Abstract
Recent studies have produced a variety of advances in the investigation of genetic similarities and differences among human populations. Here, I pose a series of questions about human population-genetic similarities and differences, and I then answer these questions by numerical computation with a single shared population-genetic dataset. The collection of answers obtained provides an introductory perspective for understanding key results on the features of worldwide human genetic variation.
Keywords: ANCESTRY, APPORTIONMENT, CLUSTERING, HETEROZYGOSITY, MICROSATELLITES, PARTITION OF VARIATION, POPULATION STRUCTURE, PRIVATE ALLELES
In this expository overview, I seek to clarify recent developments in the study of the magnitude of the genetic variabilityamong human populations. Specifically, I examine the answers to several questions about human genetic similarities and differences, all in the context of a single standardized set of samples and markers.
Are most alleles widely distributed, or are they largely confined to specific parts of the world?
Do there exist distinctive alleles for specific geographic regions that distinguish individuals in one group from those of other groups?
Of the genetic variants that exist in the human genome, how many are present within a given geographic region?
On average, how different are two individuals from the same local population, in comparison with two individuals chosen from any two populations anywhere in the world?
To what extent is it possible to determine the genetic ancestry of an individual using the alleles in his or her genome?
What events in human evolutionary history are responsible for the basic patterns of genetic similarity and difference evident in worldwide human populations?
Rather than providing a systematic review, this article offers an informal introductory perspective on the basis of work that my colleagues and I have performed with the genome-wide microsatellites of the Human Genome Diversity Project/Centre d’Etude du Polymorphisme Humain (HGDP-CEPH) Cell Line Panel (Rosenberg et al. 2002, 2003a, 2003b, 2005; Zhivotovsky et al. 2003; Ramachandran et al., 2004, 2005; Schroeder et al. 2007; Szpiech et al. 2008). Use of a shared dataset to address all of the questions eliminates the effects that such factors as differences in samples or loci can have in producing different outcomes across studies. Although we have previously reported results very similar to many of those shown, the analyses here are original, so that the same standardized dataset is used for all computations.
A dataset on autosomal microsatellite polymorphisms in human populations
The HGDP-CEPH Cell Line Panel (Cann et al. 2002; Cavalli-Sforza 2005), henceforth termed the “diversity panel,” consists of 1064 cell lines from individuals in more than 50 indigenous populations distributed worldwide (Figure 1, Table 1). For this article, the populations are divided among seven major geographic regions: sub-Saharan Africa, Europe, the Middle East, Central/South Asia, East Asia, Oceania, and the Americas.
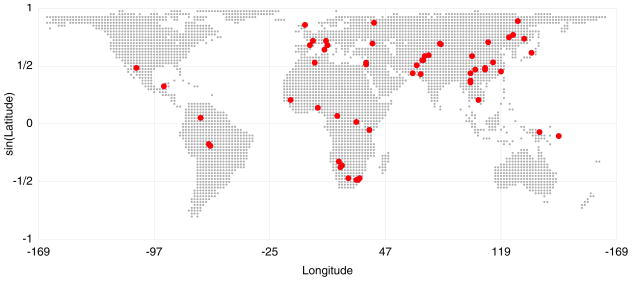
Figure 1.
Geographic locations of populations in the HGDP-CEPH Cell Line Panel. If a range of latitude and longitude coordinates was specified by Cann et al. (2002) for a given population, the population was plotted at the centroid of the range (mean of the longitudes, inverse sine of the mean of the sines of the latitudes). Except where otherwise specified, this article utilizes the exact microsatellite dataset of Rosenberg et al. (2005), a collection of 783 autosomal microsatellites in 1048 individuals from 53 populations. The map indicates 58 populations, some pairs of which overlap precisely in location, but six Bantu groups from southern Africa are grouped into a single population for the analysis. When the populations are split into regions, unless otherwise specified, the regions include sub-Saharan Africa, Europe, the Middle East (and North Africa), Central/South Asia, East Asia, Oceania, and the Americas.
Table 1.
Coordinates used in geographic analyses. Latitudes in the northern hemisphere are listed with positive values, as are longitudes in the eastern hemisphere. Additional coordinates used in some analyses include 28.39886514°S 27.6°E for Southeastern Bantu and 20.9934025°S 18.666667°E for Southwestern Bantu, respectively.
| Population | Latitude | Longitude |
|---|---|---|
| Adygei | 44 | 39 |
| Balochi | 30.49871492 | 66.5 |
| Bantu (Kenya) | −3 | 37 |
| Bantu (Southern Africa) | −25.56926433 | 24.25 |
| Basque | 43 | 0 |
| Bedouin | 31 | 35 |
| Biaka Pygmy | 4 | 17 |
| Brahui | 30.49871492 | 66.5 |
| Burusho | 36.49838568 | 74 |
| Cambodian | 12 | 105 |
| Colombian | 3 | −68 |
| Dai | 21 | 100 |
| Daur | 48.49753416 | 124 |
| Druze | 32 | 35 |
| French | 46 | 2 |
| Han | 32.26566812 | 114 |
| Han (Northern China) | 32.26566812 | 114 |
| Hazara | 33.49855601 | 70 |
| Hezhen | 47.4976192 | 133.5 |
| Italian | 46 | 10 |
| Japanese | 38 | 138 |
| Kalash | 35.99366014 | 71.5 |
| Karitiana | −10 | −63 |
| Lahu | 22 | 100 |
| Makrani | 26 | 64 |
| Mandenka | 12 | −12 |
| Maya | 19 | −91 |
| Mbuti Pygmy | 1 | 29 |
| Melanesian | −6 | 155 |
| Miao | 28 | 109 |
| Mongola | 45 | 111 |
| Mozabite | 32 | 3 |
| Naxi | 26 | 100 |
| Orcadian | 59 | −3 |
| Oroqen | 50.43389257 | 126.5 |
| Palestinian | 32 | 35 |
| Papuan | −4 | 143 |
| Pathan | 33.48700562 | 70.5 |
| Pima | 29 | −108 |
| Russian | 61 | 40 |
| San | −21 | 20 |
| Sardinian | 40 | 9 |
| She | 27 | 119 |
| Sindhi | 25.49063551 | 69 |
| Surui | −11 | −62 |
| Tu | 36 | 101 |
| Tujia | 29 | 109 |
| Tuscan | 43 | 11 |
| Uygur | 44 | 81 |
| Xibo | 43.49792973 | 81.5 |
| Yakut | 62.98287845 | 129.5 |
| Yi | 28 | 103 |
| Yoruba | 7.995094727 | 5 |
Each individual represented in the diversity panel has been genotyped for 783 microsatellite polymorphisms, spread across all 22 autosomes (Rosenberg et al. 2005). Recall that a microsatellite locus is a region of the genome in which individuals differ in their numbers of repeated copies of a basic DNA unit (Goldstein and Schlötterer 1999). Thus, for example, an individual with the DNA sequence CATCATCATCAT at a particular microsatellite has four copies of the repeated unit CAT. For each of the microsatellites we have studied, the basic repeated unit has size 2, 3, 4, or 5.
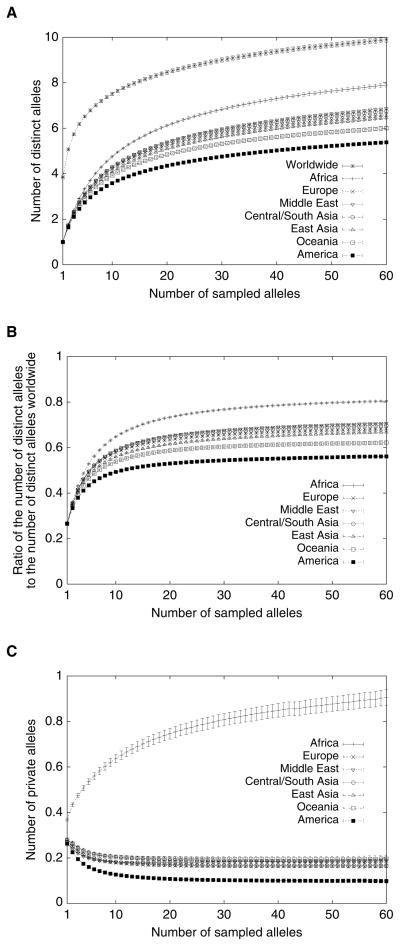
Because human microsatellites are highly variable, they provide considerable information about human genetic diversity and its geographic distribution (e.g. Bowcock et al. 1994). They tend to have at least several distinct alleles: for the 783 loci we have studied, the mean worldwide number of distinct alleles per locus is 11.94. Adjusting for the differing sample sizes of the different geographic regions in the dataset by considering subsamples of equal size from the various regions, Figure 2A shows that on average, a subsample of size 60 alleles from Africa contains ~8 distinct alleles, a subsample of size 60 from Oceania or the Americas contains ~5–6 distinct alleles, and a subsample from Europe or Asia contains ~6–7 distinct alleles. For comparison, a worldwide sample of size 420—60 from each of the seven regions—contains on average ~10 distinct alleles per locus.
Figure 2.
Mean and standard error across 783 loci of the number of distinct alleles, proportion of distinct alleles worldwide, and private alleles in geographic regions, as a function of the number of sampled alleles. (A) Number of distinct alleles. For a given locus, region, and sample size g, the number of distinct alleles averaged over all possible subsamples of g alleles from the given region is computed according to the rarefaction formula (Hurlbert 1971; Kalinowski 2004, eq. 3; Szpiech et al. 2008). (B) Proportion of alleles observed in a specific region. For a given locus, region, and sample size g, the quotient of the mean number of distinct alleles at the locus for a subsample from the region and the corresponding value for a worldwide subsample containing g alleles from each region is computed. (C) Number of private alleles. For a given locus, region, and sample size g, the number of private alleles in the region—averaging over all possible subsamples that contain g alleles each from the seven regions—is computed according to an extension of the rarefaction formula (Kalinowski 2004, eq. 4; Szpiech et al. 2008). Error bars denote the standard error of the mean across loci. In all three plots, for each sample size g, loci were considered only if their sample sizes were at least g in each geographic region.
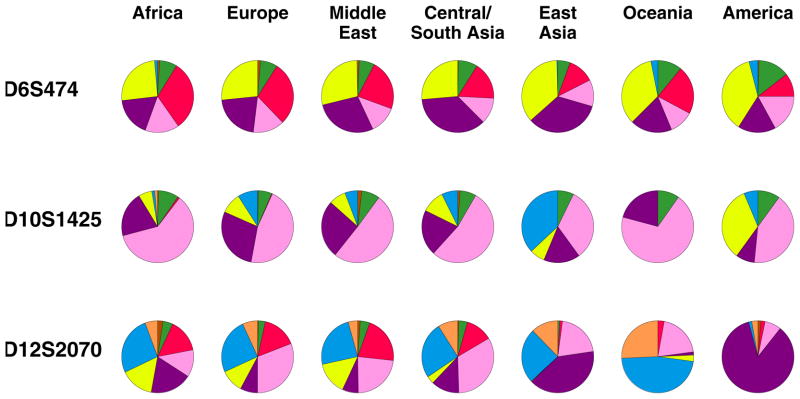
Representative microsatellite allele frequency distributions are shown in Figure 3 for three loci, each of which has exactly eight distinct alleles worldwide. The loci each have a pattern in which 3–6 of their alleles are reasonably common, and the rest are quite rare. These three loci illustrate a relatively small divergence in allele frequencies across geographic regions, a large divergence in allele frequencies across regions, and an intermediate level of divergence. For D6S474, the least diverged locus, nearly every allele has similar frequencies in all seven of the regions; for D10S1425, the locus with intermediate divergence, some but not all of the alleles have noticeable differences across regions; finally, for D12S2070, the most strongly diverged of the three loci, nearly every allele—most dramatically, the allele shown in purple—has a substantial frequency difference.
Figure 3.
Allele frequencies at three microsatellite loci. Each of the three loci has exactly eight alleles, which are displayed counterclockwise from the top of each pie chart in the following sequence of colors, proceeding in increasing order of allele size: orange, blue, yellow, purple, pink, red, green, brown. In most of the pie charts, one or more alleles is rare or absent.
Questions about human genetic variation
1. Are most alleles widely distributed, or are they largely confined to specific parts of the world?
For each of the alleles in the dataset, we can characterize its geographic distribution by identifying the locations in which it is present and those in which it is absent. Considering each of the seven regions, a given allele has one of 27-1=127 possible presence/absence distributions. If we disregard alleles that appear only once in the dataset—and that are therefore more likely than other alleles to result from genotyping errors—Table 2 gives the fractions of alleles in the dataset that have each of the possible geographic categorizations.
Table 2.
Percentages of alleles, among 8290 non-singleton alleles at 783 loci, that have given geographic distributions. Each row depicts a possible geographic distribution that an allele can possess, with an X indicating the presence of the allele in a geographic region. The percentage of all alleles with the given distribution is then indicated in the column at right. Only distributions possessed by more than 0.4% of the alleles are shown, in reverse lexical order. The remaining 94 distributions not shown together contain 10.12% of the alleles.
| Africa | Europe | Middle East | Central/ South Asia | East Asia | Oceania | America | Percent of all alleles |
|---|---|---|---|---|---|---|---|
| X | X | X | X | X | X | X | 46.60 |
| X | X | X | X | X | X | 4.98 | |
| X | X | X | X | X | X | 6.97 | |
| X | X | X | X | X | 5.09 | ||
| X | X | X | X | 1.41 | |||
| X | X | X | X | 0.52 | |||
| X | X | X | 0.55 | ||||
| X | X | X | X | 0.64 | |||
| X | X | X | 0.49 | ||||
| X | X | 0.70 | |||||
| X | X | X | X | X | 0.54 | ||
| X | X | X | X | X | 0.45 | ||
| X | X | X | X | 0.97 | |||
| X | X | X | 1.25 | ||||
| X | X | X | 0.65 | ||||
| X | X | 2.29 | |||||
| X | X | X | 0.68 | ||||
| X | X | 1.28 | |||||
| X | X | 1.34 | |||||
| X | 4.28 | ||||||
| X | X | X | X | X | X | 0.42 | |
| X | X | X | X | X | 0.63 | ||
| X | X | X | X | 0.90 | |||
| X | X | X | 0.68 | ||||
| X | X | 0.43 | |||||
| X | X | X | 0.42 | ||||
| X | X | 0.53 | |||||
| X | X | 0.48 | |||||
| X | X | 0.66 | |||||
| X | 0.64 | ||||||
| X | X | 0.78 | |||||
| X | 0.59 | ||||||
| X | 1.01 |
We can observe from Table 2 that by far the geographic distribution most prevalent among alleles, containing 46.60% of the alleles in the dataset, is presence in all seven regions. The three distributions with the next highest numbers of alleles are the categories for presence everywhere except Oceania (6.97%), everywhere except both Oceania and the Americas (5.09%), and everywhere except the Americas (4.98%). These distributions are then followed by presence only in Africa (4.28%), and presence only in Africa and the Middle East (2.29%).
Assembling the presence/absence categories with the largest numbers of alleles into a pie chart and grouping categories with small numbers of alleles, Figure 4A illustrates that most alleles tend to be found in at least two or three of the seven regions, and that relatively few alleles are private to individual regions (7.53%). Among private alleles, more than half (56.89%) are found only in Africa. This result is intensified in Figure 2C, which adjusts for sample size differences among the regions. In this figure, which considers the mean number of private alleles per locus if equal-sized subsamples are simultaneously drawn from each of the seven regions, it can be observed that on average, in a sample of size 420 containing 60 alleles from each region, a microsatellite locus has about 0.9 private alleles in Africa, and about 0.15–0.2 private alleles in each of the other regions—fewer in the Americas.
Figure 4.
Classifications of alleles by geographic distribution. The classifications are grouped by the number of regions in which alleles were found, and the distributions with the largest numbers of alleles are shown explicitly. (A) Classifications of alleles by presence or absence within regions. (B) Classifications of alleles by presence or absence within populations.
We can now answer the question posed in this section. Most alleles are widely distributed around the world, and about half of all alleles represented in the diversity panel are found in all seven geographic regions. Relatively few alleles are private to individual regions. Among the alleles that are private, more than half are found only in Africa.
2. Do there exist distinctive alleles for specific geographic regions that distinguish individuals in one group from those of other groups?
We have seen that the number of alleles that are private to individual regions is relatively small. We can now ask whether those alleles have high or low frequencies in the regions where they are found. If the frequencies of private alleles are high, these alleles could then be used as diagnostic types that could easily identify individuals as belonging to particular groups.
Considering all 624 private alleles observed more than once in the dataset, their mean estimated frequency in the region where they occur is 0.0165, with standard deviation 0.0212. Only six alleles private to a single region have frequencies greater than 0.10 in the region where they are found, and only one has frequency greater than 0.13. This allele, allele 275 at locus D9S1120, is present only in the Americas, with overall frequency 0.365. Its frequencies in the five Native American populations in the diversity panel are 0.192, 0.250, 0.300, 0.220, and 0.971 for indigenous Colombians, Karitiana, Maya, Pima, and Surui, respectively.
If we now consider all alleles in the dataset—not only the private alleles—and identify those that according to a statistic that measures ancestry information content (Rosenberg et al. 2003a) have the greatest potential to enable inferences about regional ancestry, we can see that none of these highly informative alleles has a frequency close to one in some groups but close to zero in all others (Table 3). Thus, none of the alleles is diagnostic for a particular region or group of regions. The most diverged allele has an allelic informativeness of 0.169, noticeably smaller than both 0.363, the value that would be obtained for an allele with frequency one in three of the groups and zero in the other four, and 0.278, the informativeness for an allele with frequency one in one of the seven groups and zero in all others. To visually observe the frequency differences for a locus that has an allele with strong divergence across geographic regions, note that allele 95 of locus D12S2070, shown in purple in Figure 3, has the third-highest regional ancestry information content of all 9346 alleles in the dataset.
Table 3.
The 10 alleles most informative about regional ancestry, among 9346 alleles at 783 microsatellite loci. Ancestry information content for alleles was calculated according to the In measure of Rosenberg et al. (2003a, eq. 5). Loosely speaking, according to this measure, an allele is most informative about regional ancestry if the knowledge that an individual has the allele enables accurate inferences to be made about the source population of the individual, and if the allele is sufficiently common that it enables ancestry inference for a substantial fraction of all individuals.
| Locus | Alternate name of locus | Allele size | Allelic informativeness | Allele frequency | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Africa | Europe | Middle East | Central/ South Asia | East Asia | Oceania | America | ||||
| GTTTT002P | 140 | 0.169 | 0.033 | 0 | 0.006 | 0.002 | 0.002 | 0.667 | 0 | |
| D6S1006 | ATC4D09 | 194 | 0.136 | 0.631 | 0.364 | 0.555 | 0.239 | 0.036 | 0 | 0.014 |
| D12S2070 | ATA25F09 | 95 | 0.127 | 0.187 | 0.078 | 0.072 | 0.126 | 0.403 | 0.014 | 0.852 |
| D2S2986 | 2QTEL47 | 158 | 0.124 | 0.074 | 0.029 | 0.016 | 0.071 | 0.365 | 0.569 | 0.598 |
| ATAC026P | 198 | 0.124 | 0.541 | 0.006 | 0.071 | 0.027 | 0 | 0 | 0.005 | |
| AAT258 | 145 | 0.120 | 0.070 | 0.007 | 0.046 | 0.042 | 0.021 | 0 | 0.604 | |
| GATA65E01 | 121 | 0.119 | 0.488 | 0 | 0.043 | 0.015 | 0.002 | 0 | 0 | |
| D2S441 | GATA8F03 | 135 | 0.118 | 0.045 | 0.161 | 0.106 | 0.223 | 0.213 | 0.792 | 0.755 |
| D7S1808 | GGAA3F06 | 252 | 0.115 | 0.008 | 0.019 | 0 | 0.050 | 0.305 | 0.030 | 0.519 |
| TTTA028 | 187 | 0.114 | 0.146 | 0.487 | 0.517 | 0.242 | 0.021 | 0 | 0.019 | |
The combination of these results shows that among the alleles considered, there do not exist distinctive alleles present in all members of one region but absent from individuals outside the region. While occasional alleles with large frequency differences do exist, they are unusual, and they do not typically approach the maximal possible level of divergence. As a fraction of all alleles, strongly diverged alleles are rare.
3. Of the genetic variants that exist in the human genome, how many are present in a given geographic region?
Using the values in Table 2, together with the remaining geographic distributions not shown in the table, we can calculate the fractions of alleles found in each of the geographic regions in the dataset. Considering all 8290 alleles observed more than once, 87.12% are found in Africa, 77.17% in Europe, 81.09% in the Middle East, 81.66% in Central/South Asia, 79.82% in East Asia, 57.44% in Oceania, and 60.11% in the Americas. Averaging across regions, a random region contains 74.91% of the non-singleton alleles found in the full worldwide dataset.
The quantities in Figure 2A enable us to make adjustments for the unequal sample sizes in the different geographic regions. For each region and various values of the subsample size g, Figure 2B plots the mean fraction of alleles in a randomly chosen worldwide subsample containing g alleles from each of the seven regions that are found in a random region-specific subsample of size g. Over most of the values of g considered, about 75–81% of worldwide alleles appear in Africa, 63–71% in Europe, the Middle East, Central/South Asia, or East Asia, 59–63% in Oceania, and 53–57% in the Americas. Thus, each region contains a majority of all alleles found worldwide, with the greatest fraction being observed in Africa and the smallest fraction occurring in the Americas.
4. On average, how different are two individuals from the same local population, in comparison with two individuals chosen from any two populations anywhere in the world?
On the basis of the initial analysis of protein polymorphisms performed by Lewontin (1972) and subsequent computations with other types of markers, it has often been noted that “genetic variation within populations constitutes X% of human genetic variation, and genetic variation among populations constitutes (100-X)%.” The values of X vary by study, but they generally lie in the range of 80–95% (e.g. Barbujani and Di Benedetto 2001; Brown and Armelagos 2001; Ruvolo and Seielstad 2001; Excoffier and Hamilton 2003; Long and Kittles 2003; Rosenberg et al. 2003b; Li et al. 2008).
This description of the partition of human variation suggests that the number 80–95% is the answer to a question similar to #3. However, as we have seen in the section on question #3, the fraction of alleles present in a randomly chosen geographic region is generally smaller than 80–95%, and the fraction in a randomly chosen population must be still smaller. In the literature on human genetic variation, statements about the fraction of variation within and among populations have almost always referred to the answers to questions similar to #4.
In one of the most common ways that the partitioning of human genetic variation has been conceptualized—which differs slightly from the entropy-based approach of Lewontin (1972)—populations are first classified by geographic region. A variable indicating the presence or absence of an allele in a population is expressed in an analysis-of-variance framework as the sum of terms for the mean frequency of the allele worldwide, the mean frequency of the allele in the region to which the population belongs, the mean frequency of the allele in the population, and an “error” term, which reflects within-population variation. For each distinct allele in a dataset, this linear equation is constructed for each presence/absence observation of the allele in each individual, and using analysis-of-variance techniques, the components of genetic variation are then estimated. These components correspond to the estimated fractions of the variation in the allelic indicator variable owing to variation across regions, variation across populations within regions, and “error,” or within-population variation. Estimates of these components based on the individual alleles are then combined across alleles and loci to produce an overall estimate of the genetic variance components. Some studies omit the region term in the linear model, estimating only the among-population and within-population components of genetic variation. The variance components estimation is based only on the ~0.1% of the human genome that consists of variable markers, as monomorphic markers have no variation across individuals that can be partitioned.
For our microsatellite data, we can estimate the components of genetic variation for different designs using the analysis-of-variance approach (Table 4), obtaining a similar result to previous studies, namely that in a design with two variance components, the within-population component constitutes more than 90% of human genetic variation. When we divide the populations into seven geographical regions and estimate three variance components, the within-population component is 93.9%, the among-population-within-region component is 2.4%, and the among-region component is 3.8%.
Table 4.
The partition of genetic variation. Eurasia, which denotes the combination of Europe, the Middle East, and Central/South Asia, is treated as a single region in the five-region worldwide design, but it is subdivided in the seven-region design. Variance components were estimated according to the method of Weir (1996, pp. 169–174, 184–186), assuming Hardy-Weinberg equilibrium within populations. Confidence intervals are based on 1000 bootstraps across loci.
| Sample | Number of regions | Number of populations | Variance components and 95% confidence intervals (%) | ||
|---|---|---|---|---|---|
| Within populations | Among populations within regions | Among regions | |||
| World | 1 | 53 | 94.4 (94.1, 94.6) | 5.6 (5.4, 5.9) | |
| World | 5 | 53 | 93.0 (92.7, 93.3) | 2.5 (2.4, 2.6) | 4.5 (4.3, 4.8) |
| World | 7 | 53 | 93.9 (93.6, 94.1) | 2.4 (2.3, 2.5) | 3.8 (3.5, 4.0) |
| Africa | 1 | 7 | 96.9 (96.8, 97.1) | 3.1 (2.9, 3.2) | |
| Eurasia | 1 | 21 | 98.4 (98.3, 98.5) | 1.6 (1.5, 1.7) | |
| Eurasia | 3 | 21 | 98.3 (98.1, 98.4) | 1.2 (1.1, 1.3) | 0.6 (0.5, 0.7) |
| Europe | 1 | 8 | 99.2 (99.1, 99.3) | 0.8 (0.7, 0.9) | |
| Middle East | 1 | 4 | 98.6 (98.5, 98.8) | 1.4 (1.2, 1.5) | |
| Central/ South Asia | 1 | 9 | 98.6 (98.5, 98.8) | 1.4 (1.2, 1.5) | |
| East Asia | 1 | 18 | 98.8 (98.6, 98.9) | 1.2 (1.1, 1.4) | |
| Oceania | 1 | 2 | 93.6 (93.0, 94.3) | 6.4 (5.7, 7.0) | |
| America | 1 | 5 | 88.3 (87.8, 88.7) | 11.7 (11.3, 12.2) | |
Conveniently, the estimated variance components of the allelic indicator variables, whose meaning can be difficult to interpret, are closely related to concepts that are more easily understood. First, for a variance partitioning with only two components, among-populations and within-populations, the among-population component can be viewed as an estimator of the commonly used statistic Fst, which measures the level of variation at polymorphic markers among a set of populations (the quantity θ̂ in Weir 1996, p. 169–174), and the within-population component can be seen as an estimator of 1-Fst. For a given locus, the Fst statistic can be formulated as Fst=(πt-πs)/πt, where πs is the mean probability for the locus that two alleles chosen from the same population are distinct, and πt is the mean probability that two alleles chosen from any two populations are distinct (Nei 1987, p. 162). For variance partitioning with three components, the among-region, among-population-within-region, and within-population components correspond to (πt-πr)/πt, (πr-πs)/πt, and πs/πt, respectively, where πs and πt have the same meaning as in the two-component design and πr is the mean probability of non-identity for two alleles chosen from the same region. The within-population component can be viewed as the level of genetic difference for a pair of individuals from the same population, in comparison with the level of difference between two individuals from any two populations. The among-population-within-region component then equals the excess level of difference for a pair of individuals from the same region but from different populations, and the among-region component is the excess level of difference for a pair of individuals from different regions.
The connection between variance components and probabilities of identity suggests an approach to visualizing genetic variance components in human populations. Figure 5A depicts the genome-wide distributions of pairwise differences for pairs of individuals from the same population, from the same region, and from any two arbitrarily chosen populations. In the distribution of pairwise differences for arbitrarily selected pairs, most pairs with a proportion of differing alleles above the small jump near 0.7 involve at least one individual from Africa. This result is reflected in Figure 5B, in which it can be seen that among the geographic regions, only Africa has more than a negligible probability density at values of the proportion of differing alleles above 0.7. Consistent with the general sequence of levels of diversity seen in Figure 2, both in Figure 5B and in Figure 5C, Africa has the highest mean proportion of differing alleles, followed by the Middle East, Central/South Asia, Europe, East Asia, Oceania, and the Americas (Table 5). Note that the bimodal distribution for Oceania in Figure 5B reflects the sampling of only two populations in Oceania, so that the peak to the left involves within-population pairs, and the peak to the right involves between-population pairs. Also, both the wide range in Figure 5C of the proportion of differing alleles for pairs within Native American populations and the small peaks in Oceania and Africa between 0.2 and 0.5 are consequences of the inclusion of pairs of relatives in the diversity panel, particularly in Native Americans (Rosenberg 2006).
Figure 5.
Distributions of pairwise genetic differences across 783 microsatellites. (A) Pairwise differences for pairs of individuals from the same population, pairs from the same geographic region, and pairs arbitrarily chosen from any two populations. (B) Pairwise differences for pairs of individuals from the same geographic region, separated by region. (C) Pairwise differences for pairs of individuals from the same population, with populations from the same region grouped together. The pairwise difference for a given pair of individuals was computed as one minus their proportion of shared alleles (Mountain and Cavalli-Sforza 1997). For a given pair of individuals, loci for which one or both individuals has missing data were omitted from consideration. Probability densities were estimated from pairwise genetic differences as in Venables and Ripley (1997, p. 181, rectangular kernel with parameter b).
Table 5.
The mean and standard deviation of the proportion of alleles differing between two individuals from the same geographic region, and for each region, the mean and standard deviation of the proportion of alleles differing between two individuals from the same population within the region.
| Region | Mean proportion of alleles differing between pairs of individuals | |
|---|---|---|
| Same region | Same population within a region | |
| Africa | 0.672 ± 0.025 | 0.646 ± 0.034 |
| Europe | 0.619 ± 0.013 | 0.612 ± 0.016 |
| Middle East | 0.633 ± 0.018 | 0.623 ± 0.023 |
| Central/South Asia | 0.631 ± 0.016 | 0.620 ± 0.024 |
| East Asia | 0.600 ± 0.013 | 0.590 ± 0.018 |
| Oceania | 0.587 ± 0.046 | 0.556 ± 0.050 |
| America | 0.550 ± 0.055 | 0.474 ± 0.083 |
As can be observed from Figure 5A, the mean proportion of differing alleles for arbitrary pairs of individuals, or 0.651, only slightly exceeds the mean difference for pairs from the same region, or 0.618. In turn, the mean pairwise difference within regions only slightly exceeds the mean difference for pairs from the same population, or 0.603. The fraction of the genetic differences for a random pair of individuals from anywhere in the world found in a random pair from the same population—a quantity that corresponds to the within-population component of genetic variation—equals 0.603/0.651≈0.927. The excess difference for two individuals from the same region in comparison with two individuals from the same population—which parallels the among-population-within-region component—equals (0.618–0.603)/0.651≈0.023. Finally, the excess difference for two individuals chosen from any two populations in comparison with two individuals from the same region is (0.651–0.618)/0.651≈0.050.
The variance components estimated for the division of the dataset into seven regions and 53 populations differ slightly between the analysis on the basis of pairwise differences, which obtained (0.927, 0.023, 0.050) for the three components, and that on the basis of the analysis of variance, which estimated them at (0.939, 0.024, 0.038). The differences between these estimates arise largely from differences in the nature of the estimators: the estimates in Table 4 rely on estimators that consider the different sample sizes in different populations, whereas the calculations employing Figure 5 use the graphs exactly as they appear in the figure, without sample size weights. In summary, however, the rough agreement of analysis-of-variance and pairwise-difference methods supports the general observation that the mean level of difference for two individuals from the same population is almost as great as the mean level of difference for two individuals chosen from any two populations anywhere in the world.
5. To what extent is it possible to determine the genetic ancestry of an individual using the alleles in his or her genome?
The answers to questions #1–#4 produce a view of human genetic variation in which the level of similarity among populations is relatively high, and the level of difference is low. Most alleles are widely distributed, the fraction of alleles private to individual regions is small, most populations contain most of the alleles present in the human population, and the mean genetic difference for two individuals from the same population is almost as large as that for two individuals chosen from any two populations. We will see, however, that in the accumulation of small amounts of allele frequency variation across many loci, it is possible to make inferences about individual genetic ancestry from genetic markers.
Consider one of the loci in Figure 3. If the region of origin of an individual were known, it would not be possible to predict the genotype of the individual with much accuracy. Too much variation exists within each region to enable accurate predictions: the number of alleles is too high, and the frequency of the most frequent allele is too low.
The reverse question, however, namely that of inferring the source region of an individual given his or her genotypes, begins to be tractable as the number of loci increases. Suppose an individual is known to have been sampled from one of the seven regions in Figure 3. If the genotype of the individual were known at the first locus, D6S474, little information would be obtained about the origin of the individual. For example, suppose the individual is a yellow-yellow homozygote. This genotype is reasonably common in all of the geographic regions, so that any of them could potentially be the source of the individual. If the individual is also a blue-yellow heterozygote at the second locus, D10S1425, it becomes more likely that the individual is East Asian or Native American, as the blue-yellow genotype is most common in East Asia and in the Americas. Finally, if for D12S2070 the individual is a purple-purple homozygote, it is now much more likely that the individual is Native American than East Asian. Although the combination of yellow-yellow at D6S474, blue-yellow at D10S1425, and purple-purple at D12S2070 may very well have nonzero frequency in most regions, an individual with this combination of genotypes is by far most likely to be Native American.
This example has been based only on three loci. Imagine aligning similar pie charts for 783 loci in the same manner as in Figure 3. If an individual genotype were known for all 783 loci, as long as a reasonable amount of variation in frequencies exists across regions, it would probably not be difficult to look through the 783 sets of pie charts to determine which region is the most likely source for the individual. It is also likely that a fair amount of confidence could be placed in this estimate, regardless of which multilocus genotype an individual possessed.
This type of inferential procedure is what we have performed using the clustering algorithm Structure (Pritchard et al. 2000; Falush et al. 2003), with two main differences. First, in the description above, the source regions were known in advance, so that the problem was to classify individuals on the basis of known allele frequencies. The Structure approach, however, uses an unsupervised clustering algorithm, so that the clusters to which individuals are assigned are inferred simultaneously with assignment of individual membership. Second, above it was assumed that each individual originated from a single one of the regional groups. With Structure, however, individuals can have partial membership in multiple clusters. Thus, the genome of an individual is represented as a vector of membership coefficients, with membership coefficients summing to one across clusters. The number of clusters, represented by the value of a parameter K, is selected in advance, but can be varied across independent runs of the algorithm.
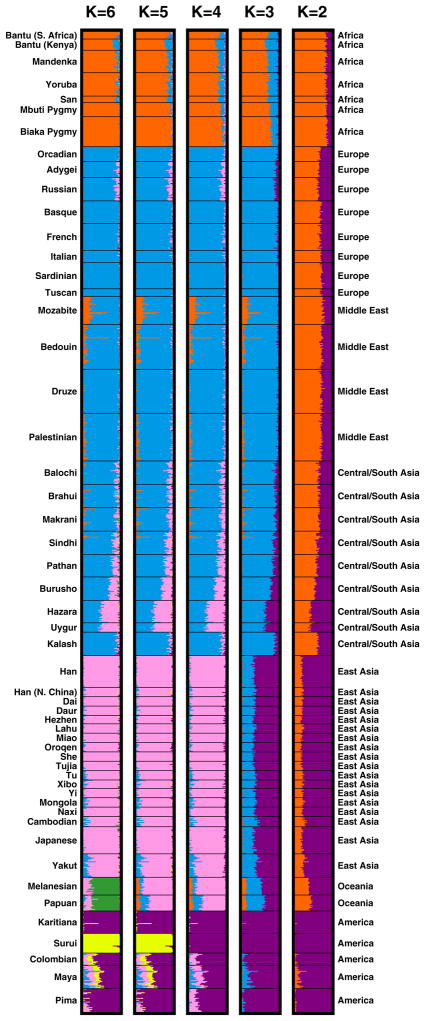
When we apply this unsupervised mixed-membership clustering approach to individual multilocus genotypes, we find that individuals from the same populations have similar membership coefficients in the inferred clusters (Figure 6). If two clusters are used, the individuals from Africa have nearly full membership in one cluster, shown in orange, and the Native Americans have nearly full membership in the other cluster, shown in purple. Moving east across Asia, the membership coefficients of individuals decrease in their similarity to those of the Africans, and they increase in similarity to those of the Native Americans.
Figure 6.
Inferred population structure for various numbers of clusters K. Each individual is represented by a thin line partitioned into K colored segments that represent the fractions of the individual’s genome estimated to belong to the K clusters. Each plot, made with Distruct (Rosenberg 2004), utilizes the highest-likelihood run among ten runs of Structure with the F model (Falush et al. 2003). Estimates were obtained from 10,000 iterations following a burn-in period of 20,000 iterations. For K=2 and K=4, all ten runs produced the same set of clusters. For K=3, three of the ten runs separated a cluster corresponding largely to East Asia and Oceania rather than one corresponding largely to Europe, the Middle East, and Central/South Asia. For K=5, the other nine runs separated a cluster corresponding to Oceania rather than one corresponding to Surui. For K=6, only one of the remaining nine runs produced the cluster corresponding to Surui. Seven of these nine runs instead separated a cluster in which many individuals from Central/South Asia—especially those from the Kalash population—had partial membership; in the ninth run, the sixth cluster partially separated the African hunter-gatherer populations (Biaka, Mbuti and San) from the other African groups.
When three clusters are used, the third cluster subdivides the orange cluster into one cluster that corresponds largely to Africans, and one that corresponds largely to individuals from Europe, the Middle East, and Central/South Asia, shown in blue. One population of note in the analysis with K=3 is the Mozabite population from northern Africa, whose individuals have mixed membership in the cluster that contains Africans and the cluster containing the populations from Europe, the Middle East, and Central/South Asia.
With K=4, a cluster corresponding to East Asia, shown in pink, separates from the purple cluster. Decreasing membership in this cluster is visible moving westward across Asia, in that populations such as Burusho, Hazara, and Uygur are estimated to have mixed membership both in the blue and pink clusters. With five clusters, the highest-likelihood replicate of the analysis separates a single Native American population, Surui from Brazil, into a distinct cluster. This result differs from our previous analyses with K=5 (Rosenberg et al. 2002, 2005), which identified a cluster corresponding to the two populations from Oceania, one from Papua New Guinea and the other from the Solomon Islands. However, only one of ten replicates here identified a Surui cluster, and the remaining nine all obtained the cluster corresponding to Oceania. With K=6, the Oceania cluster was identified in all replicates, the highest-likelihood of which also obtained the cluster corresponding to Surui. This observation is also slightly different from our previous analyses with overlapping but not identical sets of markers in the same individuals, in which the sixth cluster corresponded to the Kalash population from Pakistan (Rosenberg et al. 2002), or to a subdivision of Native Americans into more northerly and southerly populations (Rosenberg et al. 2005).
From these results, we can observe that despite the genetic similarity among populations suggested by the answers to questions #1–#4, the accumulation of information across a large number of genetic markers can be used to subdivide individuals into clusters that correspond largely to geographic regions. The apparent discrepancy between the similarity of populations in questions #1–#4 and the clustering in this section is partly a consequence of the multivariate nature of clustering and classification methods, which combine information from multiple loci for the purpose of inference, in contrast to the univariate approaches in questions #1–#4, which merely take averages across loci (Edwards 2003). Even though individual loci provide relatively little information, with multilocus genotypes, ancestry is possible to estimate at the broad regional level, and in many cases, it is also possible to estimate at the population level as well.
6. What events in human evolutionary history are responsible for the basic patterns of genetic similarity and difference evident in worldwide human populations?
The discussion of the first five questions has focused on patterns of variation observed in human populations today. This section turns to explaining these patterns using inferences that can be made about the genetic history of the human population. Suppose that the human population descends from a small ancestral group confined to a small area. Suppose also that the expansion of populations occurred by a sampling process, in which population subgroups repeatedly split off from their ancestral groups and moved short distances away. Repetition of this process of subsampling and expansion would eventually have led to habitation of a large area.
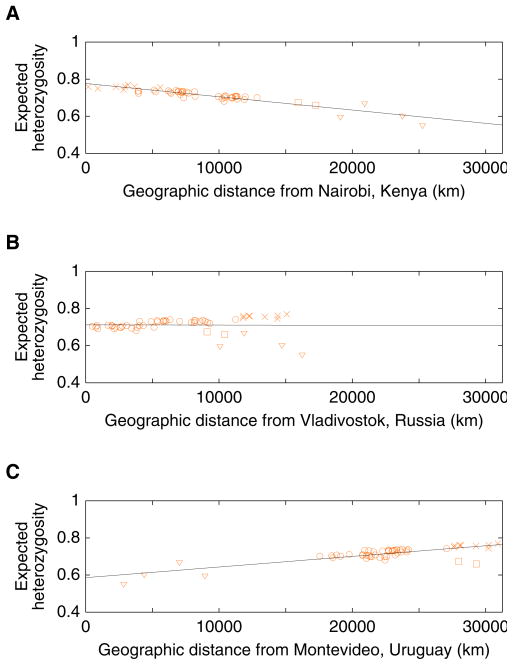
Our simulations of this serial sampling process suggest that it would produce a linear decline in levels of genetic variation, as measured by heterozygosity, with increasing geographic distance from the site of origin (Ramachandran et al. 2005; DeGiorgio et al. 2009). Considering three different locations as examples—one in Africa, one in East Asia, and one in South America—we can see in Figure 7 that a linear decline of heterozygosity occurs with distance from the location in Africa, but not with distance from each of the other points: the point in East Asia does not produce a straight line, and while the point in South America does produce a close match to a straight line, the slope of this line is positive rather than negative. These observations can potentially be explained by a serial sampling model starting from an African origin, in which South America is among the last places to have been reached during the human expansion.
Figure 7.
Heterozygosity of populations as a function of geographic distance from particular points: (A) Nairobi, Kenya (1.26666667oS 36.8°E); (B) Vladivostok, Russia (43.13333333°N 131.91666667°E); (C) Montevideo, Uruguay (34.88333333°S 56.18333333°W). For each locus and population, expected heterozygosity was computed as , where n denotes the number of alleles in the sample and pi denotes the sample frequency of distinct allele i in the population. The mean heterozygosity across loci was then computed. Geographic coordinates for populations were obtained as in Figure 1, and great-circle geographic distances between populations were computed as in Rosenberg et al. (2005), forcing paths between pairs of points to travel through the five waypoints described in Ramachandran et al. (2005). Paths to the Americas all passed through 64°N 177°E and 54°N 130°W, paths to Oceania through 11°N 104°E, and paths to Africa through 30°N 31°E; paths from Europe to Africa, the Middle East, or Oceania also passed through 41°N 28°E. As in Ramachandran et al. (2005), the Bantu samples from southern Africa were split into Southwestern Bantu (Herero, Ovambo) and Southeastern Bantu (Pedi, Sotho, Tswana, Zulu) groups, and the Surui were omitted (for the Southwestern and Southeastern Bantus, the coordinates used were the mean of the longitudes and the inverse sine of the mean of the sines of the reported locations of included individuals). The four Native American populations are marked with triangles, the two populations from Oceania with squares, and the eight sub-Saharan African populations with crosses. The remaining populations, from Europe, Asia, and northern Africa, are marked with circles. Denoting geographic distance in thousands of kilometers by D, the regression lines are H=0.770–0.00716D, H=0.712+(9.97×10−5)D, and H=0.586+0.00574D, for (A), (B), and (C), with coefficients of determination (R2) equal to 0.865, 1.16×10−4, and 0.662, respectively.
Figure 8 shows a plot of a measure of the linear fit between heterozygosity and geographic distance from a point, for points selected from around the world (excluding the Americas). The putative points of origin with the closest match to a pattern of linear decrease in heterozygosity with distance from the point all lie within Africa. Further, each point in Africa produces a better fit of the model than does any point outside of Africa, so that if the serial sampling model is sensible, the human population likely originated with a group in Africa.
Figure 8.
The fit of a linear decline of heterozygosity with increasing distance from putative geographic origins. The color of a point indicates a correlation coefficient r between expected heterozygosity and geographic distance from the point. Geographic coordinates and distances between points were obtained as in Figure 7. Excluding points in Iceland and Greenland, all points in the region shown were considered as possible origins, provided that they were both on land and on a lattice of latitudes and longitudes described by Ramachandran et al. (2005). Among the points shown, the smallest value of r (−0.932) is observed at 1.43°N 20°E.
This view of human migrations is also supported by computations of the directional “flow” of alleles for pairs of regions. For each ordered pair of geographic regions, Figure 9 shows the fraction of alleles found in the first region that are also observed in the second. Assigning each region a number of migrational steps from a putative human origin in Africa (Africa=0, Middle East=1, Europe=Central/South Asia=2, East Asia=3, Oceania=America=4), for all pairs of regions at different numbers of steps from Africa, the flow of alleles is always greater moving outward from Africa than that moving back towards Africa. In other words, the pattern of allelic presence and absence matches a history in which the gene pool of each migrating human population consisted largely of a sampling of the alleles present in its ancestral population.
Figure 9.
Schematic world map of the “flow” of microsatellite alleles. Colored boxes represent regions of the world, positioned geographically. Links entering into a geographic region indicate the percentages of distinct alleles from the geographic region found in other regions (and an edge with the number x is drawn proportionately in width to x/4–8). For example, averaging across loci, 87% of alleles observed in Europe are also observed in Africa, whereas 74% of alleles observed in Africa are also observed in Europe. More precisely, following Conrad et al. (2006), for a given locus, a sample size g, and a pair of regions A and B, the expected number α of distinct alleles that will be found in a sample of size g from region A is computed as in Figure 2A. The expected number π of distinct alleles that will be found in a sample of size g from region A but not in a sample of size g from region B is computed as in Figure 2C, as the number of alleles private to region A, averaging over all possible subsamples that contain g alleles from region A and g alleles from region B. The fraction of the alleles in region A that are found in region B is then obtained as 1-π/α. The value g=40 was used for all computations, and for a given pair of regions, only those loci with sample sizes of at least g in both regions were considered.
The serial sampling model can explain other properties of the data discussed above. We observed earlier that both the mean number of alleles found at a locus and the mean number of private alleles are greatest for African populations and smallest for populations in the Americas and Oceania, even after adjusting for sample size differences. We also saw that African groups possess a greater proportion of the alleles found in the full human population than do non-African groups, and that groups from the Americas and Oceania possess the smallest proportion of alleles. These observations are all expected if Africa was the original source of human populations, and if the populations of Oceania and the Americas trace their ancestry primarily to more recent waves of migrating human populations.
We also found that in an unsupervised cluster analysis, individuals grouped into geographical clusters largely corresponding to sub-Saharan Africa, Europe and the part of Asia west of the Himalayas, the part of Asia east of the Himalayas, Oceania, and the Americas. These observations are compatible with serial sampling, assuming that major geographic barriers such as oceans, the Sahara desert, and the Himalayas were not frequently crossed during human migrations. This reduced frequency for the traversal of major barriers would then increase the genetic similarity for individuals on the same side of a barrier relative to that of individuals on opposite sides of the barrier, with the following consequence: a discontinuity in genetic distance as a function of geographic distance would be produced for most pairs of populations on opposite side of a major barrier, in comparison with the genetic distance for pairs on the same side. This discontinuity, which is in fact observed in the diversity panel (Figure 10), would then explain the ability of clustering algorithms to identify clusters of individuals corresponding to the geographic regions bounded by the barriers that are most important. Thus, the clusters we have observed are consistent with serial sampling together with reduced permeability for major geographic barriers.
Figure 10.
Genetic and geographic distance for 630 pairs of populations in Europe, Asia, and northern Africa. All pairs involving populations sampled in these regions are plotted, except for those that involve Hazara, Kalash, or Uygur. The population pairs presented are a subset of those shown in Figure 6 of Rosenberg et al. (2005). Blue triangles indicate 324 pairs of populations on opposite sites of the Himalayas. Points marked by a red E or W indicate pairs with both populations on the east or west side of the Himalayas, respectively (153 population pairs each). The regression line based on all 630 points is Fst=0.00537+0.0023D+0.0219B, where D denotes geographic distance in thousands of kilometers (as computed for Figure 7) and B=0 for population pairs on the same side and B=1 for pairs on opposite sides. The blue and red lines illustrate the regression equation, setting B=1 and B=0, respectively. R2 equals 0.882 for the bivariate regression of Fst on B and D, and 0.659 for a univariate regression on D only. Fst genetic distance was calculated using eq. 5.3 of Weir (1996).
Discussion
Our analysis of human microsatellites supports the following main results. (1) Most genetic variants are widely distributed, with an excess present in Africa. (2) Genetic variants that distinguish individuals in one region from individuals in other regions are rare. (3) Each geographic region contains most genetic variants, with Africa possessing the largest fraction. (4) Pairs of individuals from different geographic regions tend to be only slightly more genetically different than pairs of individuals from the same region. (5) Despite the high levels of similarity across populations, the accumulation of small differences across large numbers of markers enables inference of geographic ancestry. (6) The pattern of human genetic similarities and differences can be explained as the outcome of a human expansion out of Africa via a process in which new migrating populations each carried only subsets of the variation from their parental populations, and in which major geographic barriers have historically had reduced permeability to human migration.
The design of this article, in which a single dataset has been used to answer a series of questions about human genetic similarities and differences, has supplied one viewpoint on key results in a vast collection of studies that cover many marker systems, samples, datasets, and methodological tools; the dataset has offered an approach focused in indigenous populations on highly variable markers that generally lie outside of genes and that therefore more directly reflect the history of human migrations than do loci at which natural selection has had a strong influence. While it is hoped that this article provides a point of entry into the study of genetic similarities and differences among human populations, the reader is also directed to more comprehensive reviews (e.g. Mountain 1998; Harpending and Rogers 2000; Jorde et al. 2001; Relethford 2001; Cavalli-Sforza and Feldman 2003; Tishkoff and Verrelli 2003; Jobling et al. 2004; Garrigan and Hammer 2006; Lawson Handley et al. 2007; Weaver and Roseman 2008; Barbujani and Colonna 2010; Novembre and Ramachandran 2011) for additional perspectives on the patterns of worldwide human genetic variation and their history.
Acknowledgments
I am grateful to all of my collaborators who have contributed to the work summarized in this article. M. Jakobsson, S. Mahajan, and S. Ramachandran provided assistance with the preparation of Figures 9, 6, and 8, respectively. Support has been provided by NIH grant R01 GM081441 and by a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
Literature Cited
- Barbujani G, Colonna V. Human genome diversity: frequently asked questions. Trends Genet. 2010;26:285–295. doi: 10.1016/j.tig.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Barbujani G, Di Benedetto G. Genetic variances within and between human groups. In: Donnelly P, Foley R, editors. Genes, Fossils and Behaviour. IOS Press; Cambridge: 2001. pp. 63–77. [Google Scholar]
- Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- Brown RA, Armelagos GJ. Apportionment of racial diversity: a review. Evol Anthropol. 2001;10:34–40. [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Pioffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL. The Human Genome Diversity Project: past, present and future. Nature Rev Genet. 2005;6:333–340. doi: 10.1038/nrg1596. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Feldman MW. The application of molecular genetic approaches to the study of human evolution. Nature Genet. 2003;33:S266–S275. doi: 10.1038/ng1113. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nature Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- DeGiorgio M, Jakobsson M, Rosenberg NA. Explaining worldwide patterns of human genetic variation using a coalescent-based serial founder model of migration outward from Africa. Proc Natl Acad Sci USA. 2009;106:16057–16062. doi: 10.1073/pnas.0903341106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AWF. Human genetic diversity: Lewontin’s fallacy. BioEssays. 2003;25:798–801. doi: 10.1002/bies.10315. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Hamilton G. Comment on “Genetic structure of human populations”. Science. 2003;300:1877. doi: 10.1126/science.1083411. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, Hammer MF. Reconstructing human origins in the genomic era. Nature Rev Genet. 2006;7:669–680. doi: 10.1038/nrg1941. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Schlötterer C, editors. Microsatellites: Evolution and Applications. Oxford University Press; Oxford: 1999. [Google Scholar]
- Harpending H, Rogers A. Genetic perspectives on human origins and differentiation. Annu Rev Genomics Hum Genet. 2000;1:361–385. doi: 10.1146/annurev.genom.1.1.361. [DOI] [PubMed] [Google Scholar]
- Hurlbert SH. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- Jobling MA, Hurles ME, Tyler-Smith C. Human Evolutionary Genetics: Origins, Peoples & Disease. Garland Science; New York: 2004. [Google Scholar]
- Jorde LB, Watkins WS, Bamshad MJ. Population genomics: a bridge from evolutionary history to genetic medicine. Hum Mol Genet. 2001;10:2199–2207. doi: 10.1093/hmg/10.20.2199. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST. Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv Genet. 2004;5:539–543. [Google Scholar]
- Lawson Handley LJ, Manica A, Goudet J, Balloux F. Going the distance: human population genetics in a clinal world. Trends Genet. 2007;23:432–439. doi: 10.1016/j.tig.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The apportionment of human diversity. Evol Biol. 1972;6:381–398. [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Long JC, Kittles RA. Human genetic diversity and the nonexistence of biological races. Hum Biol. 2003;75:449–471. doi: 10.1353/hub.2003.0058. [DOI] [PubMed] [Google Scholar]
- Mountain JL. Molecular evolution and modern human origins. Evol Anthropol. 1998;7:21–37. [Google Scholar]
- Mountain JL, Cavalli-Sforza LL. Multilocus genotypes, a tree of individuals, and human evolutionary history. Am J Hum Genet. 1997;61:705–718. doi: 10.1086/515510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Novembre J, Ramachandran S. Perspectives on human population structure at the cusp of the sequencing era. Annu Rev Genomics Hum Genet. 2011;12:245–274. doi: 10.1146/annurev-genom-090810-183123. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Rosenberg NA, Zhivotovsky LA, Feldman MW. Robustness of the inference of human population structure: A comparison of X-chromosomal and autosomal microsatellites. Hum Genomics. 2004;1:87–97. doi: 10.1186/1479-7364-1-2-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relethford JH. Genetics and the Search for Modern Human Origins. Wiley-Liss; New York: 2001. [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003a;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:660–671. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Response to comment on “Genetic structure of human populations”. Science. 2003b;300:1877. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Ruvolo M, Seielstad M. The apportionment of human diversity: 25 years later. In: Singh RS, Krimbas CB, Paul DB, Beatty J, editors. Thinking about Evolution: Historical, Philosophical, and Political Perspectives. Cambridge University Press; Cambridge: 2001. pp. 141–151. [Google Scholar]
- Schroeder KB, Schurr TG, Long JC, Rosenberg NA, Crawford MH, Tarskaia LA, Osipova LP, Zhadanov SI, Smith DG. A private allele ubiquitous in the Americas. Biol Lett. 2007;3:218–223. doi: 10.1098/rsbl.2006.0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech ZA, Jakobsson M, Rosenberg NA. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics. 2008;24:2498–2504. doi: 10.1093/bioinformatics/btn478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. Modern Applied Statistics with S-PLUS. 2 Springer-Verlag; New York: 1997. [Google Scholar]
- Weaver TD, Roseman CC. New developments in the genetic evidence for modern human origins. Evol Anthropol. 2008;17:69–80. [Google Scholar]
- Weir BS. Genetic Data Analysis II. Sinauer; Sunderland, MA: 1996. [Google Scholar]
- Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet. 2003;72:1171–1186. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]