Background: Phage P4 Psu protein is a capsid decoration protein with unknown structure.
Results: The first structure of Psu reveals a novel fold and a knotted dimer.
Conclusion: The V-shaped molecular architecture is important for capsid binding.
Significance: The structure of Psu will help to design peptide fragments, which can be used as drugs against the bacterial transcription machinery.
Keywords: Protein Crystallization, Structural Biology, Transcription Termination, Viral Protein, X-ray Crystallography
Abstract
Psu is a capsid decoration protein of bacteriophage P4 and acts as an antiterminator of Rho-dependent transcription termination in bacteria. So far, no structures have been reported for the Psu protein or its homologues. Here, we report the first structure of Psu solved by the Hg2+ single wavelength anomalous dispersion method, which reveals that Psu exists as a knotted homodimer and is first of its kind in nature. Each monomer of Psu attains a novel fold around a tight coiled-coil motif. CD spectroscopy and the structure of an engineered disulfide-bridged Psu derivative reveal that the protein folds reversibly and reassembles by itself into the knotted dimeric conformation without the requirement of any chaperone. This structure would help to explain the functional properties of the protein and can be used as a template to design a minimal peptide fragment that can be used as a drug against Rho-dependent transcription termination in bacteria.
Introduction
Factor-dependent transcription termination in bacteria is mediated by a hexameric RNA-dependent ATPase protein, Rho (1). This mode of transcription termination causes polarity in gene expression pattern. Psu, a polarity suppression protein, is the first reported antiterminator of Rho-specific transcription termination in bacteria, which also acts as a capsid decoration protein in bacteriophage P4 (2, 3). Protein moonlighting is a phenomenon in which a protein can perform more than one function. So, Psu can be categorized as a moonlighting protein due to its dual activities. Upon binding to the Rho hexamer, Psu reduces the ATPase activity of Rho without affecting its primary RNA binding activities (4). Furthermore, Psu interacts with P4 phage capsid protein and stabilizes P4 against heat treatment (5). However, the relation between the heat stabilization by Psu and its polarity suppression, if any, is yet to be established.
Circular dichroism studies demonstrate that Psu is predominantly an α-helical protein and size exclusion chromatography along with in vitro cross-linking experiments reveal that Psu exists as a dimer in solution (4). Extensive mutational and cross-linking studies suggests that the interaction of Psu with Rho is direct and specific, mediated by the C-terminal tail of Psu, whereas the N-terminal domain of Psu maintains the conformational integrity of the C-terminal tail (6). However, the atomic structure of Psu, its domain organization and assembly remains unknown.
Here, we report the first structure of Psu (21 kDa) consisting of a symmetric “V”-shaped homodimer. The striking feature of the dimeric interface is that it incorporates a significant portion of the polypeptide chain in a knotted conformation, which is extensive and exclusively hydrophobic in nature. Each monomer of Psu attains a novel protein fold with a long N-terminal coiled-coil motif wrapped by the C-terminal helices, whereas the central region self assembles into the knot with another monomer to form the dimer. We have validated the existence of this knotted dimeric conformation of Psu in solution by solving the structure of T123C mutant on no-Cys construct of Psu (T123C-ΔCys-Psu).4 In this mutant structure, Cys-123 from each monomer faces each other at the dimeric interface and forms a disulfide bridge under oxidizing condition. We have also demonstrated that the denatured Psu can self assemble into the knot spontaneously in solution. Furthermore, the knotted V-shaped architecture of the Psu dimer, with its inherent tensile properties makes Psu suitable to interact with the hexameric capsomere on phage capsid to block DNA leakage.
EXPERIMENTAL PROCEDURES
Preparation of WT Psu and T123C-ΔCys-Psu Mutant
The wild type Psu protein was expressed and purified by a three-step process involving a 25% ammonium sulfate precipitation in buffer containing 10 mm Tris-HCl, pH 7.9, 0.1 mm EDTA, 0.1 mm DTT, and 5% (v/v) glycerol followed by purification through Q-Sepharose (GE Healthcare) and CM Sepharose columns (GE Healthcare). Psu was eluted between 50 and 150 mm NaCl, and the eluted fractions were stored in 20 mm Tris-HCl, pH 7.9, 0.1 mm EDTA, 0.1 mm dithiothreitol, and 100 mm NaCl. This procedure yielded ∼95% pure protein. For the preparation of the T123C-ΔCys-Psu mutant, a zero-Cys Psu (ΔCys-Psu) was prepared by mutating all the cysteines (Cys-13 and Cys-117) to serine. The T123C change was made by site-directed mutagenesis on the ΔCys-Psu and was purified in a similar way as WT.
Protein Crystallization, Data Collection, and Structure Determination of Wild Type Psu and Psu T123C
The crystallization trials of Psu was carried out at 277 and 293 K. Typically, 3 ml of protein solution (5 mg/ml) was mixed with 2 ml of reservoir solution containing 7.5%(w/v) PEG 6000, 5%(v/v) glycerol, 0.5 mm DTT, and 300 mm NaCl in 0.1 m MES, pH 6.0, at 277 K. Cube-shaped crystals appeared after 7 days. For phasing, the crystals of Psu were soaked in 2 mm HgI2 in the reservoir solution for 2 h. X-ray data were collected in-house at 100 K using a MAR Research image-plate detector of diameter 345 mm and Cu,K α radiation (1.54 Å) generated by a Bruker-Nonius FR591 rotating-anode generator equipped with Osmic MaxFlux confocal optics and running at 50 kV and 90 mA. A total of 129 frames were collected with a crystal-to-detector distance of 210 mm. The exposure time for each image was 4 min, and the oscillation range was maintained at 1 degree. The data were integrated with imosflm (7) and scaled with scala in the imosflm GUI (7). Phases from the single wavelength anomalous dispersion data were calculated in Phenix Autosol (8) with a figure-of-merit of 0.48 and refinement of one Hg2+ site with an occupancy of 0.8. The density was modified after defining the protein region of the density maps as the regions with the highest local root mean square deviations with Resolve in Phenix Autosol, which produced a readily interpretable map. The initial model was built with Phenix Autobuild (8), building 142 of 190 residues and placing 91 of the side chains. The rest of the model was built manually in Coot (9) and O (10) and iterative building cycles in Phenix Autobuild. The model was refined with Phenix Refine (8). The final Rfactor was 19.21 and Rfree was 21.87, including TLS (11) with excellent stereochemistry. Density was absent for the first three residues giving a final model of 187 residues of 190. The Ramachandran plot in Molprobity (12) shows that >98% of the residues are in the most favored region with none in the disallowed region.
For crystallization of Psu T123C, the protein was dialyzed against buffer containing 50 mm Tris (pH. 8.0), 300 mm NaCl. The crystallization trials of Psu was carried out at 277 and 293 K. Typically, 3 ml protein solution (5 mg/ml) was mixed with 2 ml of reservoir solution containing 7.5% (w/v) PEG 6000, 5% (v/v) glycerol in 0.1 m HEPES, pH 7.0 at 277 K. Cube-shaped crystals appeared within a couple of days. Data were collected up to 3 Å in conditions similar to the WT Psu. For phasing, the coordinates of WT Psu was used for molecular replacement with Phaser (13) in CCP4 (14). Model building was done with coot and refinement was carried out with Phenix refine. The data processing and refinement statistics are summarized in Table 1.
TABLE 1.
Data collection, phasing, and refinement statistics
| Data collection | ||
| Protein name | Psu-WT | T123C-ΔCys-Psu |
| Space group | I422 | I422 |
| Cell dimension | ||
| a, b, c | 148.75, 148.75, 63.37 | 149.61, 149.61, 62.7 |
| α, β, γ | 90.0, 90.0, 90.0° | 90.0, 90.0, 90.0° |
| Resolution (Å)a | 47.04–2.04 (2.11–2.04) | 47.31–3.00 (3.78–3.00) |
| Rmerge | 0.057 (0.343) | 0.116 (0.463) |
| I/σ(I) | 13.4 (3.7) | 6.2 (2.0) |
| Completeness (%) | 98.5 (92.0) | 95.5 (96.4) |
| Redundancy | 4.8 (4.6) | 3.0 (3.0) |
| Refinement | ||
| Protein name | Psu-WT | T123C-ΔCys-Psu |
| Resolution (Å) | 47.04–2.04 | 47.31–3.00 |
| Rcryst/Rfree | 19.2/21.9 | 23.11/27.66 |
| No. of atoms | 1705 | 1480 |
| Protein | 1479 | 1480 |
| Ligand | 10 | |
| Water | ||
| B-factor (Å2) | 216 | |
| Average | 36.53 | 58.13 |
| Protein | 35.79 | 58.13 |
| Ligand | 31.42 | |
| Water | 40.52 | |
| Root mean square deviations | ||
| Bond length (Å) | 0.007 | 0.011 |
| Bond angles | 0.9° | 1.2° |
| Ramachandran statistics (%) | ||
| Most favored | 97.7 | 97.3 |
| Additionally allowed | 2.3 | 2.7 |
| Disallowed | 0 | 0 |
a Numbers in parentheses refer to the highest resolution shell with all data collected from a single crystal.
Structural Analysis
Average B-factors for each residue were calculated using Baverage in CCP4 (14). The oligomeric state of the protein was analyzed using the PISA web server (15). Figures were prepared using PyMOL (16). Flexible regions of the molecule were identified by Normal mode analysis with NOMAD-Ref server (17) using the elastic network model and default settings for the other parameters. Briefly, all atoms were taken for the analysis of total 16 modes with a cut-off value of 10 Å for mode calculation and 5 Å as elastic constant.
Circular Dichroism Spectroscopy
Far-UV CD spectra were acquired for protein concentrations of 1 mg/ml (0.047 mm) in 10 mm sodium phosphate (pH 7.4) at 25 °C using a BioLogic Science Instruments (France) spectropolarimeter and cuvettes of 1-mm path length. For denaturation studies, the protein was incubated in varying concentrations of guanidine chloride (GdmCl; 1, 1.5, 2, 2.5, and 3 m) at 25 °C. The protein was initially standardized for the minimum GdmCl concentration at which denaturation reaches saturation. Initial denaturation with 6, 5, 4, and 3 m gave the same CD profiles. So, maximum of 3 m GdmCl was used for performing the denaturation experiments to minimize the noise in the 200–210 nm range. The far-UV CD spectrum of each sample was acquired after 20 min of incubation. Each spectrum was recorded as an average of eight scans. In all experiments, contributions of the buffer to the spectra were subtracted, and mean residue ellipticities were determined before plotting the spectra. For renaturation studies of Psu WT and Psu T123C, the proteins at a concentration of 2 mg/ml (0.095 mm) and 1.7 mg/ml (0.08 mm), respectively, were first denatured in a reaction mixture containing 3 m GdmCl and then subsequently dialyzed against buffer without GdmCl.
Fluorescence Spectroscopy
Steady state fluorescence studies were performed using a Varian Cary Eclipse fluorescence spectrophotometer for protein concentrations of 1 mg/ml (0.047) in 10 mm sodium phosphate (pH 7.4) at 25 °C and cuvettes of 4-mm path length. For denaturation studies, the protein was incubated in varying concentrations of GdmCl (1, 1.5, 2, 2.5, and 3 m) at 25 °C for 20 min each. Fluorescence emission from Psu tryptophans were measured using an excitation at 295 nm, and the emitted intensity was recorded at 340 nm. In all experiments, contributions of the buffer to the spectra were subtracted before plotting the spectra.
Cross-linking and SDS-PAGE
For the cross-linking experiments, T123C-ΔCys-Psu was incubated with the copper phenanthroline (18) before crystallization to induce disulfide bond between the two cysteines. The disulfide bridge can be formed in the presence of copper phenanthroline only if they are quite close to each other (∼6 Å) in the tertiary/quaternary structure and are correctly oriented. The concentrations of the cross-linker and T123C-ΔCys-Psu were 5 mm and 80 μm, respectively. T123C-ΔCys-Psu and copper phenanthroline were mixed in phosphate buffer (10 mm NaH2PO4, pH 7.4) devoid of any reducing agents, and incubation was continued for 30 min at 25 °C. Non-reducing SDS sample buffer was added to the cross-linking reactions, which were subjected to SDS-PAGE (15% polyacrylamide gel).
RESULTS
Psu Has a Novel Structural Fold
The structure of Psu was solved by mercury single wavelength anomalous dispersion method at 2.04 Å (Table 1). Psu crystallizes as a dimer where each asymmetric unit accommodates one Psu monomer. The refined model of Psu includes 187 of 190 residues with unambiguous electron density (data not shown) and excellent stereochemistry (Table 1). Psu monomer adopts a “golf stick” like structure and is characterized by seven helices (α1–α7) (Fig. 1a). The N-terminal motif consists of two long antiparallel helices (“CC stem” having α1 and α2), followed by two helices involved in the dimerization (“DR knot;” α3 and α4), and finally a flexible C-terminal tail comprising three helices (“CT belt;” α5–α7), which folds back and interacts with the N-terminal helices. The polypeptide chain (135–147) that connects the helix α4 and the CT belt is oriented in a fashion such that it leaves a hole in the monomeric structure (Fig. 1a). A search of the Protein Data Bank using the DALI server (19) and PDBeFold (20) did not produce any significant match with the Psu structure designating it to be a new fold.
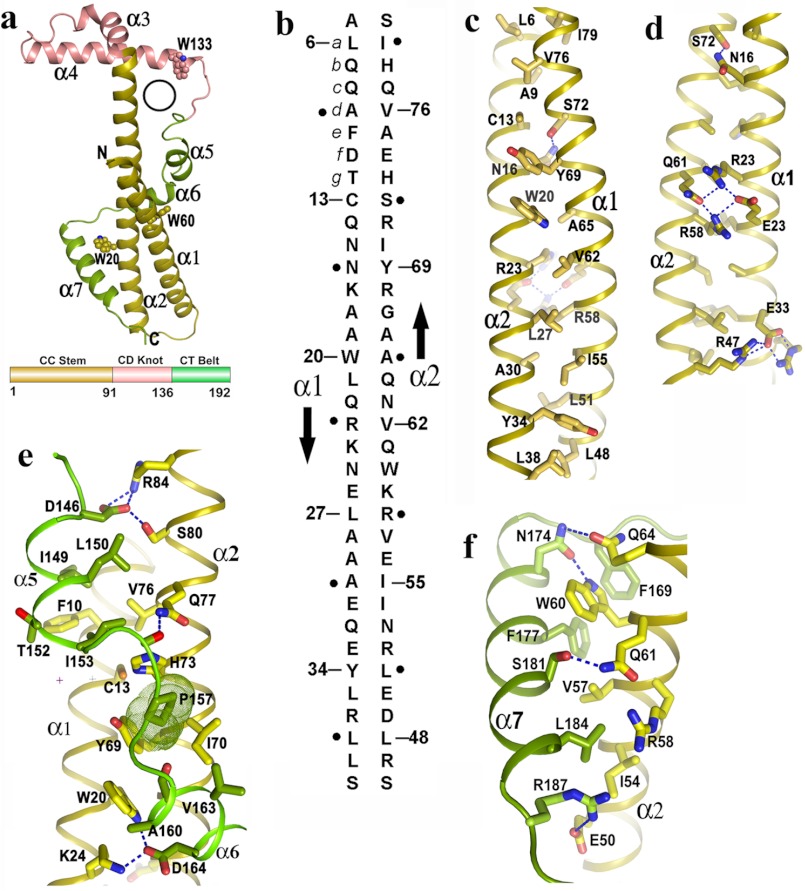
FIGURE 1.
Novel fold of Psu and stabilization of its coiled-coil region. a, schematic representation of Psu monomer resembling the structure of a golf stick, seven helices and three Trp residues are labeled. A black circle represents the hole in one monomer through which α3 of the second monomer can pass (top), topological diagram of Psu showing different regions (bottom) (same color scheme is used in c–f). b, schematic representation of antiparallel two stranded coiled-coil in Psu. The amino acid sequence of α1 and α2 are shown vertically, and the arrows indicate the chain direction. Residues at position a of each heptad repeat are numbered, whereas residues at position d are indicated as black dot. c, hydrophobic core of the CC stem formed by residues a of one helix with residues d of the other. d, view 180° rotated of c along the coiled-coil axis showing the polar/charged interactions that stabilize the CC stem. e, clamping of α5 and α6 with coiled coil. Pro-157, shown as dotted surface, packs with Cys-13, Tyr-69, Ile-57, His-73. f, interactions of α7 with α2.
A Coiled-coil Conformation at the N Terminus
The CC stem is composed of two long antiparallel helices, α1 (residues 3–39) and α2 (residues 45–91) connected by an inverse γ turn (residues 43–45) (Fig. 1a). Helices α1 and α2 form a two stranded antiparallel coiled-coil motif with five consecutive heptad repeats of amino acid residues, with the heptad positions “abcdefg” (Fig. 1b) bearing the signature of coiled-coil motifs (hxxhcxc; where “h” designates hydrophobic residues and “c” charged residues). Heptad repeat of hydrophobic residues at position “a,” hallmark of a coiled-coil motif, are observed at Leu-6, Cys-13, Trp-20, Leu-27, Tyr-34 positions of the helix α1 and at Leu-48, Ile-55, Val-62, Tyr-69, Val-76 positions of the helix α2 (Fig. 1, b and c). At the interface of the two helices, hydrophobic residues at position a of one helix lines up with the residues at position d of the other helix to form a hydrophobic core that stabilizes the coiled-coil structure (Fig. 1, b and c). Some variations are observed with respect to the proper positioning of the hydrophobic amino acids at a or d and the polar amino acids at e or g. Interestingly, these variations are used to further stabilize the coiled-coil either through formation of polar contacts or formation of salt bridge interactions. Polar/charged residues at a or d packs their hydrophobic portion in the core of the coiled-coil (Fig. 1d). For example, polar side chain of Asn-16 (d) forms hydrogen bond with Ser-72 (d) (Fig. 1d) at one end of the coiled-coil. At the middle of the motif, two positively charged residues, Arg-23 (d) and Arg-58 (d) come close to each other, and this destabilization is converted to a favorable one through salt bridge interactions by Glu-26 (e) and Gln-61 (e) (Fig. 1d). Near the inverse γ turn connector between α1 and α2, Arg-47 (e) and Arg-36 (c) make strong salt bridge interactions with Glu-33 (e) (Fig. 1d). Apart from these, there are additional violations in amino acid type, and these are engaged to make specific interactions with the CT belt residues during wrapping the coiled-coil (Fig. 1, e and f).
The CT Belt Wraps and Stabilizes the CC Stem
The C-terminal helices α5, α6, and α7 wraps the CC stem similar to a belt (Fig. 1a). During wrapping, amino acids from the CT belt makes several ionic and “knob/hole” type hydrophobic interactions with the amino acids from CC-stem. Asp-146 of helix α5 strongly interacts with Arg-84 and Ser-80 of α2, whereas residues Ile-149, Thr-152, and Ile-153 make a hydrophobic hole to pack Phe-10 (e) of α1 (Fig. 1e). Leu-150 and Ile-153 of α5 form a proper hole to accommodate the side chain of Val-76. Pro-157 is nicely packed with the amino acids from both helix α1 (Cys-13) and helix α2 (Val-76, Ile-70, and Tyr-69) (Fig. 1e). Asp-164 of α6 makes strong interactions with Lys-24 and Trp-20 of α1 (Fig. 1e). Phe-177 and Phe-169 of α7 produce an environment suitable to pack Trp-60 (f) of α2 (Fig. 1f), whereas N174 of α7 forms H-bonds with Trp-60 and Gln-64 (Fig. 1f). Leu-184 fits properly between Ile-54 (e), Leu-57 (c), and Arg-58 (d) (Fig. 1f). Arg-187 near the end of helix α7 makes a salt bridge with Glu-50 (c) of α2 (Fig. 1f). All of these interactions therefore not only reinforce the C-terminal belt in the stabilization of the coiled-coil domain but also clamp the CT belt tightly with the CC stem.
A Knotted Biological Assembly Adopted by Psu Dimer
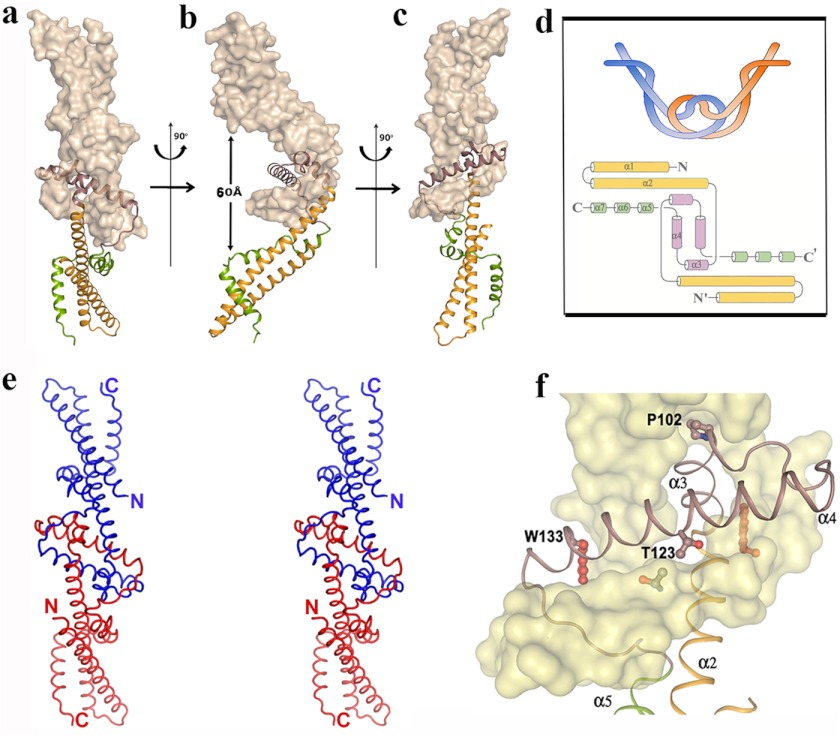
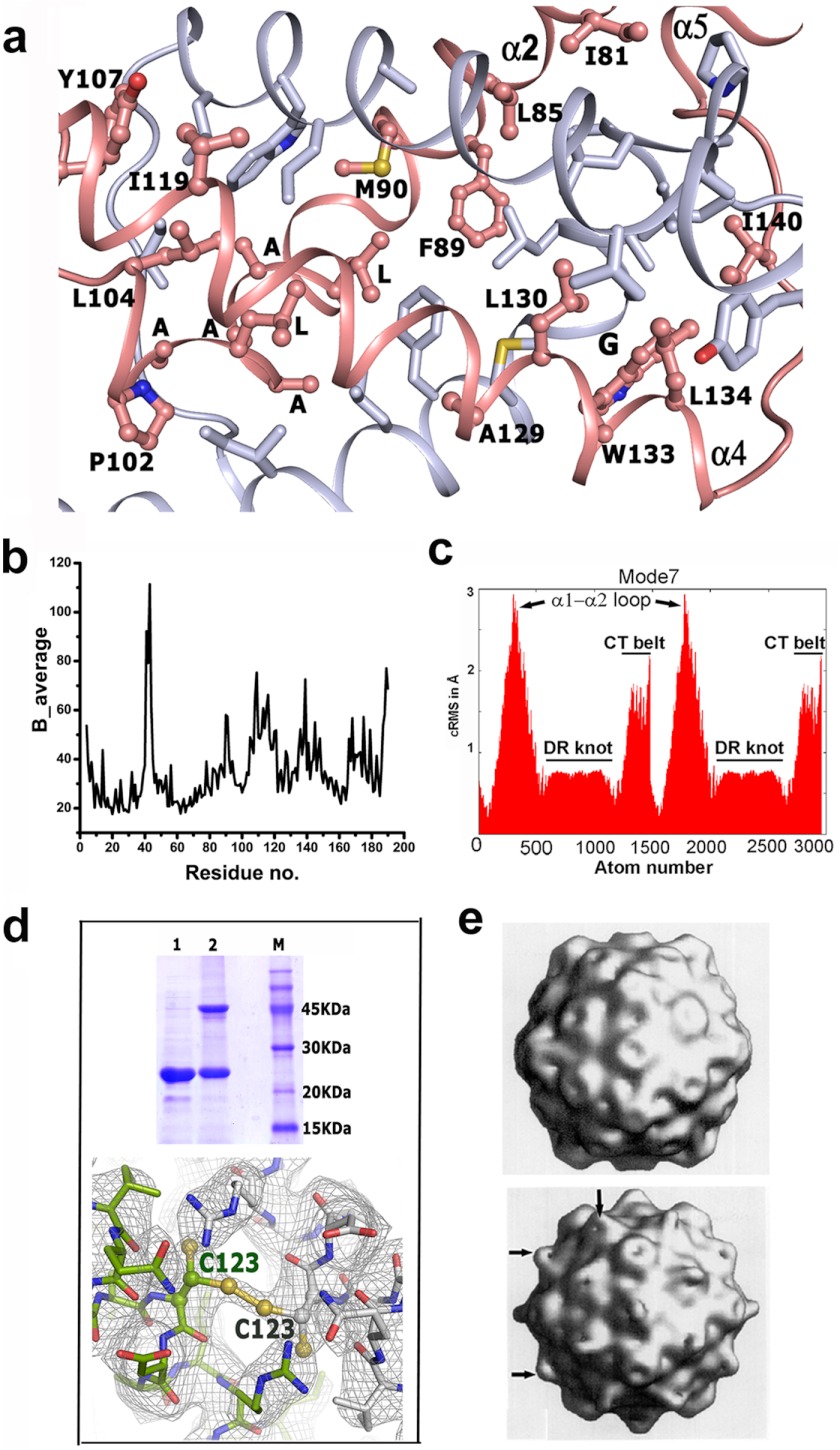
Crystal structure of Psu demonstrates a symmetric V-shaped homodimer, which is depicted in three different orientations (Fig. 2, a–c). The striking feature of this dimer is that the two monomers are knotted, which is shown schematically in Fig. 2d. This knotted dimeric self-assembly recruits part of α2, helices α3–α4, and the long loop connecting α4 to α5 from both monomers, which are related by a 2-fold at the dimeric interface. As mentioned earlier, there is a hole in the monomeric Psu structure (Figs. 1a and 2f), and during dimerization, α3 of one monomer passes through the hole of the other (Fig. 2f). In fact, the helix α2 needs to take a sharp turn of ∼80° at Gly-93 to project the short helix α3, ending at Pro-102, through the hole. At this turning point, Gly-93 faces the bulky side chain of the conserved Trp-133 of its knotting partner. Interestingly, α3 bears a stretch of highly conserved residues GXXLAAALAP in Psu homologues (Fig. 3). The region beyond Pro-102 is a short loop followed by helix α4, which is disposed in an antiparallel fashion to the α4 of the other monomer at an intersecting angle of ∼45°. The loop connecting α4 to α5 encompasses α3 of the knotting partner, and finally the CT belt wraps its own CC stem (Fig. 2f). During this process, a dimeric knot is formed that incorporates ∼25% of the polypeptide chain from each monomer. The knotted interface is devoid of any water molecule and harbors exclusively hydrophobic environment with several residues from α2, α3, and α4 (Fig. 4a). At the interface, Thr-123 lies in close proximity to the Thr-123 of its knotting partner with the 2-fold symmetry axis present in between (Fig. 2f). Presence of this 2-fold symmetry is also evident from Fig. 4a. There are only two hydrogen bonds from each monomer within this huge interface. The dimeric self assembly of Psu is also supported by PISA (15), whereupon dimer formation, each monomer buries ∼5500 Å2 surface area with a ΔGdiss value of 39 Kcal/mol. Average B-factors and normal mode analysis for the residues indicates that the knotted region is more stable than the CT belt (Fig. 4, b and c).
FIGURE 2.
Knotted dimerization of Psu. a–c, knotted dimer of Psu shown in three orientations with one monomer in surface representation and the other as a schematic. Extended V shape of the dimer and distance between the α7 helices are evident from b. d, schematic representation of the knotted dimer (top) and involvement of secondary structural elements of Psu in knot formation drawn using Topdraw (24). e, stereo view of the Psu dimer with the Cα backbone, N and C termini are labeled. f, close-up view around the knotted dimeric interface showing entrance of α3 through the hole of its knotting partner, packing of Trp-133 and Pro-102, and close proximity of two Thr-123 residues is evident.
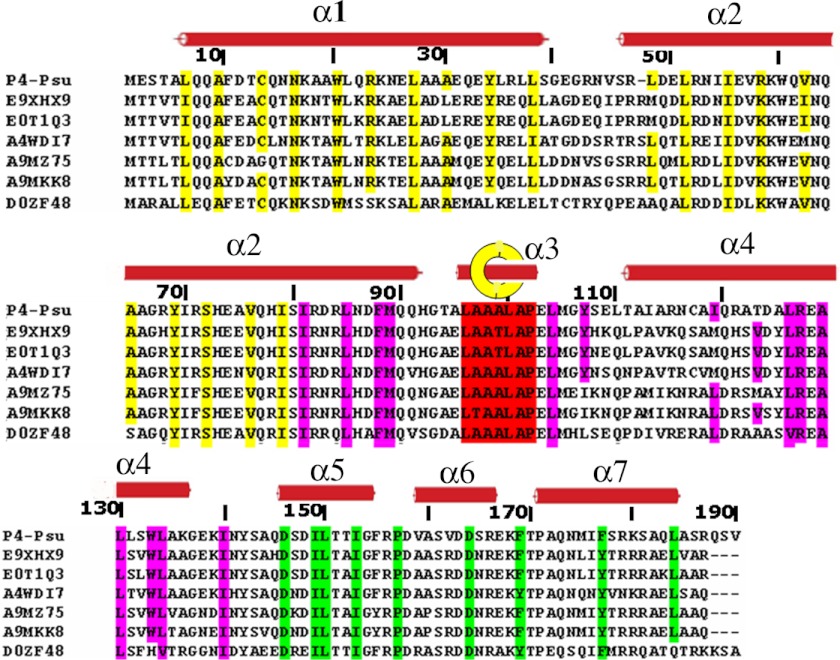
FIGURE 3.
Sequence alignment of Psu with its homologues. Important conserved residues are represented with color codes similar to those used in Fig. 1a. Secondary structural elements are overlaid on top and numbered. Yellow, coiled coil; pink, dimerization region; green, C-terminal belt; red, residues passing through the hole in the dimerization region. The yellow circle with the helix within represents the helix that passes through the knotted interface. E9XHX9, Escherichia coli TW10509; E0T1Q3, Edwardsiella tarda (strain FL6–60); E0T1Q3, Enterobacter sp. (strain 638); A4WDI7, Salmonella paratyphi B (strain ATCC BAA-1250/SPB7); A9MZ75, Salmonella arizonae (strain ATCC BAA-731/CDC346–86/RSK2980); D0ZF48, Edwardsiella tarda (strain EIB202).
FIGURE 4.
Stability of the Psu dimer from B-factor plot and normal mode analysis and validation of the Psu knot in solution. a, hydrophobic interactions at the dimeric interface. Residues interacting at the dimeric interface are shown as ball-and-stick for one monomer and labeled, whereas its knotting partner is shown as sticks and not labeled. Two monomers are related by a 2-fold that is passing through two Phe-89 and perpendicular to the paper. Residues of α3 are labeled but not numbered for clarity. Close proximity of Gly-93 and Trp-133 is evident. b, B-factor differences between residues of the Psu crystal structure. Indicated differences were calculated from the Cα main chain atoms. The B factor values of the CT belt residues are comparably higher than the rest of the molecule. c, normal mode analysis showing the low frequency mode 7 with cRMS (rmsd per residue) on the y-axis and atom number on the x-axis depicting large vibrational movements (Å) for regions α1-α2 and the CT belt regions, whereas vibrations around the knotted interface are minimum. d, formation of disulfide-linked knotted dimer of Psu in solution and in crystal structure. Top, 15% SDS-PAGE of T123C-ΔCys-Psu (lane 1), T123C-ΔCys-Psu treated with copper phenanthroline (lane 2), along with MW marker (lane 3). Bottom, T123C-ΔCys-Psu crystal structure showing the disulfide bond. e, electron micrograph of phage capsid (5) without (top) and with Psu (bottom), bulged out regions are the location of Psu (black arrow).
Psu also Adopts the Knotted Dimeric Conformation in Solution
To prove the existence of the knotted dimeric conformation in solution and not as a crystallographic artifact, we prepared a no-Cys construct of Psu (ΔCys-Psu) and then mutated its Thr-123 to Cys (hereafter termed as T123C-ΔCys-Psu). Thr-123 is located at a concave surface of Psu and just by simple association of the monomers, two Thr-123 residues cannot come in close proximity. But at the knotted dimeric interface, Thr-123 of one monomer lies in close proximity to Thr-123 of the other monomer and their side chains face each other (Fig. 2f). We reasoned that if the same dimer is formed in solution for T123C-ΔCys-Psu, then Cys-123 of the two monomers would come close in the quaternary structure and facilitate the formation of a disulfide bond. His-89 also comes close to its knotted counterpart, but His→Cys mutation may disrupt other interactions at the dimeric interface, which might be important for the dimer stability. The crystal structure of copper phenanthroline treated T23C-ΔCys-Psu showed that Cys-123 adopts two conformations of equal population, and one is able to form the disulfide bridge, validating the formation of the dimer in solution (Fig. 4d). Copper phenanthroline-treated T123C-ΔCys-Psu shows the same distribution (50% each) of the disulfide-bonded and reduced form in solution (Fig. 4d, lane 2). T123C-ΔCys-Psu dimer superposes with the Psu dimer with a root mean square deviation of 0.2 Å, implying that engineering a disulfide bond at the knotted interface does not require significant structural changes.
Dimeric Assembly of Psu on the Phage Capsid Is Similar to the Crystal Structure
A 45 Å electron micrograph reconstruction of the P4 phage capsid demonstrated that 12 pentameric and 30 hexameric capsomeres are arranged on a T = 4 lattice (Fig. 4e, top) (3). A P4+Psu reconstruction is very similar to P4 with an additional mass on top of the hexamers (Fig. 4e, bottom). This extra mass on the P4 capsid bridges across the hexamers (3) and has an extended V-shaped structure similar to the knotted dimeric assembly of Psu observed in our crystal structure (Fig. 2b). The largest holes on the P4 capsid hexamer have a diameter 50–70 Å and Psu dimer essentially covers its entire width. The shortest distance measured between the two α7 helices of our Psu dimer (Fig. 2c) is 60 Å, suggesting that Psu, particularly the two α7 helices of its dimer, are involved in the interactions with the hexameric subunits and covers up the hole to prevent DNA leakage from the capsid core.
The CT Belt Is Flexible Compared with the Relatively Rigid CC Stem
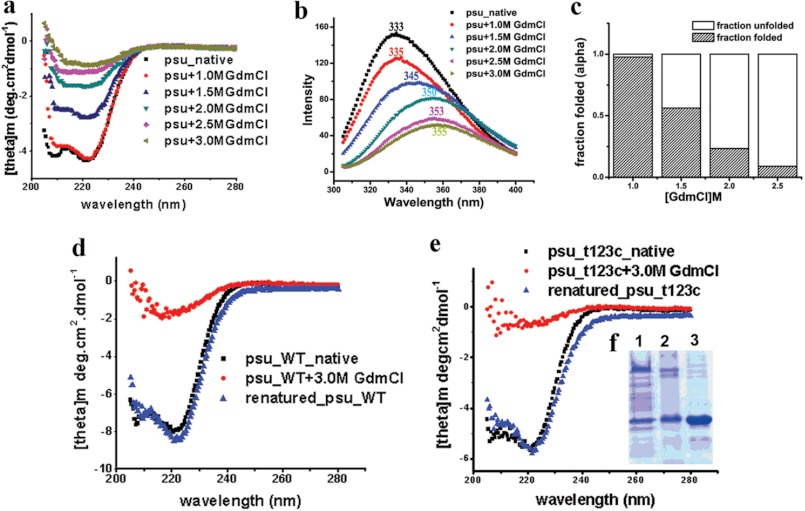
We performed CD spectroscopy in presence of GdmCl to judge the stability of different regions of Psu, especially its coiled-coil motif. The native protein gives characteristics signature of an all-α protein in CD giving minima at 208 and 222 nm, respectively (Fig. 5a), corroborating our crystal structure. Tryptophan fluorescence studies of wild type Psu shows a maxima at 333 nm, characteristics of buried tryptophan residues (Fig. 5b). Psu dimer also demonstrates that all the tryptophan residues (Trp-20, Trp-60, and Trp-133) are buried (Figs. 1a and 2f). Trp-20 and Trp-60 residing at the CC stem are obscured by the CT belt (Fig. 1a), whereas Trp-133 becomes buried during dimer formation (Fig. 2f). Unfolding of the protein with increasing concentrations of GdmCl resulted in the gradual loss of helical signature in the CD spectra (Fig. 5a; Table 2), and Psu practically loses its secondary structure at ∼2.0 m of GdmCl (Fig. 3, a and c; Table 2). Interestingly, the signature of the coiled-coil motif, indicated by the ratio of ellipiticity at 222 to 208 nm, is maintained even up to ∼2.0 m of GdmCl (Table 2), indicating that the coiled-coil part is stable until this point but unfolds as the concentration of GdmCl is increased further. A gradual decrease in the fluorescence intensity and a red-shift of the emission maxima of the protein was observed as the concentration of GdmCl is increased similar to CD experiments, indicating that the Trp residues are getting exposed to the solvent as the molecule unfolds (Fig. 5b). The major shift at 333 nm in the fluorescence spectra observed is due to the change from dimeric state to its monomer (Fig. 5b). CD experiments demonstrate that coiled-coil part of Psu is highly stable and with its unfolding whole structure disrupts. However, the relative stability of the CT belt and DR knot is not available from our CD experiments. The way the dimeric knot is formed and considering its highly hydrophobic nature and huge surface area, it is hard to believe that unfolding of the knotted region would occur before the CT belt unfolds. High B factors for the CT belt region and normal mode perturbation data (Fig. 4, b and c) (17) also supports the flexibility of the CT belt. Moreover, unfolding of the CT belt would render four of six Trp (Trp-20, Trp-60 from each monomer) residues solvent accessible, whereas only two tryptophans (Trp-133 from each monomer) would be solvent accessible when the DR knot unfolds. Considering the initial huge decrease of fluorescent intensity, we suggest that with gradual increment of denaturant CT belt of Psu unfolds first and then the DR knot, which corresponds to a transition from dimeric to monomeric form.
FIGURE 5.
Biophysical characterization of Psu knot. a, far-UV CD spectra of Psu at different GdmCl concentration. b, fluorescent emission spectra of Psu at 340 nm, using an excitation at 295 nm with concentrations of GdmCl varied similar to CD experiments. c, bar diagram showing folded fractions of Psu-WT at different GdmCl concentrations. d–e, comparison of native, denatured (3 m GdmCl) and renatured (0.26 m GdmCl) Psu-WT (d) and T123C-ΔCys-Psu (e). f, lane 1, 15% SDS-PAGE of renatured T123C-ΔCys-Psu treated with copper phenanthroline; lane 2, T123C-ΔCys-Psu treated with copper phenanthroline; lane 3, native T123C-ΔCys-Psu.
TABLE 2.
Helical content and ratio of ellipticity at θ222 and θ208 of Psu for different concentrations of GdmCl
| [GdmCl] in m | 0 | 1 | 1.5 | 2.0 |
|---|---|---|---|---|
| % Helicity | 0.901 | 0.87 | 0.55 | 0.29 |
| Θ222/θ208 | 1.03 | 1.12 | 1.69 | 1.72 |
Knotted Conformation of Psu Is Reversible and Sequence-directed
To understand the renaturation ability of Psu into the crystallographic knotted dimer, we used WT and T123C-ΔCys-Psu, denatured them using GdmCl, removed the denaturant gradually, and compared the spectroscopic properties of these proteins with the native proteins by CD spectroscopy. The CD spectra of the renatured WT and T123C-ΔCys-Psu proteins overlapped almost perfectly with their native forms (Fig. 5, d and e), indicating that the denatured proteins refolds to a structural state quite similar to their native folded forms. However, from these experiments, it is difficult to say whether the refolded proteins form the knotted structure or not. Therefore, we treated the refolded T123C-ΔCys-Psu protein with copper phenanthroline and analyzed the cross-linked products through non-reducing 15% SDS-PAGE (Fig. 5e). The gel picture shows the same profile of the protein band distribution (i.e. 50% cross-linked product) for both the renatured T123C-ΔCys-Psu and T123C-ΔCys-Psu treated with copper phenanthroline, confirming the fact that the denatured form of T123C-ΔCys-Psu upon refolding culminates to the knotted dimeric conformation. Interestingly, Psu does not require any chaperone to refold into such a complicated knotted dimer. An analysis of the primary sequence of Psu shows several patches of highly conserved sequences, which might be important in performing definite role in the overall folded structure. In particular, the conservation of coiled-coil region, sequence of α3, crucial knotting residues and the clamping residues at the CT belt (Fig. 3) are highly conserved. So, it can be proposed that the refolding of Psu to the native conformation is feasible as the sequence of the protein itself contains the necessary information for the formation of the knotted dimeric state.
DISCUSSION
The most striking feature of Psu structure is its novel fold and the knotted dimeric conformation where considerable portions of the polypeptide chain of one Psu monomer pass through the other to form the knot. Though proteins are known to form self-tying knots where the polypeptide backbone folds over itself to reach a state of lowest potential energy (21), our results show for the first time an architecture of a deeply buried knotted dimer in nature with an exclusive pattern of hydrophobic interactions over a large interface. The dimeric knot of Psu is distinctly different from domain swapping that involves the exchange of domains or structural elements between two monomers and is characterized by the presence of a hinge loop where a segment of polypeptide chain links the swapped domain to the rest of its subunit, a C-interface that occurs between domains in a monomeric subunit and an O-interface that occurs between monomers in a domain swapped dimer (22).
Why does Psu dimerize through such a complicated knot? Recently, knotted structures have gained popularity among structural biologists and material scientists due to their tensile strength and their effect on mechanical properties of polymers and biomolecules (23). Psu binds to the hexameric capsomere on the P4 capsid to prevent DNA leakage. Dimerization of Psu provides two equivalent binding sites capable of interacting with the hexameric capsomeres, whereas the knot formation can be envisaged to provide the necessary stability to withstand the shearing stress exerted by the DNA while coming out of the capsid. Interaction surface of Psu with the capsid is not yet known, but a V45F mutation was reported to abolish the antitermination activity of Psu, whereas its capsid binding ability remains unaltered (3). This observation suggests that the interaction surface of Psu with Rho and capsid are distinct. Therefore, Psu by virtue of its knotted dimerization is capable to bind both the capsid and Rho efficiently and specifically, providing the basis for its moonlighting activity.
For normal mode analysis, the first six modes were not considered as they were zero frequency modes and represented only rigid body motions. Normal mode perturbation data corresponding to mode 7 (Fig. 4c) demonstrate that the Psu dimer behaves like an arm and the knotted region acts like an elbow, allowing the dimer to swing inward and outward. During this process, the knotted region experiences minimum movements, whereas the C-terminal helix and the α1-α2 connector regions exhibit larger motions (Fig. 4c). Binding of Psu to the gpN hexameric capsomeres induces a conformational change, as the capsomeres, in their Psu-bound form, show a smaller hole compared with their Psu unbound version (5), further supporting our normal mode data.
Multiple sequence alignment of Psu-like proteins, found in strains of different species as part of cryptic prophages in their genome (6), reveals that residues involved in the packing of coiled-coil (a and d positions) are highly conserved (Fig. 3). Also, the conservation of residues forming the knot, especially the residues at α3, indicates similar dimer formation for the other Psu proteins as well. The coiled-coil region followed by the dimerization region provides an architecture around which the functional CT region is woven efficiently. Residues responsible for clamping the CT belt with the CC stem are highly conserved, which not only reduce the degrees of freedom of the CT belt but offers an interaction face of Psu oriented in the correct conformation space to bind Rho. A significant question is how this knotted dimeric conformation is attained. There may be two distinct paths to reach such a conformation. In the first mechanism, one monomer is formed and the second monomer is assembled around the first monomer. In the second mechanism, two monomers simultaneously become knotted before the folding process is complete.
The folding and unfolding of the knotted dimer by GdmCl does not exhibit any hysteresis. 123C-ΔCys-Psu refolds similar to WT Psu, and the formation of a disulfide bond in the refolded protein indicates the attainment of the same knotted structure. It would be interesting to decipher the folding energy landscape of Psu to attain the reversible knotted dimeric conformation and the role of amino acid sequence signature in this process.
Another feature of the molecule is the presence of the coiled-coil motif which is generally seen to be involved in protein-protein interactions. So, apart from providing structural rigidity to Psu, it may mediate at least in part the interaction of Psu with the Rho hexamer and the hexameric holes on the phage P4 capsid. Our structure may provide the starting point for designing a minimal fragment from the CT belt, which can be used a potential drug against the bacterial transcription machinery and would add to the existing knowledge of protein knots.
Acknowledgments
Sapna Godavarthi prepared the clone of cysteine derivative of Psu. We thank Abhijit Bhattacharya of Saha Institute of Nuclear Physics for help in data collection and Dr. Jhimli Dasgupta of St. Xavier's College (Kolkata, India) for critically reading the manuscript.
This work was supported in part by the SPGHGD project, DAE, India, Saha Institute of Nuclear Physics (to U. S.). This work was also supported by the Department of Biotechnology, India (to R. S.).
The atomic coordinates and structure factors (codes 3RX6 and 4DVD) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- T123C-ΔCys-Psu
- T123C mutant on no-Cys variant of Psu
- GdmCl
- guanidine hydrochloride.
REFERENCES
- 1. Richardson J. P., Greenblatt J. (1996) Control of RNA chain elongation and termination in Escherichia coli and Salmonella: Cellular and Molecular Biology (Neidhardt F. C., ed) pp. 822–848, American Society for Microbiology, Washington, D. C [Google Scholar]
- 2. Linderoth N. A., Calendar R. (1991) The Psu protein from bacteriophage P4 is an antitermination factor for rho-dependent transcription termination. J. Bacteriol. 173, 6722–6731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Isaksen M. L., Rishovd S. T., Calendar R., Lindqvist B. H. (1992) The polarity suppression factor of bacteriophage P4 is also a decoration protein of the P4 capsid. Virology 188, 831–839 [DOI] [PubMed] [Google Scholar]
- 4. Pani B., Banerjee S., Chalissery J., Muralimohan A., Abishek M., Loganathan R. M., Suganthan R. B., Sen R. (2006) Mechanism of inhibition of Rho-dependent transcription termination by bacteriophage P4 protein Psu. J. Biol. Chem. 281, 26491–26500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dokland T., Isaksen M. L., Fuller S. D., Lindqvist B. H. (1993) Capsid localization of the bacteriophage P4 Psu protein. Virology 194, 682–687 [DOI] [PubMed] [Google Scholar]
- 6. Pani B., Ranjan A., Sen R. (2009) Interaction surface of bacteriophage P4 protein Psu required for the complex formation with the transcription terminator Rho. J. Mol. Biol. 389, 647–660 [DOI] [PubMed] [Google Scholar]
- 7. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 10. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 11. Painter J., Merritt E. A. (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 12. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) MolProbity: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 15. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 16. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.2r3pre, Schrödinger, LLC, New York [Google Scholar]
- 17. Lindahl E., Azuara C., Koehl P., Delarue M. (2006) NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 34, W52-W56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Belogurov G. A., Vassylyeva M. N., Svetlov V., Klyuyev S., Grishin N. V., Vassylyev D. G., Artsimovitch I. (2007) Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell 26, 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krissinel E., Henrick K. (2004) Secondary-structure matching (PDBeFold), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 21. King N. P., Jacobitz A. W., Sawaya M. R., Goldschmidt L., Yeates T. O. (2010) Structure and folding of a designed knotted protein. Proc. Natl. Acad. Sci. U.S.A. 107, 20732–20737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bennett M. J., Schlunegger M. P., Eisenberg D. (1995) 3D Domain swapping: A mechanism for oligomer assembly. Protein Sci. 4, 2455–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saitta A. M., Soper P. D., Wasserman E., Klein M. L. (1999) Influence of a knot on the strength of a polymer strand. Nature 399, 46–48 [DOI] [PubMed] [Google Scholar]
- 24. Bond C. S. (2003) TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics 19, 311–312 [DOI] [PubMed] [Google Scholar]