ABSTRACT
Phage-encoded Shiga toxin (Stx) acts as a bacterial defense against the eukaryotic predator Tetrahymena thermophila. It is unknown how Stx enters Tetrahymena protozoa or how it kills them. Tetrahymena protozoa are phagocytotic; hence, Stx could gain entry to the cytoplasm through the oral apparatus or via endocytosis. We find that Stx2 can kill T. thermophila protozoa that lack an oral apparatus, indicating that Stx2 can enter these cells via endocytosis. As opposed to the lack of effect on mammalian phagocytes, Stx2 produced by bacteria encapsulated within phagocytotic vesicles is also capable of killing Tetrahymena. Addition of an excess of the carbohydrate binding subunits of Stx2 (StxB) and/or ricin (ricin B) blocks Stx2 cytotoxicity. Thus, regardless of whether Stx2 enters the cytoplasm by endocytosis or from the phagocytotic vesicle, this transport is mediated by a putative glycoconjugate receptor. Bacteriophage-mediated lysis of Stx-encoding bacteria is necessary for Stx toxicity in Tetrahymena; i.e., toxin released as a consequence of digestion of bacteria by Tetrahymena is harmless to the cell. This finding provides a rationale as to why the genes encoding Stx are found almost exclusively on bacteriophages; Stx must be released from the bacteria prior to the digestion of the cell, or it will not be able to exert its cytotoxic effect. It also suggests a reason why other bacterial exotoxins are also found only on temperate bacteriophages. Incubation of Tetrahymena with purified Stx2 decreases total protein synthesis. This finding indicates that, similar to mammalian cells, Stx2 kills Tetrahymena by inactivating its ribosomes.
IMPORTANCE
Tetrahymena is a bacterial predator and a model for mammalian phagocytosis and intracellular vesicular trafficking. Phage-encoded exotoxins apparently have evolved for the purpose of bacterial antipredator defense. These exotoxins kill mammalian cells by inactivating universally conserved factors and/or pathways. Tetrahymena and susceptible mammalian cells are killed when exposed to bacteriophage-encoded Shiga toxin (Stx). Stx toxicity in mammalian cells requires Stx binding to the globotriaosyl ceramide (Gb3) receptor, followed by receptor-mediated endocytosis (RME). We show that, similar to mammalian cells, internalized Stx inhibits protein synthesis in Tetrahymena. Although Tetrahymena lacks Gb3, our results suggest that the cytotoxic effect of Stx on Tetrahymena is apparently mediated by a receptor, thereby arguing for the existence of RME in Tetrahymena. As opposed to the case with mammalian phagocytes, Stx produced by bacteria inside Tetrahymena is cytotoxic, suggesting that these cells may represent a “missing link” between unicellular eukaryotic bacterial predators and phagocytotic mammalian cells.
Introduction
With approximately 1031 individuals, bacteriophages are likely the most abundant organisms on the planet (1, 2). Bacteriophages often bear genes encoding exotoxins (e.g., Shiga toxin [Stx], diphtheria toxin, cholera toxin, and botulinum toxin) which cause disease in mammals (3). While these phage-encoded toxins harm mammals, these phages can be prevalent where mammals are not found (4). Our earlier work demonstrated that, similar to its effect on mammalian cells, both bacterially produced and purified Stx is capable of killing Tetrahymena thermophila. That work also demonstrated that Stx is capable of functioning as part of an antipredator defense strategy, killing Tetrahymena and reducing its predation efficiency, allowing the bacteria to survive (5). Hence, susceptible mammals may not be the original or primary “targets” of this exotoxin or others.
Stx is a family of homologous AB5 exotoxins that share the same enzymatic activity (6, 7). They comprise the enzymatically active (StxA) subunit and a pentamer of receptor binding subunits (StxB), which is necessary for entry and trafficking of the toxin (8). Stx was first identified in Shigella dysenteriae serotype 1, but closely related variants are found in Escherichia coli and similar bacteria (e.g., Stx1, Stx2, Stx2c to -f) (6, 9–11). Regardless of the source, Stx holotoxin enters mammalian cells by binding a glycosphingolipid, globotriaosyl ceramide (Gb3), present on the cell surface. Gb3 is a glycosphingolipid characterized by Gal(α1–4) Gal(β1–4) Glc(β1–8) ceramide linkages (12). Recognition and binding of the Gal(α1–4) Gal linkage by StxB (12) and subsequent aggregation of bound Gb3 initiates clathrin-mediated endocytosis (CME) of Gb3-Stx complexes in susceptible mammalian cells (13). Once inside the cell, the Stx-containing endosome undergoes retrograde transport through the Golgi apparatus and into the endoplasmic reticulum (14, 15), where the StxA subunit is cleaved from the holotoxin by furin, leading to release of the StxA subunit into the cytosol. The activated Stx subsequently removes a critical adenine residue from the rRNA amino-acyl tRNA-accepting (sarcin-ricin) loop, inactivating the ribosome and eventually killing the cell (16). The position, function, and sequence of this loop are conserved among virtually all organisms, including Tetrahymena (17, 18).
Our previous investigations of Stx toxicity to Tetrahymena did not determine the mode of entry of Stx or the mechanism by which it kills the organism (5). This information could provide insight into processes related to the evolution of Stx toxicity. T. thermophila encodes all of the machinery needed for CME (19). Stx could enter Tetrahymena through the plasma membrane by receptor-mediated or nonspecific clathrin-mediated endocytosis. Importantly, these cells encode an elaborate set of Rab proteins, suggesting that they are capable of internalizing and appropriately sorting Stx-containing vesicles resulting from CME (20). Protozoa are also phagocytotic and share many features with mammalian phagocytes, especially macrophages. However, macrophages are not susceptible to killing by bacterially produced Stx (21–23), instead responding to Stx intoxication by releasing proinflammatory cytokines.
T. thermophila takes up bacteria, nutrients, and other molecules through its oral apparatus, encapsulating these in food vacuoles. Bacteria ingested by Tetrahymena remain viable in food vacuoles for a considerable length of time (24, 25). Therefore, it is formally possible that viable Stx-encoding bacteria internalized in food vacuoles by Tetrahymena are able to produce toxin that is lethal to this organism. Hence, as opposed to mammalian cells killed by Stx, lethal toxin could enter T. thermophila by either or both of two non-mutually exclusive methods, i.e., (i) plasma membrane-mediated endocytosis of cell-free toxin and/or (ii) uptake of Stx-encoding bacteria or cell-free Stx via the oral apparatus.
We showed previously that excess purified StxB specifically prevents Tetrahymena killing by purified Stx2 (5). Moreover, preliminary findings indicated that StxB is necessary for the toxicity of bacterially produced Stx to Tetrahymena (5). Since StxB is the receptor binding subunit of Stx, these findings suggested that at least part of Stx’s intoxication mechanism is mediated by the binding of Stx to a receptor. Regardless, the nature and location of this putative receptor in Tetrahymena are unclear. Moreover, it is unclear whether toxin released from internalized bacteria can also kill these organisms and/or whether this entry mechanism also involves a receptor.
Once inside mammalian cells, activated Stx inhibits protein synthesis and causes cell death (16). While Stx kills T. thermophila, we do not know if it does so in the same way as it does mammalian cells. It has been shown that Stx, though at a very low affinity, inactivates bacterial ribosomes (26), resulting in the death of bacteria that overproduce Stx. Tetrahymena ribosomes are more similar to mammalian ribosomes than bacterial ribosomes are and are therefore very likely to be inactivated by Stx.
Here we explore the mechanisms by which Stx enters Tetrahymena and how the internalized toxin kills these organisms. Our findings indicate that Stx added to or produced by bacteria in the culture medium enters Tetrahymena via a specific receptor. Our data also suggest that this putative receptor is a membrane-bound glycoconjugate. We also show that Stx produced by bacteria encapsulated within food vacuoles is capable of killing Tetrahymena. Transport of Stx produced in the food vacuoles to the cytoplasm is also apparently mediated by a glycoconjugate receptor. We find that Stx is a powerful inhibitor of protein synthesis. Although we have not defined the details of how Stx exerts its cytolethal effects, we anticipate that, similar to mammalian cells, this inhibition of protein synthesis leads to cell death.
RESULTS
Our previous results (5) showed that purified bacterially encoded Stx kills Tetrahymena (strain CU427.4). A priori, Stx could enter Tetrahymena cells via two alternative, non-mutually exclusive routes. One route entails nonspecific phagocytosis of Stx by Tetrahymena; the other involves Stx binding to a cell surface receptor, followed by receptor-mediated endocytosis. Our preliminary results suggested that purified Stx2 enters T. thermophila cells via a specific receptor and that this entry is mediated by StxB. We wished to confirm this idea. To do this, we compared the abilities of two different bacterial strains, one of which bears a wild-type 933W prophage that encodes both the A and B subunits of Stx2 (MG1655::933W) and the other of which bears a mutant 933W prophage in which the gene encoding the B subunit has been disrupted (MG1655::933WΔB) and hence encodes only the Stx2A subunit. Control immunoblot assays showed that the same amount of the StxA subunit is produced by the two strains (data not shown). When T. thermophila is cocultured with MG1655::933W, the number of T. thermophila cells is reduced by approximately 60% (Fig. 1A) compared to that of cells that are incubated in the absence of any bacteria. In contrast, the number of Tetrahymena cells increases by nearly 2-fold when they are cocultured with MG1655::933WΔB. This finding indicates that the StxA subunit alone is insufficient to intoxicate T. thermophila and that Tetrahymena killing requires both the A and B subunits. This finding is consistent with the suggestions that Stx2 enters T. thermophila cells via a specific receptor and that this entry is mediated by StxB.
FIG 1 .
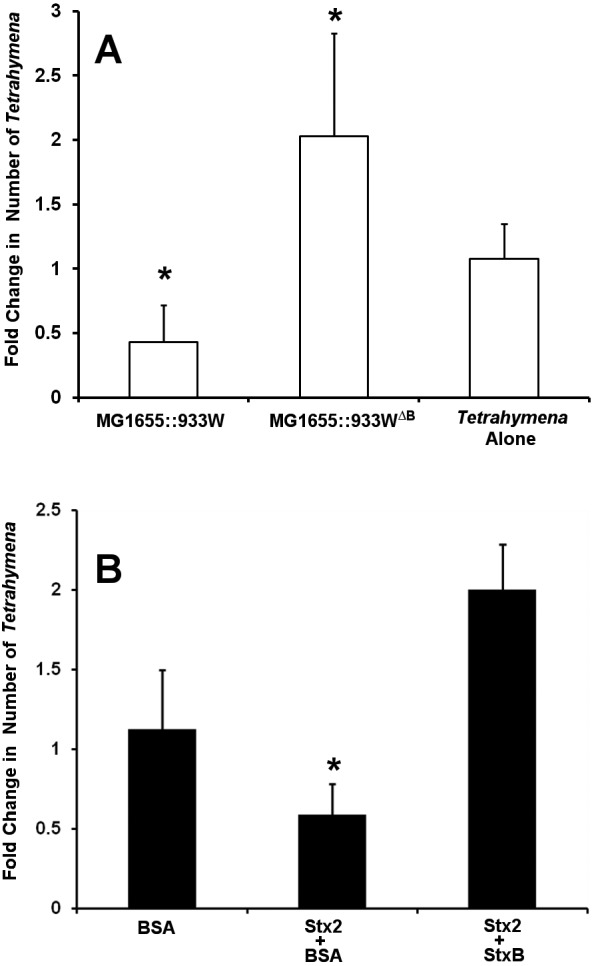
Role of StxB in Stx2-mediated Tetrahymena killing. (A) Tetrahymena cells were separately cultured in medium with no additions or containing MG1655::933W or MG1655::933WΔB for 6 h at 30°C. The number of live Tetrahymena cells was determined after 6 h as described previously (5). Data are presented as the number of live Tetrahymena cells after incubation relative to the number of live Tetrahymena cells at the start (t = 0) of incubation. Error bars represent standard deviations from three or more independent experiments, with each experiment comprising a minimum of three individual measurements *, P < 0.01 relative to the control. (B) Tetrahymena cells were axenically cultured with 40 ng/ml Stx2 holotoxin or BSA alone or 40 ng/ml Stx2 holotoxin in the presence of 2 µg/ml StxB. The number of live Tetrahymena cells was determined after 6 h. Data are presented as the number of live Tetrahymena cells after incubation relative to the number of live Tetrahymena cells at the start of incubation. Error bars represent standard deviations from three or more independent experiments, with each experiment comprising a minimum of three individual measurements. *, P < 0.01.
To further test the idea that Stx intoxication results from toxin binding to a specific receptor in T. thermophila, if such a receptor exists, we reasoned that the presence of excess StxB, the receptor binding subunit of Stx, should act as a competitor for Stx holotoxin-mediated killing of Tetrahymena, whereas a nonspecific protein would not. To do this, we incubated T. thermophila with Stx2 holotoxin in the presence or absence of StxB. Consistent with previous observations, when Tetrahymena cells are grown in the presence of purified Stx2 and an ~80-fold molar excess of BSA (bovine serum albumin) the number of viable Tetrahymena cells decreases. In contrast, when Tetrahymena cells are incubated with purified Stx2 and an ~80-fold molar excess of the StxB subunit, the number of viable Tetrahymena cells increases (Fig. 1B), indicating that StxB competes with Stx2 for entry into Tetrahymena cells Our preparation of the StxB subunit contains no proteolytic activity (data not shown). Hence, the ability of StxB, but not BSA, to protect T. thermophila from Stx2-induced killing is consistent with the idea that entry of Stx into Tetrahymena cells is mediated by a receptor. We hypothesize that each cell bears a number of these receptors, requiring an excess of StxB over Stx2 to block Stx2 binding and entry into Tetrahymena cells.
To further examine the mode of entry of Stx2 into Tetrahymena cells, we assessed the ability of Stx2 to kill Tetrahymena strain NP1, a temperature-sensitive mutant strain of T. thermophila. When grown at the permissive temperature (30°C), NP1 forms a normal oral apparatus; however, at the restrictive temperature (37°C), this strain lacks an oral apparatus (27) and consequently is unable to perform phagocytosis. This leads to the observed smaller size of NP1 at the restrictive temperature (Fig. 2). Consistent with the reported NP1 phenotype (27), at 30°C, this strain efficiently ingests bacteria, as indicated by the presence of mCherry-labeled E. coli strain MG1655 in the food vacuoles inside these cells (Fig. 2A). When grown at 37°C, NP1 does not ingest bacteria by phagocytosis, as indicated by the absence of labeled bacteria inside these cells, and does not form food vacuoles (Fig. 2B). Hence, at 37°C, the only route for compounds to enter Tetrahymena NP1 cells is via endocytosis. If Stx2 is capable of killing Tetrahymena strain NP1 at 37°C, this finding would indicate that Stx2 can enter Tetrahymena cells by endocytosis. This finding would be consistent with the suggestion that Stx2 enters Tetrahymena cells via a specific membrane-bound receptor.
FIG 2 .
Phagocytosis of fluorescent bacteria by Tetrahymena strain NP1 at permissive and restrictive temperatures. Tetrahymena cells were cocultured with EDL933 bearing a plasmid encoding the red fluorescent protein mCherry at 30°C (A) or 37°C (B). Shown are overlaid DIC/fluorescence micrographs of a ×60-magnified image of this culture. The images shown were obtained with a rhodamine red filter on a Zeiss Axio Imager Z1 microscope.
Cells of both wild-type Tetrahymena strain CU427.4 and mouthless strain NP1 increase in number when incubated for 6 h in the presence of BSA at 30°C, but NP1 grows to a higher density than does wild-type Tetrahymena under these conditions (Fig. 3A). Consistent with the cytotoxic effect of Stx shown previously (5) and in Fig. 1B, the presence of purified Stx2 inhibits the growth of both of these Tetrahymena strains (Fig. 3A) at 30°C. At 37°C, where NP1 lacks the oral apparatus, the number of viable Tetrahymena strain NP1 cells remains static when they are incubated with BSA. Under these conditions, the number of wild-type Tetrahymena cells also remains static. Incubation of NP1 with purified Stx2 at 37°C reduces the number of viable T. thermophila strain NP1 cells by approximately 60% (Fig. 3B). Under identical conditions, Stx2 also similarly decreases the viability of Tetrahymena strain CU427.4. The ability of Stx2 to kill mouthless Tetrahymena strain NP1 at the restrictive temperature indicates that Stx intoxication does not require toxin entry via phagocytosis. This finding suggests that Stx2 is able to enter Tetrahymena cells through the cell membrane. Stx’s ability to get into Tetrahymena cells through the cell membrane, combined with the finding that StxB specifically protects against Stx-mediated killing, argues that T. thermophila membranes contain a receptor for Stx.
FIG 3 .
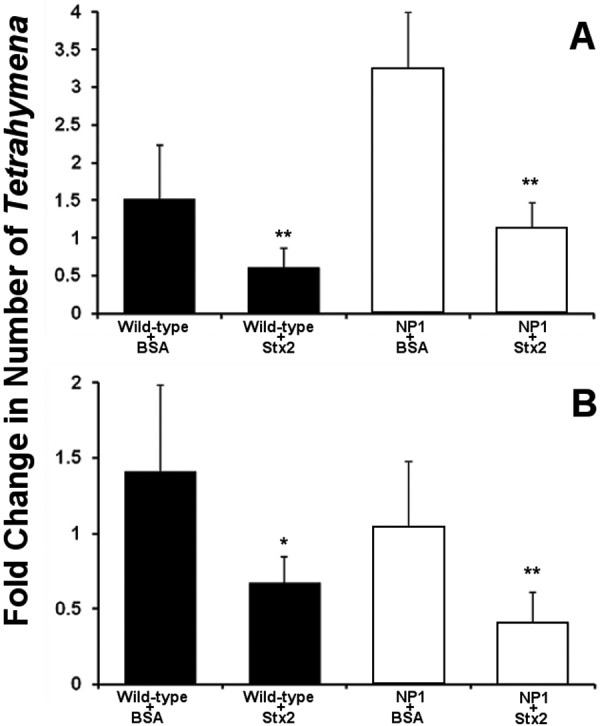
Effect of purified Stx2 on the survival of axenically growing wild-type and NP1 mutant Tetrahymena cells at various temperatures. Purified Stx2 holotoxin or BSA (40 ng/ml) was added to 104/ml Tetrahymena cells cultured as described in Materials and Methods at 30°C (A) or 37°C (B). The number of live Tetrahymena cells was determined after 6 h of incubation. Data are presented as the number of live Tetrahymena cells after incubation relative to the number of live Tetrahymena cells at the start of incubation. Error bars represent standard deviations from three or more independent experiments, with each experiment comprising a minimum of three individual measurements. *, P < 0.01; **, P < 0.001.
We wished to gain further insight into the nature of Tetrahymena’s putative Stx receptor. Since Stx2 intoxication of Tetrahymena requires its B subunit (Fig. 1A) and this subunit binds carbohydrates, we postulated that the receptor may be a glycoconjugate. To test this suggestion, we incubated Tetrahymena cells with the glycolipid binding subunit of ricin, ricin B (28), and determined whether it could protect Tetrahymena cells from Stx2-mediated killing. As shown in Fig. 1B, when incubated with 40 ng/ml Stx2, the number of viable Tetrahymena cells decreased in the presence of 2 µg/ml BSA but not in the presence of 2 µg/ml StxB. Similar to the findings with StxB, the number of viable Tetrahymena cells was unaffected by the presence of 40 ng/ml Stx2 when 2 µg/ml of ricin B was also present (Fig. 4). Hence, ricin B protects against Stx2-mediated killing. The protection of T. thermophila from Stx2-mediated killing by ricin B indicates that Stx and ricin share the same putative glycoconjugate receptor or family of receptors. This finding also implies that the putative Stx2 receptor in T. thermophila may be specific not to Stx2 but a receptor or receptor family specific to toxins that bind to similar sugar moieties.
FIG 4 .

Ricin B protects Tetrahymena from Stx2-mediated killing. Tetrahymena cells were axenically cultured with 40 ng/ml BSA, Stx2 holotoxin, or 40 ng/ml ricin B subunit alone or 40 ng/ml Stx2 holotoxin in the presence of 2 µg/ml StxB or ricin B as indicated. The number of live Tetrahymena cells was determined after 6 h. Data are presented as the number of live Tetrahymena cells after incubation relative to the number of live Tetrahymena cells at the start of incubation. Error bars represent standard deviations from three or more independent experiments, with each experiment comprising a minimum of three individual measurements. *, P < 0.01.
Release of Stx holotoxin from EDL933 or other Stx-encoding bacteria requires phage-mediated lysis of the cell—it is not secreted through any bacterial secretory machinery (29). Therefore, toxin-encoding bacteria in coculture with T. thermophila have two potential routes by which they can deliver toxin into Tetrahymena cells. One route is by bacterial lysis, releasing Stx2 outside the Tetrahymena cell, followed by its entry via endocytosis. Alternatively, bacterially derived toxin may enter Tetrahymena cells when Stx2-encoding bacteria are eaten by Tetrahymena cells and these bacteria, encapsulated within the food vacuoles, lyse, releasing toxin. We showed previously that Tetrahymena releases a factor that can stimulate SOS-dependent induction of Stx-encoding prophage (5).
We first determined whether bacterially derived Stx can enter through the cell membrane via endocytosis. To do this, mouthless Tetrahymena strain NP1 (27) was cocultured with Stx1- and Stx2-encoding EDL933 or an stx1 stx2 mutant strain, EDL933Δstx, which is identical to EDL933 except that its toxin-encoding genes are inactivated by disruption/deletion (30). The number of viable Tetrahymena NP1 cells in coculture with EDL933 decreased by approximately 40% at 30°C, the permissive temperature for the formation of the oral apparatus, and by 60% at the restrictive temperature (37°C), where NP1 lacks the oral apparatus (Fig. 5). In contrast, the number of viable NP1 cells in coculture with EDL933Δstx either increased at 30°C or remained static at 37°C (Fig. 5). The ability of EDL933 to decrease the number of NP1 cells at 37°C indicates that entry of Stx through the putative Tetrahymena cell membrane receptor is a potential route for bacterially derived Stx.
FIG 5 .
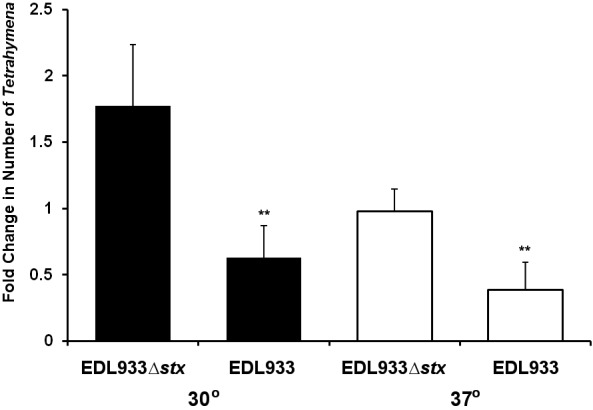
Elimination of phagocytosis does not affect killing of Tetrahymena by bacterially produced Stx. Tetrahymena strain NP1 was cocultured with EDL933 or EDL933Δstx at 30°C (black bars) or 37°C (white bars). Cultures were prepared as described in Materials and Methods. Data are expressed as n-fold changes in cell numbers at the end of a 6-h incubation relative to the beginning of the incubation. At the start of the incubation, Tetrahymena cells were present at approximately 104/ml and bacterial cells were present at 108/ml. Error bars represent standard deviations from three or more independent experiments, with each experiment comprising a minimum of three individual measurements. *, P < 0.01; **, P < 0.001.
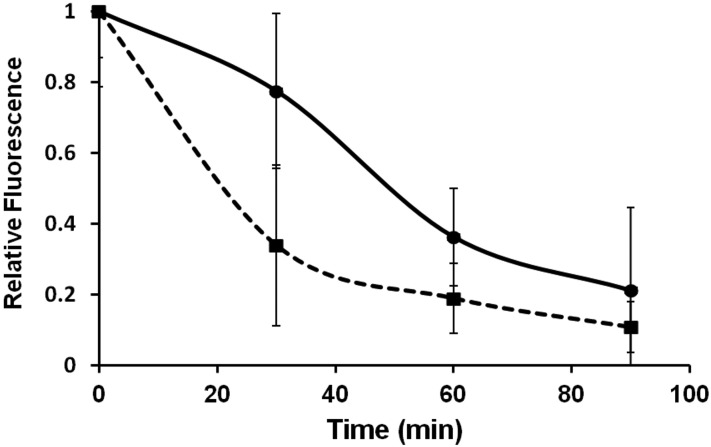
Although bacterially derived Stx apparently can enter Tetrahymena cells through the cell membrane (Fig. 5), it is possible that ingestion of toxin-encoding bacteria and the release of Stx in the food vacuoles provide an efficient route for Stx entry into T. thermophila. If this route is to be effective, some of the bacteria ingested by Tetrahymena and encapsulated in food vacuoles (Fig. 2) must remain viable as they pass through the cell. To test this idea, we followed separately the transit of fluorescently labeled EDL933 and EDL933∆stx bacteria through the cell by microscopy. We found that, consistent with previous results (31), these encapsulated bacteria transited though the cell and were expelled via the cytoproct over approximately 90 min (Fig. 6). While there were differences in the survival of EDL933 and EDL933∆stx during their passage through the cells, in both cases, only some of the ingested bacteria were digested and/or lysed during transit. At 60 min, between 20 and 40% of the labeled bacteria remained intact and at 90 min postingestion, 10 to 30% of the labeled bacteria remained intact and within Tetrahymena cells.
FIG 6 .
Persistence of intact EDL933 and EDL933Δstx in Tetrahymena. Tetrahymena cells were separately cocultured with EDL933 (•) or EDL933Δstx (▪), each of which was transformed with a plasmid expressing the red fluorescent protein mCherry. Bacteria not internalized by Tetrahymena cells were removed, and the fluorescence intensity remaining within the Tetrahymena cells was quantified at various times as described in Materials and Methods. Fluorescence intensity is expressed as a fraction of that found at t = 0. Each time point represents the average fluorescence intensity of ≥30 cells.
Since bacteria apparently survive within Tetrahymena food vacuoles, it is possible that bacterially derived Stx2 can be released from these organelles and enter the cytoplasm of Tetrahymena, causing cell death. To begin to test this idea, we designed an E. coli strain that, when transformed with an Stx2-producing plasmid, allows Stx2 to be released only when the bacteria are encapsulated within Tetrahymena. This strain, MG1655 (recA)::λcI857 bears a recA mutation and is lysogenized with bacteriophage λcI857. This phage bears a temperature sensitivity mutation in its repressor protein. Consequently, while it is able to form stable lysogens at permissive temperatures (≤30°C), this phage is unable to maintain lysogeny at temperatures above 33°C. Since any E. coli strain that contains a lysogenic prophage can be induced to grow lytically by RecA-mediated bacteriophage repressor autocleavage (32), either stochastically (33) or as a consequence of DNA damage, the presence of the recA mutation ensures that the resident prophage in this strain is essentially incapable of lytic growth at permissive temperatures. Thus, the presence of the recA mutation is required to prevent the production of Stx outside the Tetrahymena cell. Therefore, the combination of this mutation and bacteriophage λcI857 allows us to induce Stx2 production only from bacteria encapsulated within Tetrahymena cells.
We transformed MG1655 (recA)::λcI857, which does not encode Stx, with pStx, a plasmid that directs the constitutive synthesis of low levels of Stx2 holotoxin. Control immunoblot assays indicate that this plasmid produces ~4,500 molecules of intact Stx per cell (i.e., lysis of a saturated overnight culture will contain Stx2 at a concentration of 66 ng/ml). Stx2 is not secreted through any bacterial secretory machinery. Its release from the bacteria depends on phage genes that cause bacterial cell lysis (29). Consequently, at permissive temperatures, the Stx2 constitutively produced within MG1655 (recA)::λcI857/pStx cannot be released from the cell. Alternatively, at the restrictive temperature, MG1655 (recA)::λcI857/pStx lyses readily, releasing the Stx contained within the cell. Control experiments done previously have shown that in MG1655-based λcI857, lysogens are completely lysed within 30 min of incubation at restrictive temperatures (data not shown). We also transformed MG1655 (recA)::λcI857 with the empty vector for use as a control. As another control, we constructed MG1655 (recA)/pStx. Since this strain lacks an inducible prophage, Stx2 would not be released from it unless the cell is killed and lysed by some other means.
These strains, MG1655 (recA)/pStx, MG1655 (recA)::λcI857/pET17b, and MG1655 (recA)::λcI857/pStx, were separately cocultured with T. thermophila strain CU427.4 at the permissive temperature of 30°C for 30 min. Microscopic examination indicates that under these conditions, bacteria fluorescently labeled with mCherry are found in the food vacuoles of approximately 90% of the Tetrahymena cells in the culture (data not shown). Subsequently, the free bacteria were washed away by differential centrifugation as described in Materials and Methods. After washing, the cultures were shifted to 42°C and incubated for an additional 4 h.
The number of viable Tetrahymena cells cocultured with MG1655 (recA)::λcI857/pET17b doubled over the incubation period (Fig. 7). In contrast, when MG1655 (recA)::λcI857/pStx cells were cultured with Tetrahymena cells, the number of viable Tetrahymena cells decreased by 40% (Fig. 7). This result is consistent with the idea that bacterially derived Stx is capable of entering the cytosol from the food vacuoles and killing Tetrahymena cells.
FIG 7 .
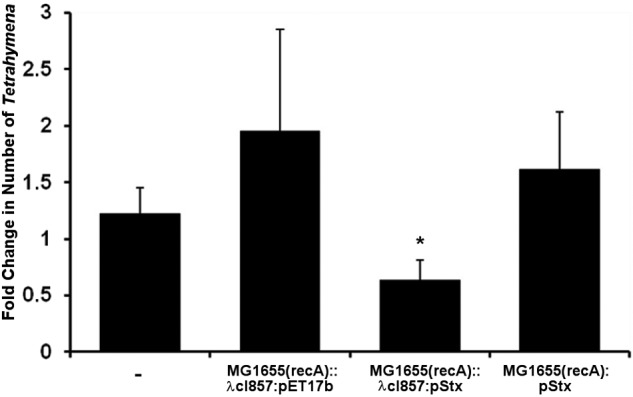
Stx produced by internalized bacteria is cytotoxic to Tetrahymena. Tetrahymena cells were cocultured with MG1655 (recA)::λcI857/pStx, MG1655 (recA)::λcI857/pET17, or MG1655 (recA)/pStx for 30 min at 30°C, uninternalized bacteria were removed by centrifugation, and the bacterium-containing Tetrahymena cells were incubated at 42°C as described in Materials and Methods. The Tetrahymena cells were enumerated subsequent to washing and again after incubation at 42°C. At the start of the incubation, Tetrahymena cells were present at approximately 104/ml. The data presented are the number of live Tetrahymena cells after incubation relative to the number of live Tetrahymena cells at the start of incubation. Error bars represent standard deviations from three or more independent experiments, with each experiment comprising a minimum of three individual measurements. *, P < 0.001.
The number of viable Tetrahymena cells incubated with MG1655 (recA)/pStx increased by 50% (Fig. 7). The only difference between this strain and MG1655 (recA)::λcI857/pStx is the absence of bacteriophage λcI857 from MG1655 (recA)/pStx. These data indicate that digestion of bacteria by Tetrahymena apparently renders the released toxin harmless to the cell and argues that bacteriophage-mediated lysis of Stx-encoding bacteria is necessary for Stx toxicity in Tetrahymena.
Taken together, the data in Fig. 5 and 7 show that bacterially produced Stx2 effectively kills Tetrahymena regardless of whether the toxin is released into the medium or produced within Tetrahymena food vacuoles. Our data suggest that Stx2 transport across the Tetrahymena plasma membrane is mediated by a glycoconjugate receptor (Fig. 1, 3, and 4). We wondered whether transport of Stx2 across the food vacuole membrane into the cytosol utilizes the same membrane receptor system. To answer this question, we incubated Tetrahymena cells with MG1655 (recA)::λcI857/pStx in the presence of either 2 µg/ml purified StxB or, as a control, 2 µg/ml BSA. The free bacteria were washed away, and the cocultures were moved to 42°C to induce Stx release into T. thermophila food vacuoles. The number of viable Tetrahymena cells in cultures incubated with BSA decreased by approximately 40%, while the number of viable Tetrahymena cells in cultures incubated with StxB increased slightly (Fig. 8). This finding shows that excess StxB in the medium taken up with bacteria protects Tetrahymena cells from bacterially produced Stx and indicates that the B subunit mediates uptake into Tetrahymena cells. This observation suggests that bacterially derived Stx2 within Tetrahymena food vacuoles enters the cell via a receptor-mediated process that may be identical to Stx2 entry from outside the cell. Hence, these two seemingly disparate modes of entry into T. thermophila apparently use the same uptake scheme.
FIG 8 .
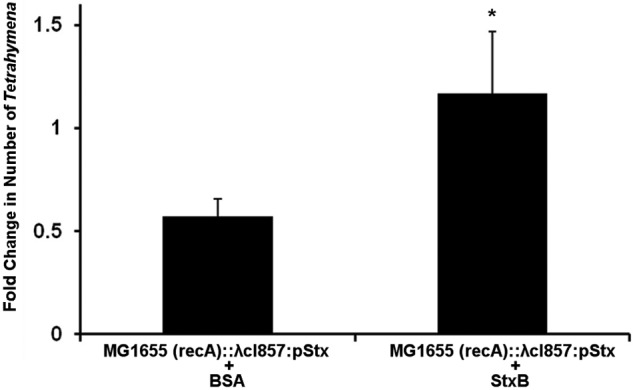
StxB decreases the cytotoxicity of Stx produced by bacteria internalized by Tetrahymena cells. Tetrahymena cells were cocultured with MG1655 (recA)::λcI857/pStx and either 2 µg/ml BSA or StxB for 30 min at 30°C, uninternalized bacteria were removed by centrifugation, and the bacterium-containing Tetrahymena cells were incubated at 42°C as described in Materials and Methods. The Tetrahymena cells were enumerated subsequent to washing and again after incubation at 42°C. At the start of the incubation, Tetrahymena cells were present at approximately 104/ml. Data are presented as the number of live Tetrahymena cells after incubation relative to the number of live Tetrahymena cells at the start of incubation. Error bars represent standard deviations from three or more independent experiments, with each experiment comprising a minimum of three individual measurements.
Having gained insight into how Stx enters T. thermophila cells, we wished to determine how this toxin kills Tetrahymena cells. Stx apparently kills susceptible mammalian cells by inactivating the ribosome and inhibiting protein synthesis (8). To determine whether purified Stx2 inhibits protein synthesis, we incubated T. thermophila with purified Stx2 and measured total protein synthesis by pulse-chase using [35S]methionine. In these experiments, we also examined the effect of paromomycin, a potent protein synthesis inhibitor in Tetrahymena cells, on [35S]methionine incorporation (34). After a 1-h incubation with Stx, Tetrahymena protein synthesis decreases 5-fold compared to that of Tetrahymena cells incubated with BSA (Fig. 9). This observation is consistent with the idea that Stx2 inhibits protein synthesis in T. thermophila. Interestingly, this decrease in [35S]methionine incorporation is nearly identical to the decrease seen when Tetrahymena cells are incubated with paromomycin (Fig. 9). This similarity indicates that Stx2 is also a potent protein synthesis inhibitor. Decreases in protein synthesis caused by both Stx and paromomycin are seen in incubation periods as short as 10 min (data not shown), indicating that both compounds act quickly to inhibit protein synthesis.
FIG 9 .
Effect of purified Stx2 on Tetrahymena protein synthesis. Tetrahymena cells (5 × 104/ml) were separately incubated with 200 ng/ml Stx2, 200 ng/ml BSA, or 0.5 mM paromomycin for 60 min and subsequently assayed for protein synthesis as described in Materials and Methods. [35S]methionine incorporation into protein was determined by scintillation counting and is represented as counts per minute per Tetrahymena cell. Error bars represent standard deviations from three or more independent experiments, with each experiment comprising a minimum of three individual measurements. *, P < 0.001.
DISCUSSION
Our earlier work demonstrated that both purified Stx and that produced by bacteria in coculture are capable of killing T. thermophila (5). Together with other findings, these observations indicated that Stx can function as part of an antipredator defense strategy, suggesting that susceptible mammals are neither the original nor the primary “targets” of this toxin. Our previous investigations did not discern the mode of entry of Stx into Tetrahymena cells and whether the mechanism by which it kills these cells was conserved between single-celled and metazoan eukaryotes. This information could provide insight into the evolution of Stx toxicity.
Stx intoxication of susceptible mammalian cells requires its specific binding to the Gb3 glycolipid receptor located on the plasma membrane. The finding that Stx kills T. thermophila under conditions where phagocytosis via the oral apparatus is blocked by mutation indicates that these organisms are able to import Stx through the plasma membrane. Tetrahymena contains all of the machinery needed for CME. CME in Tetrahymena can occur nonspecifically, via membrane recycling (20, 35–37). However, we find that Stx-mediated killing of mouthless Tetrahymena by can be specifically blocked by the addition of an excess of the receptor binding StxB subunit. It is formally possible that StxB prevents killing by “inactivating” the Stx holotoxin; however, our StxB preparation does not contain a protease or other inactivating activities (data not shown). Also, excess StxB does not appear to inactivate Stx holotoxin Gb3 binding activity (38). Importantly, we also find that Stx holotoxin killing of Tetrahymena can be blocked by the carbohydrate binding subunit of ricin, ricin B, but not by a nonspecific protein, BSA. Together, these findings suggest that Stx can be imported through the plasma membrane and this import is mediated by a receptor. Since BSA and StxB pentamers are similar in size and isoelectric point, our results are inconsistent with the idea that StxB enters cells by nonspecific CME.
It is well established that T. thermophila behavior responds in a specific and selective way to the addition of effectors (hormones, drugs) that would, at first glance, be expected to affect only cells of multicellular eukaryotes (39–41). It has been conjectured that these behaviors are mediated by ligand-receptor interactions (42). Hence, the suggestion that Stx import into these cells is mediated by a membrane-bound receptor may not be unexpected. However, in no case have ligand-mediated changes in Tetrahymena behavior been associated with the binding of these ligands to a specific receptor. While our data do not identify a specific receptor for Stx, the finding that the presence of an excess of the carbohydrate binding subunits of Stx and ricin toxin (StxB and ricin B, respectively) blocks Stx-mediated killing of Tetrahymena argues for the presence of a receptor on Tetrahymena membranes and that this receptor is a glycoconjugate.
Our analysis of Tetrahymena lipid content indicates that the glycolipid Gb3, which serves as the Stx receptor in mammalian cells, is not found in Tetrahymena (T. Hennessey, C. Lingwood, and G. Koudelka, unpublished results). Consistent with this finding, bioinformatic analysis shows that Tetrahymena does not contain a gene orthologous to that which encodes the mammalian Gb3 synthase (43, 44). Nonetheless, the suggestion that a membrane-bound glycoconjugate may mediate the uptake of bacterial exotoxins is consistent with the observation that pertussis toxin (PTX), a toxin whose import into mammalian cells is also mediated by the binding of a carbohydrate binding subunit to a membrane-bound glycoconjugate receptor, has demonstrable effects on Tetrahymena behavior (41). Therefore, while the precise identity of a receptor(s) for both Stx and PTX in Tetrahymena has yet to be determined, we hypothesize that membrane-bound glycoconjugates mediate the import of both toxins.
Both purified Stx and bacteria can potentially enter Tetrahymena cells by phagocytosis via the oral apparatus. The finding that StxB is necessary for intoxication of Tetrahymena by bacterially produced toxin and that excess StxB, but not BSA, can prevent killing by Stx produced by bacteria in food vacuoles argues that cytoplasmic import of toxin across the food vacuole membrane may be mediated by a receptor. Since the oral groove is formed by invagination of the plasma membrane, it is not surprising that the internal surface of the resulting food vacuole could contain the putative Stx receptor that is apparently located on the surface of these cells.
Tetrahymena cells are bacterivorous. Live bacteria are captured by the oral groove, encapsulated within phagosomes, and subsequently lysed, presumably as a consequence of acidification of this organelle, which occurs within 10 min postingestion, or fusion of the phagosome with lysosomes, which occurs between 20 and 40 min postingestion (45, 46). In this light, it is surprising that Stx produced by bacteria in food vacuoles is cytotoxic (Fig. 7). However, similar to the results of others (24, 25), we found that bacteria ingested by Tetrahymena cells survive in this phagosome and have a half-life of 20 to 50 min. Moreover, between 10 and 30% of the bacteria survive in the food vacuole for at least 90 min (Fig. 6). Hence, encapsulated bacteria live long enough to deliver cytolethal toxin to Tetrahymena cells.
Our previous results indicated that bacterially encoded Stx can function as part of a bacterial antipredator defense strategy (5). One question that arises from the findings that Stx produced outside the cell and imported through the cell membrane and Stx produced inside the phagosome by ingested bacteria are both cytolethal is: which route of intoxication is most relevant to the antipredator defense mechanism? We found previously that the 50% lethal dose of purified Stx is ~2.5 ng/ml, which corresponds to approximately 2 × 1010 molecules/ml. It is unclear how much Stx is produced by an individual induced bacterium; however, the amount of toxin produced by Stx-encoding bacteria is proportional to the number of phages produced (47, 48). About 150 lambdoid phages are produced per bacterium. Since Stx production is linked to the transcription of genes expressed late in the lysis pathway, it is reasonable to assume that a similar (or greater) number of molecules of toxin is released from each induced lysogen. Therefore, approximately 108 bacterial cells/ml must lyse to produce enough Stx to significantly affect the viability of Tetrahymena cells. In water, E. coli is found at ≤107 cells/ml. Thus, if Stx produced outside the cell mediates its antipredator activity, in order to effectively function as an antipredator defense, virtually all of the cells in the population would need to lyse, a clearly ineffective strategy to facilitate population survival.
Our measurements suggest that at a bacterial concentration of 108 cells/ml, each Tetrahymena cell ingests ≤500 bacteria. Tetrahymena cells reach a population of size of 104/ml in nutrient-poor medium. Hence, after the introduction of bacteria, Tetrahymena internalizes ≤5 × 106 cells/ml “prey” bacteria, leaving at least 9.5 × 107 bacterial cells/ml. While we have not precisely measured the number of internalized bacteria that are induced to produce phage or digested by Tetrahymena, and even if Tetrahymena cell densities are substantially higher than 104/ml, it is apparent that intoxication by ingested bacteria is likely the most relevant to the antipredator defense mechanism.
The results in Fig. 7 show that only toxin that is produced by bacteria bearing temperate bacteriophage is able to kill Tetrahymena cells. Stx constitutively produced in phagosomes by bacteria lacking a bacteriophage or bearing a phage that cannot be induced is not cytotoxic (Fig. 7). That is, toxin released from the bacteria as a consequence of digestion by Tetrahymena cells is not cytotoxic. These observations suggest that (i) phage induction and consequent toxin release are triggered prior to the toxin-inactivating action of bacterial digestion and (ii) the cytotoxic activity of Stx produced by bacteria in phagosomes apparently requires that production of toxin be coupled with phage-mediated lysis of the bacterial cell. Since the digestive processes that lead to bacterial lysis apparently inactivate Stx, these findings provide a rationale for why the genes encoding Stx are found almost exclusively on bacteriophages; toxin must be released from the bacteria prior to the digestion of the cell, or it will not be able to exert its cytotoxic effect. We suggest that similar reasons underlie the finding that several other bacterial exotoxins are found only on temperate bacteriophages.
Inspection of the data in Fig. 6 shows that, in otherwise isogenic strains, the half-life of bacteria bearing genes encoding Stx survive 2.5 times longer inside Tetrahymena cells than do bacteria that do not harbor these genes. This difference causes the amount of viable Stx-encoding bacteria to be 3-fold higher at 90 min than the number of Stx− bacteria. Similar findings were reported in reference 25. Hence, Stx appears to partially protect phagocytosed bacteria from digestion. Although the precise cause is likely different, this observation is similar to that made with Legionella pneumophila; i.e., strains of L. pneumophila that are pathogenic to mammals are less susceptible to digestion than are nonpathogenic strains. Apparently, pathogenic L. pneumophila prevents the food vacuoles from fusing with lysosomes (49).
With this in mind, we envision two potentially overlapping mechanisms by which Stx enhances the survival of ingested bacteria in Tetrahymena cells. In both cases, some, but not all, of the bacteria containing a prophage and toxin may be induced to produce phage and consequently toxin. According to one idea, transport of this toxin into the cytoplasm inhibits protein synthesis and therefore inhibits the expression of proteins that mediate further digestion of the food vacuole contents. Alternatively, Stx released into the cytoplasm or contained within the food vacuole alters the trafficking of the Stx-containing bacteria so that they are less efficiently targeted to lysosomes.
Similar to our observation in Tetrahymena cells, bacteria bearing genes that encode Stx2 also survive better in human macrophages than do otherwise isogenic bacterial strains bearing mutations in these genes (50). Nonetheless, despite the Stx-mediated increase in intracellular bacterial survival, human monocytes have been found to be resistant (21–23) or, at best, weakly susceptible (50) to this toxin. Apparently, the failure of this bacterially expressed Stx to efficiently kill macrophages stems from the inability of this toxin to be exported into the cytoplasm and the subsequent delivery of the vesicle containing the toxin and bacteria to lysosomes (51, 52). Hence, although the structures and mechanism of phagocytosis and vesicular transport by these two cell types appear to be highly similar (19, 53), in Tetrahymena, the toxin produced by internalized bacteria can clearly be trafficked to the cytoplasm to exert its cytolethal effect. In sensitive mammalian cells, vesicles containing Stx bypass lysosomes and the toxin is brought to the cytoplasm via retrograde transport (54–56). It remains to be determined whether Stx produced by ingested Stx-encoding bacteria uses this same or a different pathway to reach the Tetrahymena cytoplasm.
MATERIALS AND METHODS
Strains and chemicals.
EDL933 was obtained from the ATCC. EDL933∆stx, an EDL933 variant containing deletions of both the stx1 and stx2 genes (30), was a gift from Christine Miller, Institut National de la Recherche Agronomique. E. coli strain YYC7 was obtained from the CGSC. Strain MG1655(recA) was created as described earlier (57). T. thermophila strains CU427.4 and NP1, a mouthless strain (27), were obtained from the Tetrahymena Stock Center (Cornell University).
Purified Stx2 toxin and anti-Stx antibodies were obtained from Toxin Technologies (Sarasota, FL) and BEI Resources (Manassas, VA). Bacterial strains EDL933:mCherry and EDL933∆stx:mCherry were made by transforming either strain with the pmCherry vector from Clontech (Mountain View, CA).
Purification of StxB.
StxB was purified from MG1655 transformed with pSU108, a plasmid that directs the overproduction of StxB, and was a gift from Ludger Johannes. The protein was purified as described previously (56). The purified protein was stored at −80°C in phosphate-buffered saline and supplemented with 10% glycerol.
Construction of MG1655::933W and MG1655::933WΔB.
The MG1655::933W lysogen was constructed as previously described (58), using wild-type 933W bacteriophage isolated by spontaneous induction of bacterial strain EDL933. A portion of the stxB gene from this strain was subcloned into plasmid pET17b. A DNA fragment containing the gene for chloramphenicol resistance (cat) was amplified from pACYC184 and inserted into the PflMI site of the stxB gene, and the recombinant plasmid, pStxΔB, was transformed into MG1655. The MG1655::933WΔB strain was created by P1 transduction (59) of strain MG1655::933W using a lysate from MG1655/pStxΔB and selecting for chloramphenicol resistance. Disruption of the stxB gene was confirmed by PCR analysis and sequencing of PCR products derived from the relevant region of the MG1655::933WΔB chromosome.
Construction of MG1655 (recA)::λcI857 and Stx variant strains.
Bacteriophage λcI857, which has a temperature sensitivity mutation in the cI protein that makes it unable to maintain lysogeny at temperatures above 33°C (60), was purified from lysogenized strain YYC7. YYC7 was grown overnight in Luria broth (LB) at 30°C. The culture was diluted 1:50 in LB and grown to an optical density at 600 nm of 0.6. The mid-log-phase culture of YYC7 was incubated at 42°C, inducing the phage and lysing the cells. A few drops of chloroform were added to the culture, the lysate was vortexed, and the debris was removed by centrifugation at 18,000 × g. The supernatant was spotted onto a lawn of MG1655 (recA) in top agar. Plaques were scraped and streaked onto LB agar plates. Individual colonies were tested for the ability to produce phage at 42°C. The resulting strain, MG1655 (recA)::λcI857, was transformed with either pStx (5) or pET17b (EMB Biosciences).
Protection from Stx2 killing by ricin B.
Stationary-phase cultures of T. thermophila (CU427.4) were diluted 5-fold in proteose peptone plus FeCl2 and grown for 3 days at 30°C. The cells were washed twice in 10 mM Tris-HCl (pH 7.4) and suspended in the same medium. Washed cells were added to M9 plus 0.08% Na-citrate at a density of 104/ml. These cells were incubated for 6 h at 30°C with 40 ng/ml Stx2 and 2 µg/ml BSA, StxB, or ricin B. As a control, Tetrahymena cells were also separately incubated with 40 ng/ml BSA. Tetrahymena cell density was determined (5) at the beginning and end of the incubation period. Tetrahymena cells killed by Stx are not visible by Lugol staining because they are lysed. Each measurement was performed in triplicate, and the data were averaged. The data shown represent the average of nine or more replicates.
Effect of Stx2 on Tetrahymena strain NP1.
Saturated cultures of T. thermophila strains NP1 and CU427.4 were diluted 5-fold in EPP medium (27) and grown for 3 days at 37°C. The cells were washed twice in 10 mM Tris-HCl (pH 7.4) and suspended in the same medium. Washed cells were added to M9 plus 0.08% Na-citrate at a density of 104/ml. These cells were incubated for 6 h at 37°C in the presence of 40 ng/ml BSA or Stx2. As a control for growth at 37°C, the experiment was replicated at 30°C. Tetrahymena cells were enumerated as described above. Each measurement was performed in triplicate, and the data were averaged. The data represent the average of nine or more replicate experiments.
Coculture of EDL933 with Tetrahymena strains NP1 and CU427.4.
T. thermophila strains NP1 and CU427.4 were prepared as described above, except that the washed, resuspended cells were added to medium containing bacterial cells that had been previously grown to saturation at 37°C in M9 plus 0.08% glucose, washed twice in M9 plus 0.08% Na-citrate, and suspended in the same medium. The cocultures containing 104 Tetrahymena cells/ml and 108 EDL933 bacterial cells/ml were incubated for 6 h at 37°C. As a control for growth at 37°C, the experiment was replicated at 30°C. Tetrahymena cells were enumerated as stated above. Each measurement was performed in triplicate, and the data represent the average of nine or more replicate experiments.
Bacterial survival within Tetrahymena food vacuoles.
T. thermophila CU427.4 and bacterial strains EDL933:mCherry and EDL933∆stx:mCherry were prepared as described above, except that the final Tetrahymena cell density was approximately 5 × 104/ml and bacterial cultures were supplemented with 100 µg/ml ampicillin. The bacterial cells produce mCherry, a red fluorescent protein with an excitation maximum at 587 nm and an emission maximum at 610 nm (61). To allow uptake of bacteria by Tetrahymena, cocultures containing Tetrahymena cells at approximately 5 × 104/ml and bacterial cells at 108/ml were incubated for 30 min at 30°C. After 30 min, the cocultures were washed free of bacteria by 3-fold dilution in 10 mM Tris-HCl (pH 7.4) and pelleted at 1,000 rpm for 5 min. Pelleted Tetrahymena cells containing bacteria were washed twice more and resuspended in 1 ml of 50% proteose peptone with FeCl2. Bacterium-containing cells were incubated at 30°C, and samples were taken for microscopic examination every 30 min for 2 h. Samples were prepared by adding sodium azide to a final concentration of 0.1% to kill the Tetrahymena cells. Samples were loaded onto slides and inspected at ×60 magnification using differential interference contrast (DIC) and a rhodamine red filter on a Zeiss Axio Imager Z1 microscope. Images were taken using a high-resolution AxioCam MRm digital camera. Average fluorescence was quantified by analyzing fluorescent images using AxioVision version 4.8.2 (manufactured by Zeiss MicroImaging GmbH).
Killing and StxB subunit competition from within Tetrahymena food vacuoles.
Tetrahymena strain CU427.4 and bacterial strains MG1655 (recA)::λcI857/pStx, MG1655 (recA)::λcI857/pet17, and MG1655 (recA)/pStx were prepared as described above, except that bacteria were grown at 30°C. After incubation at 30°C for 30 min, the Tetrahymena cells were washed free of uninternalized bacteria as described above, except that they were resuspended in a final volume of 4 ml of M9 plus 0.08% Na-citrate. The Tetrahymena cells containing internalized bacteria were then incubated at 42°C for 6 h. MG1655 (recA)::λcI857/pet17 acts as the toxin negative control, while MG1655 (recA)/pStx acts as the phage negative control. To determine if StxB affects bacterially mediated killing of Tetrahymena, cocultures were supplemented with 2 µg/ml BSA or StxB. Tetrahymena cells were enumerated as described above. Each measurement was performed in triplicate, and the data were averaged. The data represent the average of nine or more replicate experiments.
Effect of Stx2 on Tetrahymena protein synthesis.
Tetrahymena cells were prepared as described above, except that their final density was approximately 5 × 104/ml. The Tetrahymena cells were subsequently incubated with 200 ng/ml Stx2 for 60 min. Subsequently, the Tetrahymena cells were washed twice in M9 plus 0.08% glucose and 2 µCi of [35S]methionine was added to 100 µl of Tetrahymena cells and incubated for an additional 15 min at 25°C. Next, 10 µl of 50 mM Casamino acids was added to the mixture and the mixture was incubated for an additional 5 min at 25°C. Cells were lysed by adding 50 µl of Tetrahymena lysis buffer (62) and incubating the mixture for 30 min on ice. Cellular debris was removed by centrifugation at 18,000 × g for 15 min. Total protein in the supernatant was precipitated by incubation with 12.5% trichloroacetic acid on ice for 10 min. Precipitate was collected by vacuum filtration through a glass fiber filter using a vacuum manifold and washed three times with 1 ml acetone. The total amount of labeled protein synthesized was measured by counting with a scintillation counter.
Statistical methods.
Except where indicated, the error bars shown in the figures are standard deviations of multiple (more than nine) replicate experiments. Student two-tailed, two-sample, equal-variance t tests were used to determine the significance of differences in the measured data.
ACKNOWLEDGMENT
This work was supported by a grant from the National Science Foundation (MCB-0956454) to G.B.K.
Footnotes
Citation Stolfa G, Koudelka GB. 2012. Entry and killing of Tetrahymena thermophila by bacterially produced Shiga toxin. mBio 4(1):e00416-12. doi:10.1128/mBio.00416-12.
REFERENCES
- 1. Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95:6578–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wommack KE, Colwell RR. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brüssow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Casas V, et al. 2006. Widespread occurrence of phage-encoded exotoxin genes in terrestrial and aquatic environments in southern California. FEMS Microbiol. Lett. 261:141–149 [DOI] [PubMed] [Google Scholar]
- 5. Lainhart W, Stolfa G, Koudelka GB. 2009. Shiga toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila. J. Bacteriol. 191:5116–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herold S, Karch H, Schmidt H. 2004. Shiga toxin-encoding bacteriophages—genomes in motion. Int. J. Med. Microbiol. 294:115–121 [DOI] [PubMed] [Google Scholar]
- 7. O’Brien AD, et al. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694–696 [DOI] [PubMed] [Google Scholar]
- 8. Sandvig K. 2001. Shiga toxins. Toxicon 39:1629–1635 [DOI] [PubMed] [Google Scholar]
- 9. Melton-Celsa AR, Rogers JE, Schmitt CK, Darnell SC, O’Brien AD. 1998. Virulence of Shiga toxin-producing Escherichia coli (STEC) in orally-infected mice correlates with the type of toxin produced by the infecting strain. Jpn. J. Med. Sci. Biol. 51(Suppl.):S108–S114 [DOI] [PubMed] [Google Scholar]
- 10. Schmidt H. 2001. Shiga-toxin-converting bacteriophages. Res. Microbiol. 152:687–695 [DOI] [PubMed] [Google Scholar]
- 11. Schmitt CK, McKee ML, O’Brien AD. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H- strain E32511. Infect. Immun. 59:1065–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ling H, et al. 2000. A mutant Shiga-like toxin IIe bound to its receptor Gb(3): structure of a group II Shiga-like toxin with altered binding specificity. Structure 8:253–264 [DOI] [PubMed] [Google Scholar]
- 13. Sandvig K, et al. 2002. Pathways followed by ricin and Shiga toxin into cells. Histochem. Cell Biol. 117:131–141 [DOI] [PubMed] [Google Scholar]
- 14. Johannes L, Decaudin D. 2005. Protein toxins: intracellular trafficking for targeted therapy. Gene Ther. 12:1360–1368 [DOI] [PubMed] [Google Scholar]
- 15. Sandvig K, van Deurs B. 2005. Delivery into cells: lessons learned from plant and bacterial toxins. Gene Ther. 12:865–872 [DOI] [PubMed] [Google Scholar]
- 16. Yu M, Haslam DB. 2005. Shiga toxin is transported from the endoplasmic reticulum following interaction with the luminal chaperone HEDJ/ERdj3. Infect. Immun. 73:2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Engberg J, et al. 1990. Comparison of primary and secondary 26S rRNA structures in two Tetrahymena species: evidence for a strong evolutionary and structural constraint in expansion segments. J. Mol. Evol. 30:514–521 [DOI] [PubMed] [Google Scholar]
- 18. Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334:941–948 [DOI] [PubMed] [Google Scholar]
- 19. Elde NC, Morgan G, Winey M, Sperling L, Turkewitz AP. 2005. Elucidation of clathrin-mediated endocytosis in Tetrahymena reveals an evolutionarily convergent recruitment of dynamin. PLoS Genet. 1:e52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bright LJ, Kambesis N, Nelson SB, Jeong B, Turkewitz AP. 2010. Comprehensive analysis reveals Dynamic and evolutionary plasticity of Rab GTPases and membrane traffic in Tetrahymena thermophila. PLoS Genet. 6:e1001155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramegowda B, Tesh VL. 1996. Differentiation-associated toxin receptor modulation, cytokine production, and sensitivity to Shiga-like toxins in human monocytes and monocytic cell lines. Infect. Immun. 64:1173–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tesh VL, Ramegowda B, Samuel JE. 1994. Purified Shiga-like toxins induce expression of proinflammatory cytokines from murine peritoneal macrophages. Infect. Immun. 62:5085–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Setten PA, Monnens LA, Verstraten RG, van den Heuvel LP, van Hinsbergh VW. 1996. Effects of verocytotoxin-1 on nonadherent human monocytes: binding characteristics, protein synthesis, and induction of cytokine release. Blood 88:174–183 [PubMed] [Google Scholar]
- 24. Gourabathini P, Brandl MT, Redding KS, Gunderson JH, Berk SG. 2008. Interactions between food-borne pathogens and protozoa isolated from lettuce and spinach. Appl. Environ. Microbiol. 74:2518–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steinberg KM, Levin BR. 2007. Grazing protozoa and the evolution of the Escherichia coli O157:H7 Shiga toxin-encoding prophage. Proc. Biol. Sci. 274:1921–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skinner LM, Jackson MP. 1998. Inhibition of prokaryotic translation by the Shiga toxin enzymatic subunit. Microb. Pathog. 24:117–122 [DOI] [PubMed] [Google Scholar]
- 27. Orias E, Rasmussen L. 1976. Dual capacity for nutrient uptake in Tetrahymena: IV. Growth without food vacuoles and its implications. Exp. Cell Res. 102:127–137 [DOI] [PubMed] [Google Scholar]
- 28. Wales R, Richardson PT, Roberts LM, Woodland HR, Lord JM. 1991. Mutational analysis of the galactose binding ability of recombinant ricin B chain. J. Biol. Chem. 266:19172–19179 [PubMed] [Google Scholar]
- 29. Wagner PL, et al. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44:957–970 [DOI] [PubMed] [Google Scholar]
- 30. Gobert AP, et al. 2007. Shiga toxin produced by enterohemorrhagic Escherichia coli inhibits PI3K/NF-kappaB signaling pathway in globotriaosylceramide-3-negative human intestinal epithelial cells. J. Immunol. 178:8168–8174 [DOI] [PubMed] [Google Scholar]
- 31. Görtz HD, et al. 1999. Intra- and intercellular communication systems in ciliates. Naturwissenschaften 86:422–434 [DOI] [PubMed] [Google Scholar]
- 32. Tyler JS, Mills MJ, Friedman DI. 2004. The operator and early promoter region of the Shiga toxin type 2-encoding bacteriophage 933W and control of toxin expression. J. Bacteriol. 186:7670–7679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Livny J, Friedman DI. 2004. Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Mol. Microbiol. 51:1691–1704 [DOI] [PubMed] [Google Scholar]
- 34. Palmer E, Wilhelm JM. 1978. Mistranslation in a eukaryotic organism. Cell 13:329–334 [DOI] [PubMed] [Google Scholar]
- 35. Dacks JB, Field MC. 2007. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell Sci. 120(Pt. 17):2977–2985 [DOI] [PubMed] [Google Scholar]
- 36. Nusblat AD, Bright LJ, Turkewitz AP. 2012. Conservation and innovation in Tetrahymena membrane traffic: proteins, lipids, and compartments. Methods Cell Biol. 109:141–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Turkewitz AP, Bright LJ. 2011. A Rab-based view of membrane traffic in the ciliate Tetrahymena thermophila. Small GTPases 2:222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramotar K, et al. 1990. Characterization of Shiga-like toxin I-B subunit purified from overproducing clones of the SLT-I B cistron. Biochem. J. 272:805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Csaba G, Kovács P, Pállinger E. 2008. Comparison of the insulin binding, uptake and endogenous insulin content in long- and short-term starvation in Tetrahymena. Cell Biochem. Funct. 26:64–69 [DOI] [PubMed] [Google Scholar]
- 40. Csaba G, Kovács P, Pállinger E. 2008. Effect of femtomolar concentrations of hormones on insulin binding by Tetrahymena, as a function of time. Cell Biochem. Funct. 26:205–209 [DOI] [PubMed] [Google Scholar]
- 41. Lampert TJ, Coleman KD, Hennessey TM. 2011. A knockout mutation of a constitutive GPCR in Tetrahymena decreases both G-protein activity and chemoattraction. PLoS One 6:e28022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Christensen ST, Guerra CF, Awan A, Wheatley DN, Satir P. 2003. Insulin receptor-like proteins in Tetrahymena thermophila ciliary membranes. Curr. Biol. 13:R50–R52 [DOI] [PubMed] [Google Scholar]
- 43. Coyne RS, et al. 2008. Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genomics 9:562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eisen JA, et al. 2006. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4:e286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baumert U, Vosskühler C, Tiedtke A. 1998. Formation and maturation of phagosomes labeled by magnetic micro particles: an ultrastructural study in Tetrahymena thermophila. Eur. J. Protistol. 34:291–300 [Google Scholar]
- 46. Nilsson JR. 1987. Structural aspects of digestion of Escherichia coli in Tetrahymena. J. Eukaryot. Microbiol. 34:1–6 [Google Scholar]
- 47. Johansen BK, Wasteson Y, Granum PE, Brynestad S. 2001. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology 147(Pt. 7):1929–1936 [DOI] [PubMed] [Google Scholar]
- 48. Wagner PL, Acheson DW, Waldor MK. 1999. Isogenic lysogens of diverse Shiga toxin 2-encoding bacteriophages produce markedly different amounts of Shiga toxin. Infect. Immun. 67:6710–6714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Joshi AD, Swanson MS. 2011. Secrets of a successful pathogen: Legionella resistance to progression along the autophagic pathway. Front. Microbiol. 2:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Poirier K, et al. 2008. Escherichia coli O157:H7 survives within human macrophages: global gene expression profile and involvement of the Shiga toxins. Infect. Immun. 76:4814–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dacosta B, Sansonetti P, Ryter A. 1990. Immunolabelling of Shiga toxin in macrophages infected with Shigella dysenteriae 1. Res. Microbiol. 141:543–549 [DOI] [PubMed] [Google Scholar]
- 52. Falguières T, et al. 2001. Targeting of Shiga toxin B-subunit to retrograde transport route in association with detergent-resistant membranes. Mol. Biol. Cell 12:2453–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Przyborowski L, Smajkiewicz A. 1975. Colorimetric determination of Bromisoval and carbromal. Pol. J. Pharmacol. Pharm. 27:645–648 [PubMed] [Google Scholar]
- 54. Johannes L, Tenza D, Antony C, Goud B. 1997. Retrograde transport of KDEL-bearing B-fragment of Shiga toxin. J. Biol. Chem. 272:19554–19561 [DOI] [PubMed] [Google Scholar]
- 55. Mallard F, et al. 1998. Direct pathway from early/recycling endosomes to the Golgi apparatus revealed through the study of Shiga toxin B-fragment transport. J. Cell Biol. 143:973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mallard F, Johannes L. 2003. Shiga toxin B-subunit as a tool to study retrograde transport. Methods Mol. Med. 73:209–220 [DOI] [PubMed] [Google Scholar]
- 57. Shkilnyj P, Koudelka GB. 2007. Effect of salt shock on the stability of λimm434 lysogens. J. Bacteriol. 198:3115–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arber W, et al. 1983. Lambda vol II, p 433–466 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 59. Winans SC, Elledge SJ, Krueger JH, Walker GC. 1985. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J. Bacteriol. 161:1219–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chang YY, Cronan JE., Jr 1982. Mapping nonselectable genes of Escherichia coli by using transposon Tn10: location of a gene affecting pyruvate oxidase. J. Bacteriol. 151:1279–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shaner NC, et al. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 [DOI] [PubMed] [Google Scholar]
- 62. Bolivar I, Guiard-Maffia J. 1989. Cellular localization of the SerH surface antigen in Tetrahymena thermophila. J. Cell Sci. 94:343–354 [DOI] [PubMed] [Google Scholar]