Background: Recognition of human milk glycans (HMGs) by lectins, antibodies, and pathogens is poorly understood.
Results: Microarrays of isolated HMGs exhibited specific binding to proteins and pathogens.
Conclusion: HMG microarray interrogation and novel metadata-assisted glycan sequencing provide a functional glycomics approach to discovering HMG function.
Significance: HMGs represent a potential “liquid innate immune system” that is specifically recognized by antibodies and pathogens.
Keywords: Biomarkers, Glycobiology, Glycomics, Microarray, Virus, Biomarkers for Human Embryonic Stem Cells, Functional Glycomics, Glycan Microarray, Human Milk Glycans, Virus Interactions
Abstract
Human milk contains a large diversity of free glycans beyond lactose, but their functions are not well understood. To explore their functional recognition, here we describe a shotgun glycan microarray prepared from isolated human milk glycans (HMGs), and our studies on their recognition by viruses, antibodies, and glycan-binding proteins (GBPs), including lectins. The total neutral and sialylated HMGs were derivatized with a bifunctional fluorescent tag, separated by multidimensional HPLC, and archived in a tagged glycan library, which was then used to print a shotgun glycan microarray (SGM). This SGM was first interrogated with well defined GBPs and antibodies. These data demonstrated both the utility of the array and provided preliminary structural information (metadata) about this complex glycome. Anti-TRA-1 antibodies that recognize human pluripotent stem cells specifically recognized several HMGs that were then further structurally defined as novel epitopes for these antibodies. Human influenza viruses and Parvovirus Minute Viruses of Mice also specifically recognized several HMGs. For glycan sequencing, we used a novel approach termed metadata-assisted glycan sequencing (MAGS), in which we combine information from analyses of glycans by mass spectrometry with glycan interactions with defined GBPs and antibodies before and after exoglycosidase treatments on the microarray. Together, these results provide novel insights into diverse recognition functions of HMGs and show the utility of the SGM approach and MAGS as resources for defining novel glycan recognition by GBPs, antibodies, and pathogens.
Introduction
As the natural food source for newborns, human milk provides not only all of the nutrients necessary for infants to grow and develop but also provides health benefits in early childhood (1). In addition to antibodies that greatly enhance defense of the infant against various diseases, human milk possesses a rich pool of free-reducing oligosaccharides (glycans) that generally are unique in composition to humans and differ from those in the milk of other mammals (2, 3). The mature milk is estimated to contain 5–15 g/liter free human milk glycans (HMGs)3 depending on individual lactation time and blood group type (4, 5). So far, >200 unique HMGs have been detected, and >100 have been structurally characterized (6, 7). The large number and the remarkable structural diversity of these glycans suggest that they possess multiple biological effects (7–9). The prebiotic and anti-adhesive effects are the most common functions attributed to HMGs (10, 11). Furthermore, HMGs are associated with neonatal intestinal development (12–14) and limiting risks of necrotizing enterocolitis (7).
Although the multiple functions of HMGs are poorly understood, numerous studies have shown that their functional activities are dependent on their structures. For example, sialylated HMGs inhibit cholera toxin binding (15) and leukocyte adhesion to cultured human umbilical vein endothelial cells (16). Specific fucosylated HMGs are recognized by enteropathogens, including Helicobacter pylori (17), rabbit calicivirus (18), and Norwalk virus (19). Neutral HMGs, especially H type 2 glycans, inhibit Campylobacter jejuni adherence to Hep-2 cells and intestinal mucosa (20). Although the reported in vitro and in vivo data provide important information for understanding the effect of HMGs, typical experiments with HMG utilize either a small number of defined glycans or mixtures of HMG fractions. Such limitations represent challenges in studying HMGs, where the goal is to determine the roles of specific glycans in the milk glycome and to establish the relationships between glycan structures and their biological effects. However, linking function to structures of HMGs is difficult; many HMGs are comprised of linear and branched polymers of type 1 and type 2 lactosamine, Galβ1–3GlcNAc and Galβ1–4GlcNAc, respectively, substituted with α-linked Neu5Ac and Fuc. It is difficult to assign structures by mass spectrometry alone because of isobaric and isomeric structures, and a wide variety of approaches is often required, thus hindering progress in this area (21, 22).
We and others have made extensive use of glycan microarrays with defined chemo-enzymatically derived glycans to explore glycan recognition by glycan-binding proteins (GBPs) and microorganisms (23–27). However, because glycan synthesis is difficult, only a small fraction of the predicted, large number of glycans in the human glycome (28) is available for array production. To address this limitation we developed an alternative strategy termed “shotgun glycomics” (29) in which mixtures of free glycans derived from glycoproteins and glycolipids are derivatized with a bifunctional fluorescent tag and separated by multidimensional HPLC, and individual glycans are printed as a shotgun glycan microarray (SGM). In this approach glycan structures are defined after they are identified through their recognition by a GBP or pathogen and, therefore, are potentially functionally important. Here we have applied this approach to HMGs and defined those HMGs that are individually recognized by selected antibodies and pathogens. In addition, we combined the use of mass spectrometry, recognition by defined GBPs, and exoglycosidase treatments to help provide more detailed information about specific glycan structures in an approach termed metadata-assisted glycan sequencing (MAGS). This work represents the first use of a shotgun glycomics approach to prepare a natural glycan microarray of HMG containing >100 glycans.
EXPERIMENTAL PROCEDURES
Materials
Free reducing glycans used as standards were purchased from Sigma and V-LABS, Inc. (Covington, LA). All standard chemicals were bought from Sigma and used without further purification. Human milk was purchased from the Mothers Milk Bank (Austin, TX). Asialo, biantennary N-glycan (NA2), was prepared by mild acid hydrolysis and peptide N-glycosidase F digestion of a chicken egg yolk glycopeptide prepared as described (30). 2-Amino-N-(2-aminoethyl)benzamide (AEAB) was synthesized as described previously (31). β1–3 galactosidase, β1–4 galactosidase, and α2–3 neuraminidase were obtained from New England Biolabs (Ipswich, MA); neuraminidase (Arthrobacter ureafaciens) was from Roche Applied Science; jack bean β-galactosidase was from ProZyme (Hayward, CA). HPLC solvents were purchased from ThermoFisher Scientific (Waltham, MA). An Ultraflex-II TOF/TOF system from Bruker Daltonics was used for MALDI-TOF mass spectrometry analysis of glycan conjugates. Biotinylated lectins were from Vector Laboratories (Burlingame CA), Cy5-Streptavidin and Alexa488-labeled goat anti-mouse IgG were from Invitrogen. Anti-blood group Lewis a (Lea) and anti-CD15 antibodies were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Mouse monoclonal antibodies to blood group H1 antigen (ab3355) and to Lewis b (Leb) (ab3968) were purchased from Abcam (Cambridge, MA). Anti-sialyl Lea antibody was a kind gift from Professor Nancy Louis (Emory University School of Medicine, Atlanta, GA), and the anti-type 1 chain antibody was a kind gift from Dr. Irving Weissman (Stanford University, Stanford, CA). Anti-Lex antibody was prepared in the Cummings laboratory.4 Anti-TRA-1–81 and TRA-1–60 monoclonal antibodies were purchased from Millipore (Billerica, MA). Printing of glycan arrays was performed using a Piezorray Printer (PerkinElmer Life Sciences, Waltham, MA) on NEXTERION® Slide H slides, which are N-hydroxysuccinimide-activated slides from Schott Nexterion (Schott North America, Louisville, KY), as previously described (29, 32).
Preparation of Human Milk Glycans
The procedure for isolation of HMGs is illustrated in supplemental Fig. S1A (33). Briefly, human milk was defatted by centrifugation at 6000 × g for 30 min (4 °C); skimmed milk was filtered through glass wool and mixed with 2 volumes of ethanol and allowed to stand at 4 °C overnight to precipitate the bulk of the lactose and proteins. After centrifugation, the supernatant was concentrated and fractionated with Sephadex G-25 column to fraction A, B, and C. Fraction A, enriched with glycans larger than lactose, was applied to a DEAE column equilibrated with 2 mm pyridinium acetate, yielding neutral, monosialyl, and disialyl fractions by eluting sequentially with 2, 20, and 200 mm pyridine acetate, respectively (34). The resulting fractions were lyophilized.
Glycan-AEAB Conjugation and Purification
Standard glycans and the three human milk glycan fractions were conjugated with AEAB as described previously (31). Briefly, 1–10 mg of glycan was mixed with 50–250 μl of freshly prepared 0.35 m AEAB hydrochloride salt and an equal volume of 1 m NaCNBH4 in DMSO/AcOH (v/v = 7/3). The conjugation reaction was left to proceed for 2 h at 65 °C and was stopped by the addition of 10 volumes of cold acetonitrile and allowed to stand for 30 min at −20 °C. The precipitated glycan-AEAB derivatives were collected after centrifugation at 10,000 × g for 3 min.
High Performance Liquid Chromatography
A Shimadzu HPLC CBM-20A system coupled with a UV detector SPD-20A and a fluorescence detector RF-10AXL was used for HPLC analysis and separation of glycan-AEABs. The AEAB-conjugated milk glycans were first separated by normal phase HPLC on an Agilent NH2 column (250 × 4.6 mm). The mobile phase linear gradient was from 80% acetonitrile, 10 mm ammonium acetate, pH 4.5, to 10% acetonitrile, 125 mm ammonium acetate, pH 4.5, in 160∼200 min. Individual peaks were collected and dried. Each peak collected on the NH2 column was further purified in a second dimension by reverse phase HPLC on a porous graphitized carbon column (150 × 4.6 mm) with a gradient of 15–45% acetonitrile (0.1% trifluoroacetic acid) in 30 or 15 min. The effluents were monitored by UV absorption at 330 nm and/or fluorescence at 420 nm with excitation 330 nm. LNFPIII-AEAB or lactose-AEAB was utilized as the standard for the quantification of the AEAB derivatives. The individual glycans after the second dimension chromatography were quantified, dried, and reconstituted in water at a concentration of 200 μm and stored as a HMG-tagged glycan library (TGL).
Printing of the SGM, Binding Assays, and Analysis of Microarray
The printing conditions for the SGM, binding assays, and data analysis of glycan microarrays have been previously described (31, 35, 36). Briefly, each glycan in the TGL was adjusted to 100 μm in sodium phosphate buffer (100 mm, pH 8.5) in a final volume of 10 μl and distributed into the 384-well source plate of the Piezorray printer. The glycans were printed as 0.3-nl aliquots in replicates of 5. The biotinylated lectins or unlabeled antibodies were incubated for 1–3 h on the slides. After washing, the bound lectins were detected by a secondary incubation with cyanine 5-labeled streptavidin (5 μg/ml), and the bound antibodies were detected using Alexa488-labeled goat anti-mouse IgG (5 μg/ml). The fluorescence signals were generated using a ProScanArray Scanner (PerkinElmer Life Sciences) with the excitation/emission wavelength set at 649/670 nm and 495/510 nm for cyanine 5 and Alexa488, respectively.
Virus Preparations and Binding Assays
The human parainfluenza (hPIV) and influenza A were grown in LLC-MK2 cells and MDCK cells, respectively, as previously described (37, 38). Harvested viruses were purified through a sucrose gradient centrifugation (10–60% for hPIV and 10–40% for influenza A) and resuspended in calcium-magnesium saline followed by labeling with Alexa488 succinimidyl ester (Molecular Probes, Invitrogen). For the binding assays, the slides were incubated with labeled viruses for 1 h at 4 °C and directly scanned after washing. The WT and mutant strains of minute virus of mice (MVM) were constructed and prepared as previously described (39–42). Briefly, the WT MVMp empty particles were grown in mouse A9 ouabr11 fibroblasts, MVMi mutants (MVMi-ggA and MVMi-agD) were grown in NB324K cells, and VLPs (virus-like particles) were grown in sf9 insect cells. The harvested viruses were released by repeated freezing and thawing and then purified by centrifugal sedimentation and density equilibrium gradients. The purity and integrity of prepared viruses were assessed by hemagglutination, SDS-PAGE, and electron microscopy (41).
Exoglycosidase Digestion of Glycans Printed on a HMG Subarray
On-array exoglycosidase digestions were carried out at 37 °C in buffers recommended by the suppliers of the enzymes. Enzymes were used without further purification. Microarrays were rehydrated in the digestion buffer for 5 min before enzyme addition and washed 4 times with TSM buffer (20 mm Tris-HCl, 150 mm sodium chloride, 2 mm calcium chloride, and 2 mm magnesium chloride) supplemented with 0.1% Tween 20, 4 times with TSM buffer, and 4 times with water before detection with lectins or antibodies. Reaction conditions were optimized for each exoglycosidase.
RESULTS
Preparation of the Human Milk Glycome SGM
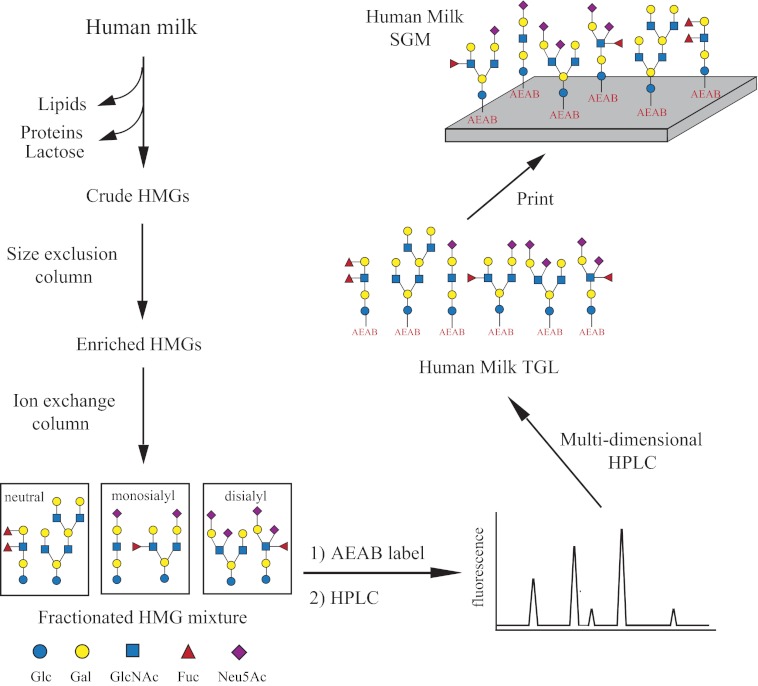
The generation of a SGM of HMGs from a single individual milk sample is illustrated in Fig. 1. To extract the human milk free glycome, the lipids, proteins, and most of the lactose were removed by centrifugation and ethanol precipitation. The glycans were obtained after size exclusion chromatography and then separated into three distinct groups (neutral, monosialyl, and disialyl) by ion-exchange chromatography (supplemental Fig. S1) (33). Adopting our shotgun glycomics approach, the glycan mixtures in each group were conjugated with AEAB and separated by two-dimensional HPLC into a total of 156 individual fractions. Selected fractions were subjected to an additional round of reverse-phase HPLC to further improve the purity and obtain 127 glycans in the TGL. The array contains 73 (57%) sialylated glycans and 54 (43%) neutral glycans. It should be noted that although the number of sialylated glycans exceeds the neutral ones, the total abundance of neutral glycans is much higher than that of sialylated ones. Analysis of the observed masses of all samples (supplemental Table S1) revealed glycans ranging in size from 2 residues (2 hexoses (Hex)), i.e. lactose, up to 12 residues (6 Hex + 4 HexNAc + 2 Fuc/Neu5Ac) and in mass from 506.337 [M+H]+ (2 Hex) to 2550 [M+H]+ (6 Hex + 4 HexNAc + 2 Neu5Ac). Of the 54 neutral glycans, >80% carry 1–3 fucose residues, whereas less than half of the sialylated glycans are fucosylated. Of the 73 sialylated glycans, 17 were purified from the monosialyl group, and 56 were from the disialyl group. However, mass results indicated that some of the samples purified from the disialyl group only carry one sialic acid. After quantification using AEAB fluorescence, the 127 glycans were adjusted to the same concentration (regardless of their abundance) and printed (replicates of n = 5) on N-hydroxysuccinimide-activated glass slides along with 11 structurally defined glycans that serve as controls for binding experiments (supplemental Table S1). Subsequently, the HMG microarray was interrogated with lectins and antibodies to evaluate whether the glycans were effectively printed.
FIGURE 1.
Schematic for generation of HMG SGM. Human milk glycans were extracted, fractionated, AEAB-conjugated (labeled with a tag), and separated. The purified fractions were quantified and printed to create a human milk glycan SGM available for studies with GBPs and microorganisms.
Preliminary Characterization of the HMGs on the SGM by Lectins and Antibodies
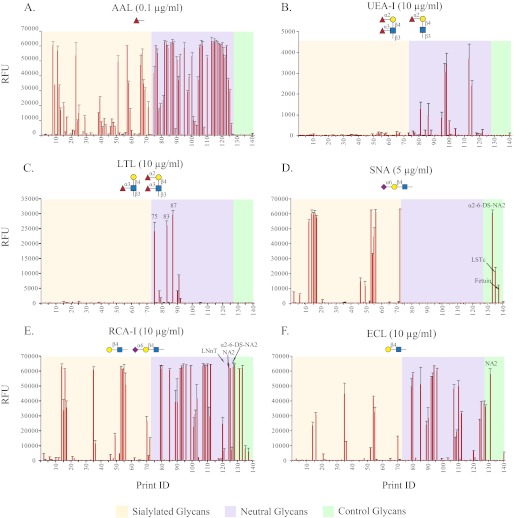
Defined glycan microarrays are used to explore the specificity of GBPs, including lectins and anti-glycan antibodies (23). In turn, GBPs with well defined binding specificity can assist in the elucidation of glycan structures. To interrogate the SGM and evaluate the structural diversity of isolated HMGs, 10 biotinylated lectins, each at several concentrations in the range of 0.001–10 μg/ml, were applied to the array. No significant binding was observed with concanavalin A, Vicia villosa lectin, Griffonia simplicifolia lectin II, and Maackia amurensis lectin I. These lectins recognize glycans containing mannose (43–45), terminal GalNAc (46), terminal GlcNAc (47), and terminal Neu5Acα2–3Galβ1–4GlcNAc (48), respectively; the absence of their binding is consistent with the lack of such structures in human milk free glycans (6). Independently the binding of these lectins to a defined glycan microarray from the CFG (v5.0) was evaluated, and the data are available online as glycan array data on the CFG website. The other six lectins, Aleuria aurantia lectin (AAL), Sambucus nigra agglutinin (SNA), Lotus tetragonolobus lectin (LTL), Ulex europaeus agglutinin I (UEA-I), Ricinus communis agglutinin I (RCA-I), and Erythrina cristagalli Lectin (ECL), exhibited binding to many of the HMGs on the SGM as discussed below, and the binding histograms showing average relative fluorescence units (RFU) for selected concentrations of each lectin are shown in Fig. 2, and the binding data are listed in supplemental Table S2.
FIGURE 2.
Plant lectins binding to human milk SGM. The human milk SGM microarray was characterized with biotinylated lectins AAL (0.1 μg/ml; A), UEA-I (10 μg/ml; B), LTL (10 μg/ml; C), SNA (5 μg/ml; D), RCA-I (10 μg/ml; E), and ECL (10 μg/ml; F). A total of 140 glycans was printed on the microarray. Glycans 1–73 are sialylated glycans (tan), 74–127 are neutral glycans (violet), and 128–140 are controls of structurally defined glycans (light green). The structures in symbols indicate the binding specificity of each lectin identified by defined glycan microarray (CFG v5.0). Considering the variation of binding affinity, the histogram shows the data at the concentration that yielded the best signal/background ratio instead of the data at the same concentration.
Three fucose binding lectins, AAL, LTL, and UEA-I, were used to reveal the fucosylated glycans on the SGM. Approximately 61 glycans (48%) showed strong binding with AAL (RFU > 10,000), which binds to terminal α-linked l-fucose in 1–2, 1–3, and 1–4 linkages (49), and an additional 24 glycans (19%) had weaker binding (RFU 1,000–8,000). Together, the fucosylated glycans recognized by AAL made up 67% of the total isolated HMGs, which is close to the percentage found by using HPLC-ChIP/MS method (21). In addition, most of the neutral glycans and about 40% of the sialylated glycans are fucosylated, in agreement with the results from glycan composition analysis based on mass (supplemental Table S1). UEA-I, which is specific for α1–2-linked fucose on a type 2 chain (50, 51), bound very weakly (<4,000 RFU) to several multifucosylated, neutral glycans (Fig. 2B), suggesting the presence of α1–2 fucose on our HMG array. However, this weak binding was presumably due to the cross-reactivity of UEA-I with other fucose-containing glycans as shown in Table 1, and supplemental Table S2 and actually not from Fucα1–2. This conclusion was supported by the observation that glycans were resistant to α1–2 fucosidase digestion in solution (data not shown). This result indicated the absence of H type 2 (Fucα1–2Galβ1–4GlcNAc) and Ley (Fucα1–2Galβ1-4(Fucα1–3)GlcNAc) structures in the SGM. LTL recognizes α1–3-linked fucose within type 2 glycans (52, 53), like Lex and Ley determinants. It showed strong binding to three neutral glycans (H-75, H-83, and H-87) and weak binding to another five glycans (Fig. 2C). The three high affinity binders likely contain terminal Lex determinants because the absence of UEA-I binding (Fig. 2B) excludes the possibility of α1–2-linked fucose.
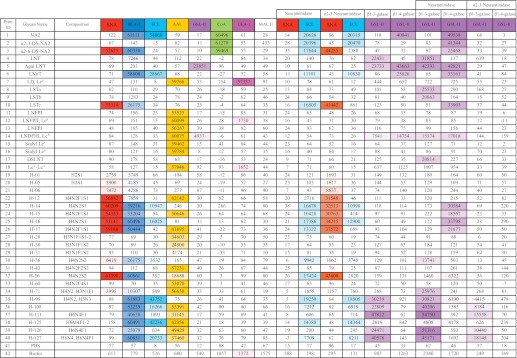
TABLE 1.
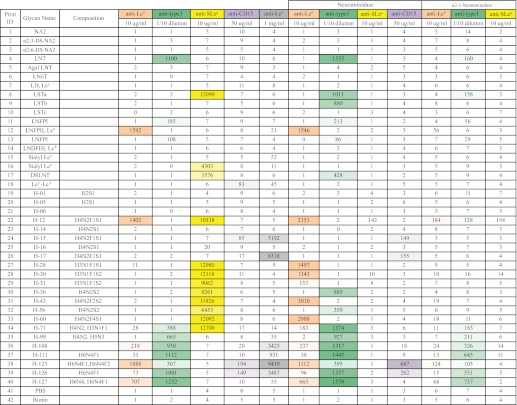
Heat map of lectin-binding data for structural analysis
A heat map summary of the binding data from analyses of the HMG subarray with different plant lectins before and after specific exoglycosidase digestion(s) is shown. A total of 40 glycans were printed on the HMG subarray, including 18 control glycans and 22 selected HMGs. The color-coded numbers highlight the binding intensity (the darker the color, the higher the binding intensity). The color scale for the Excel spreadsheet was set using 0 as minimum and 60,000 RFU as maximum. The untreated HMG subarrays were tested with eight lectins. The HMG subarrays treated with non-specific neuraminidase and α2-3-specific neuraminidase were tested with SNA and ECL. The β1–3 galactosidase (β1–3 galase)- or β1–4 galactosidase (β1–4 galase, from either New England Biolabs or Prozyme)-treated slides were tested with GSL-II. The lectin concentrations used were 10 μg/ml with the exception of AAL, which was 1 μg/ml.
SNA, which binds to the Neu5Acα2–6Galβ1–4GlcNAc determinant (23, 54), bound well to 14 (of 73) sialylated glycans on the SGM along with three control glycans (2–6-DS-NA2, LSTc, and fetuin, structures shown in supplemental Table S1), indicating the existence of α2–6-sialylated type 2 structure in 19% of the glycans (Fig. 2D). Both RCA-I and ECL recognize terminal Galβ1–4GlcNAc (55–59), but the former has much higher affinity and also binds to Neu5Acα2–6Galβ1–4GlcNAc with slightly lower affinity, whereas the latter can also bind Fucα1–2Galβ1–4Glc (60). Consistent with these features, the binding pattern of RCA-I on this SGM was similar to the combined pattern of SNA and ECL (Fig. 2, D–F) together, suggesting that ∼46% of glycans have terminal type 2 or sialylated type 2 structures. In summary, the results of lectin binding both validated the preparation of the SGM and provided significant structural information on individual HMGs.
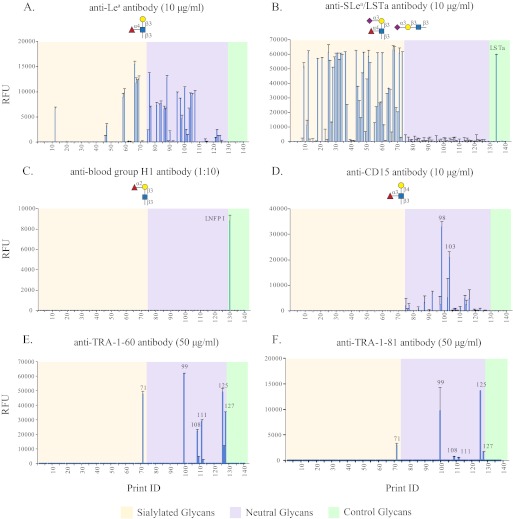
The structures of HMGs reflect the Lewis blood type and secretor status of the mothers (61, 62). Certain fucosylated glycans that were suggested to possess important biological functions (3) occur only in the milk of Lewis positive and/or Secretor-positive mothers due to the expression of FUT-3 and FUT-2, respectively (19). Because there are no lectins specific for Lewis blood group antigens, we analyzed the SGM with several blood group-related monoclonal antibodies. As shown in Fig. 3A, the mAb to Lea bound to 34 glycans, including 9 sialylated ones, with fluorescence signals in the range of 500–15,000 RFU. To reveal additional Lea-containing glycans, we interrogated the array with an anti-sialyl Lea (SLea) antibody. According to the data from the structurally defined CFG glycan microarray, this antibody also binds to LSTa (Neu5Acα2–3Galβ1–3GlcNAcβ1–3Galβ1–4Glc) moiety with lower signal. At the concentration of 10 μg/ml, 31 sialylated glycans showed binding signals higher than 30,000 RFU with the antibody, and another 19 glycans had signals >5,000 RFU (Fig. 3B). Although we could not simply assign SLea and LSTa structures by signal intensity, it is certain that many of these sialyl glycans are fucosylated based on their AAL binding (Fig. 2A) and may contain the SLea moiety. Nevertheless, the abundance of Lea-containing glycans showed that the SGM was from a Lewis positive donor. To determine the secretor status of the donor, we interrogated the SGM with anti-Leb and anti-blood group H type 1 antibodies. There was little to no binding observed with the anti-Leb antibody at any concentration (data not shown), which indicates that the milk sample is from a secretor negative donor. This finding was confirmed by assaying with anti-H1 antibody, which bound only the control LNFP I (Fucα1–2Galβ1–3GlcNAcβ1–6Galβ-4Glc) (Fig. 3C), the precursor of Leb antigen.
FIGURE 3.
Antibodies binding to human milk SGM. The human milk SGM was interrogated with antibodies: anti-Lea antibody (10 μg/ml; A), anti-SLea/LSTa antibody (10 μg/ml; B), anti-blood group H1 antibody (1:10 dilution; C), and anti-CD15 antibody (10 μg/ml; D). The microarray was also used to test the binding specificity of anti-TRA-1–60 antibody (50 μg/ml; E) and anti-TRA-1–81 antibody (50 μg/ml; F). Glycans 1–73 are sialylated glycans (tan), 74–127 are neutral glycans (violet), 128–140 are controls of structurally defined glycans (light green).
We also interrogated the array with anti-CD15 antibody, known to recognize Lex antigen. Strong binding was observed to glycans H-98 and H-103 with weaker binding to several other neutral glycans (Fig. 3D). When comparing CD15 antibody with lectin LTL, the two Lex-recognizing proteins showed distinct specificity toward different HMGs, although with some overlap. This could indicate that the recognition does not solely depend on the Lex determinant for complex glycans, and the nearby residues or branches could affect the binding.
Detection of HMGs That Are Epitopes of Anti-TRA-1 Antibodies, Specific for Human Pluripotent Stem Cells
Recently, Natunen et al. (63) predicted epitopes for mAbs anti-TRA-1–60 and anti-TRA-1–81 based on binding data from Version 4.2 of the CFG glycan microarray. These mAbs, which are specific for human pluripotent stem cells, bound only to two glycans, both containing the type 1 lactosamine epitope, Galβ1–3GlcNAcβ1–3Galβ1–4GlcNAc, on that version of the CFG glycan microarray. We further examined the two mAbs on Version 5.0 of the CFG array, which contains many multiantennary glycans with poly-N-acetyllactosamine (64) and observed strong binding by three additional glycans (supplemental Fig. S2). Importantly, these three glycans (#522, #572, and #573) are multiantennary glycans with two to three type 1 lactosamine repeats at their non-reducing ends. In addition, we observed weak but significant binding to 387, a glycan with a fucosylated type 1 lactosamine chain. Considering that HMG is a rich source of type 1 and type 2 lactosamine structures, we interrogated the SGM with the two anti-TRA-1 antibodies at several concentrations (1–100 μg/ml), and the results at 50 μg/ml are shown in Fig. 3, E and F. Consistent with the CFG array data, TRA-1–60 and TRA-1–81 share similar receptor specificity as both bind glycans H-71, H-99, H-108, H-111, H-125, and H-127. However, unlike the CFG results, which showed no significant difference in the signal intensity and binding patterns for the two mAbs at 50 μg/ml, we observed that with the SGM the signal intensity for TRA-1–60 binding was always severalfold higher than that of TRA-1–81 at the same concentration and that there are three more low affinity binders for TRA-1–60, H-109, H-112, and H-126 (Fig. 3E). To define the glycan epitope in HMG for TRA-1 antibodies, we retrieved glycans H-71, H-99, H-108, H-111, H-125, H-126, and H-127 from the TGL for further characterization. With the exception of sialylated glycan H-71, all of the TRA-1-bound glycans are neutral fucosylated structures consisting of 2–4 lactosamine repeats. We predict from previous studies (63) that these glycans possess type 1 lactosamine. Our data also indicate that the two anti-TRA-1 mAbs recognize complex glycans, as described below.
Investigation of Virus Binding to the SGM
The HMG-derived SGM provides a library of 127 naturally occurring glycans that permits us to investigate the binding properties of biologically relevant proteins and pathogens and to provide interesting insights into the potential function of HMGs. To explore the general application of this HMG-derived SGM for exploring pathogen interactions, we examined MVM and influenza virus, both of which attach to the sialic acid on the surface of their target cells at the initial stage of infection (40, 65).
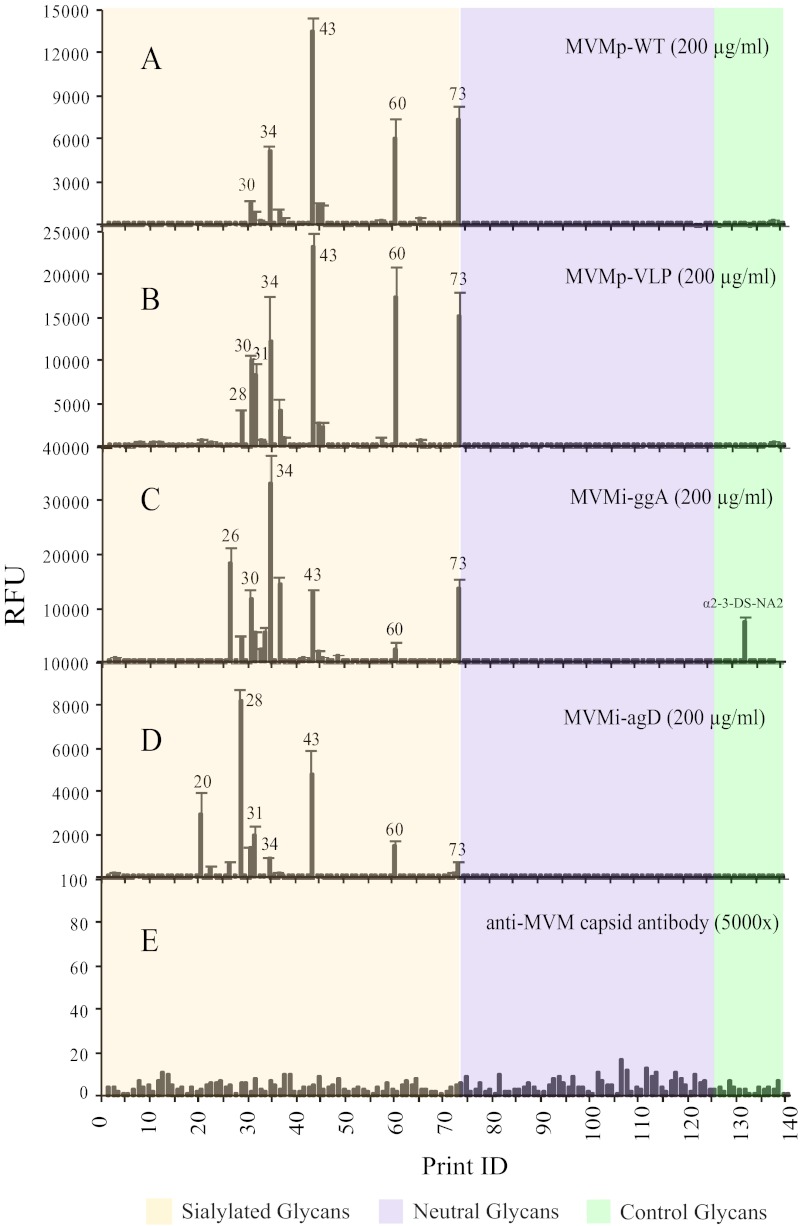
Two prototype strains, empty capsid (MVMp-WT) and virus-like particle (MVMp-VLP), and two immunosuppressive strain mutants, MVMi-agD and MVMi-ggA (39), were tested at 200 μg/ml concentration and detected by a rabbit anti-MVM capsid antibody. All of the MVM viruses recognized glycans H-30, H-31, H-34, H-43, H-60, and H-73 with each non-WT strain binding several additional glycans (Fig. 4). Interestingly, the initial MALDI data (supplemental Table S1) showed that the 6 common binders are disialylated glycans, and the lectin and antibody binding data revealed that all of the binders were also recognized by the anti-SLea/LSTa antibody, suggesting that the terminal sialyl α2–3-linked type 1 motif might be part of the binding determinant. This is a new finding compared with the previous report with CFG Glycan Array Version 3.0 (∼180 glycans), which concluded that MVMs specifically recognized α2–3-linked type 2 motif and MVMi also bound to α2–8-linked multisialylated glycans (41). These data are complementary to the data from the CFG array as there is very little overlap between the structures found on the CFG array and HMG-derived SGM. In fact, these observations suggest that the repertoire of glycan receptors of MVM is broader than originally reported, and these viruses prefer highly charged or possibly multisialylated glycans. We retrieved glycans that were bound by all MVM strains from the TGL for more detailed structural analysis discussed below.
FIGURE 4.
MVM viruses binding to human milk SGM. The binding preferences of several strains of MVM, MVMp-WT (prototype strain, empty capsid; A), MVMp-VLP (prototype strain, virus-like particles (B)), MVMi-ggA (capsid protein mutant of immunosuppressive strain, empty capsid; C), and MVMi-agD (Non-structural protein mutant of immunosuppressive strain, empty capsid; D) were evaluated on the human milk SGM. Each virus was tested at 200 μg/ml and detected by anti-MVM capsid antibody. Panel E shows that there was no background binding from the anti-MVM capsid antibody to HMG microarray. Glycans 1–73 are sialylated glycans (tan), 74–127 are neutral glycans (violet), and 128–140 are controls of structurally defined glycans (light green).
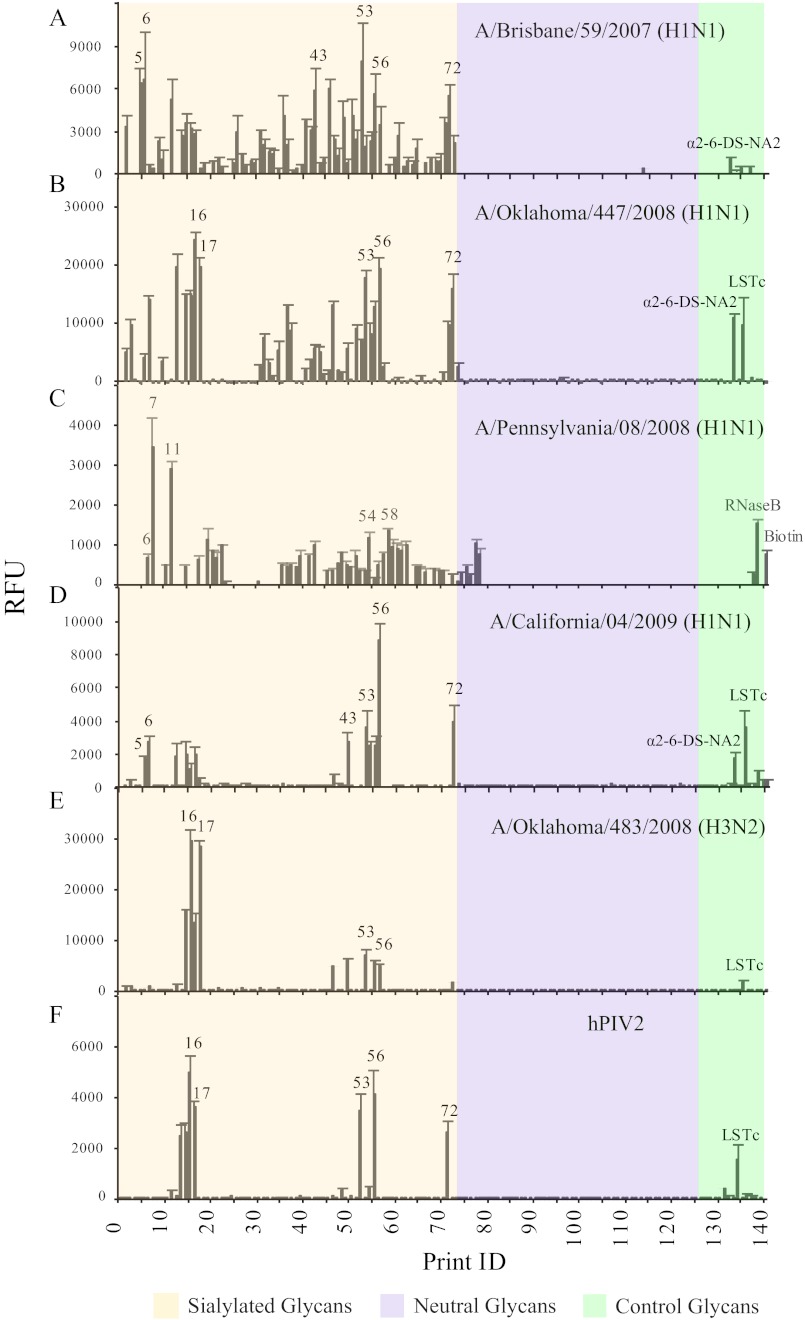
Recently, we utilized the CFG glycan microarray to determine the receptor binding properties of 2009 swine isolates along with some other strains of influenza viruses (37). Our results showed that the highest binding of H1N1 isolates was toward sialylated poly-N-acetyllactosamine structures, which are abundant among HMGs. To determine if HMGs contained natural glycans capable of binding influenza A virus, we interrogated the SGM with three seasonal human H1N1 strains (A/Brisbane/59/2007, A/Pennsylvania/08/2008, and A/Oklahoma/447/08), one human pandemic H1N1 strain (A/California/04/2009), and one H3N2 virus (A/Oklahoma/483/08) for comparison. Consistent with our previous findings, A/Brisbane isolate showed the broadest binding specificity and preferred glycans with terminal α2–6-linked sialic acid (Fig. 5A). Interestingly, the glycans recognized by A/Brisbane/59/2007 (H1N1) were the same glycans that bound SNA, which is specific for the determinant, Neu5Acα2–6Galβ1–4GlcNAc. The binding profile of A/Oklahoma/447/08 (H1N1) virus is similar to the A/Brisbane virus but displayed a much higher signal to noise ratio (Fig. 5B). When compared with binding data from lectins and antibodies, A/Oklahoma/447/08 (H1N1) displayed a clear preference for glycans with α2–6 sialic acid, binding strongly to 2–6-DS-NA2, LSTc, and all of the glycans bound by SNA. Additionally, like A/Brisbane/59/2007, several glycans were recognized by the anti-SLea/LSTa antibody, suggesting certain specificity toward α2–3 sialic acid-containing glycans. A/Pennsylvania/08/2008, which was shown to preferentially bind glycans having terminal α2–6 sialic acid when assayed on the CFG defined glycan array, differed from the other H1N1 strains, A/Oklahoma/447/08 and A/Brisbane/59/2007, in that it did not bind some of the HMGs (such as glycan H-12, H-15, H-16, H-53, and H-55) that possess the Neu5Acα2–6Galβ1–4 motif (Fig. 5C). This result suggests that the virus binding does not solely rely on the sialic acid linkage. In the case of A/California/04/2009, a human pandemic H1N1 isolate, the binding pattern overlaps with SNA (Fig. 5D), which confirms the results from CFG microarray that A/California/04/2009 has a restricted binding preference for α2–6 sialic acid-linked type 2 glycans. Furthermore, as a comparison to the H1N1 A/Oklahoma/447/08 virus, we tested the A/Oklahoma/483/08 H3N2 virus isolate (Fig. 5E) and obtained a more restricted binding pattern where all the bound glycans contain Neu5Acα2–6Galβ1–4GlcNAc structure and were recognized by the H1N1 A/Oklahoma/447/08.
FIGURE 5.
Influenza viruses binding to human milk SGM. The binding preferences of various influenza virus isolates, A/Brisbane/59/2007 H1N1 (A), A/Oklahoma/447/2008 H1N1 (B), A/Pennsylvania/08/2008 (C), A/California/04/2009 H1N1 (D), A/Oklahoma/483/2008 H3N2 (E), and hPIV2 were evaluated with HMG microarray (F). Glycans 1–73 are sialylated glycans (tan), 74–127 are neutral glycans (violet), and 128–140 are controls of structurally defined glycans (light green).
Finally, we evaluated the binding properties of three human parainfluenza viruses on the human milk SGM. It is known that these viruses require the presence of Neu5Acα2–3Galβ1-4GlcNAc motif (66). However, our M. amurensis lectin I binding data showed that this structure is not a component of the HMG. Thus, as we would predict, no binding was observed with the type 1 and 3 (hPIV1 and hPIV3) viruses. By contrast, unlike type 1 and 3, the type 2 parainfluenza virus (hPIV2) displayed a very strict preference for α2–6-sialic acid-containing glycans (Fig. 5F), with receptor specificity similar to the H1N1 strains. Overall, the virus binding experiments demonstrated that many of the HMGs might function as decoys for cell-bound receptors and that the elements in the HMGs might be found on cell-bound receptors. Glycans that bound viruses were selected for more detailed characterization as described below.
Structural Characterization of Selected HMGs
Interrogation of the partially characterized HMG-derived SGM with antibodies against biological markers and viruses demonstrated the potential to identify the receptors of GBPs, including lectins, anti-glycan antibodies, and GBPs in pathogens. Although the existing lectin/antibody binding data already provided some common features of the receptors, detailed structural analysis is necessary to relate the specific structures with biological functions. We selected relevant glycans from the human milk TGL and attempted the use of tandem mass spectrometry and/or serial enzymatic digestion (67) to decipher these structures and found that although MALDI analysis of the glycan derivatives generated excellent data and in some cases good secondary fragmentation data, more sophisticated analyses of permethylated glycan-AEAB derivatives were difficult to interpret due to the complexity of the spectra generated from partial methylation of the primary and secondary amines introduced by the AEAB. These complexities limited the detailed structural analysis of selected glycans from the TGL by MS/MS.
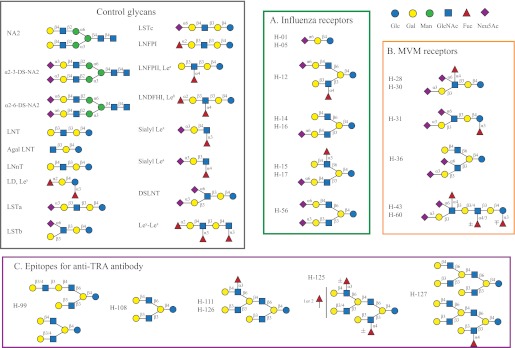
We realized, however, that our lectin-based analysis of the human milk SGM provided significant structural information in a rather high throughput format as all 127 glycans could be analyzed in a single assay. In addition we reasoned that digestion of the glycans on the microarray would be an efficient approach to do in situ structural analysis by combining specific exoglycosidase digestion with defined lectin binding. To demonstrate this, we selected a total of 22 functionally identified glycans, and their HPLC profiles and MALDI-TOF spectra are shown in supplemental Fig. S3. These structures include seven glycans bound by anti-TRA-1 antibodies (H-71, H-99, H-108, H-111, H-125, H-126, and H-127), six glycans bound by MVM (H-28, H-30, H-31, H-36, H-43, and H-60) and nine glycans bound by influenza viruses (H-1, H-5, H-6, H-12, H-14, H-15, H-16, H-17, and H-56). These glycans were printed as a separate array designated “HMG subarray” on N-hydroxysuccinimide-derivatized slides along with 18 structurally defined glycan standards. The 18 control glycans represent some typical structural motifs found in human milk, such as type 1 and type 2 glycans and Lewis blood group glycans (structures shown in Fig. 6). The results from these glycans were used to monitor the behaviors of reagents and to direct structure predictions.
FIGURE 6.
The structures of 17 defined glycans used for controls and the predicted structures of 20 selected human milk glycans. The control glycans (left panel) are listed by common names and the HMGs (panels A, B, and C) are listed by fraction names that are the same as the glycan ID on the microarray. Glycans H01-H17 and H-56 were ligands of influenza viruses, glycans H-28-H-36 were ligands of MVMs, and glycans H-99-H-127 were ligands of anti-TRA-1 antibodies. 18 control glycans were printed; however, glycans 11 and 13 were both LNFPI.
To obtain on-array sequence information, the non-reducing terminal structures of the selected glycans were first determined by screening the HMG subarray with a variety of defined lectins and antibodies (listed in supplemental Table S3A) whose specificities were defined by analysis on v5.0 of the CFG defined glycan array. We reasoned that collection of these data along with the predicted compositional data from mass spectrometry could be combined as a collection of metadata and would provide information about the specific structures of glycans that mass spectrometry alone might not easily resolve. We have termed this approach MAGS. To this end, positive/negative binding by lectins or antibodies to each glycan indicates the presence/absence of the corresponding moiety that each lectin or antibody recognizes, e.g. ECL for Galβ1–4GlcNAc, AAL for fucose, and anti-Lea antibody for Lea epitope. The binding data from multiple GBPs were analyzed in detail to assign the structures. For example, if SNA, RCA-I, and anti-type 1 chain antibody, but not ECL, showed binding toward a glycan, it would suggest that this glycan might possess a terminal Neu5Acα2–6Galβ1–4GlcNAc determinant (SNA and RCA-I positive; ECL negative) together with a terminal Galβ1-3GlcNAc determinant (ECL negative and anti-type 1 antibody-positive). These data do not distinguish between an asymmetric biantennary glycan and the presence of two glycans; however, other metadata can be associated with the individual printed glycans such as a MALDI analysis to determine the number of molecular ions and composition and the shape of the individual peak(s) during HPLC to evaluate glycan homogeneity. The next series of experiments involved the use of a group of specific exoglycosidases to treat the glycans directly on the microarray followed by interrogating with defined lectins or antibodies. The gain and loss of binding can provide composition and/or linkage information for the terminal and penultimate sugar residues. To accomplish this we optimized the reaction condition of five exoglycosidases (supplemental Table S3B) for on-array digestion, including the nonspecific neuraminidase from A. ureafaciens, the recombinant α2–3-neuraminidase from Salmonella typhimurium, the jack bean β1–4/6 galactosidase, the recombinant β1–3-galactosidase from Xanthomonas manihotis, and the recombinant β1–4-galactosidase from Bacteroides fragilis. In general we found that longer incubation times and high enzyme concentrations were necessary to achieve effective digestion when compared with the reactions in solution. The binding results after enzymatic treatment can be divided into two categories, loss and gain. The loss of signal after digestion confirms the prediction from the positive signals before digestion, as in the case of neuraminidase treatment, where the loss of SNA or anti-SLea antibody binding confirms the presence of a specific sialic acid linkage. Similarly, β-galactosidase digestion confirms a type 1 or type 2 chain structure. For the type 1 chain, the binding of anti-type 1 chain antibody is specifically diminished by β1–3-galactosidase treatment, whereas β1–4-galactosidase has no effect. On the contrary, the binding of ECL to a type 2 chain is lost only after β1–4-galactosidase digestion. Furthermore, the type 1 and type 2 structures can also be distinguished by the gain of G. simplicifolia lectin II binding after β1–3/4-galactosidase digestion removes a terminal Gal revealing a terminal GlcNAc. Beyond the single enzymatic treatment, we also conducted sequential digestion involving desialylation first with neuraminidase or α2–3-specific neuraminidase followed by specific β-galactosidase treatment. This set of experiments is particularly useful for sialylated glycans and HMGs that are comprised of many isomers of type 1 and type 2 linear and branched lactosamines and poly-N-acetyllactosamines. Finally, all of the collected metadata including molecular ions, fragmentation MS or MS/MS data, and the behavior on ion exchange chromatography and HPLC are combined to provide predictions of structures that were not possible with MALDI-TOF and MALDI-TOF/TOF analyses alone.
Using this MAGS approach, we were able to predict the structural moieties and most of the linkages for 20 of the 22 HMGs as shown in Fig. 6. The structures of H-06 and H-71 were not proposed because these glycans appeared to be mixtures. The binding data are compiled in supplemental Table S4, and the lectin binding and specific antibody binding data are summarized in Tables 1 and 2, respectively. The detailed description of the logic used to predict the structure of each glycan is provided in the supplemental material. Six pairs of glycans were found to be the same structure (H1/H5, H14/H16, H15/H17, and H28/H30) or have the same general structures (H43/H60 and H111/H126) based on HPLC profiles, MALDI analysis, and binding data. This is due to the overlap of glycans in the fractions obtained during the multi-dimensional chromatography. In addition, several samples were contaminated with minor impurities. Nevertheless, the correlation of proposed structures with the function defined by antibody and virus binding revealed interesting findings. The five influenza virus receptors, including monosialylated H-01/05, H12, H14/16, H15/17, and disialylated H-56 all contain the Neu5Acα2–6Galβ1–4Glc/GlcNAc moiety as indicated by SNA, RCA-1, ECL, neuraminidase, and β1–4 galactosidase data (Table 1). Except for the sialyl lactose (H01/05), the other four structures are biantennary glycans with one type 2 chain branch. It appears that the other branch can be diverse structures, as we observed the presence of Lea, Lex, type 2 chain, and sialylated type 1 chain. The on-array structural analysis also revealed common features for the MVM receptors. Mostly relying on antibodies (anti-SLea/LSTa and anti-type 1) and exoglycosidases (specific and unspecific neuraminidase) data (Table 2), we proposed that the four disialyl structures all carry an α2–3-sialylated type 1 chain with an additional sialic acid attached to the GlcNAc in α2–6 linkage (Neu5Acα2–3Galβ1–3(Neu5Acα2–6)GlcNAc). It is possible that the disialyl LNT motif is one of the recognition determinants for MVM. Similar to influenza virus, modifications such as fucosylation and branching on this motif did not block the virus recognition. Furthermore, our results together with the CFG data indicated that the recognition of MVMs is beyond the sialic acid as the viruses did not bind to all the multisialylated glycans. In the case of anti-TRA-1 antibodies, the sialylated binder H-71 was found to have relatively low purity, and thus its structure was not elucidated. The other six binders (H-99, H-108, H-111, H-125, H-126, and H-127) are neutral complexed glycans, especially for the latter four, which are multi-branched structures. Although we did not obtain all of the linkage information for these large glycans, we found that all the receptors contain the common motif: type 1 lactosamine epitope. Although H-99 is a relatively simple lactosamine glycan, similar to the structure identified from CFG array, the others contain an additional type 2, Lea or Lex branch, and it seems that these extra branches do not prevent the binding of the antibodies. The structures corresponding to abbreviated glycans are defined in Fig. 6 and supplemental Table S4.
TABLE 2.
Heat map of antibody-binding data for structural analysis
A heat map summary of the binding data from analyses of the HMG subarray with different defined antibodies before and after specific exoglycosidase digestion(s) is shown. A total of 40 glycans were printed on the HMG subarray, including 18 control glycans and 22 selected HMGs. The color-coded numbers highlight the binding intensity (the darker the color, the higher the binding intensity). The color scale for the Excel spreadsheet was set using 0 as minimum and 1,500 RFU as maximum for anti-Lea, anti-type 1 and anti-CD15 antibodies and 0 as minimum and 10,000 RFU as maximum for anti-SLea and Lex antibodies. The untreated HMG subarrays were tested with five antibodies. The HMG subarrays treated with non-specific neuraminidase were interrogated with anti-Lea, anti-type 1, anti-SLea, anti-CD15, and the HMG-subarrays treated with α2-3-specific neuraminidase were interrogated with anti-Lea, anti-type 1, and anti-SLea.
DISCUSSION
Human milk glycans have been studied for many decades, and many components have been purified and structurally characterized. It is recognized that HMGs play multiple biological roles, and probably each function is directly correlated with specific structures of individual glycans (6, 8). However, there has been no efficient method for functional glycomic analyses of HMGs in terms of their recognition and interactions with other molecules. Using a shotgun glycomics approach, we isolated a human milk free glycan glycome and generated a corresponding glycan microarray that allowed us to investigate the function of individual glycans in a high-throughput format. The binding patterns of defined lectins and antibodies to the human milk SGM revealed the structural diversity of the isolated glycome. The human milk TGL, which represents a permanent repository for HMG analyses, is enriched in sialylated glycans, type 1 and type 2 glycans, and Lea-containing glycans but lacks α1–2-linked fucosylated glycans, such as H type 1 and Leb structures. These data indicate that the milk sample used for preparation of the tagged glycan library was from a Lewis positive individual and confirm that the individual was a secretor negative donor. We chose to use milk from a secretor negative donor in this first study, as it would limit the diversity of glycans available for interrogation. In further studies we are preparing human milk SGMs from donors of other genotypes. It is clear from our studies that analyses of glycans of individual milk samples on microarrays using specific blood group antibodies may represent an alternative strategy to the existing mass-based (68) or HPAEC-based methods (4) for screening the genotype of milk donors.
The human milk SGM described here provides a source of purified glycans that is conveniently available as an archived resource for interrogation with antibodies, pathogenic microorganisms, and potentially live cells for evaluating functions of individual structures. In this approach, once a biologically relevant structure is identified, many of its structural features can be described by collecting metadata in a database for the entire shotgun array. If more detailed analysis, i.e. actual sequence and definition of complete structure, are required, the preliminary structural features will be invaluable for analysts using other physical techniques such as mass spectrometry and NMR. For example, monoclonal antibodies TRA-1–60 and TRA-1–81 are widely used to identify the biomarker for human pluripotent stem cells. These antibodies were generated using cells as antigens, and they were originally thought to be directed against keratan-sulfate proteoglycan (69). In recent studies using the defined glycan array from the CFG (Version 4.2), Natunen et al. (63) identified type 1 lactosamine as an epitope for TRA-1–60 and TRA-1–81 antibodies. Here, we extended those observations to additional structures that were identified first as glycans recognized by these antibodies followed by determining their structure using metadata collected on these specific glycans on the shotgun subarrays. Our results revealed the presence of ligands for the anti-TRA-1 antibodies in the human milk glycome and showed that the two stem cell marker antibodies recognize complicated large glycans that possess the reported minimum recognition moiety, type 1 lactosamine. Although there is no obvious correlation between the stem cell marker antibodies and the function of HMGs, we demonstrated that human milk SGM is a glycan library useful for investigating the binding specificity of proteins due to the diversity and complexity of HMGs and especially due to its enrichment of blood group related epitopes, which are the receptor of various proteins and microorganisms. Another recent example is the newly generated anti-stage-specific embryonic antigen-5 monoclonal antibody, which was found to only recognize H type 1 glycans (70) that are characteristic of HMGs.
Inhibition of pathogen binding is considered the essential immune protection provided to the infant by human milk. It is believed that certain HMGs act as the receptor analogues or decoy receptors to bind to pathogens and thus prevent their attachment to cell targets (3). The established human milk SGM provides a unique platform to study the interaction between pathogens and milk oligosaccharides. To demonstrate the ability of the human milk SGM for evaluating the binding specificity of pathogens, we tested MVM and influenza virus, both known to recognize sialic acids, which are enriched in the human milk SGM. It was interesting that our study revealed that MVMs specifically bound to α2–3-linked multisialyl glycans, which are a category of novel receptors that are not present on the CFG microarray. Although there is no connection between the HMG function and MVMs, these results could provide additional insight into the receptor preferences of MVMs, which were previously found to recognize α2–8-linked multisialylated glycans and sialyl Lex moieties on the CFG microarray (41). Combining the study from both glycan arrays, it is clear that MVMs have a unique preference for certain multisialylated oligosaccharides, like disialyl LNT glycans we identified in this paper.
To expand our study to human influenza viruses beyond the synthetic CFG microarray (37) and the sialylated glycan microarray (71), we interrogated the human milk SGM with several influenza strains. Similar to previous findings, the viruses have a strong preference toward α2–6-sialylated glycans, but not all the α2–6-sialylated glycans were recognized by the viruses, which once again suggested that the recognition of influenza viruses are not limited only to the terminal glycans. One common feature for the receptors is that they all contain an α2–6-linked sialic acid to a type 2 chain. Overall, our virus experiments demonstrated that the human milk SGM is useful for studying pathogen binding properties and presumably possesses receptors for pathogens associated with infant diseases, which will be the focus of future studies.
A major tool used in the current work was the combination of metadata from mass spectrometry, lectin, and antibody binding and on-array enzymatic degradation of glycans in the technique we termed MAGS. This approach can complement the use of mass spectrometry and other approaches and provides key information important in predicting glycan structure. Development of MAGS partly arose from our needs to identify the recognition motifs and even underlying glycan structures required for better understanding the interactions of glycans with proteins and microorganisms. Compared with the low cost and high efficiency of nucleic acid and protein sequencing analyses, glycan sequence analysis is very difficult and often requires highly sophisticated instrumentation and skilled interpretation. Although progress in mass spectrometry has significantly advanced the structural analysis of glycans, there is a need to develop more high-throughput and fast glycan sequencing methods that only require small amounts of samples. In this work here, we initiated the on-array glycan sequencing method of MAGS and chose human milk SGM as the working model.
Using lectins and antibodies whose binding specificities are known, we collected binding data from original glycans and enzyme-treated glycans. Various sugar moieties and linkage information were deduced from analyzing all of the data. Extremely minute amounts of HMGs were used, and many individual glycans were analyzed simultaneously on a single microarray. These studies demonstrated the successful use of the on-array glycan sequencing method and also showed that this technique is currently limited only by the availability of lectins and antibodies with defined binding specificities as well as specific exoglycosidases. For example, there is no reagent that can differentiate the branch point (β1–3/6) to the core lactose/lactosamine, although the current literature indicates that the lactosamine unit at this β1–6 branch point is a type 2 structure (21). Except for neuraminidase, the enzymes used showed relative low activity on the slides compared to the activity in solution and thus required longer incubation time and relatively large amounts of enzymes (supplemental Table S3B). Clearly, the analyses described here could be extended by the use of specific α-l-fucosidases, but these studies were limited by their availability and cost from commercial sources. Nevertheless, using the on-array sequencing analysis, we were able to successfully identify many of the structural features important for antibody binding and the receptors for several viruses in a high throughput format.
Collecting milk glycan structures and having methods to rapidly identify milk glycans is of course useful and can generate interesting data on the metabolic changes of specific milk glycans but generally do not provide direct information on glycan function. Having glycans immobilized on a microarray for interrogation by biologically relevant GBPs or microorganisms and retrievable from a TGL provides a true approach to functional glycomic analysis to accelerate discovery of HMG function. As an added feature, by collecting metadata on each glycan associated with a SGM as the array is interrogated with both defined GBPs and GBPs of interest to reveal glycan function, these arrays of immobilized glycans can be structurally characterized in a high throughput format. As more defined GBPs and highly purified exoglycosidases become available, this MAGS approach could be miniaturized and automated as a high throughput analytical tool capable of sequencing thousands of glycans simultaneously.
Acknowledgments
We thank Shelly Gulati (University of Oklahoma Health Sciences Center) for help on influenza virus preparations and Dr. Jamie Heimburg-Molinaro (Emory University School of Medicine) for manuscript editing and review.
This work was supported, in whole or in part, by National Institutes of Health Grants GM62116 (to the Consortium for Functional Glycomics) and RO1 GM085448 (to D. F. S.). This work was also supported by National Science Foundation Grant MCB 0718948 (to M. A.-M.) and United States Department of Health and Human Services Contract HHSN266200700006C (to the NIAID Centers of Excellence for Influenza Research and Surveillance).

This article contains supplemental descriptions of structural predictions of selected HMGs, Tables S1–S4, and Figs. S1–S3.
M. Mandalasi, N. Dorabawila, D. F. Smith, J. Heimburg-Molinaro, R. D. Cummings, A. K. Nyame, unpublished observations.
- HMG
- human milk glycan
- SGM
- shotgun glycan microarray
- GBP
- glycan-binding protein
- AEAB
- 2-amino-N-(2-aminoethyl)-benzamide
- Hex
- hexose
- HexNAc
- N-acetylhexosamine
- Fuc
- fucose
- AAL
- A. aurantia lectin
- SNA
- S. nigra agglutinin
- LTL
- L. tetragonolobus lectin
- UEA-I
- U. europaeus agglutinin I
- RCA-I
- R. communis agglutinin I
- ECL
- E. cristagalli Lectin
- RFU
- relative fluorescence units
- Lea
- Lewisa
- Leb
- Lewisb
- Lex
- Lewisx
- Ley
- Lewisy
- CFG
- Consortium for Functional Glycomics
- MVM
- minute virus of mice
- hPIV
- human parainfluenza virus
- MAGS
- metadata-assisted glycan sequencing
- VLP
- virus-like particle
- TGL
- tagged glycan library
- Neu5Ac
- N-acetylneuraminic acid (sialic acid).
REFERENCES
- 1. Schack-Nielsen L., Michaelsen K. F. (2007) Advances in our understanding of the biology of human milk and its effects on the offspring. J. Nutr. 137, 503S–510S [DOI] [PubMed] [Google Scholar]
- 2. Newburg D. S. (2005) Innate immunity and human milk. J. Nutr. 135, 1308–1312 [DOI] [PubMed] [Google Scholar]
- 3. Newburg D. S., Ruiz-Palacios G. M., Morrow A. L. (2005) Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 25, 37–58 [DOI] [PubMed] [Google Scholar]
- 4. Thurl S., Munzert M., Henker J., Boehm G., Müller-Werner B., Jelinek J., Stahl B. (2010) Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br. J. Nutr. 104, 1261–1271 [DOI] [PubMed] [Google Scholar]
- 5. Thurl S., Henker J., Siegel M., Tovar K., Sawatzki G. (1997) Detection of four human milk groups with respect to Lewis blood group dependent oligosaccharides. Glycoconj. J. 14, 795–799 [DOI] [PubMed] [Google Scholar]
- 6. Kobata A. (2010) Structures and application of oligosaccharides in human milk. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86, 731–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bode L. (2009) Human milk oligosaccharides. Prebiotics and beyond. Nutr. Rev. 67, S183–S191 [DOI] [PubMed] [Google Scholar]
- 8. Bode L. (2006) Recent advances on structure, metabolism, and function of human milk oligosaccharides. J. Nutr. 136, 2127–2130 [DOI] [PubMed] [Google Scholar]
- 9. Espinosa R. M., Taméz M., Prieto P. (2007) Efforts to emulate human milk oligosaccharides. Br. J. Nutr. 98, S74–S79 [DOI] [PubMed] [Google Scholar]
- 10. Sela D. A., Mills D. A. (2010) Nursing our microbiota. Molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 18, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. German J. B., Freeman S. L., Lebrilla C. B., Mills D. A. (2008) Human milk oligosaccharides. Evolution, structures, and bioselectivity as substrates for intestinal bacteria. Nestle Nutr. Workshop Ser. Pediatr. Program 62, 205–218; discussion 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Donovan S. M. (2009) Human milk oligosaccharides. The plot thickens. Br. J. Nutr. 101, 1267–1269 [DOI] [PubMed] [Google Scholar]
- 13. Kuntz S., Kunz C., Rudloff S. (2009) Oligosaccharides from human milk induce growth arrest via G2/M by influencing growth-related cell cycle genes in intestinal epithelial cells. Br. J. Nutr. 101, 1306–1315 [DOI] [PubMed] [Google Scholar]
- 14. Kuntz S., Rudloff S., Kunz C. (2008) Oligosaccharides from human milk influence growth-related characteristics of intestinally transformed and non-transformed intestinal cells. Br. J. Nutr. 99, 462–471 [DOI] [PubMed] [Google Scholar]
- 15. Idota T., Kawakami H., Murakami Y., Sugawara M. (1995) Inhibition of cholera toxin by human milk fractions and sialyllactose. Biosci. Biotechnol. Biochem. 59, 417–419 [DOI] [PubMed] [Google Scholar]
- 16. Bode L., Kunz C., Muhly-Reinholz M., Mayer K., Seeger W., Rudloff S. (2004) Inhibition of monocyte, lymphocyte, and neutrophil adhesion to endothelial cells by human milk oligosaccharides. Thromb. Haemost. 92, 1402–1410 [DOI] [PubMed] [Google Scholar]
- 17. Borén T., Falk P., Roth K. A., Larson G., Normark S. (1993) Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262, 1892–1895 [DOI] [PubMed] [Google Scholar]
- 18. Ruvoën-Clouet N., Ganière J. P., André-Fontaine G., Blanchard D., Le Pendu J. (2000) Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo-blood group family. J. Virol. 74, 11950–11954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marionneau S., Ruvoën N., Le Moullac-Vaidye B., Clement M., Cailleau-Thomas A., Ruiz-Palacois G., Huang P., Jiang X., Le Pendu J. (2002) Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122, 1967–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruiz-Palacios G. M., Cervantes L. E., Ramos P., Chavez-Munguia B., Newburg D. S. (2003) Campylobacter jejuni binds intestinal H(O) antigen (Fucα1, 2Galβ1, 4GlcNAc) and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 278, 14112–14120 [DOI] [PubMed] [Google Scholar]
- 21. Ninonuevo M. R., Park Y., Yin H., Zhang J., Ward R. E., Clowers B. H., German J. B., Freeman S. L., Killeen K., Grimm R., Lebrilla C. B. (2006) A strategy for annotating the human milk glycome. J. Agric. Food Chem. 54, 7471–7480 [DOI] [PubMed] [Google Scholar]
- 22. Niñonuevo M. R., Lebrilla C. B. (2009) Mass spectrometric methods for analysis of oligosaccharides in human milk. Nutr. Rev. 67, S216–S226 [DOI] [PubMed] [Google Scholar]
- 23. Smith D. F., Song X., Cummings R. D. (2010) Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods Enzymol. 480, 417–444 [DOI] [PubMed] [Google Scholar]
- 24. Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C. H., Paulson J. C. (2004) Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rillahan C. D., Paulson J. C. (2011) Glycan microarrays for decoding the glycome. Annu. Rev. Biochem. 80, 797–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Paz J. L., Seeberger P. H. (2012) Recent advances and future challenges in glycan microarray technology. Methods Mol. Biol. 808, 1–12 [DOI] [PubMed] [Google Scholar]
- 27. Liu Y., Childs R. A., Palma A. S., Campanero-Rhodes M. A., Stoll M. S., Chai W., Feizi T. (2012) Neoglycolipid-based oligosaccharide microarray system. Preparation of NGLs and their noncovalent immobilization on nitrocellulose-coated glass slides for microarray analyses. Methods Mol. Biol. 808, 117–136 [DOI] [PubMed] [Google Scholar]
- 28. Cummings R. D. (2009) The repertoire of glycan determinants in the human glycome. Mol. Biosyst. 5, 1087–1104 [DOI] [PubMed] [Google Scholar]
- 29. Song X., Lasanajak Y., Xia B., Heimburg-Molinaro J., Rhea J. M., Ju H., Zhao C., Molinaro R. J., Cummings R. D., Smith D. F. (2011) Shotgun glycomics. A microarray strategy for functional glycomics. Nat. Methods 8, 85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seko A., Koketsu M., Nishizono M., Enoki Y., Ibrahim H. R., Juneja L. R., Kim M., Yamamoto T. (1997) Occurence of a sialylglycopeptide and free sialylglycans in hen's egg yolk. Biochim. Biophys. Acta 1335, 23–32 [DOI] [PubMed] [Google Scholar]
- 31. Song X., Xia B., Stowell S. R., Lasanajak Y., Smith D. F., Cummings R. D. (2009) Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem. Biol. 16, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song X., Lasanajak Y., Olson L. J., Boonen M., Dahms N. M., Kornfeld S., Cummings R. D., Smith D. F. (2009) Glycan microarray analysis of P-type lectins reveals distinct phosphomannose glycan recognition. J. Biol. Chem. 284, 35201–35214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kobata A., Ginsburg V., Tsuda M. (1969) Oligosaccharides of human milk. I. Isolation and characterization. Arch. Biochem. Biophys. 130, 509–513 [DOI] [PubMed] [Google Scholar]
- 34. Smith D. F., Zopf D. A., Ginsburg V. (1978) Fractionation of sialyl oligosaccharides of human milk by ion-exchange chromatography. Anal. Biochem. 85, 602–608 [DOI] [PubMed] [Google Scholar]
- 35. Heimburg-Molinaro J., Song X., Smith D. F., Cummings R. D. (2011) Preparation and analysis of glycan microarrays. Curr. Protoc. Protein Sci. Chapter 12, Unit 12.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song X., Heimburg-Molinaro J., Smith D. F., Cummings R. D. (2012) Glycan microarrays. Methods Mol. Biol. 800, 163–171 [DOI] [PubMed] [Google Scholar]
- 37. Bradley K. C., Jones C. A., Tompkins S. M., Tripp R. A., Russell R. J., Gramer M. R., Heimburg-Molinaro J., Smith D. F., Cummings R. D., Steinhauer D. A. (2011) Comparison of the receptor binding properties of contemporary swine isolates and early human pandemic H1N1 isolates (Novel 2009 H1N1). Virology 413, 169–182 [DOI] [PubMed] [Google Scholar]
- 38. Kumari K., Gulati S., Smith D. F., Gulati U., Cummings R. D., Air G. M. (2007) Receptor binding specificity of recent human H3N2 influenza viruses. Virol. J. 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. D'Abramo A. M., Jr., Ali A. A., Wang F., Cotmore S. F., Tattersall P. (2005) Host range mutants of Minute Virus of Mice with a single VP2 amino acid change require additional silent mutations that regulate NS2 accumulation. Virology 340, 143–154 [DOI] [PubMed] [Google Scholar]
- 40. López-Bueno A., Rubio M. P., Bryant N., McKenna R., Agbandje-McKenna M., Almendral J. M. (2006) Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J. Virol. 80, 1563–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nam H. J., Gurda-Whitaker B., Gan W. Y., Ilaria S., McKenna R., Mehta P., Alvarez R. A., Agbandje-McKenna M. (2006) Identification of the sialic acid structures recognized by minute virus of mice and the role of binding affinity in virulence adaptation. J. Biol. Chem. 281, 25670–25677 [DOI] [PubMed] [Google Scholar]
- 42. Cotmore S. F., Hafenstein S., Tattersall P. (2010) Depletion of virion-associated divalent cations induces parvovirus minute virus of mice to eject its genome in a 3′-to-5′ direction from an otherwise intact viral particle. J. Virol. 84, 1945–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brewer C. F., Bhattacharyya L. (1986) Specificity of concanavalin A binding to asparagine-linked glycopeptides. A nuclear magnetic relaxation dispersion study. J. Biol. Chem. 261, 7306–7310 [PubMed] [Google Scholar]
- 44. Baenziger J. U., Fiete D. (1979) Structural determinants of concanavalin A specificity for oligosaccharides. J. Biol. Chem. 254, 2400–2407 [PubMed] [Google Scholar]
- 45. Naismith J. H., Field R. A. (1996) Structural basis of trimannoside recognition by concanavalin A. J. Biol. Chem. 271, 972–976 [DOI] [PubMed] [Google Scholar]
- 46. Tollefsen S. E., Kornfeld R. (1983) The B4 lectin from Vicia villosa seeds interacts with N-acetylgalactosamine residues α-linked to serine or threonine residues in cell surface glycoproteins. J. Biol. Chem. 258, 5172–5176 [PubMed] [Google Scholar]
- 47. Lyer P. N., Wilkinson K. D., Goldstein L. J. (1976) An N-acetyl-d-glycosamine binding lectin from Bandeiraea simplicifolia seeds. Arch. Biochem. Biophys. 177, 330–333 [DOI] [PubMed] [Google Scholar]
- 48. Wang W. C., Cummings R. D. (1988) The immobilized leukoagglutinin from the seeds of Maackia amurensis binds with high affinity to complex-type Asn-linked oligosaccharides containing terminal sialic acid-linked α-2,3 to penultimate galactose residues. J. Biol. Chem. 263, 4576–4585 [PubMed] [Google Scholar]
- 49. Kochibe N., Furukawa K. (1980) Purification and properties of a novel fucose-specific hemagglutinin of Aleuria aurantia. Biochemistry 19, 2841–2846 [DOI] [PubMed] [Google Scholar]
- 50. Matsumoto I., Osawa T. (1969) Purification and characterization of an anti-H(O) phytohemagglutinin of Ulex europeus. Biochim. Biophys. Acta 194, 180–189 [DOI] [PubMed] [Google Scholar]
- 51. Pereira M. E., Kisailus E. C., Gruezo F., Kabat E. A. (1978) Immunochemical studies on the combining site of the blood group H-specific lectin 1 from Ulex europeus seeds. Arch. Biochem. Biophys. 185, 108–115 [DOI] [PubMed] [Google Scholar]
- 52. Pereira M. E., Kabat E. A. (1974) Specificity of purified hemagglutinin (lectin) from Lotus tetragonolobus. Biochemistry 13, 3184–3192 [DOI] [PubMed] [Google Scholar]
- 53. Yan L., Wilkins P. P., Alvarez-Manilla G., Do S. I., Smith D. F., Cummings R. D. (1997) Immobilized Lotus tetragonolobus agglutinin binds oligosaccharides containing the Le (x) determinant. Glycoconj. J. 14, 45–55 [DOI] [PubMed] [Google Scholar]
- 54. Shibuya N., Goldstein I. J., Broekaert W. F., Nsimba-Lubaki M., Peeters B., Peumans W. J. (1987) The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(α2–6)Gal/GalNAc sequence. J. Biol. Chem. 262, 1596–1601 [PubMed] [Google Scholar]
- 55. Green E. D., Brodbeck R. M., Baenziger J. U. (1987) Lectin affinity high-performance liquid chromatography. Interactions of N-glycanase-released oligosaccharides with Ricinus communis agglutinin I and Ricinus communis agglutinin II. J. Biol. Chem. 262, 12030–12039 [PubMed] [Google Scholar]
- 56. Baenziger J. U., Fiete D. (1979) Structural determinants of Ricinus communis agglutinin and toxin specificity for oligosaccharides. J. Biol. Chem. 254, 9795–9799 [PubMed] [Google Scholar]
- 57. Iglesias J. L., Lis H., Sharon N. (1982) Purification and properties of a d-galactose/N-acetyl-d-galactosamine-specific lectin from Erythrina cristagalli. Eur. J. Biochem. 123, 247–252 [DOI] [PubMed] [Google Scholar]
- 58. Kaladas P. M., Kabat E. A., Iglesias J. L., Lis H., Sharon N. (1982) Immunochemical studies on the combining site of the d-galactose/N-acetyl-d-galactosamine specific lectin from Erythrina cristagalli seeds. Arch. Biochem. Biophys. 217, 624–637 [DOI] [PubMed] [Google Scholar]
- 59. Kaladas P. M., Kabat E. A., Shibata S., Goldstein I. J. (1983) Immunochemical studies on the binding specificity of the blood group Leb specific lectin Griffonia simplicifolia IV. Arch. Biochem. Biophys. 223, 309–318 [DOI] [PubMed] [Google Scholar]
- 60. Svensson C., Teneberg S., Nilsson C. L., Kjellberg A., Schwarz F. P., Sharon N., Krengel U. (2002) High resolution crystal structures of Erythrina cristagalli lectin in complex with lactose and 2′-α-l-fucosyllactose and correlation with thermodynamic binding data. J. Mol. Biol. 321, 69–83 [DOI] [PubMed] [Google Scholar]
- 61. Grollman E. F., Kobata A., Ginsburg V. (1969) An enzymatic basis for Lewis blood types in man. J. Clin. Invest. 48, 1489–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kobata A. (2000) A journey to the world of glycobiology. Glycoconj. J. 17, 443–464 [DOI] [PubMed] [Google Scholar]
- 63. Natunen S., Satomaa T., Pitkänen V., Salo H., Mikkola M., Natunen J., Otonkoski T., Valmu L. (2011) The binding specificity of the marker antibodies Tra-1–60 and Tra-1–81 reveals a novel pluripotency associated type 1 lactosamine epitope. Glycobiology 21, 1125–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Marionneau S., Le Moullac-Vaidye B., Le Pendu J. (2002) Expression of histo-blood group A antigen increases resistance to apoptosis and facilitates escape from immune control of rat colon carcinoma cells. Glycobiology 12, 851–856 [DOI] [PubMed] [Google Scholar]
- 65. Varki A. (2008) Sialic acids in human health and disease. Trends Mol. Med. 14, 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Amonsen M., Smith D. F., Cummings R. D., Air G. M. (2007) Human parainfluenza viruses hPIV1 and hPIV3 bind oligosaccharides with α2–3-linked sialic acids that are distinct from those bound by H5 avian influenza virus hemagglutinin. J. Virol 81, 8341–8345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kannicht C., Grunow D., Lucka L. (2008) Enzymatic sequence analysis of N-glycans by exoglycosidase cleavage and mass spectrometry. Detection of Lewis X structures. Methods Mol. Biol. 446, 255–266 [DOI] [PubMed] [Google Scholar]
- 68. Blank D., Gebhardt S., Maass K., Lochnit G., Dotz V., Blank J., Geyer R., Kunz C. (2011) High-throughput mass finger printing and Lewis blood group assignment of human milk oligosaccharides. Anal. Bioanal. Chem. 401, 2495–2510 [DOI] [PubMed] [Google Scholar]
- 69. Badcock G., Pigott C., Goepel J., Andrews P. W. (1999) The human embryonal carcinoma marker antigen TRA-1–60 is a sialylated keratan sulfate proteoglycan. Cancer Res. 59, 4715–4719 [PubMed] [Google Scholar]
- 70. Tang C., Lee A. S., Volkmer J. P., Sahoo D., Nag D., Mosley A. R., Inlay M. A., Ardehali R., Chavez S. L., Pera R. R., Behr B., Wu J. C., Weissman I. L., Drukker M. (2011) An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat. Biotechnol. 29, 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Song X., Yu H., Chen X., Lasanajak Y., Tappert M. M., Air G. M., Tiwari V. K., Cao H., Chokhawala H. A., Zheng H., Cummings R. D., Smith D. F. (2011) A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J. Biol. Chem. 286, 31610–31622 [DOI] [PMC free article] [PubMed] [Google Scholar]