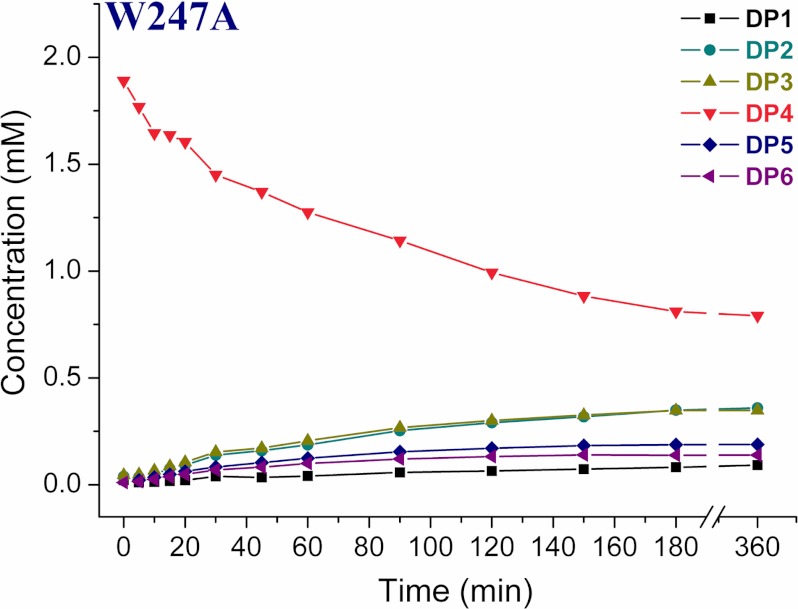
Exchange of Trp-247, at the solvent-exposed region, to Ala resulted in the reduced hydrolytic ability (Fig. 4 and supplemental Fig. S7). The formation of TG products was slow at the early time points, which increased at later time points and persisted up to 6 h. W247A produced DP2 and DP3 as the major end products. The proportion of DP1 to DP6 products at the end of 6 h of incubation reactions was 4.3, 18.2, 18.1, 42.3, 9.8, and 7.2%, respectively.
FIGURE 4.
Product profile of the mutant W247A. HPLC quantification profile of both hydrolytic and TG products up to 360 min. The reaction mixture contained 2 mm DP4 substrate and 350 nm W247A, pH 5.6, at 40 °C. The reaction products were analyzed by isocratic HPLC. Products were quantified from respective peak areas of products by using standard calibration curves of CHOS ranging from DP1 to DP6.
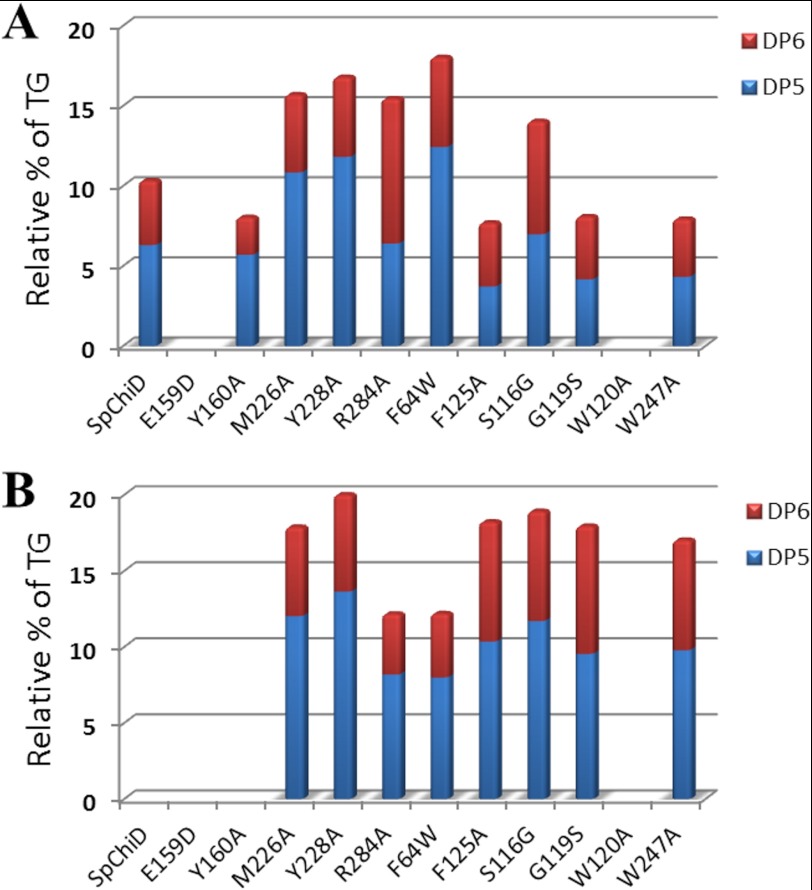
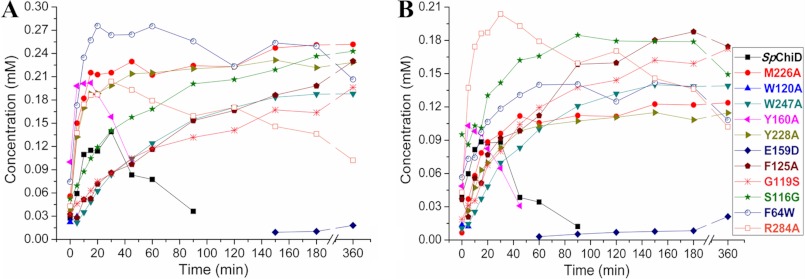
The specific activity and the extent of TG activity for all the mutants were compared with SpChiD (supplemental Fig. S8) and are listed in Table 1. The specific activity decreased for mutants E159D/F64W, E159D, F125A, G119S, S116G, and W247A and significantly increased for Y160A, F64W, Y228A, R284A, M226A, with a maximum of an 8-fold increase for W120A. However, F125A, G119S, S116G, R284A, and W247A produced more of DP5 and DP6 compared with GlcNAc. F64W in 60 min (12.9%) and other mutants M226A (12.0%), Y228A (13.6%), F125A (10.4%), and S116G (11.7%) in 6 h produced double the concentration of DP5 compared with SpChiD (6%) at 30 min (Fig. 5). However, almost double the concentration of DP6 was generated by G119S (8.4%), F125A (7.8%), and W247A (7.2%) at 6 h compared with SpChiD, which produced only 4% at 30 min (Fig. 5). The changes in proportion of TG products (DP5 and DP6) produced by all 11 single mutants at the respective time intervals were quantified and compared against the native SpChiD (Fig. 6, A and B).
TABLE 1.
Relative specific activity and TG activity of SpChiD and its mutants towards DP4 substrate
+ indicates minimum; − indicates nil; +++ indicates high, and * indicates regained.
| Name of the enzyme | Relative % of specific activity | Relative TG activity |
|---|---|---|
| SpChiD | 100 | + |
| E159D | 49.8 | − |
| Y160A | 151.3 | + |
| M226A | 204.9 | +++ |
| Y228A | 168.9 | +++ |
| R284A | 190.5 | +++ |
| W120A | 830.6 | − |
| F64W | 160.7 | +++ |
| G119S | 72.8 | +++ |
| S116G | 74.9 | +++ |
| F125A | 71.8 | +++ |
| W247A | 75.8 | +++ |
| E159D/F64W | 5.7 | * |
FIGURE 5.
Quantifiable TG products (DP5 and DP6) accumulated by SpChiD and its mutants. Reaction mixtures containing 2 mm DP4 and 350 nm of each SpChiD and its indicated mutants were incubated separately for different time periods from 0 to 360 min at 40 °C. Products were quantified from respective peak areas by using standard calibration curves of CHOS. Comparative profiles show the quantity of DP5 and DP6 TG products produced at 30 min (A) and 6 h (B).
FIGURE 6.
Comparison of quantifiable TG products. DP5 (A) and DP6 (B) accumulated at different time intervals upon incubation of 2 mm DP4 with SpChiD and its variants. Reaction was carried up to 6 h, pH 5.6, at 40 °C. Product quantification was done by a linear correlation between peak area and concentration of oligosaccharides in standard samples.
DISCUSSION
S. proteamaculans 568, a member of family Enterobacteriaceae, was isolated as a root endophyte from Populus trichocarpa (30). The Carbohydrate Active enZYme database (31) showed that at least eight genes in the genome sequence of S. proteamaculans 568 could be potentially involved in chitin turnover. We have cloned and characterized SpChiA, SpChiB, SpChiC, and SpChiD from S. proteamaculans 568 (20, 29). SpChiD had much less sequence identity (<25% and only 23% identity with human chitotriosidase) with other well characterized chitinases, except with chitinase II from K. pneumoniae for which the enzyme characterization was not available.
SpChiD was the first characterized bacterial family 18 processive endochitinase with a single catalytic domain. Among the characterized single domain chitinases, SpChiD showed significant TG activity with a potential to synthesize CHOS with DP up to 13 (20). SpChiD therefore attained importance as a unique hyper-TG chitinase produced by an endophytic bacterium. The enormous potential of SpChiD to modulate chitin substrates may have implication in the plant-microbe interaction. The chitinase-catalyzed TG activity provides a potentially useful tool for synthesis of CHOS and also the neoglycoproteins (18, 19).
TG could be improved by reducing the effective concentration of water, performing the TG reactions at high concentrations of substrate, and adding large excesses of the acceptor molecule (12). The product association or product release was the rate-determining step for chitinase-catalyzed degradation of insoluble chitin or soluble chitosan, respectively (32). Moreover, the deglycosylation step may be the rate-determining step in hydrolysis/TG (14). Because the TG products are kinetically controlled, monitoring of the reaction is necessary to ensure maximum yields (12). But in this study there was no temporal difference between the hydrolytic and TG activities of SpChiD. As both hydrolytic and TG activities start simultaneously at 0 min (20), even at very low enzyme and substrate concentrations (supplemental Fig. S4) the mutational approach could be most appropriate for improving the TG activity. Very few mutational studies were carried out to improve the TG activity in terms of extended time of TG reaction and the quantity of TG products produced by a chitinase (14, 15, 17). This study provides deeper insight into the three different possibilities of targeting SpChiD to improve TG activity as shown below.
Residues at the Catalytic Center
Among the five different amino acid substitutions made at the catalytic center, three showed increased TG activity. Met-226 and Tyr-228, present on the β6 strand of the TIM barrel, with their side chains in proximity to the catalytic triad DXDXE, have the potential to interact with the sugar molecule during catalysis. The residue Met-226 is not directly involved in catalysis but is an important supporting residue for the catalytic activity of SpChiD. Substitution of Met-243 to Glu in family 18 chitinase from Aspergillus fumigatus showed that the mutant M243E lost the TG activity toward p-nitrophenyl-(N-acetylglucosamine)2 but not with DP4 substrate (33). Docking studies revealed that M243E forms a hydrogen bond with backbone oxygen of −1 sugar instead of a hydrophobic stack between Met-243 and the +1 subsite sugar moiety. However, in this study, substitution of Met-226 with Ala decreased the hydrophobic interactions needed for the usual pace of reaction, which probably gives a chance for the oxazolinium intermediate to stay longer at the catalytic center. Moreover, it was evident that a chitin-chitosan hybrid polysaccharide could be synthesized via chitinase-catalyzed polymerization of an oxazoline derivative of a GlcN β(1→4) GlcNAc monomer. Monomer was designed as a transition state analog substrate for chitinase catalysis, which belongs to the glycoside hydrolase family 18 (34). Therefore, we speculate that the increased stay of substrate/intermediate resulted in increased TG activity both in terms of quantity of TG products formed and the duration of TG activity of the mutant M226A. It was proposed that the Tyr-214 in ChiB of S. marcescens is conserved and stabilizes the transition state (35). Y228A, where similar kind of interactions were possible with the phenolic hydroxyl group of Tyr-228, also improved the TG activity. The mutants M226A and Y228A accommodate smaller side chains on the β6 strands and therefore possess a larger cavity that revealed the possibility of increasing the TG activity possibly by decreasing the rate of reaction. Quantifiable TG products DP5 and DP6 were detected in proportions 12.0 and 5.8% by M226A and 13.6 and 6.8% by Y228A, although it was only 6.3 and 4% by SpChiD.
Arg-284 located on the β7 strand of the TIM barrel, with its side chain protruding towards the active site, forms ion pair interactions with Asp-229 in its free state. It was observed from the crystal structure of chitinase in complex with sugar (PDB code 1D2K, 1E15, and 1EDQ), that Arg-284 also forms ion pairs with the substrate at the catalytic center. Based on these two observations, we mutated Arg-284 to Ala and caused perturbations in the ion pair-forming ability of SpChiD with the incoming DP4 substrate. Such subtle changes at the catalytic center resulted in a gradual increase in the proportion of TG products DP5 (6.4%) and DP6 (9.0%) for up to 30 min. But the proportion of DP5 and DP6 products was reversed by the end of 150 min with 9.8 and 6.2%, respectively, and continued until the end of 6 h (8.2% of DP5 and 4.0% of DP6). Shifting towards increased synthesis of the DP5 product by Arg-284 could be due to the dynamic ion pair interactions with the substrate or within the protein, which remains to be examined.
The chitinase B from S. marcescens catalytic residue Asp-142 is in the “down” position, interacting with Asp-140 for family 18 chitinases in their free state. Upon substrate binding, Asp-142 moves to the “up” position and interacts with the substrate and Glu-144 (35). Such dynamic changes in all three catalytic residues play a crucial role during chitin degradation. As the carboxyl group of Glu-204 could not be substituted by Asp in chitinase A1 of Bacillus circulans W12, it was concluded that the relative disposition of the carboxyl group of Glu-204 to substrate was critical for the catalytic activity (36). In this study, Glu-159 located on the β4 strand of the TIM barrel of SpChiD was mutated to Asp, which resulted in substantial loss of both hydrolytic and TG activities, suggesting that the length of the side chain in Glu-159 was not only important for hydrolysis but also for TG activity. Because of a subtle change with the removal of a single –CH2 group, the mutant E159D had a significant difference in the activity. The adjacent residue Tyr-160 substituted with Ala reduced the time of TG to 45 min, compared with 90 min with SpChiD. The DP4 substrate was utilized rapidly within 60 min as compared with 120 min with SpChiD. The results indicated that the phenolic hydroxyl group in the Tyr-160 was important for an extended TG activity up to 90 min. The mutant Y160A acquired subtle changes at the catalytic center that favored hydrolysis but not TG.
Residues at the Catalytic Groove
We have selected five different amino acid residues in the catalytic groove for point mutations. Phe-64 with Trp, Gly-119 with Ser, Ser-116 with Gly, and Phe-125 and Trp-120 with Ala were replaced. Phe-64 was located on the β2 strand of the TIM barrel, and its substitution with Trp resulted in increased synthesis of TG products up to 60 min (12.9% of DP5 and 6.6% of DP6), later channeled in a gradual decrease up to 6 h (8.0% of DP5 and 4.2% of DP6). Although the Phe and Trp residues confer aromaticity, the bulky side chain (increased surface area) of Trp facilitates more interactions with the incoming substrate (14). The Trp also might assist in correct positioning and guiding of the sugar molecule into the active site (37). The increased TG by F64W for 60 min and then a gradual decrease may be because the sugar molecules with higher DP will possibly have more interactions with the Trp residue. When F64W is coupled with E159D, the double mutant E159D/F64W regained activity by three times in substrate utilization. The hydrolysis decreased to a greater extent for E159D/F64W, but produced a DP5 product as efficient as SpChiD at 30 min (Fig. 3B). In E159D, the relative positioning of the carboxylate group of Glu was perturbed by replacement with Asp. However, the increased aromatic surface area at the catalytic groove in F64W conversion could overcome the loss of function of E159D to a greater extent at least on TG activity.
The residues Gly-119 and Trp-120 were present on the β3 strand of TIM barrel very close to the catalytic center. Both of these residues were thought to maintain the architecture of the catalytic groove and to a major extent maintained by Trp-120 with its bulky side chain. As these two residues are directly involved in threading of the chitin molecules toward the catalytic center, point mutations were planned to identify the key residue for TG in the chitin path and also to increase the TG. To achieve this, Trp-120 was mutated to Ala and Gly-119 was replaced with Ser. Exchange of Trp-97 to Ala in ChiB of S. marcescens significantly increases the rate of hydrolysis (38), and removal of such a residue resulted in the loss of TG activity (17, 14). W120A had lost the ability of TG after 5 min, and it almost completely degraded the DP4 substrate by 30 min. The very low TG activity at the early time intervals (0–5 min) could be due to the strong inherent TG activity of the SpChiD, but it was lost completely when the trafficking of water molecules along with CHOS increased through the groove, because of the lack of guiding the aromatic residue at 120th position. This proved that residue Trp-120 was also involved in the direct interactions with the sugar molecules. However, increased TG activity was observed with the mutant G119S. There were pre-existing stacking interactions shown by Trp-120 with the substrate, which further increased with the adjacent Ser-OH group. Increased interactions at the positions 119 and 120 in turn increased the TG activity of the G119S mutant.
For an efficient TG to occur, enzyme should have an active site architecture that disfavors correct positioning of the hydrolytic water molecule and/or favors binding of incoming carbohydrate molecules, through strong interactions in the aglycon subsites (14). Ser-93 in ChiB from S. marcescens stabilizes the first Asp (Asp-140) in the DXDXE motif, when the second Asp (Asp-142) turns away from Asp-140 to Glu-144 and the oxazoline intermediate (35, 39). The second Asp (Asp-142) was shown to be important for TG activity (14). The QM/MM calculations (40) show that Asp-142 is important in the transition state and intermediate stabilization. The mutation D142N yielded an enzyme with high TG activity (14). It was proposed that active site electrostatics may change by this mutation and that this may affect the catalyzing water. In line with these observations, we have mutated Ser-116 → Gly. The residue Ser-116 is present on the β3 strand of the TIM barrel with its side chain traversing the void between the two Asp residues of the catalytic triad. Mutant S116G resulted in the decreased hydrolytic ability with a concomitant increase in TG activity. The residue Phe-125 was present at the α3 helix of the TIM barrel with its side chain eclipsing the DXDXE region from the bottom side of the active site. This was the conserved aromatic residue found in the family 18 chitinases, whose side chain forms π-π and CH-π interactions with Trp-158 located between Asp-157 and Glu-159 of the catalytic triad. The mutant F125A has a shortage of the aromatic side chain because the ascribed interactions could be disturbed. These subtle changes in the interactions resulted in increased TG activity.
Residue at the Solvent-accessible Region
Trp-247 is on the turn region next to the β6 strand of the TIM barrel. Soluble substrates like CHOS were assumed to enter the substrate binding cleft from various directions, whereas a chitin chain from crystalline chitin enters the binding site only from the edge of the cleft (41). There was no significant effect on hydrolysis of the DP5 substrate with the mutants Y56A and W53A, but the hydrolyzing activity against β-chitin microfibrils decreased in chitinase A1 from B. circulans. However, in this study the decreased rate of hydrolysis followed by increased TG activity was observed for the mutant W247A. This aromatic residue with the bulky side chain could stack against the pyranoside rings, and its replacement to Ala altered the inherent activity of the SpChiD. This clearly indicates that the residue Trp-247 has an important role in the activity of SpChiD toward soluble CHOS substrates like DP4. There is no correlation between the kcat for hydrolysis and apparent TG activity. It is the other factors such as positioning and activation of the catalytic water and optimal aligning of the acceptor that are likely to play a role (14).
TG received considerable attention for the production of CHOS with longer DP. To synthesize adequate quantities of longer chain CHOS with biological activity, there is a definite need for exploitation of TG activity. We conclude that the residues at the catalytic center and groove are the better targets to improve TG activity. The improved SpChiD variants are useful in developing a process for the production of longer chain CHOS.
Acknowledgments
We thank the Department of Science and Technology, Government of India, Funds for Infrastructure in Science and Technology, Level II support to the Department of Plant Sciences. We gratefully acknowledge the support from University Grants Commission in the form of Centre for Advanced Studies in Life Sciences, and Department Biotechnology, Government of India, Centre for Research and Education in Biology and Biotechnology to the School of Life Sciences. We also thank the support given under Department of Science and Technology-Funds for Infrastructure in Science and Technology Level II and University Grants Commission-Centre for Advanced Studies in Chemistry to the School of Chemistry, and University Grants Commission-supported University with Potential for Excellence (Phase II) to the University of Hyderabad for the infrastructural support. We thank Prof. B. M. Moerschbacher and Dr. Nour Eddine El Gueddari for suggestions during revision of the manuscript.

This article contains supplemental Figs. S1–S8 and Table S1.
- CHOS
- chitooligosaccharide
- Sp ChiD
- chitinase D from S. proteamaculans
- GH
- glycosyl hydrolase
- TG
- transglycosylation
- DP
- degree of polymerization
- PDB
- Protein Data Bank
- Ni-NTA
- nickel-nitrilotriacetic acid
- TIM
- triose-phosphate isomerase.
REFERENCES
- 1. Minke R., Blackwell J. (1978) The structure of α-chitin. J. Mol. Biol. 120, 167–181 [DOI] [PubMed] [Google Scholar]
- 2. Gardner K. H., Blackwell J. (1975) Refinement of structure of β-chitin. Biopolymers 14, 1581–1595 [DOI] [PubMed] [Google Scholar]
- 3. Kim S. K., Rajapakse N. (2005) Enzymatic production and biological activities of chitosan oligosaccharides (COS). A review. Carbohydr. Polym. 62, 357–368 [Google Scholar]
- 4. Vander P., V rum K. M., Domard A., Eddine El Gueddari N., Moerschbacher B. M. (1998) Comparison of the ability of partially N-acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves. Plant Physiol. 118, 1353–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cabrera J. C., Messiaen J., Cambier P., Van Cutsem P. (2006) Size, acetylation, and concentration of chitooligosaccharide elicitors determine the switch from defence involving PAL activation to cell death and water peroxide production in Arabidopsis cell suspensions. Physiol. Plant 127, 44–56 [Google Scholar]
- 6. Kendra D. F., Hadwiger L. A. (1984) Characterization of the smallest chitosan oligomer that is maximally antifungal to Fusariumsolani and elicits pisatin formation in Pisum sativum. Exp. Mycol. 8, 276–281 [Google Scholar]
- 7. Barber M. S., Bertram R. E., Ride J. P. (1989) Chitin oligosaccharides elicit lignification in wounded wheat leaves. Physiol. Mol. Plant Pathol. 34, 3–12 [Google Scholar]
- 8. Moerschbacher B., Kogel K. H., Noll U., Reisener H. J. (1986) An elicitor of the hypersensitive lignification response in wheat leaves isolated from the rust fungus Pucciniagraminis f. sp. tritici. I. Partial purification and characterization. Z. Naturforsch. 41, 830–838 [Google Scholar]
- 9. Gotthardt U., Grambow H. J. (1992) Near-isogenic wheat suspension cultures. Establishment, elicitor induced peroxidase activity, and potential use in the study of host/pathogen interactions. J. Plant Physiol. 139, 659–665 [Google Scholar]
- 10. Liu T., Liu Z., Song C., Hu Y., Han Z., She J., Fan F., Wang J., Jin C., Chang J., Zhou J. M., Chai J. (2012) Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164 [DOI] [PubMed] [Google Scholar]
- 11. Sakai K., Nanjo F., Usui T. (1990) Production and utilization of oligosaccharides from chitin and chitosan. Denpun. Kagaku. 37, 79–86 [Google Scholar]
- 12. Williams S. J., Withers S. G. (2000) Glycosyl fluorides in enzymatic reactions. Carbohydr. Res. 327, 27–46 [DOI] [PubMed] [Google Scholar]
- 13. Ly H. D., Withers S. G. (1999) Mutagenesis of glycosidases. Annu. Rev. Biochem. 68, 487–522 [DOI] [PubMed] [Google Scholar]
- 14. Zakariassen H., Hansen M. C., Jøranli M., Eijsink V. G., Sørlie M. (2011) Mutational effects on transglycosylating activity of family 18 chitinases and construction of a hyper transglycosylating mutant. Biochemistry 50, 5693–5703 [DOI] [PubMed] [Google Scholar]
- 15. Aronson N. N., Jr., Halloran B. A., Alexeyev M. F., Zhou X. E., Wang Y., Meehan E. J., Chen L. (2006) Mutation of a conserved tryptophan in the chitin-binding cleft of Serratia marcescens chitinase A enhances transglycosylation. Biosci. Biotechnol. Biochem. 70, 243–251 [DOI] [PubMed] [Google Scholar]
- 16. Fukamizo T., Goto S., Torikata T., Araki T. (1989) Enhancement of transglycosylation activity of lysozyme by chemical modification. Agric. Biol. Chem. 53, 2641–2651 [Google Scholar]
- 17. Taira T., Fujiwara M., Dennhart N., Hayashi H., Onaga S., Ohnuma T., Letzel T., Sakuda S., Fukamizo T. (2010) Transglycosylation reaction catalyzed by a class V chitinase from cycad, Cycas revolut., A study involving site-directed mutagenesis, HPLC, and real time ESI-MS. Biochim. Biophys. Acta 1804, 668–675 [DOI] [PubMed] [Google Scholar]
- 18. Li C., Huang W., Wang L. X. (2008) Chemo-enzymatic synthesis of N-linked neoglycoproteins through a chitinase-catalyzed transglycosylation. Bioorg. Med. Chem. 16, 8366–8372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan S. Q., Huang W., Wang L. X. (2012) Remarkable transglycosylation activity of glycosynthase mutants of endo-D, an endo-β-N-acetylglucosaminidase from Streptococcus pneumoniae. J. Biol. Chem. 287, 11272–11281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Purushotham P., Podile A. R. (2012) Synthesis of long chain chitooligosaccharides by a hypertransglycosylating processive endochitinase of Serratia proteamaculans 568. J. Bacteriol. 194, 4260–4271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi J., Blundell T. L., Mizuguchi K. (2001) FUGUE. Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310, 243–257 [DOI] [PubMed] [Google Scholar]
- 22. Sashidhar B., Inampudi K. K., Guruprasad L., Kondreddy A., Gopinath K., Podile A. R. (2010) Highly conserved Asp-204 and Gly-776 are important for activity of the quinoprotein glucose dehydrogenase of Escherichia coli and for mineral phosphate solubilization. J. Mol. Microbiol. Biotechnol. 18, 109–119 [DOI] [PubMed] [Google Scholar]
- 23. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 24. Bowie J. U., Lüthy R., Eisenberg D. (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253, 164–170 [DOI] [PubMed] [Google Scholar]
- 25. Lüthy R., Bowie J. U., Eisenberg D. (1992) Assessment of protein models with three-dimensional profiles. Nature 356, 83–85 [DOI] [PubMed] [Google Scholar]
- 26. Ramachandran G. N., Ramakrishnan C., Sasisekharan V. (1963) Stereochemistry of polypeptide chain configurations. Mol. Biol. 7, 95–99 [DOI] [PubMed] [Google Scholar]
- 27. Ke S. H., Madison E. L. (1997) Rapid and efficient site-directed mutagenesis by single-tube “megaprimer” PCR method. Nucleic Acids Res. 25, 3371–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neeraja C., Moerschbacher B., Podile A. R. (2010) Fusion of cellulose binding domain to the catalytic domain improves the activity and conformational stability of chitinase in Bacillus licheniformis DSM13. Bioresour. Technol. 101, 3635–3641 [DOI] [PubMed] [Google Scholar]
- 29. Purushotham P., Sarma P. V., Podile A. R. (2012) Multiple chitinases of an endophytic Serratia proteamaculans 568 generate chitin oligomers. Bioresour. Technol. 112, 261–269 [DOI] [PubMed] [Google Scholar]
- 30. Taghavi S., Garafola C., Monchy S., Newman L., Hoffman A., Weyens N., Barac T., Vangronsveld J., van der Lelie D. (2009) Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl. Environ. Microbiol. 75, 748–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henrissat B., Davies G. (1997) Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644 [DOI] [PubMed] [Google Scholar]
- 32. Zakariassen H., Eijsink V. G., Sørlie M. (2010) Signatures of activation parameters reveal substrate-dependent rate determining steps in polysaccharide turnover by a family 18 chitinase. Carbohydr. Polym. 81, 14–20 [Google Scholar]
- 33. Lü Y., Yang H., Hu H., Wang Y., Rao Z., Jin C. (2009) Mutation of Trp137 to glutamate completely removes transglycosyl activity associated with the Aspergillus fumigatus AfChiB1. Glycoconj. J. 26, 525–534 [DOI] [PubMed] [Google Scholar]
- 34. Makino A., Kurosaki K., Ohmae M., Kobayashi S. (2006) Chitinase-catalyzed synthesis of alternatingly N-deacetylated chitin. A chitin-chitosan hybrid polysaccharide. Biomacromolecules 7, 950–957 [DOI] [PubMed] [Google Scholar]
- 35. Synstad B., Gåseidnes S., Van Aalten D. M., Vriend G., Nielsen J. E., Eijsink V. G. (2004) Mutational and computational analysis of the role of conserved residues in the active site of a family 18 chitinase. Eur. J. Biochem. 271, 253–262 [DOI] [PubMed] [Google Scholar]
- 36. Watanabe T., Kobori K., Miyashita K., Fujii T., Sakai H., Uchida M., Tanaka H. (1993) Identification of glutamic acid 204 and aspartic acid 200 in chitinase A1 of Bacillus circulans WL-12 as essential residues for chitinase activity. J. Biol. Chem. 268, 18567–18572 [PubMed] [Google Scholar]
- 37. Norberg A. L., Dybvik A. I., Zakariassen H., Mormann M., Peter-Katalinić J., Eijsink V. G., Sørlie M. (2011) Substrate positioning in chitinase A, a processive chito-biohydrolase from Serratia marcescens. FEBS Lett. 585, 2339–2344 [DOI] [PubMed] [Google Scholar]
- 38. Krokeide I. M., Synstad B., Gåseidnes S., Horn S. J., Eijsink V. G., Sørlie M. (2007) Natural substrate assay for chitinases using high-performance liquid chromatography. A comparison with existing assays. Anal. Biochem. 363, 128–134 [DOI] [PubMed] [Google Scholar]
- 39. van Aalten D. M., Komander D., Synstad B., Gåseidnes S., Peter M. G., Eijsink V. G. (2001) Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc. Natl. Acad. Sci. U.S.A. 98, 8979–8984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jitonnom J., Lee V. S., Nimmanpipug P., Rowlands H. A., Mulholland A. J. (2011) Quantum mechanics/molecular mechanics modeling of substrate-assisted catalysis in family 18 chitinases. Conformational changes and the role of Asp142 in catalysis in ChiB. Biochemistry 50, 4697–4711 [DOI] [PubMed] [Google Scholar]
- 41. Watanabe T., Ariga Y., Sato U., Toratani T., Hashimoto M., Nikaidou N., Kezuka Y., Nonaka T., Sugiyama J. (2003) Aromatic residues within the substrate-binding cleft of Bacillus circulans chitinase A1 are essential for hydrolysis of crystalline chitin. Biochem. J. 376, 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]