Background: Kinesin-5 motors are important for formation and maintenance of the bipolar mitotic spindle.
Results: ATP binding triggers coupled conformational changes of kinesin-5 specific structural elements in the microtubule-bound motor domain.
Conclusion: Kinesin-5 mechanochemistry is tuned to its cellular functions.
Significance: Subnanometer resolution structure determination of microtubule-bound kinesin-5s and kinetics experiments reveal the molecular basis of their motor properties and of drug inhibition.
Keywords: Fluorescence, Kinesin, Kinetics, Microtubules, Mitosis, Molecular Motors, Cryoelectron Microscopy, Structural Biology
Abstract
Kinesin-5 is required for forming the bipolar spindle during mitosis. Its motor domain, which contains nucleotide and microtubule binding sites and mechanical elements to generate force, has evolved distinct properties for its spindle-based functions. In this study, we report subnanometer resolution cryoelectron microscopy reconstructions of microtubule-bound human kinesin-5 before and after nucleotide binding and combine this information with studies of the kinetics of nucleotide-induced neck linker and cover strand movement. These studies reveal coupled, nucleotide-dependent conformational changes that explain many of this motor's properties. We find that ATP binding induces a ratchet-like docking of the neck linker and simultaneous, parallel docking of the N-terminal cover strand. Loop L5, the binding site for allosteric inhibitors of kinesin-5, also undergoes a dramatic reorientation when ATP binds, suggesting that it is directly involved in controlling nucleotide binding. Our structures indicate that allosteric inhibitors of human kinesin-5, which are being developed as anti-cancer therapeutics, bind to a motor conformation that occurs in the course of normal function. However, due to evolutionarily defined sequence variations in L5, this conformation is not adopted by invertebrate kinesin-5s, explaining their resistance to drug inhibition. Together, our data reveal the precision with which the molecular mechanism of kinesin-5 motors has evolved for force generation.
Introduction
Cell division is orchestrated by the bipolar spindle. This structure is organized through the coordinated production of sustained force from opposing microtubule (MT)2 motors, and it is crucial for accurate chromosome segregation. Although these mitotic motors have evolved specific enzymologies to generate and respond to this force, the molecular basis for such specific adaptations is largely unknown. One of these motors is kinesin-5 (K5), which is important for the formation and maintenance of bipolar spindles (1). K5s oligomerize through coiled-coil interactions to form homotetramers, with pairs of motor domains (MDs) at either end of dumbbell-shaped molecules. This organization is perfectly designed for the cross-linking and sliding of adjacent MTs that contribute to the bipolar architecture of the spindle (1, 2). Dimers of K5 within the tetramer accomplish this process by taking ATP-driven steps along the MT lattice toward their plus-ends (3, 4).
The force-generating capacity of kinesins originates in their MDs, which contain both MT- and ATP-binding sites. Conserved structural elements in the nucleotide-binding site, including the P-loop, and the switch I and switch II loops, undergo coordinated conformational changes in response to ATP binding and hydrolysis (Fig. 1A). These structural changes are transmitted to other regions of the MD to control MT affinity (5, 6). In plus-end kinesins, such as K5s, the conformation of the nucleotide-binding site also controls the orientation of the neck linker (NL), a 14–18-amino acid sequence at the MD C terminus. In the case of the transport motor kinesin-1 (K1), the NL directs motor-generated movement by docking along the MD in response to ATP binding, a structural change that directs the other, tethered head of the K1 dimer toward the plus-end of the MT (7, 8). In K1s, the MD N terminus is also essential for motility because its proximal portion, or cover strand (CS), interacts with the NL, together called the cover neck bundle (CNB), which has been modeled as a β-sheet. CNB formation is hypothesized to be important in reinforcing NL movement, because NL docking alone is not sufficient to account for force transmission and motility (9, 10).
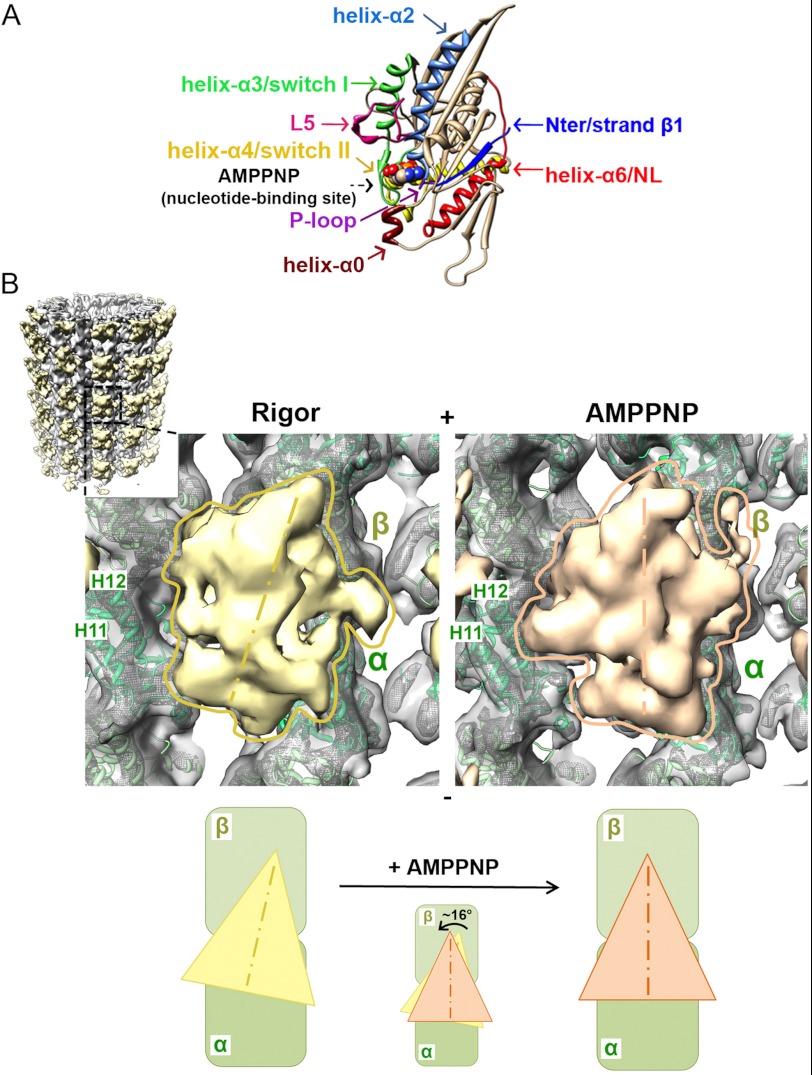
FIGURE 1.
Cryo-EM reconstructions of human K5 MD before and after ATP binding. A, crystal structure of arrowhead-shaped K5 MD bound to AMPPNP viewed with the MT-binding surface at the back. The mechanochemical elements in the MD that sense and respond to bound nucleotide are color-coded and labeled with arrows, with AMPPNP shown in space-fill representation (PDB entry 3HQD (20)). B, cryo-EM reconstructions of MT-bound human K5 MD in rigor (left) and AMPPNP (right) states. The inset shows the whole rigor reconstruction of MT-bound K5 MD, whereas the main panels show a single K5 MD bound to αβ-tubulin dimer (surface contoured at 1.1 σ and mesh contoured at 4 σ, with green ribbon docked) in rigor (yellow) and AMPPNP (orange). Helices H11 and H12 in α-tubulin are labeled. The schematic below illustrates the rotation of the K5 MD that occurs when AMPPNP binds. The MT plus-end is toward the top of this and all subsequent figures.
Despite their conserved features, kinesin MDs with different cellular physiologies show significant variability in several structural elements. This suggests that kinesins modify these regions in order to respond to the specific functional demands placed upon them. One of the most variable of these structural elements is loop 5 (L5), which interrupts the helix-α2 and is close to the P-loop, switch I, and switch II. It is among the most highly variable sequence segments in the kinesin superfamily, ranging from 7 residues in vertebrate kinesin-7 to 18 residues in vertebrate K5 (11) (Fig. 1A). Several recent studies indicate that L5 regulates the ATPase cycle of K5 by modulating ATP binding (12), ADP release (13), microtubule binding (12), and coupling to the NL (14). Moreover, L5 is the binding site of small molecule inhibitors specific to vertebrate K5s (15) that allosterically inhibit ATPase activity and directional movement (16, 17), induce spindle collapse, and arrest mitosis, (18) and have been investigated in cancer clinical trials (19). Together, these data raise fundamental questions about the molecular mechanism by which K5-specific structural elements control the motor's mechanochemistry.
A battery of biophysical, biochemical, and structural techniques has been used to address these important questions. Structures solved using x-ray crystallography have provided residue-by-residue views of the human K5 MD, including highlighting potential conformational variations in the K5 NL and L5 (11, 15, 20, 21). In parallel, spectroscopic measurements have provided dynamic data concerning the timing and regulation of conformational changes within the MD and have been correlated with K5 enzyme kinetics (12, 14, 22). The K5 crystal structures have formed the basis of molecular interpretations of these dynamic data. Unfortunately, these structures are necessarily solved in the absence of MTs, which are crucial partners for the K5 ATPase. This makes it very difficult to directly correlate the nucleotide bound in a kinesin MD crystal structure with the functional state of the motor that is captured (23–26).
To address these deficits, we have combined two complementary methods to determine the structures of the human K5 MD in discrete nucleotide states and the relative timing of conformational changes in relation to nucleotide binding. The first, cryoelectron microscopy (cryo-EM) and image reconstruction, is ideally suited to examine the structure of large complexes and has allowed us to determine the MT-bound structures of human K5 MD at successive steps in its ATPase cycle. Although prior intermediate resolution (∼25–30 Å) cryo-EM studies of human K5-MT complexes provided initial insight into the function of these motors (27, 28), the movements of individual components of the MD could not been visualized. Our subnanometer resolution reconstructions now enable us to visualize coupled, nucleotide-dependent conformational transitions of human K5 structural elements, including those in L5, the NL, and the N terminus. They also enable comparison with a previous reconstruction at equivalent resolution of the Drosophila melanogaster K5 KLP61F (29), which sheds light on the lack of inhibition of KLP61F by human K5-specific drugs (30). The second methodology, transient state kinetics utilizing a series of distance-sensitive probes in the MD, has allowed us to complement these static “snapshots” of the K5 MD structure with dynamic information in order to determine the relative timing of structural changes that occur during the motor ATPase cycle. Our work allows us to reconcile a number of prior inconsistencies in the literature (12, 14, 31, 32), and it reveals the structural basis of K5 force generation and function-specific tuning of its conserved ATPase.
EXPERIMENTAL PROCEDURES
Cloning, Expression, Purification, and Labeling of K5 Constructs
A cysteine-light human K5 motor construct (residues 1–367) with a single cysteine on the neck linker (V365C) was generated, expressed, and purified using a C-terminal His6 tag as described previously (12, 31). We also generated three additional cysteine-containing Eg5 (residues 1–367) mutants in a pET-21a vector for bacterial expression. The first of these is a construct with a single cysteine in L5 (T126C) for undecagold labeling. The other two contain two cysteines and have been used to measure the kinetics of NL docking (V256C,V365C) and CS movement (A2C,V256C) during the ATPase cycle by means of distance-sensitive spectroscopy. The A2C,V256C and V256C,V365C constructs were labeled with tetramethyl rhodamine 5′-maleimide (TMR) by incubation for 24 h with a 10-fold molar excess of TMR over MD, followed by gel filtration on Sephadex G25 to remove unbound label. Labeling stoichiometries of 1.7–1.9 were achieved for both constructs (data not shown). The MT-activated kcat for TMR-labeled V256C,V365C was 5.1 ± 1.6 s−1, whereas that for TMR-labeled A2C,V256C was 4.5 ± 0.9 s−1, both reasonably close to the corresponding value for wild type (33). Additionally, labeling with the FRET donor/acceptor pair AEDANS-DDPM was accomplished by incubating the motor domain construct with 0.2 mol eq of 1,5-IAEDANS for 24 h, followed by incubation with a 10-fold molar excess of DDPM. Excess label was removed by gel filtration on Sephadex G25 (PD10, GE Healthcare). This resulted in a 1:5 to 1:10 AEDANS/DDPM molar ratio, as determined by absorbances at 336 and 440 nm.
An undecagold cluster conjugated to a maleimide linker was used to covalently label single cysteines at specific sites in the K5 motor domain using a similar strategy as was applied to K1 (7, 34). The gold label, provided by Dr. Dan Safer (University of Pennsylvania), was activated with a 100-fold molar excess of N-methoxycarbonylmaleimide in 0.5 m sodium bicarbonate for 90 min on ice, and the reaction was stopped by adding water (50 ml of water for every 1 ml of reaction). The mixture was then applied to CM-Sepharose resin (Sigma) and eluted into labeling buffer (10 mm Hepes, pH 7, 25 mm potassium acetate, 1 mm magnesium acetate, 1 mm EGTA, 0.1 mm ATP) supplemented with 0.2 m potassium acetate. The protein was exchanged into labeling buffer supplemented with 1 mm tris(2-carboxyethyl)phosphine (Sigma) using a PD10 desalting column (Amersham Biosciences). Immediately prior to mixing with a 10-fold molar excess of activated gold, the T126C and V365C constructs were exchanged into labeling buffer (T126C) or labeling buffer supplemented with 50 mm imidazole, pH 7 (V365C), both without tris(2-carboxyethyl)phosphine. After an 8-h incubation at room temperature, labeled proteins were recovered by nickel affinity chromatography. Concentrations of eluted protein and attached gold (determined by absorbance at 420 nm) gave labeling efficiencies of 50% for T126C and 64% for V365C.
Electron Microscopy and Image Processing
MTs were polymerized using bovine brain tubulin (Cytoskeleton, Inc.). The tubulin, at a final concentration of 5 mg/ml, was incubated with 80 mm PIPES, pH 6.8, 5 mm MgCl2, 1 mm EGTA, 5 mm GTP for 2 h at 37 °C. Paclitaxel (1 mm; Calbiochem) in dimethyl sulfoxide was added, followed by a further 2-h incubation at 37 °C. Unlabeled T126C was dialyzed into 80 mm PIPES, pH 6.8, 5 mm MgCl2, 1 mm EGTA before use. MTs (2 μm) and MD (20 μm) were incubated at room temperature with either 5 mm AMPPNP (Jena Biosciences GmbH) for 90 min or with apyrase (1 unit/ml (Sigma), to remove bound nucleotides) for 10 min. Subsequently, 3.5-μl samples were applied to glow-discharged C-flat holey carbon grids (400-mesh; Protochips Inc.), at 24 °C and 100% humidity, followed by automated blotting and plunging into a liquid ethane bath (Vitrobot, FEI Co.). Low dose images were collected on a Tecnai F20 FEG microscope (FEI Co.) operating at 200 kV using a Gatan CT3500 Cryotransfer System (Gatan, Inc). Micrographs were recorded at ×50,000 nominal magnification, 0.8–2.4 μm defocus, on Eastman Kodak Co. SO-163 films and digitized (SCAI scanner, Carl Zeiss, Inc.) to a final sampling of 1.4 Å/pixel.
Segments along 13-protofilament MTs were manually picked using Boxer (35) (helix option). Three-dimensional reconstructions were generated using a previously described custom single particle procedure (36). For each MT, the orientation of the seam was determined by projection matching using a B lattice 13-protofilament MT decorated with K1 motor domain as reference. Two rounds of reference alignment and reconstruction using custom SPIDER (37) scripts and five refinement rounds using FREALIGN (38), taking into account pseudohelical symmetry and including full CTF correction based on parameters determined by CTFFIND3 (39), were performed. The resolution of the final reconstructions for the rigor and AMPPNP states was determined by two methods: 9.5 and 9.7 Å from the Fourier Shell Correlation 0.5 criterion (40) or 10.3 and 10.4 Å from Rmeasure (41) for rigor and AMPPNP, respectively (supplemental Fig. S1 and Table S1). The maps were sharpened using a band pass filter between 20-9.7 Å and 20-9.5 Å. Asymmetric units of the rigor (EMD-2078) and AMPPNP (EMD-2077) reconstructions are deposited in the Electron Microscopy Data Bank.
The same reconstruction procedure was applied to the data sets of gold-labeled T126C (T126C-Au) or V365C (V365C-Au) treated with apyrase or AMPPNP. Gold densities were clearly visible in the four reconstructions (supplemental Table S1 and Fig. S2). An asymmetric unit of each of these reconstructions is deposited in the Electron Microscopy Data Bank: gold_loop5/AMPPNP, EMD-2152; gold_loop5/rigor, EMD-2079; gold_NL/AMPPNP, EMD-2081; gold_NL/rigor, EMD-2080. To extract gold-specific densities, differences between scaled gold-labeled and unlabeled reconstructions were calculated using SPIDER (37) and IMAGIC-5 (42) packages, and low pass and/or median filters were applied to the difference maps to reduce residual noise.
Atomic Model Building
UCSF Chimera (43) was used for rigid body fitting of atomic structures in the cryo-EM reconstructions. The crystal structure of αβ-tubulin dimer (PDB entry 1JFF (44)) was fitted as a rigid body. The quality of the fit was assessed by cross-correlation between the experimental and simulated maps (supplemental Table S2). The four different human K5 MD crystal structures (PDB entries 3HQD (20), 1II6 (11), 1Q0B (15), and 2WOG (21)) were fitted as rigid bodies, and cross-correlation values were calculated (supplemental Table S2). The N-terminal residues 6–16 were added to the docked K5 MD-AMPPNP crystal structure (PDB entry 3HQD), using the Chimera function commands “addaa” and “minimize,” in order to generate a model of the MD that accounts for the EM density in the presence of AMPPNP. Among 250 different conformations of L5 (residues 119–131), produced with the tool ”Model loops with Modeler” in UCSF Chimera, a subset of 10 conformations with the top scores for stereochemistry and accounting for the EM density were selected and subjected to flexible fitting within the K5 MD. The final K5 MD-AMPPNP atomic model was refined by flexible fitting in Flex-EM (45) (supplemental Table S2). The server RIBFIND (46) was used to find the MD rigid bodies for the first iteration of flexible fitting in order to avoid overfitting. Subsequently, individual secondary structural elements were defined as independent rigid bodies in the final iteration. The atomic model for the rigor state was created as follows. 1) The coordinates of residues 16–204, 236–265, and 310–365 from K5 MD-ADP crystal structure (PDB entry 1II6), which account for the K5 MD core and the NL, and residues 205–235 and 266–309 from K5 MD-AMPPNP (PDB entry 3HQD), which account for helix-α3/switch I and helix-α4/switch II, were used to generate a chimeric K5 MD. 2) Different conformations of L5 (residues 119–131) were generated as described above, and the top 10 conformations were selected and added to the chimeric MD. The final chimeric MD with the modeled L5 conformation was refined using Flex-EM (supplemental Table S2). The atomic models are deposited in the Protein Data Bank: AMPPNP, 4AQV; rigor, 4AQW.
Kinetic Methodologies
Pre-steady-state measurements of binding of 2′dmT to the catalytic site and of ATP-induced movement of the NL and the CS were performed on a KinTek SF-2004 stopped flow spectrophotometer with an instrument dead time of 1.2 ms. Binding of 2′dmT was monitored by exciting the mant fluorophore at 295 nm through FRET from MT and MD tryptophans and observing the emission at 90º to the incident beam through a 450-nm broadband pass filter. For experiments using the AEDANS-DDPM donor acceptor pair, the donor (AEDANS) was excited at 332 nm, and a 500-nm broadband pass filter was used to monitor changes in AEDANS fluorescence emission. We had previously reported (31) that the limiting anisotropies for fluorescent probes on the Eg5 neck linker are <0.25, which implies that the error in determining κ2 is between 2 and 10% (47).
For experiments monitoring changes in TMR dimer formation, the fluorophore was excited at 520 nm, and a 590-nm long pass filter was used to monitor changes in TMR emission. Motor-MT complexes were formed prior to stopped-flow experiments by removing unbound nucleotide through gel filtration (PD10), followed by the addition of 0.2 unit/ml apyrase. All experiments were carried out in 100 mm KCl, 25 mm HEPES, 2 mm MgCl2, 1 mm EGTA, pH 7.50, at 20 °C. For experiments using AEDANS-DDPM FRET or TMR dimer formation, MD/MT stoichiometries were typically 1:5–7, using an [MT] of 5–7 μm. We had previously shown that MD/MT stoichiometric ratios from 1:4 to 1:15 gave similar kinetic results (31, 48).
RESULTS
Subnanometer Resolution Reconstructions of the Human K5 MD from Cryo-EM
We collected low dose cryo-EM images of human K5 MD bound to taxol-stabilized MTs in the presence of apyrase (to remove nucleotide) and AMPPNP, a non-hydrolyzable ATP analog. Using single-particle analysis, rigor (nucleotide-free) and AMPPNP structures were reconstructed at 9.5 and 9.7 Å resolution, respectively (Fourier shell correlation criterion 0.5; supplemental Table S1 and Fig. S1). We used a single cysteine mutant (T126C) in a cysteine-free background to generate these reconstructions. We found that the MT-activated ATPase of T126C is within the range reported for wild type and cysteine light K5 MD (T126C kcat = 5.6 ± 0.2 s−1 compared with 5.5–8.3 s−1 (31, 33)). We also monitored the transient kinetics of ATP binding by mixing a range of concentrations of 2′dmT with complexes of MD plus MTs in the stopped flow and monitoring the resulting fluorescence increase, corresponding to binding of 2′dmT to the active site. The rate constant for both wild type and T126C constructs showed similar hyperbolic dependence on [2′dmT] (supplemental Fig. S3), defining maximum rate constants and apparent 2′dmT dissociation constants of 56 ± 19 s−1 and 83 ± 47 μm, respectively, for wild type, and 55 ± 11 s−1 and 82 ± 28 μm, respectively, for the T126C mutant.
In each reconstruction, the characteristic arrowhead-shaped K5 MD points toward the MT plus-end and binds every αβ-tubulin dimer, the building block of the underlying MT lattice (Fig. 1B). The biggest difference between the two reconstructions is that the majority of the K5 MD is rotated counterclockwise by ∼16° in the AMPPNP state, such that its tip points toward the MT plus-end (Fig. 1B). Previous moderate resolution (25–30 Å) cryo-EM studies of K5 showed no such rotation upon binding ATP analogues (27, 28). However, at these lower resolutions, it has been demonstrated that it would not technically be possible to visualize such movements (49). In all cryo-EM studies of plus-end kinesins at resolutions comparable with ours (∼10 Å or better), counterclockwise rotation was seen similar to what we report here for K5s (25, 49). Such a rotation is postulated to result from a rearrangement of conserved loops in the catalytic site in response to ATP binding (25, 49). This rearrangement is structurally amplified, causing the bulk of the MD to rotate around a static contact with the MT (6). Moreover, our reconstructions show that, in addition to MD rotation, large conformational changes also occur throughout the MD when AMPPNP binds.
Generation of Pseudoatomic Models of MT-bound K5 MD in Rigor and AMPPNP States
We generated pseudoatomic models of the K5-MT complex before and after AMPPNP binding. As expected, coordinates of the αβ-tubulin dimer (PDB entry 1JFF) fit well into the MT portion of our reconstructions, validating the quality of our structures (Fig. 1B and supplemental Table S2). To evaluate the K5 MD conformation in each reconstruction, we compared rigid body fits of each of the four different conformations of the human K5 MD captured in crystal structures (supplemental Table S2). The crystal structure of human K5 MD in complex with AMPPNP (PDB entry 3HQD) yielded the best fit into the AMPPNP reconstruction, as evaluated both by the highest correlation value (supplemental Table S2) and by a visual match of the two structures. This structure was therefore used as a starting model for further refinement, discussed below. On the other hand, there is no nucleotide-free K5 crystal structure available, and no clear best fit to the rigor K5-MT reconstruction was obtained with any of the available crystal structures, with particular mismatches observed at the nucleotide-binding site, L5, and the NL (data not shown). Therefore, chimeric coordinates were generated using the AMPPNP (PDB entry 3HQD) and ADP (PDB entry 1II6) crystal structures that matched the rigor reconstruction visually and by computational optimization (Fig. 2, A–C, and supplemental Table S2). Such an approach is frequently taken in cryo-EM reconstructions and in particular was necessary for full interpretation of previous subnanometer reconstructions of MT complexes with K1 (25, 50). As described under “Experimental Procedures,” we generated a set of rigor model structures from these chimeric coordinates containing computationally generated, energy-minimized conformations of L5 that approximately matched the EM density. Both MD docked coordinates were subject to flexible fitting (supplemental Table S2).
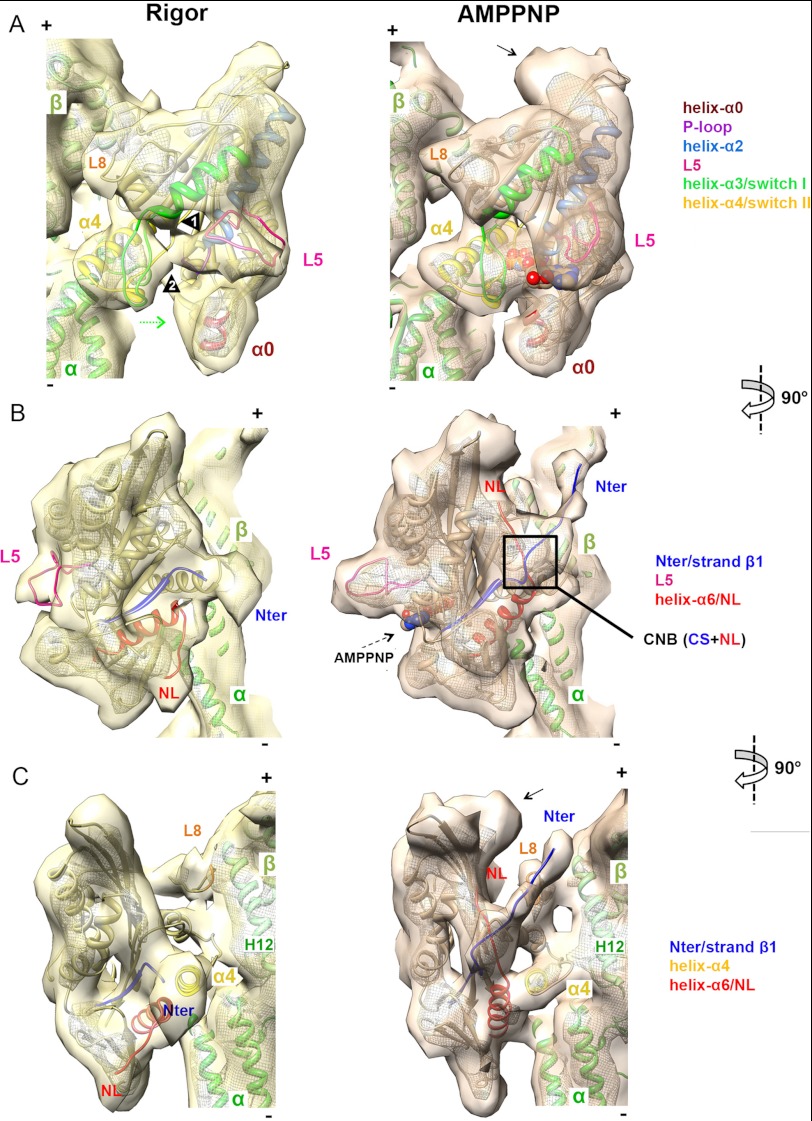
FIGURE 2.
Pseudoatomic models of nucleotide-dependent conformational changes of MT-bound K5. A–C, orthogonal views of a single αβ-tubulin dimer with K5 MD in rigor (left, in yellow) and AMPPNP (right, in orange) reconstructions, contoured as in Fig. 1B. The docked pseudoatomic coordinates are color-coded according to the key. A, view of the nucleotide-binding pocket. Arrowheads 1 and 2 indicate L5 interaction with helix-α3 and switch I, respectively. The curved density at the plus-end MD tip, not accounted for by docked coordinates in the AMPPNP reconstruction (indicated by an arrow), probably corresponds to additional NL residues and part of the histidine tag of our construct; it is only visible when the NL is docked and was previously observed in a K3-AMPPNP MT-bound reconstruction with an equivalent affinity tag (49). B, view of the front face of the K5 MD. C, the view of the NL clearly shows NL docking and ordering of the N terminus.
Binding of Nucleotide Triggers Conformational Changes throughout the K5 MD
Comparison of our rigor and AMPPNP reconstructions, aided by the docked coordinates, clarified the mechanisms by which structural changes are initiated in MT-bound K5 when it binds to ATP. In both structures, density corresponding to helix-α4 forms the main static contact with the MT wall, binding at the intra-tubulin dimer interface (Fig. 2A). K5 loop 8 also extends to the MT surface, but its contacts with helices H11 and H12 of β-tubulin shift slightly as the MD rotates upon AMPPNP binding (Fig. 2, A and C).
In the rigor reconstruction, the empty nucleotide-binding pocket is relatively open. Helix-α4 forms the back of the open active site, whereas the switch I and II loops appear retracted toward the MT binding surface, although, even at this resolution, it is not possible to model their conformations precisely. By contrast, in the AMPPNP reconstruction, the fully occupied nucleotide-binding site is more compact. This indicates that the switch I and II loops have closed (Fig. 2A, left panel, green arrow) to coordinate the bound nucleotide, as shown by the good match of the docked crystal structure in complex with AMPPNP to this region (Fig. 2A). This closure of the switch I and II loops causes rotation of the K5 MD around the static helix-α4 contact with the MT. Thus, the fundamentals of this so-called “seesaw” mechanism of force transduction, shown to apply to both K1 and K3 motors (5, 6), holds true for K5s as well.
Nucleotide-dependent Conformational Changes of the K5 L5
Comparison of the reconstructions and docked coordinates in the vicinity of the nucleotide-binding site highlight the nucleotide-dependent conformational changes in K5 L5 (Fig. 2A). In the AMPPNP reconstruction, L5 corresponds to the protruding density above the closed active site (Fig. 2, A and B). In this ATP-like state, the match between the archlike fold of our model and our reconstruction is very good (Fig. 2 and supplemental Table S2). However, there is a large conformational difference in L5 in the absence of nucleotide, indicated by the lack of the protruding density in the rigor reconstruction (Fig. 2, A and B). Instead, L5 flattens in the direction of both the MT surface and the short helix-α0 at the bottom of the MD and lies at the entrance of the active site. Density in the region of L5 is connected with the base of helix-α3 (Fig. 2A, arrowhead 1) and possibly with switch I at the nucleotide-binding site (arrowhead 2). Although our reconstruction is not detailed enough to reveal the exact conformation of L5, one such computationally generated conformer that approximately matches the EM density is shown in Fig. 2.
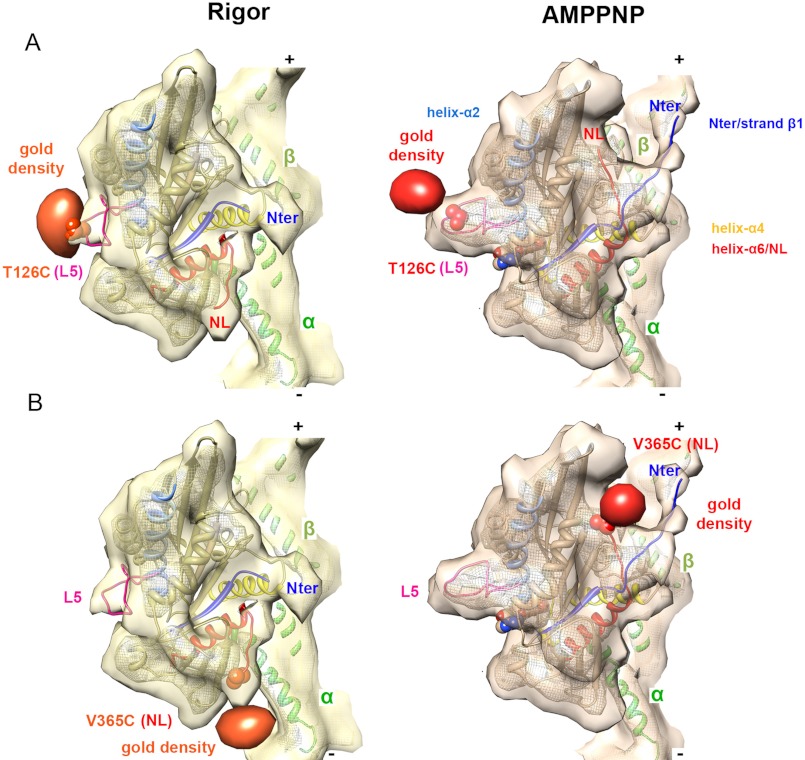
To confirm this dramatic reorientation of L5, we covalently attached an electron-dense undecagold cluster to the engineered cysteine in this loop (T126C-Au (34)) and calculated reconstructions of gold-labeled K5 MD. We first determined that the presence of the gold cluster did not perturb either the global structure of K5 MD itself or the nucleotide-dependent closure of the nucleotide-binding site and resulting MD rotation (supplemental Fig. S2). This validates our approach, used previously for elucidating the K1 stepping mechanism (7), and shows that the gold labels enable us to specifically track nucleotide-dependent conformational changes of L5.
The reconstructions of gold-labeled protein have additional density in the region of L5 compared with unlabeled K5 MD (supplemental Fig. S2). Calculation of the differences between the reconstructions in the presence and absence of gold label allowed more precise localization of the gold cluster in each nucleotide state. In the AMPPNP reconstruction, the position of the gold cluster confirms that the protruding density above the closed active site corresponds to L5 (Fig. 3A and supplemental Fig. S2). By contrast, the L5 gold label in the rigor reconstruction has shifted toward the nucleotide-binding site, consistent with our pseudoatomic modeling. These data confirm that AMPPNP binding moves L5 from a position proximal to the catalytic site to a more distant position on the MD surface (Fig. 3A). The position of the gold densities also validates the L5 pseudoatomic models for both rigor and AMPPNP states.
FIGURE 3.
Site-specific gold labels track nucleotide-dependent conformational changes. A, density corresponding to a gold cluster covalently attached to L5 (via T126C shown in a space-filling representation) is superimposed on the K5-MD reconstructions in rigor and AMPPNP and confirms the reorientation of L5 when AMPPNP binds. Gold difference maps in orange (rigor) and red (AMPPNP) are contoured at 2.6 and 2.4 σ, respectively. B, density corresponding to a gold cluster covalently attached to the NL (via V365C shown in a space-filling representation) is superimposed on the K5-MD reconstructions in rigor and AMPPNP and confirms the ordered-to-ordered transition of the NL toward the MT plus-end when AMPPNP binds. Gold difference maps in orange (rigor) and red (AMPPNP) are contoured at 1.6 and 2.4 σ, respectively.
Nucleotide-dependent Conformational Changes in the K5 NL
In K1s and K3s, ATP binding causes the NL to undergo a structural transition, from a state where it is flexible and unstructured to one where it is docked along the MD toward the MT plus-end and structured (5, 6). This docking is part of the allosteric communication mechanism between MDs in kinesin dimers that enables them to take multiple steps along the MT lattice, a property called processivity. However, it has been unclear whether the same nucleotide-dependent changes of NL structure also occur for K5s, given that 1) the K5 NL sequence is longer and non-conserved compared with K1s and K3s; 2) K5 dimer constructs show variable processivity compared with K1s (3, 4, 51); 3) the K5 NL has been captured in an unusual, immobilized conformation in crystal structures in the presence of ADP (11, 21); and 4) the K5 NL may be relatively ordered in rigor as well as in the presence of nucleotide, suggesting that nucleotide-dependent conformational changes in the NL in K5s are significantly different from what is seen for K1s (14, 31).
Although the K1 NL is not visible in rigor reconstructions of MT-bound motor (7, 36), density corresponding to the NL in K5 can be readily visualized both before and after nucleotide binding, consistent with prior spectroscopic studies (31). In particular, we find that the K5 NL is structured in rigor, where it points toward the MT minus-end (Fig. 2, B and C). This orientation was originally seen in a K5 crystal structure solved in the presence of ADP (11) or ADP plus S-trityl-l-cysteine bound to L5 (21), both of which represent a nucleotide state different from that of our rigor reconstruction. We interpret this finding as further evidence of the structural uncoupling that can occur between nucleotide occupancy and motor conformation in the absence of MTs. The close match between our rigor reconstruction and this NL conformation supports the idea that in rigor, the MT-bound K5 NL is structured and directed toward the MT minus-end.
In contrast, the conformation of the K5 NL in our AMPPNP reconstruction matches the K5 NL conformation in the AMPPNP crystal structure and, like K1, is seen to be docked along the MD toward the plus-end (Fig. 2, B and C). Although the distal portion of the K5 NL contains a well conserved proline (Pro-363) that might be predicted to influence NL conformation (11), the presence of this geometry-constraining residue does not appear to influence the fundamental process of NL docking along the MD upon binding to AMPPNP (7, 20). It also highlights that the crystal structure of the K5 MD in AMPPNP, generated in the absence of MTs, correlates closely with many aspects of the MT-bound AMPPNP motor that we observe in our reconstructions.
To validate our interpretation of these nucleotide-dependent NL movements, we covalently attached a gold cluster to a single reactive cysteine in the NL (V365C-Au) (12). We calculated reconstructions of this gold-labeled protein (supplemental Fig. S2), determined that there were no detectable structural perturbations in the gold-labeled MD, and identified the position of the gold in difference maps (Fig. 3B). The gold density matches the location of the NL in our docked models very well, and it emphasizes the large reorientation of ∼120°, from minus-end-directed to plus-end-directed, that the K5 NL undergoes upon binding AMPPNP (Figs. 2 (B and C) and 3B) (14, 31).
In a recent study, it was shown that an EPR spin probe on the NL was immobilized in both rigor and ATP-like states (14). The authors assumed that this spectroscopic finding corresponded structurally to a plus-end orientation, and they concluded that NL docking in K5, unlike the case for K1, occurs with ADP release. However, our ability to directly visualize the K5 NL orientation in rigor and ATP now allows us to conclude that 1) although the K5 NL is immobilized in rigor, consistent with the EPR studies, it is directed toward the MT minus-end, and 2) as with K1s, NL docking in K5 occurs with ATP binding. Our reconstructions reveal a large movement of an ordered K5 NL when ATP binds (Figs. 2 and 3), in which it swings from a minus- to a plus-end direction. This ATP-dependent ordered-to-ordered structural transition distinguishes the K5 NL from that for K1, which has previously been shown to undergo a disordered-to-ordered transition (25, 31, 48).
Evidence for CNB Formation in K5
A relatively unexplored aspect of the K5 motor mechanism is the role of its 19-residue N terminus. In our AMPPNP reconstruction, validation of the docked NL conformation by the location of the gold label enables us to infer that the nearby elongated density, also pointing toward the MT plus-end, probably corresponds to the K5-specific N terminus, partially modeled in Figs. 2 and 3.
That the docked NL and the N terminus both point toward the MT plus-end in our AMPPNP reconstruction suggests that CNB formation occurs in K5s as it does in K1s (9, 10). This is consistent with the observation of ATP-dependent correlated movements of the N terminus and NL in K5s (12). In contrast, the minus-end-directed NL conformation in rigor prevents formation of the CNB, supported by our finding that the N terminus is disordered in this state (Figs. 2 (B and C) and 3B). These data serve to highlight that CNB formation is likely to be a conserved aspect of kinesin motility and force generation across plus-end kinesin families.
Spectroscopic Probes Can Detect NL and CS Movement
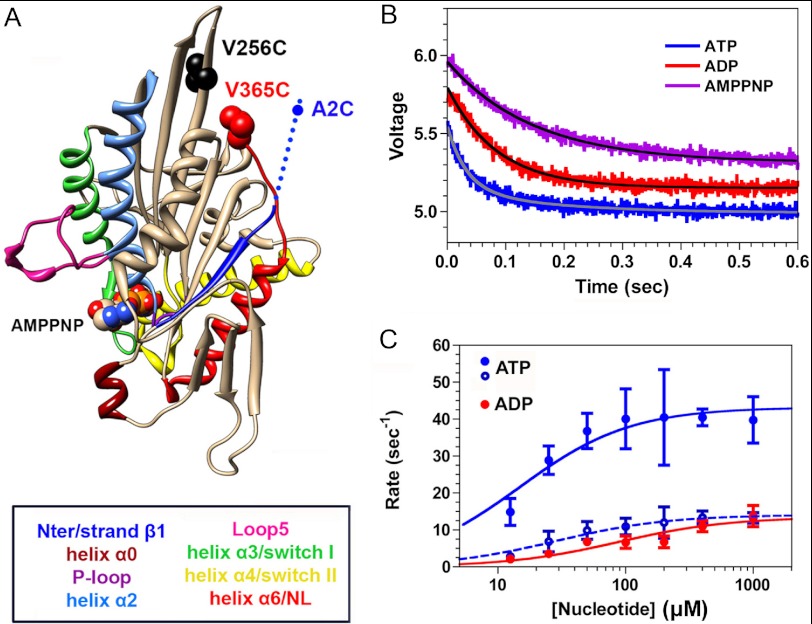
Although our cryo-EM reconstructions provide new information about how nucleotide binding alters the structure of the K5 MD, they do not allow us to determine the timing of these movements. This requires transient kinetic experiments, using spectroscopic probes that are sensitive to position. In our prior study (12), we attached a fluorescent donor (FlAsH) and a non-fluorescent acceptor (QSY7) to the MD N terminus and the C terminus of the NL, respectively, of a K5 MD-MT complex. Mixing with ATP induced a rise in fluorescence, indicating a separation of NL from CS, that followed a lag of ∼20 ms, corresponding to a rate constant of 40–50 s−1 (12). By contrast, mixing with ADP produced a rise in fluorescence without a lag. We argued on the basis of these findings that 1) ATP binding induces the CS and NL to “dock” as a unit with a rate constant of ∼40–50 s−1, corresponding to the lag phase; 2) the NL and CS subsequently separate with formation of a weak MT-binding conformation, producing a fluorescence enhancement; and 3) the relative orientation of the CS and NL in rigor and in ADP are different from each other. We interpreted these findings by assuming that the N terminus was flexible and randomly oriented throughout the ATPase cycle and that any changes in FRET efficiency reflected movements of the NL alone. However, our cryo-EM reconstructions clearly show that both the NL and CS reorient themselves in response to nucleotide, and they underscore the importance of measuring the kinetics of CS and NL movements individually. In order to accomplish this, we generated two double cysteine K5 MD mutants, A2C,V256C and V256C,V365C. In both, we placed one cysteine within the β-core of the MD (V256C, near the plus-end tip (Fig. 4A)), because this residue does not appear to change orientation with nucleotide binding and can therefore serve as a stable frame of reference. We placed the other cysteine at either the N terminus (A2C) or the C terminus of the NL (V365C) (Fig. 4A). These mutations did not appear to have any appreciable effect on the MT-activated ATPase kcat (data not shown).
FIGURE 4.
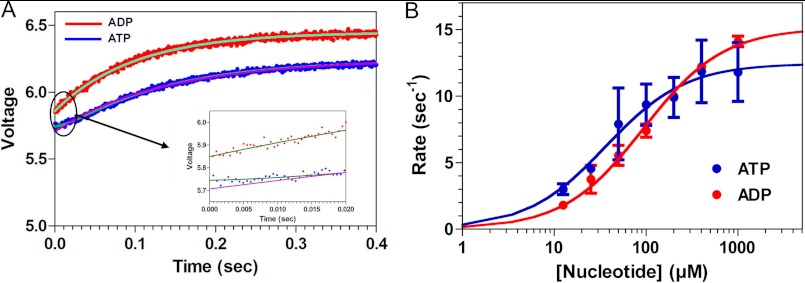
Kinetics of nucleotide-induced movement of the NL measured by TMR quenching. A, location of the inserted cysteines. B, mixing a rigor complex of 4 μm TMR-labeled V256C,V365C plus 20 μm MTs with 4 mm ATP (blue jagged curve) in the stopped flow produces a decrease in fluorescence intensity over time that fits a double exponential rate equation (solid gray curve), whereas the corresponding experiments with 4 mm ADP (red jagged curve) and 4 mm AMPPNP (magenta jagged curve) fit a single exponential rate process (solid black curves). The transient for AMPPNP has been offset by +0.2 V, whereas that for ATP has been offset by −0.2 V in order to allow the three transients to be distinguished. C, for both ATP and ADP, the value of the rate constants varied with [nucleotide]. Fitting of the rate constants for the ATP-induced transients to a hyperbolic isotherm yields maximum values of 43.2 ± 3.5 and 14.1 ± 1.4 s−1 for the fast (filled circles) and slow phases (unfilled circles), respectively. The corresponding hyperbolic fitting for ADP yields a maximum rate constant of 13.5 ± 1.7 s−1. Each point and bar represents the mean of four independent measurements ± 1 S.D. Conditions were as follows: 100 mm KCl, 25 mm HEPES, 2 mm MgCl2, 1 mm EGTA, pH 7.50, 20 °C.
We used a distance-sensitive spectroscopic approach with high signal/noise ratio to measure the kinetics of NL and CS movements, involving the static quenching of TMR fluorescence through dimer formation. This method takes advantage of the ability of TMR to dimerize weakly in solution, a process that can be markedly enhanced if the TMR fluorophores are attached to structural elements within a protein that approach each other to within the 10–20 Å necessary to achieve static quenching (52). TMR dimer formation produces a blue shift in the absorption spectrum and a >20-fold reduction in fluorescence emission. The energy transfer efficiency from FRET varies continuously with the degree of separation donor and acceptor fluorophores, and at the ensemble level, it provides a measure of the mean distance between these fluorophores. By contrast, TMR quenching is “all or none,” and fluorescence intensity at the ensemble level measures the fractional population of protein molecules whose attached TMR fluorophores can approach each other within the necessary distance to achieve quenching. We had previously used this method to monitor the separation of the K1 neck linkers at both the ensemble and single molecule levels during the ATPase cycle (8, 53). As demonstrated in supplemental Fig. S4, the resulting absorption spectrum for a V256C,V365C preparation in the absence of nucleotide and MTs demonstrates a 518/555 nm absorbance ratio of 1.19, which implies that ∼85% of the TMR probes are dimerized in this state (solid red curve). Unfolding the MD with 6 m guanidine prevents any intradomain interactions and produces a TMR absorption spectrum characteristic of monomer (supplemental Fig. S4, dashed blue curve). Similar results were seen for the A2C,A256C construct (data not shown). This implies that the NL of a MT-free, rigor MD is close to the motor core, contrary to what we are observing by cryo-EM in our rigor MD-MT complexes (Figs. 2 and 3). This suggests that binding of a rigor MD to the MT induces undocking of the NL from the motor core. Furthermore, it predicts that mixing TMR-labeled, rigor V256C,V365C with a 5-fold molar excess of MTs in the stopped flow should separate the two TMR fluorophores from each other and enhance the fluorescence emission. This is confirmed in supplemental Fig. S5, which illustrates that MT binding unquenches TMR fluorescence.
Kinetics of Nucleotide-induced NL Movement
We mixed a complex of TMR-labeled V256C,V365C plus MTs with 4 mm ATP, ADP, or AMPPNP in the stopped flow, and the resulting transients are depicted in Fig. 4B. In each case, they consisted of a reduction in TMR fluorescence, implying that nucleotide binding moves the NL in close proximity to position 256 in the motor core (Fig. 4A). However, in the case of ATP (Fig. 4B, blue), the transient was biphasic, with the faster phase constituting 60–70% of the total signal amplitude. By contrast, the transients produced by mixing with ADP (Fig. 4B, red) or AMPPNP (magenta) were monophasic. Over a range of ATP and ADP concentrations, the rate constants derived from the biexponential and monoexponential rate equations, respectively, varied hyperbolically with [nucleotide] (Fig. 4C), defining maximum rates of 43.2 ± 3.5 and 14.1 ± 1.4 s−1 for ATP (blue) and 13.5 ± 1.7 s−1 for ADP (red). The slower of the two rate constants for ATP and the rate constant for ADP are still faster than those for nucleotide-induced MT dissociation (8.5–8.7 s−1) (12).
We confirmed these findings by labeling the V256C,V365C construct with the FRET pair 1,5-IAEDANS (5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1 sulfonic acid; fluorescent donor) and DDPM (fluorescence quencher), forming a nucleotide-free complex with MTs, mixing in the stopped flow with ATP or ADP, and monitoring the change in AEDANS donor fluorescence. As with our experiments with TMR, we observed a fluorescence decrease that was monophasic for ADP and biphasic for ATP, implying that the mean distance between AEDANS and DDPM had shortened (supplemental Fig. S6A). As a control, we repeated these experiments with the double cysteine construct labeled only with donor (AEDANS) and found that mixing with nucleotide had little effect on the AEDANS fluorescence emission (data not shown). The rate constants varied hyperbolically with [nucleotide], and the extrapolated maximum rate constants (47.2 ± 3.0 and 14.4 ± 1.7 s−1 for ATP (blue) and 12.4 ± 0.4 s−1 for ADP (red) are similar to what we observed with TMR (supplemental Fig. S6B).
Our results suggest that ATP-induced movement of the NL toward the motor core is a two-step process that precedes MT dissociation. The first step occurs at roughly the same rate as ATP binding to the catalytic site in K5 (31) and reorients the NL from a rigor orientation to what is seen in the presence of AMPPNP (Fig. 3). The second step occurs at the same rate as the strong-to-weak transition in K5 (31), a process that precedes dissociation from the MT. By contrast, mixing with AMPPNP produces a single phase in the fluorescence transient (Fig. 4B), as would be expected for this non-hydrolyzable ATP analog. The slower rate constant for this process (6.7 ± 0.1 s−1) compared with ATP is consistent with its >10-fold lower affinity for the catalytic site of kinesins (54). Furthermore, our finding that ADP binding also quenches TMR fluorescence suggests that, unlike K1, the NL of K5 remains docked during the transitions from ATP to ADP states, so long as it is bound to the MT, and that the orientation of the K5 NL in the presence of ADP is different from that in rigor (7).
Kinetics of Nucleotide-induced CS Movement
We used the same approach described above with our A2C,V256C construct in order to measure the movement of the CS relative to the motor core. TMR labeling of this construct also was associated with an absorption spectrum consistent with TMR dimerization (data not shown). Mixing a complex of 4 μm TMR-labeled A2C,V256C plus 20 μm MTs with ATP or ADP in the stopped flow produced a rise in TMR fluorescence (Fig. 5A), implying that binding of either nucleotide induces the CS to move away from the motor core. In the case of ATP, fitting of the observed fluorescence transient (jagged blue curve) to a single exponential term was inadequate (smooth magenta curve in Fig. 5A and inset) and required two exponential terms, one corresponding to a lag phase associated with a rate constant of ∼50 s−1 and the second corresponding to a rise in fluorescence (smooth cyan curve; highlighted in the inset on a shortened time scale). This is qualitatively very similar to what we had previously observed using FRET donor/acceptor pairs on the NL and N terminus (12). The rate constant for the rising phase of the transient varied hyperbolically with [ATP], defining a maximum rate of 10.2 ± 0.3 s−1 (Fig. 5B, blue). This extrapolated rate constant is nearly identical to what we had measured previously using a FRET donor/acceptor pair on the N terminus and NL (12). In contrast, adequate fitting of the ADP transient (Fig. 5A, jagged red curve) could be accomplished with a single exponential process (Fig. 5A, smooth green curve). The rate constant for this process varied hyperbolically with [ADP] (Fig. 5B, red), defining a maximum rate of 13.8 ± 0.9 s−1, also nearly identical to what we had previously measured for ADP-induced separation of the CS and NL (12).
FIGURE 5.
Kinetics of nucleotide-induced movement of the CS. A, a double cysteine K5 construct (A2C,V256C) was labeled with TMR, and a rigor complex of 4 μm TMR-labeled V256C,V365C plus 20 μm MTs was mixed in the stopped flow with ATP (blue jagged curve) or ADP (red jagged curve). For ATP, the resulting fluorescence transient consisted of an increase in fluorescence, implying a separation of the CS from residue 256 in the motor core. However, fitting of this transient to a single exponential rate equation (smooth magenta curve) is inadequate at short time intervals (black oval), and this is illustrated in the inset. Adequate fitting required a double exponential rate equation, which assumes the presence of a lag (smooth cyan curve, illustrated on an expanded scale in the inset). By contrast, the corresponding transient produced by mixing with ADP (red jagged curve) could be adequately fit with a single exponential process (smooth green curve, illustrated on an expanded scale in the inset). B, for both ATP and ADP, the rate constant for the rising phase varied hyperbolically with nucleotide concentration, defining maximum rate constants of 10.2 ± 0.3 s−1 for ATP (blue) and 13.8 ± 0.9 s−1 for ADP (red). Each point and bar represents the mean of four independent measurements ± 1 S.D. Conditions were as follows: 100 mm KCl, 25 mm HEPES, 2 mm MgCl2, 1 mm EGTA, pH 7.50, 20 °C.
These results imply that the TMR fluorophores are detecting a change in CS orientation that occurs with or after ATP hydrolysis. To test this further, we mixed a complex of 1 μm TMR-labeled A2C,V256C plus 5 μm MTs with either 4 mm ATP, ADP, or AMPPNP. Results are depicted in supplemental Fig. S7. Mixing ATP (blue) and ADP (red) again produced a fluorescence increase with rate constants in the 10–14 s−1 range. By contrast, mixing with the non-hydrolyzable ATP analog AMPPNP produced no appreciable fluorescence change over the same time course.
By combining our current results with our earlier study (12), we can make several conclusions. First, in a rigor K5-MT complex, the N terminus is flexible and can interact at least transiently with the motor core. This is consistent with our cryo-EM reconstructions of rigor K5-MT complex, which indicate that this region is unstructured. It may also explain why the fluorescence of a rigor, TMR-labeled A2C,V256C-MT is quenched (Fig. 5). Second, ATP binding induces a coordinated docking of the NL and CS along the surface of the motor core at a rate of 40–50 s−1. Third, ATP hydrolysis and formation of a weak MT binding state causes the CS to peel away from the motor core and break its interaction with the NL. This latter effect might in turn be expected to lead to a reorientation of the NL and could account for the slower phase that we observe of ATP-induced changes in TMR fluorescence (Fig. 4B). However, even in the weak MT-binding state that is induced by ATP hydrolysis or by ADP binding, our spectroscopic results argue that the NL is still in a forward orientation, because we still observe quenching of TMR and AEDANS fluorescence. The loss of an interaction with the CS might be expected to reduce NL stability and could explain the enhanced flexibility of the NL noted in earlier EPR studies in the presence of ADP (14).
DISCUSSION
The model of the spindle as “mitotic muscle” (55) captures the ensemble nature of coordinated force generation by many motors that act to establish spindle organization and drive its function. K5s are central players in mitosis because they slide spindle MTs and sustain cross-bridges between them in the face of sustained opposing force from minus-end-directed motors, including ncd and dynein (56). Given that the enzymology and structure of molecular motors are shaped by the physiologic requirements placed on them, it seems likely that the molecular properties of the K5 MD must be very different from those of a transport motor, such as K1. Our data show the structural basis for some of the adaptations made by the K5 MD in order to serve these requirements.
Cryo-EM of the K5 MD Reveals Coupled Conformational Changes in Response to ATP Binding
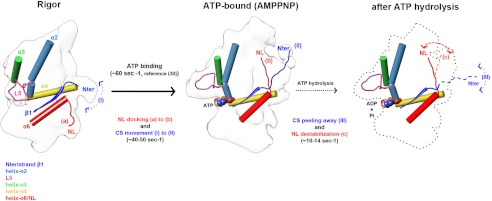
We have used the ATP analog AMPPNP in conjunction with subnanometer cryo-EM structure determination to identify the structural consequences of ATP binding. This work demonstrates that the local conformational changes induced by ATP binding, including closure of the switch I and II loops around the active site, trigger a set of further conformational changes that are propagated throughout the MD (supplemental Movie S1). These include 1) retraction of L5 away from the catalytic site, 2) a counterclockwise motion of the K5 MD on the MT lattice, 3) a large swing of a structured K5 NL toward the MT plus-end, and 4) the formation of a K5 CNB between the NL and N-terminal CS. How do our structural findings relate to K5 function?
K5 L5 Conformation Transmits Nucleotide Occupancy
Consistent with prior EPR and molecular dynamics studies (12, 14, 57), our data argue that L5 can control nucleotide binding by direct steric effects (supplemental Fig. S8) as well as indirectly, through allosteric effects on the dynamics of switch I. The proximity of L5 to the nucleotide-binding site in our rigor structure suggests that the negatively charged L5 and the negatively charged phosphates of ATP might generate an electrostatic repulsion that blocks ATP binding. In particular, the displacement of L5 away from the nucleotide-binding site to a conformation protruding from the surface of the MD when AMPPNP is bound could correspond to a previously described isomerization that rate-limits ATP binding compared with K1 and which also correlates with NL docking (Figs. 2 and 3 and supplemental Movie S1) (14, 31, 58). The proximity of L5 to the nucleotide-binding site provides a means of communication between the two sites, such that ADP release would result in collapse of unliganded L5 toward the nucleotide-binding pocket. Specifically, the side chain of L5 residue Glu-118 hydrogen-bonds with the sugar moiety of an ATP analog (20) or with ADP in the presence of a small molecule inhibitor (15, 59). Mutation of Glu-118 affects nucleotide affinity and catalytic efficiency (59, 60), suggesting that it could act as a sensor of active site occupancy. Intriguingly, such nucleotide binding regulation by a family-specific insert is reminiscent of the proposed role of “insert 1” in myosin VI, which occupies a position with respect to the nucleotide binding site that is structurally equivalent to L5 (61).
Molecular Basis of K5 Drug Inhibition Mechanism
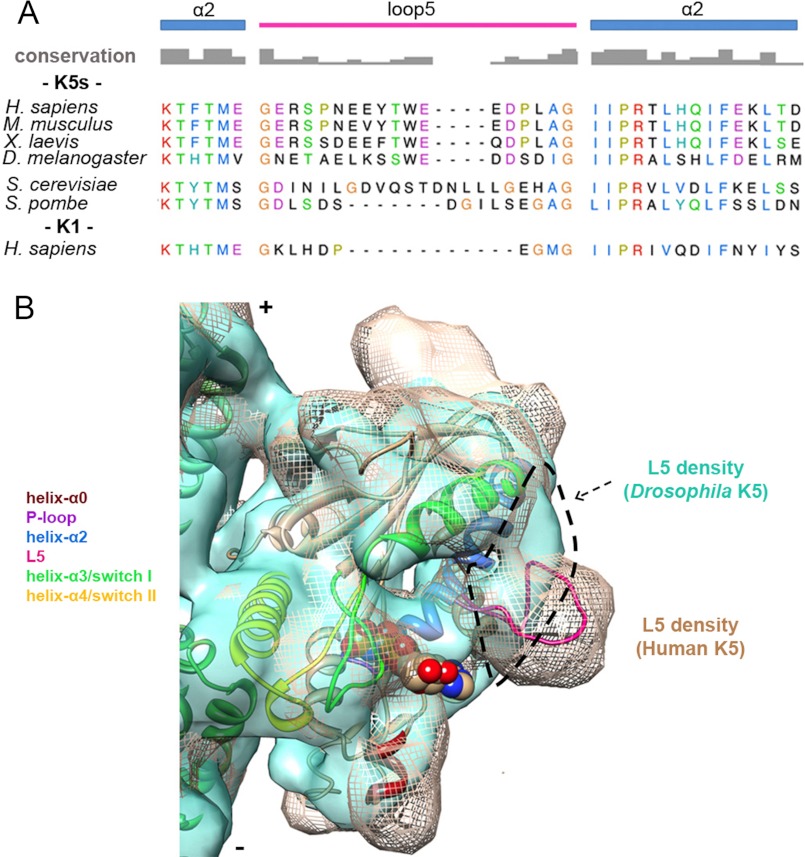
The length and content of L5 are extremely well conserved in vertebrate K5s (11, 20, 30) (Fig. 6A). Its conformation in human K5 is constrained by Pro-121 and Pro-131. Mutating these residues to alanine affects MT and ATP binding (12), and this provides further support for the argument that the conformation of L5 plays a critical role in vertebrate K5 nucleotide exchange (30). However, although L5 in the Drosophila K5 homologue, KLP61F, is the same length as in vertebrate K5s, these conserved prolines are missing. This may explain why, when compared with our AMPPNP reconstruction of human K5, L5 in MT-bound KLP61F-AMPPNP adopts a very different, flattened conformation against helix-α3 (29) (Fig. 6B). The conformation of L5 that we observe in our human K5 AMPPNP reconstruction is extremely similar to that in the presence of allosteric small molecule inhibitors (15) (supplemental Fig. S9). This suggests that these allosteric inhibitors bind in an L5 pocket that forms in the course of the vertebrate K5 ATPase cycle rather than through an induced fit mechanism (16, 59). We propose that small molecule binding stabilizes this conformation, thereby blocking motor function. Our structural comparison (Fig. 6B) explains why KLP61F is insensitive to drug inhibition (22, 30). Drug sensitivity can be imposed on KLP61F if the human L5 sequence is spliced into its MD, emphasizing the unusual modularity of this loop (22).
FIGURE 6.
Comparison of K5s L5 sequence and conformation. A, alignment of helix-α2/L5 sequences from K5s (Homo sapiens P52732, 111–149; Mus musculus Q6P9P6, 110–148; Xenopus laevis P28025, 104–142; D. melanogaster P46863, 109–147; Saccharomyces cerevisiae P28742, 137–189; Schizosaccharomyces pombe P24339, 155–200) and K1 (H. sapiens P33176, 91–121) generated using T-coffee (63). Secondary structure elements and amino acid conservation are indicated. Among these sequences, identical or similar residues are shown with the same color, whereas non-conserved residues are in black. B, cryo-EM reconstruction of MT-bound Drosophila K5 MD (KLP61F)-AMPPNP (light green surface, 0.5 σ, Electron Microscopy Data Bank entry 1604 (29)) superposed on the human K5 MD-AMPPNP structure (mesh, 1.1 σ). Our pseudoatomic AMPPNP MD model is fitted in the mesh map. The dashed outline delineates loop 5 density in KLP61F reconstruction showing the very different conformations of L5 in these K5s.
Coupled Movement of NL and CS during Force Generation
Our reconstructions show that the movement of L5 in response to AMPPNP binding correlates with a ratchet-like reorientation of the K5 NL toward the MT plus-end. Although there is a lack of consensus in the literature about the extent of K5 processivity (3, 4, 51), our current results provide a structural framework in which to consider different ideas concerning K5 mechanochemistry.
First, the observation of CNB formation when AMPPNP binds and, thus, in K5 force generation helps to explain the results of protein engineering experiments that showed that the K1 and K5 CNB components are not functionally interchangeable (i.e. the N terminus and NL of individual MDs have co-evolved and must be paired to generate wild-type motility) (62). CNB formation also has a significant regulatory effect on K5 nucleotide affinity and motor ATPase (32, 62), further highlighting the numerous allosteric effects transmitted across the K5 MD (59). The disorder of the N terminus in the K5-AMPPNP crystal structure (20) further emphasizes the essential role of the MT in full K5 function.
Our kinetic studies, using distance-sensitive spectroscopic probes, support the argument that the CS and NL function in a coordinated manner. Combined with our prior work (12), the current study supports our proposal that ATP binding induces coupled movement of the CS and NL together, followed by separation of the CS from the NL with formation of a weak MT binding state. One concern is that our results with TMR dimer quenching could be influenced by the fact that in solution, TMR fluorophores have a weak affinity for each other (Kd ∼600 μm (52)). The presence of such a weak interaction might bias a forward orientation for the NL and the CS in our labeled preparations, and it could explain why, in the absence of MTs, the TMR fluorescence of labeled V256C,V365C and A2C,V256C is quenched (supplemental Fig. S4). However, the kinetics of ATP and ADP-induced changes in NL orientation, as measured by TMR quenching, are very similar to those using AEDANS-DDPM, where such interprobe interactions do not occur (supplemental Fig. S6). Furthermore, binding of V256C,V365C to the MT unquenches the TMR fluorescence (supplemental Fig. S5). Together, these findings imply that any weak stabilizing effect of TMR dimer formation in the unbound MD is overcome by the effect of MT binding, which appears to enforce an orientation of the NL consistent with our cryo-EM reconstructions.
Our current results shed new light on structural and kinetic factors that impact on K5 mechanochemistry and distinguish it from that of K1s (Fig. 7). The K5 NL is ordered before and after ATP binds to the MT-attached K5 MD, and K5 NL docks toward the MT plus-end at the same rate as ATP binding. In contrast, the N terminus is mobile in rigor but becomes more ordered when ATP binds and the CS forms the CNB in association with the NL. Our kinetic data show that whereas the K5 CS is released as ATP hydrolysis proceeds, the NL remains docked, a significant difference compared with K1s and probably having an impact on K5 dimer stepping. This could correlate with L5 retaining its position above the nucleotide binding site while nucleotide is bound (14), pointing again to a strong regulatory role for L5 in overall K5 activity.
FIGURE 7.
Structural and kinetic schematic of K5 MD mechanochemistry. Left, when the K5 MD is bound to MTs in the rigor state, L5 is proximal to the nucleotide binding pocket, the NL (red) is ordered and directed toward the MT minus end (a), and the N terminus is disordered and interacts flexibly with the MD (I). Middle, ATP binds (∼60 s−1 (12)) and induces closure of the nucleotide binding site and MD rotation; L5 is displaced from the nucleotide binding pocket. The NL reorients and docks toward the MT plus-end (b) coupled with ordering of the N terminus (∼40–50 s−1), also toward the MT plus-end (II), enabling formation of the CNB. Right, following ATP hydrolysis, the N terminus becomes disordered again (III), whereas the NL remains oriented toward the plus-end (c) although slightly destabilized (∼10–14 s−1). MD outlines and docked secondary structure elements are based on EM reconstructions. The orientation of the NL and N terminus after ATP hydrolysis revealed by the kinetics data (Figs. 4 and 5) are represented by the dotted lines. The unknown structure of the MD is indicated by the dotted outline.
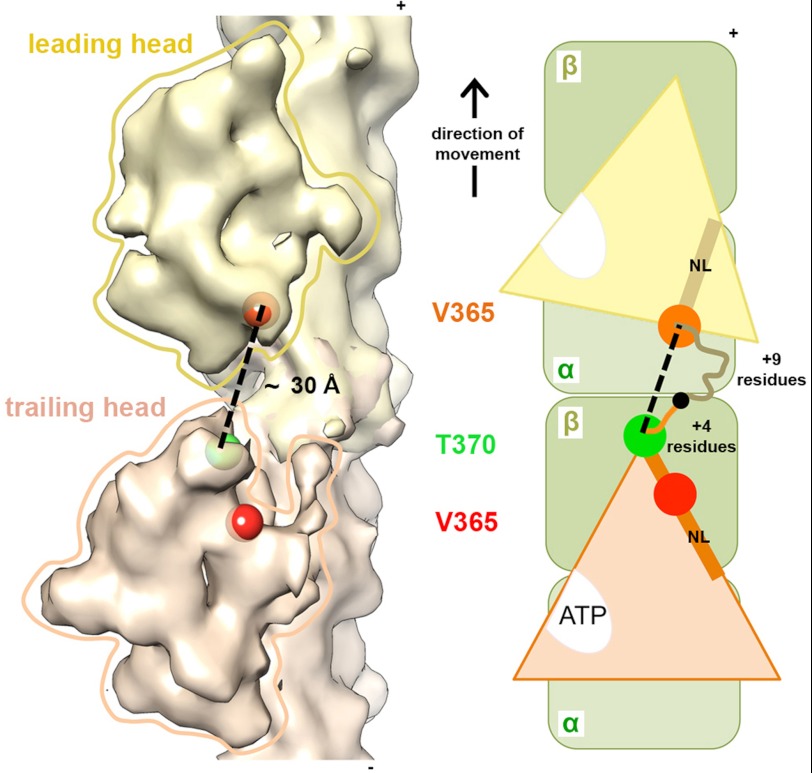
Finally, our results also provide additional data on the controversial subject of K5 processivity (3, 4, 51); a model of a hypothetical K5 dimer based on our reconstructions, with the leading head in a rigor conformation and the trailing head in an AMPPNP-like conformation, supports the idea that the 18-residue K5 NL would be expected to reduce intradimer tension, limit K5 processivity, and presumably define the mode of K5 force generation (Fig. 8). The added consideration of the NL remaining structured in ADP will further perturb dimer stepping mechanisms. Understanding how these structural aspects of K5 mechanochemistry manifest themselves in the functioning spindle will be an important challenge for future studies.
FIGURE 8.
Model of a putative K5 dimer. A, based on our reconstructions and pseudoatomic models, we generated a model of a K5 dimer with two MDs bound to adjacent αβ-tubulin dimers along an MT protofilament, with the leading head in rigor (yellow) and the trailing head in AMPPNP (orange); the positions of Val-365 in each MD are indicated with red (trailing head) and orange (leading head) spheres. Modeling using a K1 crystal structure with a full-length NL docked (PDB 1MKJ) indicates that 5 additional residues span from Val-365 to the plus-end tip of the MD, and therefore, the green sphere indicates the position of the K5 Thr-370 residue. The center-to-center distance (dashed line) between Thr-370 (trailing) and Val-365 (leading) measured ∼30 Å (using Chimera). B, schematic of the putative dimer with the NL of each MD depicted as a rod and the position of residues Val-365 and Thr-370 indicated by circles. K5 NLs contain 4 residues between Thr-370 and the putative dimerization point (indicated by the black circle) in the trailing head and 9 residues between Val-365 and the dimerization point in the leading head.
Acknowledgments
We thank Elena Orlova and members of the Birkbeck EM group for helpful discussions and Dr. Dan Safer for undecagold.
This work was supported, in whole or in part, by National Institutes of Health Grant GM102875. This work was also supported by Biotechnology and Biological Sciences Research Council Grant BB/H005137/1.

This article contains supplemental Tables S1 and S2, Figs. S1–S9, and Movie S1.
The atomic coordinates and structure factors (codes 4AQV and 4AQW) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MT
- microtubule
- 2′dmT
- 2′-deoxy-3′-mant-ATP
- CS
- cover strand
- DDPM
- N-[4-(dimethylamino)-3,5-dinitrophenyl]maleimide
- K1, K3, and K5
- kinesin-1, -3, and -5, respectively
- L5
- loop 5
- MD
- motor domain
- NL
- neck linker
- TMR
- tetramethyl rhodamine 5′ maleimide
- AMPPNP
- 5′-adenylyl-β,γ-imidodiphosphate
- CNB
- cover neck bundle
- PDB
- Protein Data Bank
- AEDANS
- 5-(((2-amino)ethyl)amino)naphthalene-1-sulfonic acid.
REFERENCES
- 1. Ferenz N. P., Gable A., Wadsworth P. (2010) Mitotic functions of kinesin-5. Semin. Cell Dev. Biol. 21, 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kapitein L. C., Peterman E. J., Kwok B. H., Kim J. H., Kapoor T. M., Schmidt C. F. (2005) The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114–118 [DOI] [PubMed] [Google Scholar]
- 3. Valentine M. T., Fordyce P. M., Krzysiak T. C., Gilbert S. P., Block S. M. (2006) Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol. 8, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Düselder A., Thiede C., Schmidt C. F., Lakämper S. (2012) Neck-linker length dependence of processive kinesin-5 motility. J. Mol. Biol. 423, 159–168 [DOI] [PubMed] [Google Scholar]
- 5. Kikkawa M., Sablin E. P., Okada Y., Yajima H., Fletterick R. J., Hirokawa N. (2001) Switch-based mechanism of kinesin motors. Nature 411, 439–445 [DOI] [PubMed] [Google Scholar]
- 6. Sindelar C. V. (2011) A seesaw model for intermolecular gating in the kinesin motor protein. Biophys. Rev. 3, 85–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rice S., Lin A. W., Safer D., Hart C. L., Naber N., Carragher B. O., Cain S. M., Pechatnikova E., Wilson-Kubalek E. M., Whittaker M., Pate E., Cooke R., Taylor E. W., Milligan R. A., Vale R. D. (1999) A structural change in the kinesin motor protein that drives motility. Nature 402, 778–784 [DOI] [PubMed] [Google Scholar]
- 8. Clancy B. E., Behnke-Parks W. M., Andreasson J. O., Rosenfeld S. S., Block S. M. (2011) A universal pathway for kinesin stepping. Nat. Struct. Mol. Biol. 18, 1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hwang W., Lang M. J., Karplus M. (2008) Force generation in kinesin hinges on cover-neck bundle formation. Structure 16, 62–71 [DOI] [PubMed] [Google Scholar]
- 10. Khalil A. S., Appleyard D. C., Labno A. K., Georges A., Karplus M., Belcher A. M., Hwang W., Lang M. J. (2008) Kinesin's cover-neck bundle folds forward to generate force. Proc. Natl. Acad. Sci. U.S.A. 105, 19247–19252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Turner J., Anderson R., Guo J., Beraud C., Fletterick R., Sakowicz R. (2001) Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J. Biol. Chem. 276, 25496–25502 [DOI] [PubMed] [Google Scholar]
- 12. Behnke-Parks W. M., Vendome J., Honig B., Maliga Z., Moores C., Rosenfeld S. S. (2011) Loop L5 acts as a conformational latch in the mitotic kinesin Eg5. J. Biol. Chem. 286, 5242–5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Waitzman J. S., Larson A. G., Cochran J. C., Naber N., Cooke R., Jon Kull F., Pate E., Rice S. E. (2011) The loop 5 element structurally and kinetically coordinates dimers of the human kinesin-5, Eg5. Biophys. J. 101, 2760–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larson A. G., Naber N., Cooke R., Pate E., Rice S. E. (2010) The conserved L5 loop establishes the pre-powerstroke conformation of the kinesin-5 motor, eg5. Biophys. J. 98, 2619–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan Y., Sardana V., Xu B., Homnick C., Halczenko W., Buser C. A., Schaber M., Hartman G. D., Huber H. E., Kuo L. C. (2004) Inhibition of a mitotic motor protein. Where, how, and conformational consequences. J. Mol. Biol. 335, 547–554 [DOI] [PubMed] [Google Scholar]
- 16. Cochran J. C., Gatial J. E., 3rd, Kapoor T. M., Gilbert S. P. (2005) Monastrol inhibition of the mitotic kinesin Eg5. J. Biol. Chem. 280, 12658–12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwok B. H., Kapitein L. C., Kim J. H., Peterman E. J., Schmidt C. F., Kapoor T. M. (2006) Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat. Chem. Biol. 2, 480–485 [DOI] [PubMed] [Google Scholar]
- 18. Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L., Mitchison T. J. (1999) Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971–974 [DOI] [PubMed] [Google Scholar]
- 19. Sarli V., Giannis A. (2008) Targeting the kinesin spindle protein. Basic principles and clinical implications. Clin. Cancer Res. 14, 7583–7587 [DOI] [PubMed] [Google Scholar]
- 20. Parke C. L., Wojcik E. J., Kim S., Worthylake D. K. (2010) ATP hydrolysis in Eg5 kinesin involves a catalytic two-water mechanism. J. Biol. Chem. 285, 5859–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaan H. Y., Ulaganathan V., Hackney D. D., Kozielski F. (2010) An allosteric transition trapped in an intermediate state of a new kinesin-inhibitor complex. Biochem. J. 425, 55–60 [DOI] [PubMed] [Google Scholar]
- 22. Liu L., Parameswaran S., Liu J., Kim S., Wojcik E. J. (2011) Loop 5-directed compounds inhibit chimeric kinesin-5 motors. Implications for conserved allosteric mechanisms. J. Biol. Chem. 286, 6201–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vale R. D., Milligan R. A. (2000) The way things move. Looking under the hood of molecular motor proteins. Science 288, 88–95 [DOI] [PubMed] [Google Scholar]
- 24. Sindelar C. V., Budny M. J., Rice S., Naber N., Fletterick R., Cooke R. (2002) Two conformations in the human kinesin power stroke defined by x-ray crystallography and EPR spectroscopy. Nat. Struct. Biol. 9, 844–848 [DOI] [PubMed] [Google Scholar]
- 25. Sindelar C. V., Downing K. H. (2010) An atomic-level mechanism for activation of the kinesin molecular motors. Proc. Natl. Acad. Sci. U.S.A. 107, 4111–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kikkawa M. (2008) The role of microtubules in processive kinesin movement. Trends Cell Biol. 18, 128–135 [DOI] [PubMed] [Google Scholar]
- 27. Hirose K., Henningsen U., Schliwa M., Toyoshima C., Shimizu T., Alonso M., Cross R. A., Amos L. A. (2000) Structural comparison of dimeric Eg5, Neurospora kinesin (Nkin) and Ncd head-Nkin neck chimera with conventional kinesin. EMBO J. 19, 5308–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krzysiak T. C., Wendt T., Sproul L. R., Tittmann P., Gross H., Gilbert S. P., Hoenger A. (2006) A structural model for monastrol inhibition of dimeric kinesin Eg5. EMBO J. 25, 2263–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bodey A. J., Kikkawa M., Moores C. A. (2009) 9-Angstrom structure of a microtubule-bound mitotic motor. J. Mol. Biol. 388, 218–224 [DOI] [PubMed] [Google Scholar]
- 30. Maliga Z., Mitchison T. J. (2006) Small-molecule and mutational analysis of allosteric Eg5 inhibition by monastrol. BMC Chem. Biol. 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rosenfeld S. S., Xing J., Jefferson G. M., King P. H. (2005) Docking and rolling, a model of how the mitotic motor Eg5 works. J. Biol. Chem. 280, 35684–35695 [DOI] [PubMed] [Google Scholar]
- 32. Zhao Y. C., Kull F. J., Cochran J. C. (2010) Modulation of the kinesin ATPase cycle by neck linker docking and microtubule binding. J. Biol. Chem. 285, 25213–25220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cochran J. C., Sontag C. A., Maliga Z., Kapoor T. M., Correia J. J., Gilbert S. P. (2004) Mechanistic analysis of the mitotic kinesin Eg5. J. Biol. Chem. 279, 38861–38870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Safer D. (1999) Undecagold cluster labeling of proteins at reactive cysteine residues. J. Struct. Biol. 127, 101–105 [DOI] [PubMed] [Google Scholar]
- 35. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN. Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 36. Sindelar C. V., Downing K. H. (2007) The beginning of kinesin's force-generating cycle visualized at 9-Å resolution. J. Cell Biol. 177, 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. (1996) SPIDER and WEB. Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 38. Grigorieff N. (2007) FREALIGN. High-resolution refinement of single particle structures. J. Struct. Biol. 157, 117–125 [DOI] [PubMed] [Google Scholar]
- 39. Mindell J. A., Grigorieff N. (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 [DOI] [PubMed] [Google Scholar]
- 40. van Heel M. (1987) Similarity measures between images. Ultramicroscopy 21, 95–100 [Google Scholar]
- 41. Sousa D., Grigorieff N. (2007) Ab initio resolution measurement for single particle structures. J. Struct. Biol. 157, 201–210 [DOI] [PubMed] [Google Scholar]
- 42. van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 43. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 44. Löwe J., Li H., Downing K. H., Nogales E. (2001) Refined structure of αβ-tubulin at 3.5 Å resolution. J. Mol. Biol. 313, 1045–1057 [DOI] [PubMed] [Google Scholar]
- 45. Topf M., Lasker K., Webb B., Wolfson H., Chiu W., Sali A. (2008) Protein structure fitting and refinement guided by cryo-EM density. Structure 16, 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pandurangan A. P., Topf M. (2012) Finding rigid bodies in protein structures. Application to flexible fitting into cryoEM maps. J. Struct. Biol. 177, 520–531 [DOI] [PubMed] [Google Scholar]
- 47. Dale R. E., Eisinger J., Blumberg W. E. (1979) The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys. J. 26, 161–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rosenfeld S. S., Jefferson G. M., King P. H. (2001) ATP reorients the neck linker of kinesin in two sequential steps. J. Biol. Chem. 276, 40167–40174 [DOI] [PubMed] [Google Scholar]
- 49. Kikkawa M., Hirokawa N. (2006) High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J. 25, 4187–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li M., Zheng W. (2012) All-atom structural investigation of kinesin-microtubule complex constrained by high-quality cryo-electron-microscopy maps. Biochemistry 51, 5022–5032 [DOI] [PubMed] [Google Scholar]
- 51. Shastry S., Hancock W. O. (2010) Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr. Biol. 20, 939–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Geoghegan K. F., Rosner P. J., Hoth L. R. (2000) Dye-pair reporter systems for protein-peptide molecular interactions. Bioconjug. Chem. 11, 71–77 [DOI] [PubMed] [Google Scholar]
- 53. Toprak E., Yildiz A., Hoffman M. T., Rosenfeld S. S., Selvin P. R. (2009) Why kinesin is so processive. Proc. Natl. Acad. Sci. U.S.A. 106, 12717–12722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rosenfeld S. S., Rener B., Correia J. J., Mayo M. S., Cheung H. C. (1996) Equilibrium studies of kinesin-nucleotide intermediates. J. Biol. Chem. 271, 9473–9482 [DOI] [PubMed] [Google Scholar]
- 55. McIntosh J. R., Hepler P. K., Van Wie D. G. (1969) Model for mitosis. Nature 224, 659–663 [Google Scholar]
- 56. Sharp D. J., McDonald K. L., Brown H. M., Matthies H. J., Walczak C., Vale R. D., Mitchison T. J., Scholey J. M. (1999) The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144, 125–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harrington T. D., Naber N., Larson A. G., Cooke R., Rice S. E., Pate E. (2011) Analysis of the interaction of the Eg5 Loop5 with the nucleotide site. J. Theor. Biol. 289, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cochran J. C., Gilbert S. P. (2005) ATPase mechanism of Eg5 in the absence of microtubules. Insight into microtubule activation and allosteric inhibition by monastrol. Biochemistry 44, 16633–16648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim E. D., Buckley R., Learman S., Richard J., Parke C., Worthylake D. K., Wojcik E. J., Walker R. A., Kim S. (2010) Allosteric drug discrimination is coupled to mechanochemical changes in the kinesin-5 motor core. J. Biol. Chem. 285, 18650–18661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sheth P. R., Basso A., Duca J. S., Lesburg C. A., Ogas P., Gray K., Nale L., Mannarino A. F., Prongay A. J., Le H. V. (2009) Thermodynamics of nucleotide and inhibitor binding to wild-type and ispinesib-resistant forms of human kinesin spindle protein. Biochemistry 48, 11045–11055 [DOI] [PubMed] [Google Scholar]
- 61. Pylypenko O., Song L., Squires G., Liu X., Zong A. B., Houdusse A., Sweeney H. L. (2011) Role of insert-1 of myosin VI in modulating nucleotide affinity. J. Biol. Chem. 286, 11716–11723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kalchishkova N., Böhm K. J. (2008) The role of kinesin neck linker and neck in velocity regulation. J. Mol. Biol. 382, 127–135 [DOI] [PubMed] [Google Scholar]
- 63. Notredame C., Higgins D. G., Heringa J. (2000) T-Coffee. A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]