ABSTRACT
The interaction of Candida albicans with macrophages induces the production of interleukin 1β (IL-1β) through inflammasome activation in a process that is required for host survival. C. albicans hypha formation has been linked to IL-1β production, but the question of whether hyphae are sufficient to trigger IL-1β production has not been examined directly. To address this question, a C. albicans library of 165 transcription factor deletion mutants was screened for strains with altered IL-1β production by lipopolysaccharide (LPS)-primed J774 cells, a murine macrophage-like cell line. Eight mutants with decreased and two mutants with increased IL-1β secretion were identified. In addition, 12 mutants with previously identified morphology deficits were found to induce IL-1β secretion to levels similar to those of the wild type. Examination of the morphology of both low and normal IL-1β-inducing mutants in macrophages revealed that two mutants (upc2Δ/upc2Δ and ahr1Δ/Δ mutants) were indistinguishable from the wild type with respect to morphology yet induced low levels of IL-1β; conversely, the ndt80Δ/Δ mutant was deficient for hypha formation but induced levels of IL-1β similar to those of the wild type. Transcription factor mutants deficient for IL-1β secretion also caused markedly lower levels of macrophage lysis. Similarly, the ability of a mutant to cause macrophage lysis was independent of its ability to form hyphae. Taken together, our observations indicate that the physical formation of hyphae is not sufficient to trigger IL-1β secretion or macrophage lysis and suggest that other mechanisms, such as pyroptosis, a caspase-1-dependent response to intracellular pathogens, may play a role in the interaction of macrophages with C. albicans.
IMPORTANCE
The ability of Candida albicans to transition from yeast to filamentous cells plays an important and complex role in pathogenesis. Recent results from a number of investigators indicate that the host responds to yeast and hyphal C. albicans differently. For example, a C. albicans mutant unable to form hyphae also fails to induce interleukin 1β (IL-1β) secretion from macrophages. We have identified C. albicans transcription factor mutants that have decreased IL-1β secretion but retain the ability to form hyphae in response to macrophages. In addition, these mutants cause significantly less macrophage lysis. These observations indicate that the physical presence of the hyphal structure in the macrophage is not sufficient to trigger IL-1β secretion nor does it cause physical lysis of the cell. Our data indicate that characteristics of hyphae separate from its physical morphology are responsible for triggering the release of IL-1β release and causing macrophage lysis. Since these observations are inconsistent with some current models, alternative mechanisms for the interaction of C. albicans with macrophages must be considered.
Observation
Candida albicans is both an important component of the human microflora and a common cause of human fungal disease, particularly in people with altered immune function. C. albicans infections manifest as either superficial mucosal diseases such as vaginitis and oropharyngeal mucositis or invasive diseases involving the bloodstream, kidneys, and other organ systems (1). The mechanisms that mediate the transition of C. albicans from a harmless commensal organism to a potentially lethal pathogen are the subject of intense current interest with relevance not only to fungal pathogenesis but also to the pathogenesis of opportunistic infections in general. A homeostatic relationship exists between C. albicans and the human host defense system (2). When organisms begin to invade tissue, the host must mount an early and effective immune response to clear the organism and maintain C. albicans as a commensal. For C. albicans to transition from a commensal to a pathogen, the host response must be insufficient and/or the organism must alter its physiology to adapt to its new niche.
An important change in C. albicans that is associated with pathogenesis is its ability to transition from an ovoid yeast form to a filamentous form (hyphae and pseudohyphae) (3). Although C. albicans mutants that are unable to undergo normal morphogenesis are frequently avirulent, the role of morphogenesis in C. albicans pathogenesis is complex and remains incompletely understood (4). One aspect of morphogenesis that has been the subject of intense interest is its role in host recognition. A number of studies have shown that macrophages, dendritic cells, and epithelial cells recognize yeast and filamentous C. albicans differently and, accordingly, trigger distinct inflammatory responses (2). For example, recent reports suggest that yeast cells do not trigger the production of interleukin 1β (IL-1β) by phagocytes, whereas cells that form filaments induce IL-1β production through inflammasome activation (5–7). The importance of IL-1β signaling in protecting the mammalian host from invasive C. albicans infection has been clearly demonstrated by the fact that mice deficient in IL-1β had decreased survival and increased fungal burdens relative to wild-type mice (7, 8).
The production of IL-1β is regulated by a two-step process: first, an NF-κB-dependent pathway triggers transcription of the pro-IL-1β gene; second, pro-IL-1β is processed by caspase-1 to produce mature IL-1β (9). In monocytes and macrophages, transcription of pro-IL-1β mRNA appears to be induced by the interaction of C. albicans with pathogen recognition receptors such as Toll-like receptor 2, dectin-1, and mannose receptor (7, 10). With respect to the second step, a number of labs have reported that, in response to C. albicans, processing of pro-IL-1β by caspase-1 is mediated by the NLRP3 (Nod-like receptor family, pyrin domain containing 3) inflammasome (5, 7, 11). Furthermore, the NLRP3 inflammasome appears to be required to prevent the systemic dissemination of mucosal infection (7). In contrast, one report has suggested that processing of pro-IL-1β is carried out by constitutively active caspase-1 (10), thus bypassing the need for inflammasome activation. Recently, it has also been reported that activation of inflammasomes by the noncanonical caspases-8 and 11 (also known as caspase-4) occurs in response to fungi, Gram-negative bacteria, and mycobacteria (12).
Three groups have reported experiments indicating that C. albicans activation of the NLRP3 inflammasome and subsequent production of mature IL-1β in macrophages is dependent on the ability of the organism to undergo the morphological transition from the yeast form to filaments after ingestion by the macrophage (5–7). C. albicans organisms lacking both EFG1 and CPH1, two partially redundant transcriptional regulators required for hyphal morphogenesis, are locked in the yeast form and fail to induce IL-1β production. Interestingly, fully formed hyphal elements are also inefficient inducers of IL-1β. Joly et al. and Lewis et al. have speculated that the latter observation can be explained by the finding that the hyphal C. albicans form is relatively resistant to phagocytosis, which may be required for initiation of the second signal (5, 13). Recently, proteins and lipids secreted by C. albicans have been shown to contribute to IL-1β production. Specifically, the addition of purified secreted aspartyl proteases Sap1, Sap2, Sap3, or Sap6 to human monocytes induced IL-1β secretion (14). Interestingly, Sap proteins associated with yeast (Sap2) and hyphae (Sap3) induced IL-1β (14). In addition, C. albicans mutants lacking farnesol synthesis genes induced lower levels of IL-1β mRNA expression than the wild type did (15). These observations suggest that factors other than morphogenesis may contribute to IL-1β production.
Based on these considerations, an important, but unresolved, question is whether the physical presence of C. albicans hyphal structures is sufficient to trigger IL-1β production. Prior to our work, the conclusion that hyphae were required for IL-1β production by macrophages was based on studies of a single afilamentous mutant, the efg1Δ/efg1Δ cph1Δ/Δ mutant. To more fully explore the role of morphogenesis in IL-1β production, we screened a publically available collection of 165 C. albicans transcription factor deletion mutants (16) for those that altered IL-1β production in lipopolysaccharide (LPS)-primed J774 cells, a well-characterized mouse macrophage-like cell line. We used macrophages that were primed with LPS to exogenously trigger the first signal, a technique that is commonly used to focus on signals that trigger inflammasome activation (5, 7). The strains in the mutant collection had been previously screened for morphology defects in vitro; therefore, we could correlate IL-1β production with altered morphology.
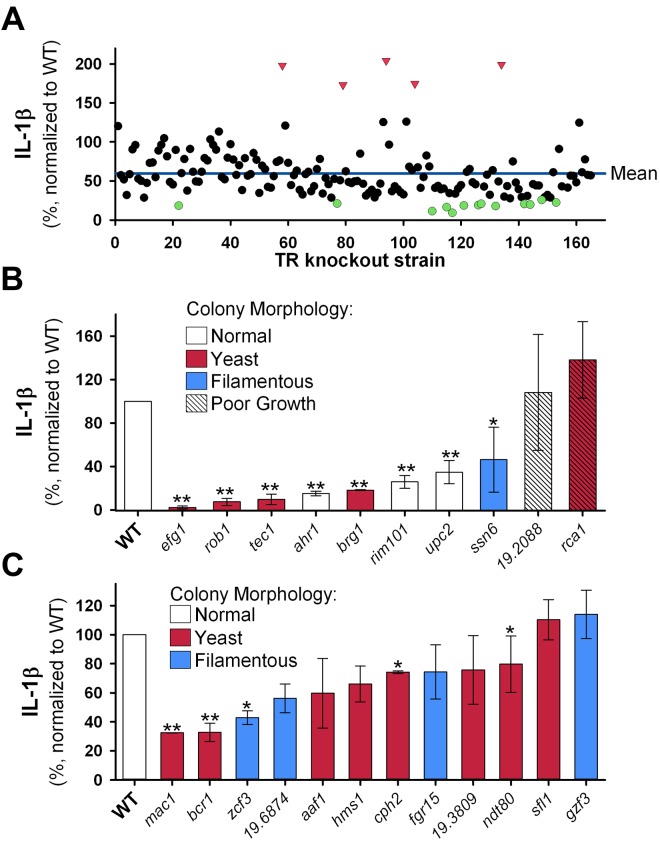
As shown in Fig. 1A, the majority (92%) of the mutants triggered lower IL-1β production than did the congenic reference strain (SN250 [4]; for simplicity, we will refer to this strain as the “wild type”). The overall mean IL-1β level for all of the mutants was 59.4% (normalized to the amount induced by the wild-type strain). Therefore, we defined a mutant as triggering “low” IL-1β production if it induced IL-1β concentrations that were at least 1 standard deviation below the mean for the mutant collection; this corresponded to a cutoff value of 25.8%. Because the mean IL-1β production for the collection of mutants was substantially below that of the wild-type strain, an IL-1β level that was 1 standard deviation above the mean was actually below the wild-type level. Thus, we defined mutants as “high” inducers of IL-1β production if they induced IL-1β levels greater than or equal to 2 standard deviations above the mean (126.6%).
FIG 1.
Production of IL-1β in response to Candida albicans transcription factor mutants does not correlate with morphology. A library containing 165 C. albicans transcription factor deletion mutants was individually screened for their ability to trigger IL-β production. Each mutant was grown overnight in yeast extract-peptone-dextrose (YPD) medium, washed, counted using a Countess (Invitrogen/ Life Technologies, Carlsbad, CA) automated hemocytometer, and diluted in phosphate-buffered saline (PBS) to a uniform organism density. J774 cells were primed by culture for 2 h in medium containing 50 ng/ml lipopolysaccharide (LPS) (E. coli O111:B4; Calbiochem/EMD Millipore, Billerica, MA), after which the C. albicans inoculum was added to microtiter plate wells at a multiplicity of infection of 2:1. After coculture for 5 h, the supernatants were removed, and the concentration of IL-1β was determined by enzyme-linked immunosorbent assay (ELISA) (eBiosciences, San Diego, CA). To allow comparison of IL-1β levels from different assays, the wild-type (WT) strain was included in each assay and was used to normalize results. (A) IL-1β induction for each mutant is plotted as a percentage of the level of the wild-type strain. The values for mutants that triggered an IL-1β level greater than 2 standard deviations (SDs) above the mean (red triangles) and the values for mutants that triggered an IL-1β level less than 1 SD below the mean (green circles) are indicated. (B) All mutants that induced “low” or “high” levels of IL-1β were retested. The mean level of IL-1β induced by each mutant over all subsequent assays is represented by the height of the bar; error bars indicate the standard deviations (n ≥ 3; range, 3 to 8). The color of the bar represents the colony morphology on Spider medium or medium containing serum, as reported in Homann et al. (13). Each mutant was grown and assayed independently; therefore, one-sample Student’s t tests were used to compare each strain to the WT. Values that are significantly different from the value for the WT are indicated by asterisks as follows: *, P < 0.05; **, P < 0.001. (C) A subset of mutants that triggered normal levels of IL-1β and that were reported to have abnormal colony morphology were retested. The results are presented and analyzed as for panel B, but with n ≥ 2 and n ranging from 2 to 7.
Of the 14 strains identified in the initial screen as having decreased IL-1β levels, 8 were confirmed to induce <25% of wild-type IL-1β production on repeat testing (Fig. 1B). Importantly, the efg1Δ/Δ mutant, which is related to the mutant previously shown by Joly et al. (5) to induce very low IL-1β production, was identified as triggering low IL-1β production, thus validating our screening assay. In the primary screen, 5 mutants induced increased IL-1β production, 2 of which, the rca1Δ/Δ mutant and the 19.2088Δ/Δ mutant, were initially confirmed on retesting. We noticed significant assay-to-assay variability for the ssn6Δ/Δ, rca1Δ/Δ, and 19.2088Δ/Δ mutants (Fig. 1B). All three mutants grow as aggregates of yeast or pseudohyphae in liquid YPD (yeast extract-peptone-dextrose) culture, making it difficult to accurately adjust the density of the fungal inoculum. Because multiplicity of infection significantly affects the IL-1β production, we excluded these three strains from further analysis.
The phenotypic characteristics, including morphogenesis, of the strains in the transcription factor deletion mutant collection were comprehensively analyzed in vitro by Homann et al. (16). One of the 7 mutants identified as inducing low levels of IL-1β, the upc2Δ/Δ mutant, had normal morphology in all of the morphological assays used by Homann et al. (16). Two additional mutants, the ahr1Δ/Δ and rim101Δ/Δ mutants, had normal morphogenesis on both Spider medium at 37°C and serum-containing medium (two of the most commonly used hypha-inducing conditions) but had abnormal colony morphology on media that are infrequently used to study filamentation (16). Thus, a significant portion of the mutants that were unable to trigger normal IL-1β production are predicted to have normal morphogenesis on the basis of colony morphology.
Because we also found a number of mutants with altered colony morphology that induced levels of IL-1β similar to the level induced by the wild type, we retested a set of these mutants; on retesting, 12/14 mutants induced levels of IL-1β that were “normal” according to our screening thresholds (Fig. 1C). The two strains that did not have normal IL-1β levels on repeat testing were strains that had abnormal morphology in liquid culture in YPD and thus were difficult to accurately adjust for organism density. Of the 12 strains that had “normal” levels of IL-1β on retesting, five strains, the mac1Δ/Δ, bcr1Δ/Δ, zcf3Δ/Δ, cph2Δ/Δ, and ndt80Δ/Δ mutants, induced levels of IL-1β that were significantly different from those induced by the wild type on statistical analysis. Two of these strains, the cph2Δ/Δ and ndt80Δ/Δ mutants, induced levels of IL-1β that differ only slightly from the levels induced by the wild type (74% and 80% of wild type, respectively). In contrast, the mac1Δ/Δ, bcr1Δ/Δ, and zcf3Δ/Δ mutants induced IL-1β levels that were 2- to 3-fold lower than those induced by the wild-type strain. The fact that these mutants were not designated “low” inducers of IL-1β in our initial screen relates to our screening strategy. We used a stringent cutoff in order to minimize detection of mutants that induced relatively normal levels of IL-1β. On the basis of these data, we estimate that the screening strategy has a false-negative rate of up to 25%. However, our goal was not to identify the entire set of mutants that induce low levels of IL-1β but to correlate morphology with IL-1β induction.
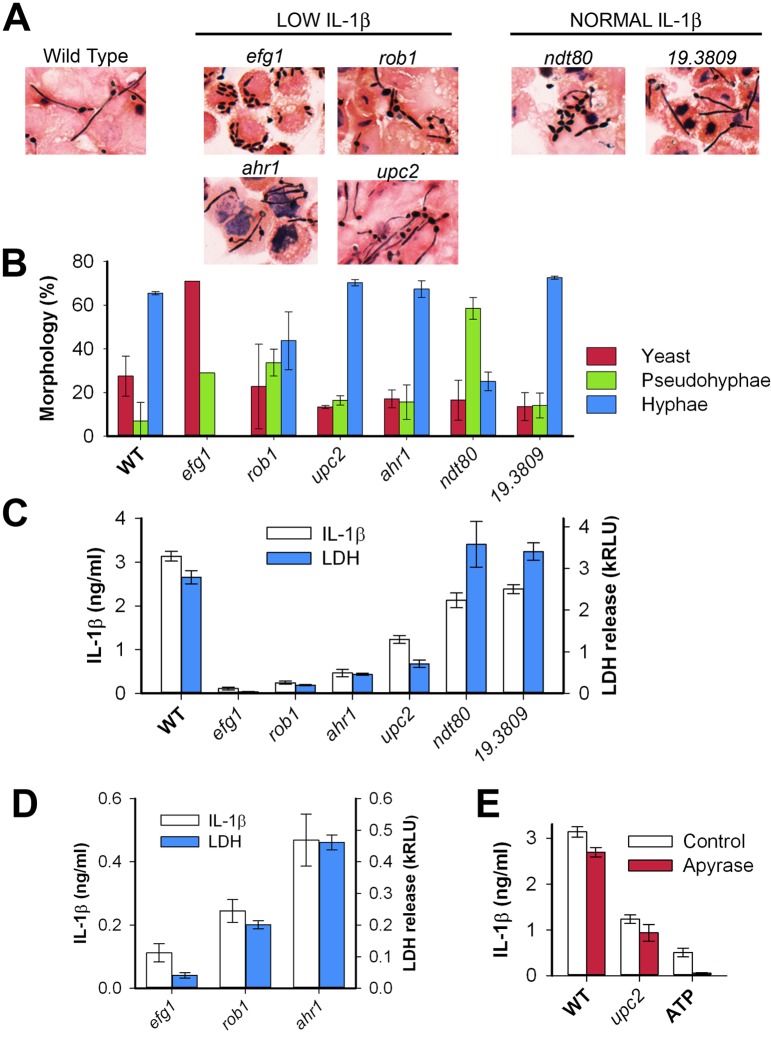
Our data suggest that the ability of C. albicans to undergo filamentation does not correlate with its ability to induce IL-1β production. To further investigate the relationship between morphogenesis and IL-1β production, we examined the ability of the mutants in Fig. 1 to undergo morphogenesis under the tissue culture conditions used for these assays (liquid medium, 37°C, 5% CO2, 1% serum). In liquid tissue culture medium at 37°C and 5% CO2, 18/19 mutants formed filaments within 2 h; the efg1Δ/Δ mutant was the only one that failed to form any filaments (data not shown). Fungal morphogenesis is highly condition dependent, and therefore, we cocultured all of the mutants that were low inducers of IL-1β and a set of normal/high inducing mutants with J774 cells and then examined the C. albicans morphology by bright-field microscopy of both live cultures (data not shown) and fixed, Gram-stained samples. Photomicrographs of six representative mutants (4 low inducing and 2 normal inducing) are presented in Fig. 2A. We then quantified the production of yeast, pseudohyphae, and hyphae by using standard criteria (Fig. 2B). The majority of wild-type cells form hyphae when exposed to macrophages, whereas efg1Δ/Δ cells form essentially no hyphae. Other than the efg1Δ/Δ mutant, all mutants tested were capable of forming filaments that were at least as long as the width of a typical host cell within 2 h of inoculation.
FIG 2.
Macrophage production of IL-1β in response to Candida albicans correlates with macrophage lysis independent of organism morphology. The morphology of the transcription factor mutants within LPS-primed macrophages was analyzed by coculturing C. albicans mutants with J774 macrophages in 8-well chamber slides (Millipore), using the conditions described in the legend to Fig. 1, except that the coculture time was 2 h. After coculture, the wells were washed with water to remove unbound organisms, the chambers were removed, and the slides were Gram stained (Harleco/EMD Millipore). (A) Photomicrographs of representative fields for the indicated mutants are presented. (B) The distribution of morphotypes for each mutant was determined by light microscopy of Gram-stained slides (n = 100 per replicate). The color of the bar indicates the morphotype. The height of the bar represents the mean result; error bars indicate the standard deviations for two replicates. (C) The ability of mutants to induce IL-1β was correlated with their ability to lyse macrophages by coculturing C. albicans mutants with J774 macrophages for 5 h, as in Fig. 1, and dividing the supernatant into two samples. One sample was used to measure IL-1β (as in Fig. 1); the other sample was used to measure macrophage lysis, as indicated by the release of lactate dehydrogenase (LDH) (CytoTox-ONE homogeneous membrane integrity assay; Promega Corp., Madison, WI). The height of the bar represents the mean level of IL-1β or LDH from a representative assay, while error bars indicate the standard deviation (n = 3). Each strain was tested in at least three assays (range, 3 to 7 assays). kRLU, thousands of relative light units. (D) Rescaled plot of IL-1β production and LDH release data from panel C for efg1, rob1, and ahr1 mutants to illustrate correlation between IL-1β production and LDH release. (E) The effect of extracellular ATP on IL-1β production was analyzed by performing macrophage-C. albicans cocultures, as in Fig. 1, except that either apyrase or PBS was added immediately prior to inoculation to a final concentration of 2.5 U/ml (Sigma-Aldrich, St. Louis, MO). For a control for apyrase activity, ATP was added to LPS-primed macrophages (without exposure to C. albicans) to a final concentration of 1 mM. The height of the bar indicates the mean level of LDH release in a representative assay; the error bars indicate the standard deviations (n = 3). Three independent assays produced equivalent results.
Examination of the data in Fig. 2B reveals a striking set of comparisons. The formation of hyphae upon exposure to macrophages is identical to that of the wild type for the ahr1Δ/Δ, upc2Δ/Δ, and 19.3809Δ/Δ mutants, yet the arh1Δ/Δ and upc2Δ/Δ mutants induce low levels of IL-1β, while the 19.3809Δ/Δ mutant is similar to the wild type (Fig. 1B and 2C). In contrast, the distribution of morphotypes for the ndt80Δ/Δ mutant is markedly altered relative to the wild type, but it induces IL-1β levels that are within 20% of the level induced by the wild type. Furthermore, the rob1Δ/Δ mutant, which like the ndt80Δ/Δ mutant has an increased ratio of pseudohyphal cells relative to hyphal cells, stimulates 13-fold-lower IL-1β than the wild-type strain does. Of all the mutants we have tested, only the efg1Δ/Δ mutant is completely unable to form hyphae on exposure to macrophages. Thus, it is possible that formation of some level of hyphae is still necessary for IL-1β formation. However, our data strongly suggest that the simple physical presence of hyphae is not sufficient for IL-1β production.
Because we found that IL-1β production does not appear to correlate with morphogenesis, we examined the effect of the low-IL-1β-inducing mutants on other measures of macrophage-C. albicans interaction. Using previously reported methods (17), we did not observe any differences in phagocyte uptake among the mutants shown in Fig. 2B (data not shown), suggesting that differences in phagocytosis of the mutants is unlikely to explain the variation in IL-1β production. C. albicans is thought to evade macrophage responses by undergoing filamentation within the phagolysosome which, in turn, leads to physical rupture of the macrophage (18). Therefore, we tested whether the low-IL-1β-inducing mutants caused less phagocyte lysis using a standard lactate dehydrogenase (LDH) cytotoxicity/lysis assay. As shown in Fig. 2C, wild-type cells induced substantial LDH release, whereas the low-IL-1β-inducing mutants caused dramatically lower levels of LDH release. Importantly, despite the fact that the upc2Δ/Δ and ahr1Δ/Δ mutants form morphologically normal filaments within macrophages, they cause approximately 5-fold-less damage to phagocytes. In contrast to the strains that induce low levels of IL-1β production, the ndt80Δ/Δ mutant, which induces normal IL-1β production, is deficient in filament formation yet causes LDH release essentially identical to that of the wild type. Furthermore, the rob1Δ/Δ mutant, which displays a morphotype profile similar to that of the ndt80Δ/Δ mutant, induces low levels of LDH release. As with IL-1β production, the ability of a mutant to trigger LDH release/macrophage lysis appears to be disconnected from its ability to undergo normal morphogenesis. This is consistent with the findings of McKenzie et al., who found that C. albicans strains deficient in cell surface glycosylation are able to form hyphae within macrophages but also fail to cause macrophage lysis (17). Thus, we have found that C. albicans morphology does not correlate with LDH release, nor does it predict C. albicans-induced macrophage IL-1β production. Furthermore, LDH release correlates well with IL-1β production as highlighted by comparing the efg1Δ/Δ, rob1Δ/Δ, and ahr1Δ/Δ mutants (Fig. 2D). These findings are inconsistent with the standard model for hypha-induced damage of macrophages and suggest that other mechanisms mediate C. albicans damage to phagocytes.
Because IL-1β production can be triggered by extracellular ATP (19), one explanation for our observations is that macrophage lysis results in ATP release, which, in turn, activates IL-1β production in the remaining host cells through a paracrine-type mechanism. To test this possibility, we assayed IL-1β production and macrophage damage in response to the mutant strains in the presence and absence of apyrase, an enzyme that degrades extracellular ATP (19). We found that apyrase blocks IL-1β production (Fig. 2D) and cell toxicity (data not shown) triggered by exogenously added ATP, but it does not affect production of IL-1β or cell lysis in response to either wild-type or mutant C. albicans. Thus, the production of IL-1β in response to these strains is not due to the release of extracellular ATP.
We have found that mutants with virtually identical filamentation patterns, such as the ndt80Δ/Δ and rob1Δ/Δ mutants, or the wild type and ahr1Δ/Δ and upc2Δ/Δ mutants, have drastically different levels of IL-1β production and macrophage lysis. Interestingly, there is a striking overlap between the mutants that we identified as inducing low IL-1β/macrophage lysis and the recently described transcriptional regulatory network that controls biofilm formation (20). In their description of this network, Nobile et al. (20) identified six transcription factors that function as master regulators of biofilm formation, EFG1, BRG1, TEC1, ROB1, BCR1, and NDT80. Of these six regulators, the efg1Δ/Δ, brg1Δ/Δ, tec1Δ/Δ, and rob1Δ/Δ mutants were identified in our original screen as triggering low IL-1β production. The bcr1Δ/Δ strain failed to meet our screening cutoff for designation as “low” IL-1β inducing (≤25.8%), however, our screening strategy was designed to exclude strains that trigger normal IL-1β production, not to identify all strains with low production. In fact, the bcr1Δ/Δ screening result was 27.6%, or 3.6-fold-lower than that of the wild type. Therefore, we included the bcr1Δ/Δ strain in subsequent assays and found that it had significantly lower levels of IL-1β production than the wild-type strain (32.7%; P < 0.001 by one-sample t test). Thus, five of the six master regulators of biofilm formation lack the ability to induce wild-type levels of IL-1β. In contrast, we found that the ndt80Δ/Δ mutant triggers normal IL-1β production; it is interesting to note that out of all of the regulators in the biofilm regulatory network, Ndt80p has a substantially higher number of target genes that are independent of the network, suggesting that its function may be somewhat distinct from the others (20).
As the field of C. albicans biology and pathogenesis has matured, the role of hyphal morphogenesis in these processes has become better understood and is now appreciated to be highly complex. In their report describing the activation of IL-1β production by C. albicans, Joly et al. qualified their observation that hyphae were required for this production by indicating that the filaments themselves may not be required (5). Our data support a model in which the physical formation of a filament is not sufficient to trigger IL-1β release or even to mediate macrophage lysis. Rather, these data suggest that specific C. albicans factors are responsible for triggering IL-1β production and macrophage lysis. An intriguing potential mechanism that provides an attractive explanation of our observations is that C. albicans triggers pyroptosis, a caspase-1-mediated programmed cell death process that causes macrophages to produce IL-1β and lyse in response to intracellular pathogens (21, 22). Although pyroptosis has not been previously described for the interaction of a fungal pathogen with macrophages, it is consistent with our data. Alternatively, macrophage secretion of IL-1β and lysis may represent two independent consequences of its interaction with C. albicans, e.g., inflammasome activation and necrosis. Although additional studies will be required to elucidate these mechanisms, our work clearly establishes that morphogenesis, in and of itself, is not sufficient to fully explain this important aspect of C. albicans host-pathogen interaction.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (1R01AI098450-01A1 [to D.J.K.]) and in part by the University of Rochester CTSA award from the NIH National Center for Research Resources and the National Center for Advancing Translational Sciences of the National Institutes of Health (KL2 RR024136, ULI RR02160, and UL1 TR000042, to M.W.).
Footnotes
Citation Wellington M, Koselny K, and Krysan DJ. 2012. Candida albicans morphogenesis is not required for macrophage interleukin 1β production. mBio 4(1):e00433-12. doi:10.1128/mBio.00433-12.
REFERENCES
- 1. Moran GP, Coleman D, Sullivan D. 2012. An introduction to the medically important Candida species, p. 11–25 In Calderone R, Clancy CJ, Candida and candidiasis, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 2. Cheng SC, Joosten LA, Kullberg BJ, Netea MG. 2012. Interplay between Candida albicans and the mammalian innate host defense. Infect. Immun. 80:1304–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Whiteway M, Oberholzer U. 2004. Candida morphogenesis and host-pathogen interactions. Curr. Opin. Microbiol. 7:350–357 [DOI] [PubMed] [Google Scholar]
- 4. Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42:590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Joly S, et al. 2009. Candida albicans hyphae formation triggers activation of the Nlrp3 inflammasome. J. Immunol. 183:3578–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng SC, et al. 2011. The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. J. Leukoc. Biol. 90:357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hise AG, et al. 2009. An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe 5:487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vonk AG, et al. 2006. Endogenous interleukin (IL)-1 alpha and IL-1 beta are crucial for host defense against disseminated candidiasis. J. Infect. Dis. 193:1419–1426 [DOI] [PubMed] [Google Scholar]
- 9. Franchi L, Muñoz-Planillo R, Núñez G. 2012. Sensing and reacting to microbes through the inflammasomes. Nat. Immunol. 13:325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van de Veerdonk FL, et al. 2009. Bypassing pathogen-induced inflammasome activation for the regulation of interleukin-1beta production by the fungal pathogen Candida albicans. J. Infect. Dis. 199:1087–1096 [DOI] [PubMed] [Google Scholar]
- 11. Gross O, et al. 2009. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature 459:433–436 [DOI] [PubMed] [Google Scholar]
- 12. Gringhuis SI, et al. 2012. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1β via a noncanonical caspase-8 inflammasome. Nat. Immunol. 13:246–254 [DOI] [PubMed] [Google Scholar]
- 13. Lewis LE, et al. 2012. Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 8:e1002578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pietrella D, et al. 2010. The inflammatory response induced by aspartic proteases of Candida albicans is independent of proteolytic activity. Infect. Immun. 78:4754–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ghosh S, et al. 2010. Candida albicans cell wall components and farnesol stimulate the expression of both inflammatory and regulatory cytokines in the murine RAW264.7 macrophage cell line. FEMS Immunol. Med. Microbiol. 60:63–73 [DOI] [PubMed] [Google Scholar]
- 16. Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5:e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McKenzie CG, et al. 2010. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 78:1650–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vázquez-Torres A, Balish E. 1997. Macrophages in resistance to candidiasis. Microbiol. Mol. Biol. Rev. 61:170–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Piccini A, et al. 2008. ATP is released by monocytes stimulated with pathogen-sensing receptor ligands and induces IL-1beta and IL-18 secretion in an autocrine way. Proc. Natl. Acad. Sci. U. S. A. 105:8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nobile CJ, et al. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bergsbaken T, Fink SL, Cookson BT. 2009. Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7:99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miao EA, Rajan JV, Aderem A. 2011. Caspase-1-induced pyroptotic cell death. Immunol. Rev. 243:206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]