Abstract
A widely used approach for assessing genome instability in plants makes use of somatic homologous recombination (SHR) reporter lines. Here, we review the published characteristics and uses of SHR lines. We found a lack of detailed information on these lines and a lack of sufficient evidence that they report only homologous recombination. We postulate that instead of SHR, these lines might be reporting a number of alternative stress-induced stochastic events known to occur at transcriptional, posttranscriptional, and posttranslational levels. We conclude that the reliability and usefulness of the somatic homologous recombination reporter lines requires revision. Thus, more detailed information about these reporter lines is needed before they can be used with confidence to measure genome instability, including the complete sequences of SHR constructs, the genomic location of reporter genes and, importantly, molecular evidence that reconstituted gene expression in these lines is indeed a result of somatic recombination.
SOMATIC HOMOLOGOUS RECOMBINATION REPORTER LINES: LACK OF SUFFICIENT EVIDENCE
Exciting recent advances have illustrated that genetic and epigenetic changes in plant genomes have important roles in adaptation to biotic and abiotic stresses (reviewed in Boyko and Kovalchuk, 2011; Waterworth et al., 2011). The data from these studies indicate that heritable adaptation to stresses is much faster than we have otherwise expected from Mendelian laws. Stress-induced epigenetic changes in the plant genome allow plants to adapt to stress rapidly. It has been suggested that such epigenetic changes may be inherited for at least the following generation (Boyko and Kovalchuk, 2010). A large number of stresses, including those that do not damage DNA directly, are also known to increase genome instability. The data obtained mostly from the somatic homologous recombination (SHR) reporter lines suggests that repair of such damage seems to increase the frequency not only of meiotic but also SHR events (reviewed in Boyko and Kovalchuk, 2011; Waterworth et al., 2011). A number of research articles in this area suggest that increases in homologous recombination (HR) frequencies may be a programmed response to accelerate evolutionary adaption and generate new resistance traits, enabling greater genomic plasticity in plants in response to adverse environmental conditions (Molinier et al., 2006; Boyko and Kovalchuk, 2011; Yao et al., 2011). However, we question the validity of these conclusions since these data are obtained almost exclusively using the SHR reporter lines that are not adequately characterized in the published literature.
Detecting HR or any genome rearrangements in somatic plant cells is a difficult task. Peterhans et al. (1990) and Lebel et al. (1993) published the first technique that allowed the detection of SHR at a single genomic locus using hemi- or homozygous pairs of deletion derivatives of the neomycin phosphotransferase (nptII) gene (Peterhans et al., 1990; Lebel et al., 1993). HR within the overlapping parts of the nptII gene restored the function, and the resulting kanamycin resistance was used for scoring recombination frequency. The recombination events were confirmed by the appearance of a characteristic 1245bp EcoRV fragment detected in all (over 40) kanamycin-resistant clones tested (Lebel et al., 1993). Using this technique, the authors found that the rate of spontaneous recombination was around 10−6. Ionizing radiation, mitomycin C, and heat shock markedly increased the frequency of intrachromosomal recombination (two- to ninefold). The method required generation of mesophyll protoplasts and culturing them in the presence of an antibiotic. It has not been adopted since tissue culture is known to modify chromatin drastically (Miguel and Marum, 2011) and such modification along with selection pressure could lead to HR at any time during culturing and thus would not necessarily represent the original HR events.
This system subsequently was modified by switching to assayable marker genes, such as uidA (β-GLUCURONIDASE [GUS]) (Swoboda et al., 1994) and LUCIFERASE (LUC) (Ilnytskyy et al., 2004; Molinier et al., 2004), eliminating the need for selection and tissue culturing. Reporter lines containing LUC were shown to be more sensitive than GUS in recombination assays. Analysis of several independent reporter lines carrying these marker genes indicated that the average somatic recombination frequency detected with the LUC transgene was ∼10-fold higher (10−5) in Arabidopsis thaliana and tobacco (Nicotiana tabacum) plants compared with GUS recombination frequencies (Ilnytskyy et al., 2004). Using such GUS- or LUC-based SHR reporter lines, it was shown that abiotic and especially biotic stresses lead to enhanced somatic recombination (Kovalchuk et al., 1998, 2001a, 2001b, 2003; Lucht et al., 2002; Filkowski et al., 2004; Ilnytskyy et al., 2004; Molinier et al., 2004, 2005, 2006; Schuermann et al., 2005). Using such a system, it was also discovered that plants can remember a stress perceived in one generation for several following generations. Stress (UV-C or flagellin) applied to Arabidopsis plants increased levels of HR that persisted in several subsequent, untreated generations (Molinier et al., 2006). Most recently, it was shown that volatile signals from stressed plants also trigger an increase in genome instability in neighboring unstressed plants (Yao et al., 2011).
However, based on the published literature, we are not convinced that differences in reconstitution of split marker gene expression in such reporter lines necessarily result from SHR. In our opinion, there is no bona fide evidence that recovery of gene expression from the split marker gene is correlated with recombination. Lacking in the published literature is a complete description of the lines, including the constructs, sequences of the vectors, the genomic location and sequence of the transgenic locus, and, especially critical, the linker DNA used between the two partial copies of the reporter gene.
We found two reports that showed recovery of marker gene expression to be correlated with recombination in one or two cases that were analyzed by DNA gel blot analysis (Swoboda et al., 1994; Molinier et al., 2004). The work by Swoboda et al. (1994) is one of the most frequently cited studies for HR in SHR lines. These authors showed results of a single DNA gel blot analysis from only one GUS SHR line. In order to have sufficient tissue for DNA isolation and DNA gel blot analyses, these authors took samples only from leaf sections expressing the reconstituted gene in large areas instead of just spots and then cultured these cells to increase cell mass. At that time, this was one of the best methods available to produce evidence that recombination took place. However, Swoboda et al. (1994) provided no further information for how many large GUS-expressing sectors were subjected to DNA gel blot analysis. Most importantly Swoboda et al. (1994) did not examine the individual blue spots expressing the reconstituted GUS gene in their analyses. The work of Molinier et al. (2004) is another highly cited article for the proof of SHR. These authors did not use spots of reconstituted LUC expression. Instead, to obtain sufficient DNA material for blotting, they performed DNA gel blot analysis only on the progeny of plants showing reconstituted LUC gene expression and uniformly expressing the transgene (five out of 1,000,000 plants screened expressed the transgene uniformly). DNA gel blot analysis of these five plants confirmed that HR took place in each plant (Molinier et al., 2004). However, since these plants were the progeny of the original SHR reporter lines, recombination could have happened during meiosis and therefore would not represent somatic events observed in leaves (Ries et al., 2000). After exhaustive study of relevant literature, we failed to find a publication definitively showing that each spot of gene expression on leaves is a result of true HR. However, SHR reporter lines continue to be used in numerous publications without any further data showing that recombination actually takes place. Instead, researchers have relied on conclusions reached from the earlier publications with limited molecular data, obtained either from leaf sections expressing the reconstituted gene in large areas instead of just spots, followed by callus culturing to increase cell mass and DNA gel blot analysis on DNA isolated from these tissues (Swoboda et al., 1994) or from the sexual progeny of SHR lines having uniform transgene expression (Molinier et al., 2004). However, these articles do not supply proof positive that marker gene expression in spots of somatic plant cells is the result of true recombination. It appears rather that such spots are simply assumed to be the result of bona fide HR. We believe that this assumption may require revision, especially considering scientific understanding of numerous possible events that might operate and lead to other means of marker gene expression in these lines in the absence of HR.
STRESS-INDUCED STOCHASTIC EVENTS: ALTERNATIVE EXPLANATIONS?
We postulate that besides SHR, a number of alternative events occurring at the transcriptional, posttranscriptional, and posttranslational levels could lead to reconstituted marker gene expression in the SHR reporter lines (Figure 1). Furthermore, these processes could be induced under stress or pathogen infection or combinations of these and other yet unknown factors. We term these phenomena stress-induced stochastic (SIS) events. SIS events can be seen as errors or untypical processes from an overloaded defense system. We consider HR as one type of SIS event. Here, we describe other SIS events that could lead to recovery of split gene expression in reporter lines.
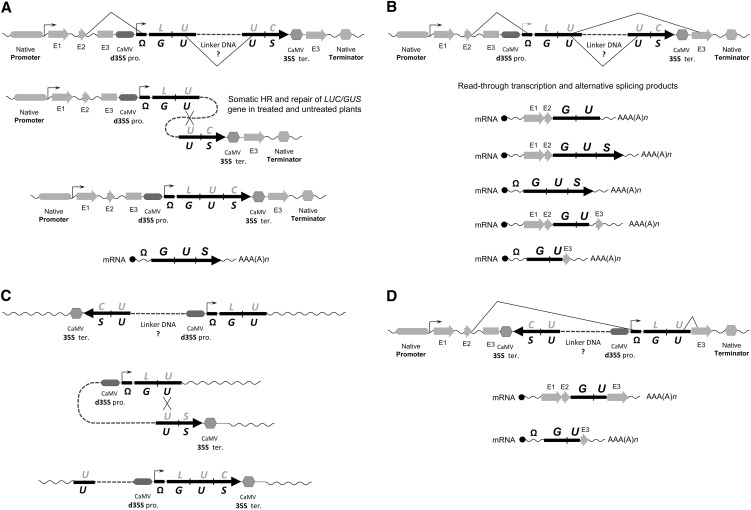
Figure 1.
SIS Events, Such as Read-Through Transcription and Alternative Splicing, Could Lead to Reconstitution of Functional GUS/LUC Protein.
The HR mechanism believed to be involved in reconstitution of split partially overlapping marker genes oriented as direct (A) or inverted (C) repeats. An illustrated alternative means of how reconstitution of such marker genes organized as direct (B) or inverted (D) repeats could take place. In the case of (B) and (D), marker gene expression is proposed to be the result of read-through transcripts initiated either from the flanking plant gene sequences or from the introduced test construct. Alternative splicing of these read-through transcripts would lead to elimination of linker DNA along with some parts of the duplicated marker transcript. These events could generate mRNA species that could be translated into fully functional reconstituted marker enzymes. As described in the text, the flanking plant sequences as well as the cryptic splice donor and acceptor sites located in the omega leader sequence and the GUS/LUC coding sequences are important for generation of functional transcripts and enzymes. Also shown are chimeric transcripts containing both plant gene and marker gene sequences predicted to be generated under conditions that regulate the native plant gene expression. Additionally, it is proposed that such plant transcripts that are fused to the partial marker gene can complement the missing activity or structure of the translated protein for its native enzymatic activity.
Cryptic Introns and Promoters in LUC or GUS Fragments
It is conceivable that both firefly LUC and bacterial GUS genes have cryptic introns that might be recognized and spliced out of the transgene transcripts containing two partial halves of the marker gene, leading to intact GUS or LUC coding sequence without any linker sequence in the mRNA (Figure 1). For example, an 84-nucleotide cryptic intron was previously detected in the GFP (for green fluorescent protein) coding sequence and was shown to be efficiently recognized and excised from its transcripts (Haseloff et al., 1997). Removing this cryptic intron from GFP in transgene constructs improved expression and detection substantially. Splicing of bacterial genes in higher eukaryotes was predicted to be a more common phenomenon than previously recognized (Lorbach et al., 1998). Low expression of Bacillus thrungiensis insecticidal crystal protein (encoded by cry) in plants led to the discovery that cry IA(b) precursor mRNA carries at least three regions that are recognized as introns, and point mutations in the 5′ splice site of the most distal intron allow high accumulation levels of the full-length mRNA (van Aarssen et al., 1995). Even bacterial terminators have been found to supply splice sites in a transgenic T-DNA insertion line that showed read-through transcription, skipping the terminator by splicing it out in various ways (Ülker et al., 2008).
Recently, it was discovered that firefly luc, which is used in many SHR reporter lines, contains a cryptic promoter as well (Vopalensky et al., 2008). Attempts to fine map the transcription start site in the coding sequence failed as these authors discovered that several transcription initiation sites were present within the LUC coding region, leading to many different transcripts. This increases the possibilities for the generation of various transgene transcripts that could have a role in the reconstitution of the LUC expression. For example, stress conditions could lead to modified plant responses, including but not limited to alternative splicing (Wang and Brendel, 2006)
Intron Donor or Acceptor Sites in the Leader Sequence
In generating the SHR lines, so far only 35S cauliflower mosaic virus (CaMV) promoters were used that contained either leader sequences having 29 amino acids of the open reading frame V of CaMV (Swoboda et al., 1994) or the omega (Ω) enhancer of Tobacco mosaic virus (Gorbunova et al., 2000) (Figure 1). These fragments are believed to enhance transgene protein production up to a level necessary to visualize clonal GUS (Schultze et al., 1990) or LUC spots. It is possible that these leader sequences supply splice donor or acceptor sites. Viral leader and gene sequences are shown to contain multiple splice donor and acceptor sites (Viaplana et al., 2001). Alternative splicing was also shown to be an essential part of the CaMV replication cycle (Kiss-Laszlo et al., 1995). Alternative RNA splicing is well documented in animal DNA viruses and retroviruses (Pongoski et al., 2002; Akusjarvi and Stevenin, 2003). If such a splice acceptor is present, for example, in the viral leader sequence used in gene constructs, it could lead to conditional reconstitution of LUC or GUS mRNA under a control of an endogenous stress-inducible plant promoter (Figures 1B and 1D).
Read-Through Transcription
Due to the presence of the 35S promoter, promoter(s) in the linker DNA, and promoters flanking the transgene integration site, it is expected that several transcripts in both sense and antisense directions might be generated. These transcripts could also lead to read-through transcripts due to stress or weak terminators, and conditional generation of such read-through transcripts and their alternative splicing could lead to functionally reconstituted LUC or GUS mRNA. Theoretically, this could lead to some rare LUC or GUS expression events during untreated conditions but an increased number of spots during stress treatments due to an enhanced responsiveness of the plant promoter to stress and increased errors that the transcription and splicing machineries make under stress conditions. Alternatively, splicing of the 35S promoter from a conditional read-through transcription originating from a stress-regulated flanking plant gene could be sufficient to induce complementation of LUC or GUS expression (Figure 1). Another observation about the CaMV 35S promoters used in SHR lines is that they appear to be stress responsive. Boyko et al. (2010b) showed that acute exposure to various types of stresses results in the transient change of the 35S promoter–driven transgene expression, whereas chronic exposure to stress does not lead to any significant changes. Additionally, it is known that most terminators used in plant science, such as NOS, 35S, and G7, are somewhat leaky and can lead to read-through transcription (Rose and Last, 1997; Luo and Chen, 2007; Ülker et al., 2008) or even supply cryptic intron splice sites (Ülker et al., 2008). Since the sequence details are missing from publications that make use of the SHR lines, it has not been possible to analyze whether these sequences could have stochastic- or pathogen-induced promoter, terminator, or intron activities.
Trans-Splicing of Linked or Unlinked GUS/LUC Transcripts
Trans-splicing is a spliceosome-directed specific joining of exons from two discontiguous primary transcripts. Trans-splicing can occur between two different transcripts of the same gene, transcripts of different genes, intergenic regions, or even those transcripts whose coding sequences are located on different chromosomes (Lasda and Blumenthal, 2011). Trans-splicing was proposed to be the mechanism of repair for two different Giardia lamblia genes, namely, Dynein heavy chain (Kamikawa et al., 2011) and Heat shock protein 90 (Kamikawa et al., 2011; Nageshan et al., 2011). In G. lamblia, these genes occur in pieces that are transcribed separately. The partial transcripts are brought together by formation of secondary structures (paired RNAs) that can form between the separate pre-mRNAs leading to removal of some parts (intron-like sequences), thus generating full-length mRNAs. Many examples of trans-splicing have also been identified in plant chloroplasts and mitochondria (reviewed in Bonen, 1993). Interestingly, trans-splicing was also suggested to be involved in the mRNA maturation of a nuclear rice (Oryza sativa) gene named SPK, a calcium-dependent seed-specific protein kinase (Kawasaki et al., 1999). The coding sequence of the SPK mRNA was shown to be divided into two regions located on different chromosomes in the rice genome sharing some overlapping sequence region. It was suggested that the SPK mRNA is derived from these separately transcribed RNAs involving a process like trans-splicing that joins them. Another convincing case of trans-splicing was demonstrated using a transgenic approach in Drosophila melanogaster for the mod (mdg4) gene, which allowed the authors to follow trans-splicing from two different chromosomal locations (Dorn et al., 2001).
Identification and analysis of a large number of chimeric transcripts in yeast, fly, mouse, and human led Li et al. (2009) to speculate that chimeric transcripts could also be generated via a transcriptional slippage model (Li et al., 2009). This model proposes that as a pre-mRNA molecule is being transcribed, it dissociates in some cases from the template strand and then the short homologous sequences at its 3′ end of pre-mRNA misaligns with the short homologous sequences at another position of the same locus or another locus. A chimeric RNA can then be generated if the transcription process continues on the new template. Since transcriptional slippage does not involve the spliceosome and is independent of introns, generation of chimeric transcripts relies on only short homologous sequences in both transcribed DNA units. The mechanistic details of the trans-splicing or the transcriptional slippage are not well understood, but there are striking similarities between these cases described and the constructs used in SHR reporter lines. Therefore, it is possible that split and partial transcripts of marker genes could also be trans-spliced leading to full-length normal transcripts and reconstitution of marker gene expression without any recombination of DNA.
Slipped-Strand Mispairing
Variation in gene expression can be generated by slipped-strand mispairing followed by RNA synthesis by the RNA polymerase through the mispaired regions but skipping the unpaired DNA in the loop. Illegitimate (out of register) base pairing in regions of repetitive DNA during replication, coupled with inadequate DNA mismatch repair systems, can produce deletions or insertions of repeat units (Levinson and Gutman, 1987). However, if these mispairings are not repaired, RNA polymerase could skip the bulging region containing one of the repeats and the linker DNA during the transcription. Such a template switch would precisely eliminate the linker DNA region and duplications in the marker gene from the RNA transcript.
Split Protein Complementation
Interactions of two split and independently made polypeptides containing parts of the GUS or LUC protein could produce a functional protein, in a similar manner to split-LUC assays, where LUC fragments are split into partly overlapping (19–amino acid–long overlap) constructs (N-LUC [amino acids 1 to 416]; C-LUC [amino acids 398 to 550]) (Luker et al., 2004; Gehl et al., 2011). N- and C-terminal subfragments of proteins could become functional if they are brought closer together by interacting proteins that are fused to them. Such reconstitution of split reporter proteins are used in investigations of protein–protein interactions, such as bimolecular fluorescence complementation, which uses split GFP, yellow fluorescent protein, or their derivatives (Bhat et al., 2006), split-ubiquitin assays (Johnsson and Varshavsky, 1994), and split-LUC assays (Luker et al., 2004; Gehl et al., 2011).
Two transcripts leading to partial polypeptides could result from read-through transcription originating from the 35S promoter used in these constructs, continuing into the linker sequence and terminating somewhere in the linker, or from a promoter located in the linker or even a partially duplicated region of the LUC gene. The second transcript might be initiated somewhere in the linker DNA or even in the second LUC fragment. Studies on the firefly luc gene, for example, showed that this gene contains a cryptic promoter (Vopalensky et al., 2008). Similarly, depending on the locus structure, the number of T-DNA insertions in the transgene locus, and their orientations, there might be hybrid transcripts containing partly interrupted plant gene transcripts and partly transgene encoded transcripts. Such transcripts might lead to translation and generation of reconstituted LUC expression due to the presence of interacting plant protein sequences in the hybrid polypeptides. Alternatively, plant transcripts could be produced that fuse the partial marker gene to a polypeptide that can complement the missing activity or structure of the translated protein, allowing its native enzymatic activity. Furthermore, such an interrupted plant gene might be pathogen or stress inducible; therefore, such hybrid transcripts would only be generated upon pathogen attack, stress, or other unknown conditions.
Intein-Mediated Protein Splicing
Inteins are internal protein elements that self-excise from their host protein and catalyze ligation of the flanking sequences (exteins) with a peptide bond (reviewed in Elleuche and Poggeler, 2010). The products of the protein splicing process are two stable proteins, the mature protein and the intein. By analogy to pre-mRNA introns and exons, the segments are called intein, for internal protein sequence, and extein, for external protein sequence. Protein splicing can also occur in trans. In this case, the intein is separated into N- and C-terminal domains, which are synthesized as separate components, each joined to an extein. The intein domains reassemble and link the joined exteins into a single functional protein. Protein splicing was first observed in yeast (Hirata et al., 1990; Kane et al., 1990; Xu et al., 1993) and later also in a wide range of organisms, including bacteria, archaea, plants, and humans.
Yang et al. (2003) have shown that if a split GUS gene is expressed as fusion protein together with a split intein coding sequences in plants, the resulting partial polypeptides can come together to form an active intein, which is then able to cleave itself out from the polypeptide and generate perfectly reconstituted GUS enzyme (Yang et al., 2003). Since the sequence of the linker DNA used in the SHR lines is not available in the published literature, we cannot determine whether or not, upon transcription and translation, these DNA sequences could act as intein. This scenario is also possible for hybrid polypeptides originating from a plant gene and LUC/GUS transgene. Since in some publications, such as Yao et al. (2011), neither the sequence of the linker DNA nor the plant locus where the transgene is located is given, we cannot determine whether these DNA sequences might act as inteins in reconstitution of functional LUC/GUS enzymes upon transcription and translation.
T-DNA Integration Locus
Transgene copy number and position in the genome are critical aspects of the SHR lines, as shown in earlier experiments with such reporter lines, indicating that there are significant differences between lines (Kovalchuk et al., 2000; Filkowski et al., 2004; Molinier et al., 2004). The constructs used in generation of SHR lines carry more than two-thirds of the coding sequences of LUC or GUS in split fragments. Our analyses of published LUC and GUS protein structures indicate that key catalytic domains of both of these enzymes are likely located in both split fragments (Conti et al., 1996; Wallace et al., 2010). Unfortunately we cannot fully be certain because in every publication only the size of the overlapping LUC or GUS fragments is given and not the exact sequences or the positions. Insertion of the transgene into a plant locus could lead to complementation of missing parts of the mRNA leading to functional enzyme. Therefore, analysis of several different lines is necessary to get the most reliable data. For example, Yao et al. (2011) used only one line (line 15D8) for their analysis. This is understandable for the crosses with the Arabidopsis hormone signaling mutants. However, no further information was provided with respect to why this line was chosen, the number of transgene copies, comomplete sequence of the constructs, and their positions in the plant genome.
Unknown Biological Factors Affecting Transcription or Translation
Since the plants tested are not in completely sterile conditions, it is conceivable that microbes (pathogenic or nonpathogenic) in the growth rooms could infect plant cells, and this infection could lead to a chain of events finally reconstituting GUS or LUC activity. This can result from pathogen effectors interfering with the normal transcription and translation in plant cells to weaken the plant immune system (da Cunha et al., 2007).
LINKER DNA: A CRITICAL ELEMENT THAT HAS BEEN OVERLOOKED
As discussed throughout this article, there are many possible explanations for the reconstitution of LUC/GUS gene expression in the SHR reporter lines. There appears to be little to no possibility of tracing the vector sequences used for generating these lines from the published literature, as they originated, in part, from unpublished vectors or discontinued products. The linker sequences cloned between two overlapping copies of the truncated GUS or LUC gene could have unexpected effects on reconstitution of these marker genes. In most cases the information about these linker DNA sequences is missing, incomplete, or possibly inaccurate. For example, Yao et al. (2011) reported that they employed a 500 bp noncoding DNA fragment as linker. However, after consulting the references cited, we found that the linker sequence used in their study in fact appears to be much larger and contains the bar gene cassette providing resistance to phosphinothricin (Gorbunova et al., 2000). Van der Auwera et al. (2008) used mannopine synthase promoter and enhancer elements as the linker sequence but they considered this information and sequence details irrelevant to their study (Van der Auwera et al., 2008). This region was simply assumed not to influence marker genes and their reconstitution. We suggest that there is no such thing as nonfunctional DNA, so the linker DNA used in these constructs could function as promoter, terminator, intron, etc., depending on conditions. The resistance gene promoter in these studies appears to be a 1' promoter that is derived from the bidirectional 1'2' promoter (Velten et al., 1984), but it is unclear which version of the promoter is present. Furthermore, the terminator in the transgene construct could support our hypothesis of alternative SIS events leading to reconstitution of gene expression, depending on the orientation of the bar gene cassette in relation to the LUC fragments, yet the identity of the terminators is also unclear from the literature. Importantly, the identity of the promoter and terminator used for the bar cassette could have a profound effect on the interpretation of research results. Some promoters are bidirectional or can become bidirectional depending on the sequence context surrounding them (Xie et al., 2001).
REPORTER LINES WITH INVERTED PARTIAL REPEATS
Reporter lines containing the partial marker gene repeats in either direct or indirect orientations have been used in analyses of SHR in plants (Figure 1). For example, a study using two transgenic lines carrying a single-copy transgene in either direct (line 1406) or inverted (line 1415) repeats of GUS fragments in Arabidopsis showed that UV-C, bleomycin, and xylanase all increased somatic recombination frequency, and the orientation of GUS fragments did not influence the recombination frequency (Molinier et al., 2005). Another study using similar single-copy lines carrying the GUS gene fragments in direct (A11) or inverted (A651) orientations in Arabidopsis showed that background spontaneous recombination frequency was much higher in line A11 compared with A651 (1.90 versus 0.21 spots, respectively) (Ilnytskyy et al., 2004). However, the fold induction upon UV-C treatment was higher in line A651 compared with A11 (4.4- versus 2.1-fold induction, hence leading to 4 versus 0.9 total spots, respectively). Lines A11 and A651 were also used in another study designed to measure the effect of pathogen- and stress-induced salicylic acid on somatic recombination frequency in Arabidopsis (Lucht et al., 2002). Curiously, however, only line A651 containing the GUS fragments in inverted orientation showed a significant induction of HR upon external application of the salicylic acid analogs 2,6-dichloroisonicotinic acid (INA) and benzothiadiazole onto seedlings. These authors concluded that chemical inducers of the salicylic acid–dependent pathogen response pathway stimulate HR in recombination reporter transgenes of different structure and at different positions in the plant genome (Lucht et al., 2002).
These observations suggest that even single-copy lines carrying the split marker gene in the same or opposite orientations can give different background- and treatment-induced recombination frequencies. This emphasizes the influence of transgene locus in the plant genome as well as the dependence of this system on environmental conditions. The SIS events described mainly for the direct repeats of split marker genes are equally applicable to the reporter lines containing such genes in inverted orientations in reconstitution of marker gene and enzyme functions.
POORLY CHARACTERIZED SHR LINE 1445
Arabidopsis Columbia-0 recombination reporter line 1445 containing partial inverted repeats of the GUS gene has been used by several groups as it is one of the best lines reporting biotic stress–related events (Molinier et al., 2006; Durrant et al., 2007; Wang et al., 2010; Song et al., 2011). Since this line was claimed to be homozygous and carries a single copy of the reporter construct (Molinier et al., 2006), we sought to determine from the published literature the location of the transgene in the Arabidopsis genome to determine the structure of the locus and possible effects of the flanking sequences on reconstitution of GUS expression under biotic stress. We found out that the information on the reporter line 1445 is confusing, incomplete, and in some cases contradictory.
The articles reporting the use of this line cite one or more of the following sources: Tinland et al. (1994), Gherbi et al. (2001), Fritsch et al. (2004), or Lucht et al. (2002), yet none of these cited articles offers any detailed information on this line. Therefore, the origin of this line is unclear, along with the details of transgene copy number and location. In addition, there is conflicting information on the location of the reporter construct in the Arabidopsis genome. Sun et al. (2008), referencing Gherbi et al. (2001) as the source of the line 1445, state that it has a single copy of the inverted split GUS transgene and mention that the transgene is located on chromosome 5 in position 8633790 in the Arabidopsis genome. By contrast, in another study employing line 1445, Pecinka et al. (2009) cite Gherbi et al. (2001) and Tinland et al. (1994) and report the location of the transgene in chromosome 2 position 14424870. Therefore, we suggest that this line may have two copies of the reporter construct in its genome.
CONFLICTING RESULTS FROM SHR REPORTER LINES
Recent studies making use of the various SHR lines have produced some conflicting results. For example, the transgenerational effect of stress on genome rearrangements in plants observed by Molinier et al. (2006) could not be reproduced by Pecinka et al. (2009). However, Boyko et al. (2010a) were able to detect such an effect, but only in the first generation of progeny. Similarly, in contrast with Molinier et al. (2005) and Yao et al. (2011), who found a seven- or threefold increase in recombination frequency, respectively, as a result of UV-C irradiation in Arabidopsis, Van der Auwera et al. (2008) did not find a significant effect of UV-C in their SHR lines containing GUS recombination reporter (Van der Auwera et al., 2008). Furthermore, in contrast with earlier results (Kovalchuk et al., 2001b; Boyko et al., 2005), Van der Auwera et al. (2008) reported that the presence of heavy metals, such as lead or cadmium ions, a heat shock of 50°C, or growth at elevated temperatures and increased daylength had no measurable effect on the mutation or recombination frequencies. In contrast with the reported 1.5-fold to sevenfold increased recombination level in two tested Arabidopsis lines as a reaction to spraying plants with the salicylic acid analogs INA and benzothiadiazole (Lucht et al., 2002), or the results of Yao et al. (2011) who used methyl salicylate, Van der Auwera et al. (2008) did not find a significant difference when they added sodium salicylate to the growth medium (Van der Auwera et al., 2008). As mentioned before, the transgenic Arabidopsis line A11 having a single copy of the GUS reporter gene in direct orientation also failed to show an induction of recombination frequency upon treatment with SA analog INA (Lucht et al., 2002). In contrast with the observations of Yao et al. (2011), addition of methyl jasmonate in growth medium did not increase recombination frequency in the experiments of the Van der Auwera et al. (2008). At least for the treatments with plant defense–inducing hormones, Van der Auwera et al. (2008) suggested differences in the application of these hormones to the tested plants as a possible explanation, but for the other treatments, they could offer no reasons to why their data differed from other published results.
POSSIBLE SOLUTIONS
Refined Tests for HR: Laser-Assisted Microdissection
The detection of SHR events is difficult because of the extremely low rates of occurrence. However, we believe that single somatic cells expressing SHR marker genes could be dissected and their DNA and RNA content analyzed to obtain the necessary molecular proof that reconstituted marker gene expression in the SHR lines is the result of SHR as opposed to other SIS events. Only with such analyses we will be able to determine whether recombination or other nonrecombination-related processes occurring at the transcriptional, posttranscriptional, translational, or posttranslational level give rise to reporter expression and the relative frequency of SHR and SIS events. One such analysis could be achieved with the use of laser-assisted microdissection (LAM). LAM is a powerful tool for isolating specific tissues, cell types, and even organelles from sectioned biological specimens for the extraction of RNA, DNA, or protein (Emmert-Buck et al., 1996; Day et al., 2005). LAM is generally applicable to all cells that can be histologically identified and enables biologists to isolate discrete cell populations in a routine manner. It has also been successfully adapted for use with plant tissues (Day et al., 2005). This technique should be applicable to the dissection of histochemically stained and fixed GUS-expressing cells. In addition, PCR and RT-PCR assays using various primer combinations on the DNA and RNA isolated from marker gene–expressing sectors could be performed. Sequencing of these PCR products and comparison to the vector DNA sequence would indicate whether or not recombination within the PCR-amplified region occurred. Similarly, RT-PCR assays using various primer combinations and sequencing of the amplification products would help to identify transcripts containing the marker gene as well as other flanking regions. Repetition of these analyses for a sufficient number of cells from treated and untreated plants would indicate the fraction of cells in which true recombination occurred and the fraction, if any, that experienced alternative possible SIS events.
Supportive Tests for Genome Instability
Several DNA damaging agents, such as ionizing radiation or clastogenic chemicals, trigger double-strand DNA breaks and problems in mitotic cell division. These cells can be detected by analyzing anaphase bridge formation between two dividing cells (Gisselsson, 2008). Chromatin fibers that are broken and fused to wrong ends lead to formation of continuous strings of chromatin stretching from one pole of the anaphase to the other in dividing cells. This is best detected in anaphase and is a relatively easy assay in dividing tissue. Comparing the anaphase bridge formation frequency in treated and untreated plants would be informative in determining whether these treatments and, most importantly, the secondary volatiles produced from the infected plants, can cause increased anaphase bridge formation in the untreated bystander plants. Furthermore, the terminal deoxynucleotidyl transferase dUTP nick end labeling assay would also be informative in this aspect (Gavrieli et al., 1992). The terminal deoxynucleotidyl transferase dUTP nick end labeling method can be used to detect DNA fragmentation by labeling the terminal fragmented end with fluorescent nucleotides in vivo. This technique has been used in plants to measure HR (Dubois et al., 2011). Quantitative RT-PCR or RNA gel blot assays for analysis of transcriptional activity of DNA repair genes would also strengthen the conclusions. The use of PCR has been problematic for the detection of rare SHR events because of the high potential for artifacts generated from PCR errors and jumping PCR. Single-molecule PCR may prove helpful in this regard (Kraytsberg and Khrapko, 2005; Lloyd et al., 2012). The increasing availability and ease of large-scale sequencing should also help to provide definitive answers in the near future.
CONCLUSIONS
Recombination reporter lines have been instrumental for two decades in studying DNA repair mechanisms and understanding how plants might cope with environmental stresses. The data generated from these lines has lead researchers to theorize exciting and far-reaching biological claims on the contribution of environmental stresses to genome evolution, such as the recent article by Yao et al. (2011) suggesting that volatile signals released from stressed plants trigger an increase in genome instability in neighboring unstressed plants. This interesting work and other hypotheses that have been put forth in this field await confirmation. In light of our review of the published literature, we have become skeptical about the conclusions reached in experiments making use of SHR reporter lines, as most of these lines are not adequately characterized, many details are missing, and there is a lack of sufficient evidence that HR truly has occurred.
We urge researchers making use of SHR lines to take the lead and supply the scientific community detailed information about these reporter lines, the SHR constructs, and their complete sequences. The location of the reporter genes in plant genomes should now also be determined as it might have unexpected influence on marker gene expression and other related events. Most importantly, molecular evidence should be provided to show whether these reporter lines indicate only somatic recombination events or other SIS events that may be occurring at the transcriptional and posttranscriptional levels. As we also tried to demonstrate, single transgenic SHR lines are not sufficient to draw conclusions from experiments; hence, several other characterized reporter lines as well as supporting experiments and methodologies are necessary to substantiate conclusions.
Acknowledgments
We thank Ales Pecinka (Max Planck Institute for Plant Breeding Research, Cologne, Germany) for his critical reading of the article and helpful suggestions. We also thank Nancy Eckardt for editing the article and providing helpful suggestions for its organization. We thank Mahmudur Rahman, a former Ph.D. student in the Plant Molecular Engineering Group, for literature searches related to trans-splicing and protein splicing as well as contributions to the discussions.
AUTHOR CONTRIBUTIONS
B.U. conceived the idea for the commentary, studied the relevant literature, wrote the article, and prepared the figure. C.H.M., T.B., S.T., B.C., A.C.O., K.W.B., and L.F. contributed to discussions, studied the relevant literature, and edited the article. All authors read and approved the final version of the article.
References
- Akusjärvi G., Stévenin J. (2003). Remodelling of the host cell RNA splicing machinery during an adenovirus infection. Curr. Top. Microbiol. Immunol. 272: 253–286 [DOI] [PubMed] [Google Scholar]
- Bhat R.A., Lahaye T., Panstruga R. (2006). The visible touch: In planta visualization of protein-protein interactions by fluorophore-based methods. Plant Methods 2: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L. (1993). Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J. 7: 40–46 [DOI] [PubMed] [Google Scholar]
- Boyko A., Blevins T., Yao Y., Golubov A., Bilichak A., Ilnytskyy Y., Hollunder J., Meins F., Jr, Kovalchuk I. (2010a). Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS ONE 5: e9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A., Filkowski J., Kovalchuk I. (2005). Homologous recombination in plants is temperature and day-length dependent. Mutat. Res. 572: 73–83 [DOI] [PubMed] [Google Scholar]
- Boyko A., Kovalchuk I. (2010). Transgenerational response to stress in Arabidopsis thaliana. Plant Signal. Behav. 5: 995–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A., Kovalchuk I. (2011). Genome instability and epigenetic modification—Heritable responses to environmental stress? Curr. Opin. Plant Biol. 14: 260–266 [DOI] [PubMed] [Google Scholar]
- Boyko A., Molinier J., Chatter W., Laroche A., Kovalchuk I. (2010b). Acute but not chronic exposure to abiotic stress results in transient reduction of expression levels of the transgene driven by the 35S promoter. New Biotechnol. 27: 70–77 [DOI] [PubMed] [Google Scholar]
- Conti E., Franks N.P., Brick P. (1996). Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure 4: 287–298 [DOI] [PubMed] [Google Scholar]
- da Cunha L., Sreerekha M.V., Mackey D. (2007). Defense suppression by virulence effectors of bacterial phytopathogens. Curr. Opin. Plant Biol. 10: 349–357 [DOI] [PubMed] [Google Scholar]
- Day R.C., Grossniklaus U., Macknight R.C. (2005). Be more specific! Laser-assisted microdissection of plant cells. Trends Plant Sci. 10: 397–406 [DOI] [PubMed] [Google Scholar]
- Dorn R., Reuter G., Loewendorf A. (2001). Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc. Natl. Acad. Sci. USA 98: 9724–9729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois E., Córdoba-Cañero D., Massot S., Siaud N., Gakière B., Domenichini S., Guérard F., Roldan-Arjona T., Doutriaux M.P. (2011). Homologous recombination is stimulated by a decrease in dUTPase in Arabidopsis. PLoS ONE 6: e18658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant W.E., Wang S., Dong X. (2007). Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc. Natl. Acad. Sci. USA 104: 4223–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleuche S., Pöggeler S. (2010). Inteins, valuable genetic elements in molecular biology and biotechnology. Appl. Microbiol. Biotechnol. 87: 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Buck M.R., Bonner R.F., Smith P.D., Chuaqui R.F., Zhuang Z., Goldstein S.R., Weiss R.A., Liotta L.A. (1996). Laser capture microdissection. Science 274: 998–1001 [DOI] [PubMed] [Google Scholar]
- Filkowski J., Kovalchuk O., Kovalchuk I. (2004). Dissimilar mutation and recombination rates in Arabidopsis and tobacco. Plant Sci. 166: 265–272 [Google Scholar]
- Fritsch O., Benvenuto G., Bowler C., Molinier J., Hohn B. (2004). The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol. Cell 16: 479–485 [DOI] [PubMed] [Google Scholar]
- Gavrieli Y., Sherman Y., Ben-Sasson S.A. (1992). Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119: 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C., Kaufholdt D., Hamisch D., Bikker R., Kudla J., Mendel R.R., Hänsch R. (2011). Quantitative analysis of dynamic protein-protein interactions in planta by a floated-leaf luciferase complementation imaging (FLuCI) assay using binary Gateway vectors. Plant J. 67: 542–553 [DOI] [PubMed] [Google Scholar]
- Gherbi H., Gallego M.E., Jalut N., Lucht J.M., Hohn B., White C.I. (2001). Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2: 287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D. (2008). Classification of chromosome segregation errors in cancer. Chromosoma 117: 511–519 [DOI] [PubMed] [Google Scholar]
- Gorbunova V., Avivi-Ragolski N., Shalev G., Kovalchuk I., Abbo S., Hohn B., Levy A.A. (2000). A new hyperrecombinogenic mutant of Nicotiana tabacum. Plant J. 24: 601–611 [DOI] [PubMed] [Google Scholar]
- Haseloff J., Siemering K.R., Prasher D.C., Hodge S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata R., Ohsumk Y., Nakano A., Kawasaki H., Suzuki K., Anraku Y. (1990). Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 265: 6726–6733 [PubMed] [Google Scholar]
- Ilnytskyy Y., Boyko A., Kovalchuk I. (2004). Luciferase-based transgenic recombination assay is more sensitive than beta-glucoronidase-based. Mutat. Res. 559: 189–197 [DOI] [PubMed] [Google Scholar]
- Johnsson N., Varshavsky A. (1994). Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. USA 91: 10340–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawa R., Inagaki Y., Tokoro M., Roger A.J., Hashimoto T. (2011). Split introns in the genome of Giardia intestinalis are excised by spliceosome-mediated trans-splicing. Curr. Biol. 21: 311–315 [DOI] [PubMed] [Google Scholar]
- Kane P.M., Yamashiro C.T., Wolczyk D.F., Neff N., Goebl M., Stevens T.H. (1990). Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H(+)-adenosine triphosphatase. Science 250: 651–657 [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Okumura S., Kishimoto N., Shimada H., Higo K., Ichikawa N. (1999). RNA maturation of the rice SPK gene may involve trans-splicing. Plant J. 18: 625–632 [DOI] [PubMed] [Google Scholar]
- Kiss-László Z., Blanc S., Hohn T. (1995). Splicing of cauliflower mosaic virus 35S RNA is essential for viral infectivity. EMBO J. 14: 3552–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk I., Kovalchuk O., Arkhipov A., Hohn B. (1998). Transgenic plants are sensitive bioindicators of nuclear pollution caused by the Chernobyl accident. Nat. Biotechnol. 16: 1054–1059 [DOI] [PubMed] [Google Scholar]
- Kovalchuk I., Kovalchuk O., Hohn B. (2000). Genome-wide variation of the somatic mutation frequency in transgenic plants. EMBO J. 19: 4431–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk I., Kovalchuk O., Hohn B. (2001a). Biomonitoring the genotoxicity of environmental factors with transgenic plants. Trends Plant Sci. 6: 306–310 [DOI] [PubMed] [Google Scholar]
- Kovalchuk I., Kovalchuk O., Kalck V., Boyko V., Filkowski J., Heinlein M., Hohn B. (2003). Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 423: 760–762 [DOI] [PubMed] [Google Scholar]
- Kovalchuk O., Titov V., Hohn B., Kovalchuk I. (2001b). A sensitive transgenic plant system to detect toxic inorganic compounds in the environment. Nat. Biotechnol. 19: 568–572 [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y., Khrapko K. (2005). Single-molecule PCR: an artifact-free PCR approach for the analysis of somatic mutations. Expert Rev. Mol. Diagn. 5: 809–815 [DOI] [PubMed] [Google Scholar]
- Lasda E.L., Blumenthal T. (2011). Trans-splicing. Wiley Interdiscip. Rev. RNA 2: 417–434 [DOI] [PubMed] [Google Scholar]
- Lebel E.G., Masson J., Bogucki A., Paszkowski J. (1993). Stress-induced intrachromosomal recombination in plant somatic cells. Proc. Natl. Acad. Sci. USA 90: 422–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G.A. (1987). Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4: 203–221 [DOI] [PubMed] [Google Scholar]
- Li X., Zhao L., Jiang H., Wang W. (2009). Short homologous sequences are strongly associated with the generation of chimeric RNAs in eukaryotes. J. Mol. Evol. 68: 56–65 [DOI] [PubMed] [Google Scholar]
- Lloyd A.H., Wang D., Timmis J.N. (2012). Single molecule PCR reveals similar patterns of non-homologous DSB repair in tobacco and Arabidopsis. PLoS ONE 7: e32255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorbach E., Wang Z., Dröge P. (1998). RNA splicing of bacterial genes in eukaryotes. Biol. Chem. 379: 1355–1358 [DOI] [PubMed] [Google Scholar]
- Lucht J.M., Mauch-Mani B., Steiner H.Y., Metraux J.P., Ryals J., Hohn B. (2002). Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat. Genet. 30: 311–314 [DOI] [PubMed] [Google Scholar]
- Luker K.E., Smith M.C., Luker G.D., Gammon S.T., Piwnica-Worms H., Piwnica-Worms D. (2004). Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. USA 101: 12288–12293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Chen Z. (2007). Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19: 943–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel C., Marum L. (2011). An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. J. Exp. Bot. 62: 3713–3725 [DOI] [PubMed] [Google Scholar]
- Molinier J., Oakeley E.J., Niederhauser O., Kovalchuk I., Hohn B. (2005). Dynamic response of plant genome to ultraviolet radiation and other genotoxic stresses. Mutat. Res. 571: 235–247 [DOI] [PubMed] [Google Scholar]
- Molinier J., Ries G., Bonhoeffer S., Hohn B. (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Ries G., Zipfel C., Hohn B. (2006). Transgeneration memory of stress in plants. Nature 442: 1046–1049 [DOI] [PubMed] [Google Scholar]
- Nageshan R.K., Roy N., Hehl A.B., Tatu U. (2011). Post-transcriptional repair of a split heat shock protein 90 gene by mRNA trans-splicing. J. Biol. Chem. 286: 7116–7122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A., Rosa M., Schikora A., Berlinger M., Hirt H., Luschnig C., Mittelsten Scheid O. (2009). Transgenerational stress memory is not a general response in Arabidopsis. PLoS ONE 4: e5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhans A., Schlüpmann H., Basse C., Paszkowski J. (1990). Intrachromosomal recombination in plants. EMBO J. 9: 3437–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongoski J., Asai K., Cochrane A. (2002). Positive and negative modulation of human immunodeficiency virus type 1 Rev function by cis and trans regulators of viral RNA splicing. J. Virol. 76: 5108–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries G., Heller W., Puchta H., Sandermann H., Seidlitz H.K., Hohn B. (2000). Elevated UV-B radiation reduces genome stability in plants. Nature 406: 98–101 [DOI] [PubMed] [Google Scholar]
- Rose A.B., Last R.L. (1997). Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J. 11: 455–464 [DOI] [PubMed] [Google Scholar]
- Schuermann D., Molinier J., Fritsch O., Hohn B. (2005). The dual nature of homologous recombination in plants. Trends Genet. 21: 172–181 [DOI] [PubMed] [Google Scholar]
- Schultze M., Hohn T., Jiricny J. (1990). The reverse transcriptase gene of cauliflower mosaic virus is translated separately from the capsid gene. EMBO J. 9: 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Durrant W.E., Wang S., Yan S., Tan E.H., Dong X. (2011). DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell Host Microbe 9: 115–124 [DOI] [PubMed] [Google Scholar]
- Sun X., Zhang Y., Yang S., Chen J.Q., Hohn B., Tian D. (2008). Insertion DNA promotes ectopic recombination during meiosis in Arabidopsis. Mol. Biol. Evol. 25: 2079–2083 [DOI] [PubMed] [Google Scholar]
- Swoboda P., Gal S., Hohn B., Puchta H. (1994). Intrachromosomal homologous recombination in whole plants. EMBO J. 13: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland B., Hohn B., Puchta H. (1994). Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus. Proc. Natl. Acad. Sci. USA 91: 8000–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B., Peiter E., Dixon D.P., Moffat C., Capper R., Bouché N., Edwards R., Sanders D., Knight H., Knight M.R. (2008). Getting the most out of publicly available T-DNA insertion lines. Plant J. 56: 665–677 [DOI] [PubMed] [Google Scholar]
- van Aarssen R., Soetaert P., Stam M., Dockx J., Gosselé V., Seurinck J., Reynaerts A., Cornelissen M. (1995). cry IA(b) transcript formation in tobacco is inefficient. Plant Mol. Biol. 28: 513–524 [DOI] [PubMed] [Google Scholar]
- Van der Auwera G., Baute J., Bauwens M., Peck I., Piette D., Pycke M., Asselman P., Depicker A. (2008). Development and application of novel constructs to score C:G-to-T:A transitions and homologous recombination in Arabidopsis. Plant Physiol. 146: 22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten J., Velten L., Hain R., Schell J. (1984). Isolation of a dual plant promoter fragment from the Ti plasmid of Agrobacterium tumefaciens. EMBO J. 3: 2723–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaplana R., Turner D.S., Covey S.N. (2001). Transient expression of a GUS reporter gene from cauliflower mosaic virus replacement vectors in the presence and absence of helper virus. J. Gen. Virol. 82: 59–65 [DOI] [PubMed] [Google Scholar]
- Vopálenský V., Masek T., Horváth O., Vicenová B., Mokrejs M., Pospísek M. (2008). Firefly luciferase gene contains a cryptic promoter. RNA 14: 1720–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B.D., Wang H., Lane K.T., Scott J.E., Orans J., Koo J.S., Venkatesh M., Jobin C., Yeh L.A., Mani S., Redinbo M.R. (2010). Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330: 831–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.B., Brendel V. (2006). Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 103: 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Durrant W.E., Song J., Spivey N.W., Dong X. (2010). Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc. Natl. Acad. Sci. USA 107: 22716–22721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth W.M., Drury G.E., Bray C.M., West C.E. (2011). Repairing breaks in the plant genome: the importance of keeping it together. New Phytol. 192: 805–822 [DOI] [PubMed] [Google Scholar]
- Xie M., He Y., Gan S. (2001). Bidirectionalization of polar promoters in plants. Nat. Biotechnol. 19: 677–679 [DOI] [PubMed] [Google Scholar]
- Xu M.Q., Southworth M.W., Mersha F.B., Hornstra L.J., Perler F.B. (1993). In vitro protein splicing of purified precursor and the identification of a branched intermediate. Cell 75: 1371–1377 [DOI] [PubMed] [Google Scholar]
- Yang J., Fox G.C., Jr, Henry-Smith T.V. (2003). Intein-mediated assembly of a functional beta-glucuronidase in transgenic plants. Proc. Natl. Acad. Sci. USA 100: 3513–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Danna C.H., Zemp F.J., Titov V., Ciftci O.N., Przybylski R., Ausubel F.M., Kovalchuk I. (2011). UV-C-irradiated Arabidopsis and tobacco emit volatiles that trigger genomic instability in neighboring plants. Plant Cell 23: 3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]