Abstract
Marker-transgene–dependent lines of Arabidopsis thaliana measuring somatic homologous recombination (SHR) have been available for almost two decades. Here we discuss mechanisms of marker-gene restoration, comment on results obtained using the reporter lines, and stress how caution must be applied to avoid experimental problems or false interpretation in the use of SHR reporter lines. Although theoretically possible, we conclude that explanations other than SHR are unlikely to account for restoration of marker gene expression in the SHR lines when used with appropriate controls. We provide an overview of some of the most important achievements obtained with the SHR lines, give our view of the limitations of the system, and supply the reader with suggestions on the proper handling of the SHR lines. We are convinced that SHR lines are and will remain in the near future a valuable tool to explore the mechanism and influence of external and internal factors on genome stability and DNA repair in plants.
The germline in plants is formed only during late development; therefore, any change in DNA sequence occurring in somatic tissue during the lifetime can be passed on to the next generation. Thus, it had been a long-standing wish for plant scientists to possess an assay to quantify somatic homologous recombination (SHR) in whole plants. The large number of repeated sequences in plant genomes was expected to be strictly controlled so as to avoid continuous duplications and deletions. In 1994, the first marker-transgene–dependent lines of Arabidopsis thaliana measuring SHR were published (Swoboda et al., 1994). Numerous articles on the influence of genetic, environmental, developmental, and other parameters on the frequency of SHR, discovered through the use of these Arabidopsis SHR reporter lines, have since been published. In the adjacent commentary by Ülker et al. (2012), the validity of the published plant lines as testers for SHR has been brought into question. Here we respond to the suggested alternative explanations for marker-gene restoration, comment on results obtained using the reporter lines, and stress how caution must be applied to avoid experimental problems or false interpretation in the use of SHR lines. The focus of Ülker et al. (2012) is based on alternative explanations of how marker gene expression might be achieved in transgenic plant lines harboring nonfunctional overlapping parts of marker genes; therefore, we first evaluate the arguments brought forward by these authors. We then give a short overview of some of the most important achievements obtained with the SHR lines. Finally, we give our view of the limitations of the system and supply the reader with suggestions on proper handling of the SHR lines.
IS THERE ANY INDICATION THAT MARKER GENE RESTORATION IS NOT CAUSED BY HOMOLOGOUS RECOMBINATION?
Originally, Arabidopsis plants were produced that carried transgenes with overlapping parts of a β-glucuronidase (GUS) gene in direct or inverted orientation. Only after restoration of the complete open reading frame (ORF) could the GUS marker be expressed (Swoboda et al., 1994). SHR was expected to occur infrequently, and thus only a few cells at most were expected to express the reconstituted marker gene. Indeed, only rare events, visible as blue spots after staining, were found (example in Figure 1). DNA gel blot analyses of callus derived from GUS-positive blue spot tissue and from surrounding (GUS-negative) tissue showed that the former, and not the latter, contained the recombined chromosomal GUS gene (Swoboda et al., 1994). The concept of using a transgenic chromosomal reporter gene that is restored by homologous recombination was not new—it had been used previously in countless studies in yeast and animal cells (Paques and Haber, 1999) and even in plant cell culture and cotyledons (Peterhans et al., 1990). The novelty was that recombination could be directly detected in planta, making the assay an attractive and easily applicable system for analysis of SHR during development. Staining for the activity of the enzyme GUS could be performed during the complete life cycle and in all tissues of the plant. Adaptation of this system to the luciferase marker meant the assay could even be used in a noninvasive way.
Figure 1.
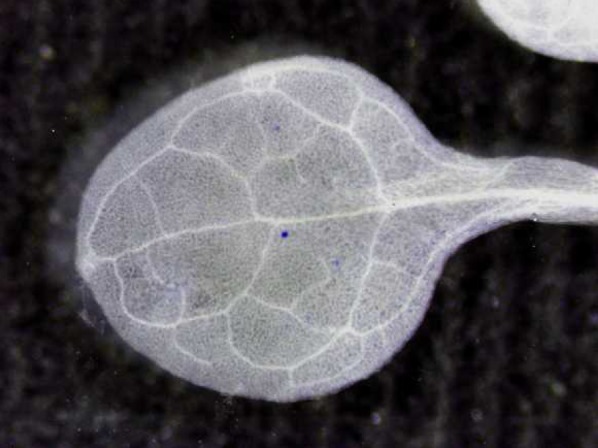
Leaf of a Seedling of a Transgenic SHR Line after Histochemical Staining.
The blue sectors are caused by expression of the GUS gene, which results from restoration by HR (Schuermann et al., 2009). (Figure courtesy of David Schürmann.)
Ülker et al. (2012) question whether the restoration of the marker is indeed caused by homologous recombination (HR) and they raise doubts about the molecular proof supplied. However, various lines of argument in our opinion make any of the alternative explanations brought forward by Ülker and colleagues unlikely. In the following, we shortly discuss the five most important ones.
1. The Stochastic Pattern of Events
The sectors visualized are products of stochastic events, in the order of 10−6 events per genome. These can be regarded as spontaneous yes or no decisions for SHR. These events can be induced or repressed by genetic or environmental means (see below), resulting in a change in the frequency of sectors. If any of the alternative explanations of Ülker et al. (2012) were true, the result would be expected to be an extremely slight to slight change in marker gene expression in all cells or at least in tissue in which these alternative gene expression changes are taking place. This would result in an extremely pale blue or rather invisible coloration, but definitely not in spots or sectors, which represent clonal populations of constitutive high-level GUS expression.
2. The Influence of the Orientation of the Truncated Part of the Marker Gene
In most of the alternative explanations suggested by Ülker and colleagues, the mechanism for the correction of gene activity depends on the direct, parallel orientation of the truncated versions of the marker gene. In many experiments reported in the literature, inverted repeats of the two parts of the marker gene were also used, with similar conclusions for the experiments in question. Such inverted orientation means that a transcript through the locus would only have one of the two truncated GUS copies in sense orientation. This would thus preclude explanations based on splicing and translation. Only the process of trans-splicing, one of the possible alternative explanations given by the authors, cannot be dismissed, except that, in our opinion, the other arguments given above and below exclude this possibility as well. However, experiments analyzing the mechanisms of recombination-dependent rescue of DNA replication blocks deliberately called for the comparison of data from the direct and inverted recombination targets (Schuermann et al., 2009).
3. The Induction of SHR by Double-Strand Breaks and DNA Damaging Agents
For DNA to recombine efficiently, a double-strand break (DSB) in at least one of the recombination partners is required. Experimental introduction of site-specific DSBs in target DNA by the use of the endonucleases HO or I-SceI increased the efficiency of SHR by up to two orders of magnitude (Chiurazzi et al., 1996; Orel et al., 2003). Enzymatic DSB induction also led to tremendous improvements in gene targeting (Puchta et al., 1996; Fauser et al., 2012). The fact that introduction of DSBs in the recombination partner(s) strongly increased the frequency of HR, including gene targeting, clearly speaks in favor of a mechanism that can be explained solely by HR. In addition, transposition-induced DSBs in recombination targets strongly induced SHR in Arabidopsis and tobacco (Nicotiana tabacum) (Shalev and Levy, 1997; Xiao and Peterson, 2000). Furthermore, the introduction of DNA breaks in a nontargeted fashion, as for instance with γ irradiation stemming from the exploded Chernobyl reactor (Kovalchuk et al., 1998) or by DNA damage–inducing agents, such as UV radiation, methyl methane sulfonate, mitomycin C, cisplatin, and others, were found to lead to increased rates of HR (Puchta et al., 1995; Ries et al., 2000). It is not evident how these agents would contribute to restoration of marker-gene expression via any pathway suggested by Ülker et al. (2012).
4. The Genetics of Marker Gene Restoration
Over the years, many genes, mostly from Arabidopsis, have been found to influence marker gene restoration frequencies in the SHR lines. Almost all are known to be involved in the process of HR (for a detailed discussion, see below) and are difficult to be viewed as influencing splicing, read-through transcription, or protein reconstruction on the protein level, as suggested by Ülker and colleagues.
5. The Molecular Proof of HR
During the past decades, the measurement of HR of marker genes has been a standard assay used in hundreds of studies in all kinds of organisms and cell lines, from bacteria to human. Proof for the restoration of the respective maker gene was mostly supplied by DNA gel blots (as mentioned above); PCR analysis would have produced artifacts during the amplification of sequences originating from molecules with overlaps. Single cells carrying a recombination event, as evidenced by GUS staining, were cloned and propagated in cell culture to obtain the required amount of DNA for blotting. For propagation of recombined material in the in planta system, leaf sectors expressing the restored GUS gene were detected by short nondestructive staining, excised, and cultured. Restoration of the GUS marker by HR could be proven by DNA gel blot analysis of plants, each carrying one of five different recombination traps (Swoboda et al., 1994; Orel et al., 2003; Molinier et al., 2004). In an independent study in tobacco, a negative selectable marker was inserted between direct overlapping parts of the GUS gene. Here, in all eight independent cases tested, it could be demonstrated that after DSB induction by I-SceI, restoration of the marker gene expression was linked to HR (Siebert and Puchta, 2002). Also, tobacco plants carrying overlapping truncated versions of a kanamycin resistance gene yielded molecular proof for in planta HR (Peterhans et al., 1990; Lebel et al., 1993). Thus, sufficient molecular evidence was supplied that supported the conclusion that disrupted marker genes carrying overlapping sequences are indeed restored by HR in planta. Ülker et al. (2012) also argue that in the in planta system, recombination might have occurred in tissue culture only after excision of the blue sectors and not in planta. This argument can be refuted by the fact that, in such a case, the recombination frequency in tissue culture would have to be several orders of magnitude higher than in planta, which clearly is not the case (Peterhans et al., 1990; Lebel et al., 1993). Formally, we cannot exclude the possibility that other kinds of tissue culture–induced mutation might lead to restoration of marker gene activity, but experimental data for such a mechanism are lacking.
Thus, the arguments provided by Ülker and colleagues contrast sharply with the wealth of information that the use of the recombination reporter lines has provided. It is, of course, possible that other unknown factors influence the behavior of the recombination substrates such that a fully functional gene is restored, but such a mechanism will have to be explained at the molecular level. Ülker et al. (2012) also propose that sequences from the plant genome might supply the complementary information, thus restoring the intactness of the transgene. Again, this possibility is extremely unlikely, especially because many different genomic locations of the integrated T-DNAs carrying various constructs yielded recombination-proficient transgenes. Moreover, several different linkers have been used in various constructs, excluding a general role of a specific linker sequence in HR, which has also been suggested by Ülker and colleagues as another alternative for restoration of the function of a split marker gene.
MULTIPLE VARIATIONS OF THE SHR SETUP
Soon after the first publication of the SHR system (Swoboda et al., 1994), various modified recombination traps were constructed and used in Arabidopsis and tobacco (Puchta et al., 1995; Gherbi et al. 2001). The recombination markers also were successfully used in rice (Oryza sativa) (S. Toki, personal communication). Apart from the GUS gene as marker, the luciferase gene was used, permitting noninvasive and sensitive assays of recombination (Kovalchuk et al., 2003; Ilnytskyy et al., 2004). Equally important is the development of specific recombination lines, the use of which allowed answering specific questions about the mechanism of SHR. A system was set up in Arabidopsis to demonstrate ectopic recombination by restoration of a GUS gene after transposition out of the marker-gene of the transposable element Dissociater (Ds) (Shalev and Levy, 1997). To test whether small heterologies within homologous sequences might influence the efficiency of recombination (homoeologous recombination), GUS-based recombination substrates that contained defined numbers of mismatches within the overlap were set up. The presence of mismatches indeed reduced recombination frequencies (Li et al., 2004; Opperman et al., 2004). It became clear that the sister chromatids play an important role in SHR; therefore, a system was established that permitted the analysis of interchromatid recombination (Molinier et al., 2004). Different mechanisms exist by which a DSB can be repaired by the use of intrachromosomal homologous sequences in somatic cells, (reviewed in Puchta, 2005). Single-strand annealing (SSA) between tandem repeats can occur in a nonconservative way such that the intervening sequence is lost. On the other hand, by synthesis-dependent strand annealing (SDSA), a sequence present in close proximity to homologous sequences can be used as template for repair of a DSB in a conservative way (gene conversion) without changing the donor. A useful modification was the insertion of the rare-cutting yeast-derived I-SceI endonuclease target sequence into the truncated and partially duplicated GUS gene. The use of this assay system allowed the measurement of the efficiency of the SSA and SDSA pathways specifically in plants. It could be demonstrated definitively that, in somatic plant cells, SSA is more efficient than SDSA (Orel et al., 2003). This system, which because of the high rates of break-induced SHR is much easier to use than the classical one, was also used for the recent demonstration of the involvement of small RNAs in DSB repair in plants (Wei et al., 2012).
In plants, most DSBs are repaired by nonhomologous end joining (Siebert and Puchta, 2002; Puchta, 2005). However, the reporter transgenes are built in such a way that in contrast with HR, nonhomologous end joining theoretically can lead to restoration of the marker only in a small minority of cases. This is demonstrated in recent work wherein we showed that HR factors involved in strand exchange are indeed required for efficient restoration of the marker in the SDSA lines (Roth et al., 2012).
An important application of the SHR tester lines was their use in screens of mutated Arabidopsis lines carrying recombination targets. Agrobacterium tumefaciens–mediated mutation was used to produce large populations of tagged lines. Examples of identification of genes involved in positive or negative influences on SHR include the genes encoding the chromatin remodelling factor INO80, centrin2, and DNA polymerase δ (Fritsch et al., 2004; Schuermann et al., 2005; Liang et al., 2006; Schuermann et al., 2009). Without the convenient transgenic SHR tester lines, the search for such genes would not have been possible.
ELUCIDATING THE ROLE OF MULTIPLE FACTORS IN SHR IN PLANTS
In addition to screens, specific mutants suspected to be involved in SHR could be analyzed upon crossing the mutant Arabidopsis lines with tester lines. It is far beyond this article to list all genes that have been tested with these assay systems (for an earlier review, see Schuermann et al., 2005); therefore, we will focus only on a few important and recent ones. In general, one can discriminate between two classes of genes that were characterized by the SHR assay: those contributing to the frequency of HR and those in the absence of which recombination is enhanced.
In different eukaryotic organisms, in both somatic and meiotic cells, several proteins involved in strand exchange have conserved functions in common steps of HR. In Arabidopsis, defects detected by the SHR assay could be correlated with meiotic defects in RAD51C and BRCA2 mutants (Abe et al., 2005; Seeliger et al., 2012). In addition, the use of SHR lines also permitted the identification of proteins involved in processing of recombination intermediates or in postreplicative repair (Mannuss et al., 2010).
On the other hand, several different classes of genes involved in hyperrecombination were also characterized. Helicases and their interaction partners are involved in the suppression of certain HR reactions in Arabidopsis, as is known for other eukaryotic organisms as well. Insertion mutants of RECQ4A, RMI1, Top3alpha, and FANCM revealed a hyperrecombination phenotype (Hartung et al., 2007; Hartung et al., 2008; Knoll and Puchta, 2011; Knoll et al., 2012). A similar phenotype of hyperrecombination was also seen in plants with reduced expression of the gene coding for the catalytic subunit of the DNA polymerase δ (Schuermann et al., 2009), indicating that replication stress enhances HR. Loss of Chromatin assembly factor1 (CAF-1), a heterotrimeric complex involved in the reconstitution of S-phase chromatin, induced a strong hyperrecombination phenotype in Arabidopsis (Endo et al., 2006; Kirik et al., 2006).
INDUCING RECOMBINATION BY EXTERNAL STRESSES
An in planta system was especially suitable to measure whether various environmental stresses applied to a plant would induce genome instability. It was no surprise that application of genotoxins, such as x-rays or methyl-methane-sulfonate, cross-linking agents, and UV irradiation (all factors that induce DNA damage), led to elevated levels of SHR (Puchta et al., 1995). SHR plants were used successfully to document radioactive pollution after the Chernobyl accident, probably as a result of the DNA breaking activity of Cs137-mediated γ radiation (Kovalchuk et al., 1998).
Even more interestingly and quite unexpectedly, stresses not known to cause DNA damage could indeed induce recombination. This was demonstrated for high salinity (Puchta et al., 1995) but also pathogen attack (Lucht et al., 2002; Kovalchuk et al., 2003). The application of flagellin, a bacterial-derived elicitor of plant defense, was shown to enhance recombination significantly (Molinier et al., 2006). These and more recent articles on environmentally induced changes in SHR, too many to list in this contribution, made and make us aware that such studies have a strong effect on our understanding of evolution.
WHERE ARE THE LIMITS?
There is no question that each experimental system has its limitations, and the better we are aware of these, the better we can apply the system. The decisive limitation of the SHR reporter system is the detection of recombination events, because this depends on the expression of the restored marker. Our concerns are thus in direct contradiction to the ones raised by Ülker et al. (2012). Whereas we have no doubts that all sectors are indeed caused by HR, any modulation of expression level might lead to changes in the detection level of the recombination events in the plants. We must assume that not all recombination events are detectable under all circumstances. Especially small sectors, which contain less activity of the restored protein, will only become visible after extensive staining—if at all.
Most recombination traps used at present are built on 35S promoters. Although this promoter is regarded as ubiquitous in expression under certain stresses, the expression might differ to a certain extent (Qin et al., 1994); thus, an uncritical use of the system in stress studies without appropriate controls might lead to artifacts.
If the influence of a specific stress factor on genome stability is to be tested, we suggest checking by quantitative RT-PCR (qRT-PCR) whether the stress itself is enhancing transcription of the SHR marker gene. Specifically, transcription of the 5′ part of the recombination target transgene before recombination must be controlled, but not transcription of the restored marker gene. Only with the former approach can transcriptional changes of the marker be detected independently of recombination. Only if the RNA level of the marker is not changed on application of the stress factor can one be sure that recombination is indeed induced directly.
As mentioned by Ülker et al. (2012), controversial results have been published using different SHR lines in different laboratories. Growth conditions might indeed differ significantly between different laboratories, thus possibly resulting in different stress responses depending on growth conditions. Therefore, it is advisable to take minor environmentally increased HR values for granted only if they can be reproduced in another laboratory. However, even this may be inconclusive, because unknown, untested growth conditions may also vary from laboratory to laboratory.
Another understandable concern is that the marker could be silenced in single cells, parts of a plant, or even the complete plant. It has long been known that transgene promoters might be inactivated progressively over generations (Kilby et al., 1992). More related to the present discussion, natural direct repeats have been shown to induce gene silencing (Kinoshita et al., 2007). Often, insertion mutants in genes to be tested for influence on SHR also contain 35S promoters (sometimes even a silenced GUS gene) within their T-DNAs. Stacking of T-DNAs carrying 35S promoters might induce silencing (Daxinger et al., 2008). The situation might be especially problematic when double or triple mutants are produced by crossing. In case the influence of a mutant background on HR is being tested, it is advisable to check by qRT-PCR whether the expression of the marker gene itself is not altered in the mutant background. A further important control is the complementation of the mutant with the natural gene, which should result in wild-type SHR frequencies. In principle, qRT-PCR should also be applied in experiments in which different mutant backgrounds are combined to exclude any cross-effect on the expression of the marker.
The recombination traps contain direct or inverted repeats; therefore, epigenetic changes may be induced by chance during propagation. Over the years, many of these lines have been propagated in different laboratories under various conditions. In addition, the conditions of seed storage may influence the properties of a plant line. After multiple rounds of selfing, the recombination line 11 (described in Swoboda et al., 1994) lost activity of the hygromycin resistance gene, which resided between the two truncated parts of the GUS gene (Holger Puchta, unpublished data). This can be taken as indication of a silencing phenomenon, in which case such plants should no longer be used for assays. It is advisable therefore to ensure that a line to be used for experiments in fact exhibits recombination sectors. Another reason for concern is, as in any other plant work, the use of different plant ecotypes as experimental and control lines. Different ecotypes might differ slightly in the expression of DNA repair genes, which might lead to certain differences in the use of different DNA repair pathways. Upon crossing of the reporter and mutant lines, test lines carrying the mutation to be analyzed as well as control lines, missing this mutation, should be isolated from the segregating population.
One study reported that after induction of recombination using SHR lines, increased levels of HR persisted in the subsequent, untreated generations, suggesting the involvement of an epigenetic effect (Molinier et al., 2006). In other studies, such effects could hardly be reproduced or were found to persist only to the next generation (Pecinka et al., 2009; Boyko et al., 2010) Thus, although transgenerational effects on SHR frequency might occur, they most likely are not a general response to abiotic stress and may be induced by special growth conditions.
Another important point concerns the number of recombination events necessary to produce statistically relevant data. Normally, a clonal population of seedlings is analyzed (30 to 100 plantlets). Depending on the SHR line and treatment/background applied, one can detect frequencies of less than one event per plant to up to hundreds of sectors per seedling. Therefore, the sample sizes must be adjusted to secure statistically significant levels of recombination events of both the control sample and the experimental sample. Moreover, independent biological replicates performed with different seed batches at different times are required to sustain reproducibility. These are general principles of good scientific experimentation.
FURTHER INFORMATION ABOUT SHR LINES
Because it was not standard practice at the time, the early SHR lines were not characterized in such detail as one would expect from current studies. In the intervening years since publication of the lines, the insertion sites in the Arabidopsis genome have been provided by several workers; Table 1 provides the respective information for the most commonly used lines. For the more recently established lines (Orel et al., 2003), Figure 2 provides a simplified annotation that highlights the relevant sections of the constructs. Complete annotated sequences of the transgenes are available in GenBank (accession numbers provided below). As mentioned by Ülker et al. (2012), conflicting results on the T-DNA insertion site(s) of line 1445 have indeed been reported by different groups (Sun et al., 2008; Pecinka et al., 2009). The construction of the respective T-DNA is described by Tinland et al. (1994), and the transgenic line carrying this T-DNA was used in the study of Fritsch et al. (2004) but was neither described nor used by Gherbi et al. (2001). We would discourage workers from using line 1445 unless they are willing to characterize it in detail before use.
Table 1.
Integration Site of Commonly Used SHR Lines
| SHR line | Ecotype | Reference | Insertion site (chromosome site or Atg no.) | Determined by |
| N1IC4 651 | C24 | Puchta et al. (1995) | Chr 5_10455625 | D. Tian; F. Hartung |
| N1DC1 11 | C24 | Swoboda et al. (1994) | Chr 2_11765197 | D. Tian |
| 1406 | Col | Gherbi et al. (2001) | Chr 3_15367918 | D. Tian |
| IC9C | Col | Molinier et al. (2004) | At5g25050 nt1767/nt1812 | F. Hartung |
| DGU.US-1a | Col | Orel et al. (2003) | At3g21080 | S. Dukowic-Schulze |
| DU.GUS-8a | Col | Orel et al. (2003) | At5g20870 | S. Dukowic-Schulze |
| IU.GUS.-8a | Col | Orel et al. (2003) | At1g72710 | S. Dukowic-Schulze |
T-DNA structure of reporter constructs shown in Figure 2; complete sequences available in GenBank (see Accession Numbers).
Figure 2.
T-DNA Structure of HR Reporter Constructs.
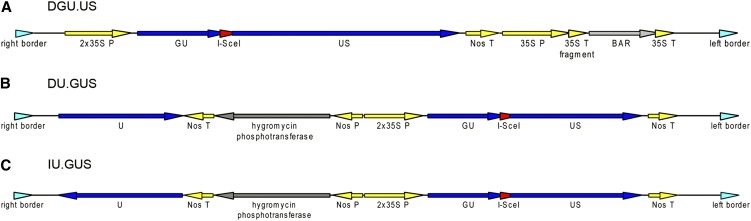
(A) Line DGU.US contains two fragments of the GUS ORF with homologous parts (GU, US; blue) in direct orientation that are separated by a recognition site of the megaendonuclease I-SceI (red). Expression of the recombined complete GUS gene is under the control of a 2x35S promoter and a Nos terminator (yellow). Downstream, a phosphinothricin acetyltransferase ORF (BAR; gray) is flanked by a 35S promoter and a 35D terminator (yellow). 3′ of the 35S promoter, there is a fragment of 35S terminator (yellow) that does not affect BAR selection.
(B) In line DU.GUS, two fragments of the GUS ORF without homology to each other (GU, US; blue) in direct orientation are separated by a recognition site of the megaendonuclease I-SceI (red). Upstream, a central fragment of the GUS ORF homologous to both GU and US is located (U; blue). Expression of the recombined complete GUS gene is under the control of a 2x35S promoter and a Nos terminator (yellow). Between the U and GU fragments, a hygromycin phosphotransferase ORF (gray) under the control of a Nos promoter and Nos terminator (yellow) is located.
(C) Line IU.GUS is constructed similarly to line DU.GUS. The central GUS ORF fragment (U; blue) is in an inverted orientation here.
All constructs are flanked by left and right border sequences (turquoise).
Conclusions
Taking all necessary controls into account, we are convinced that SHR lines are and will remain in the near future a valuable tool for exploring the mechanism and influence of external and internal factors on genome stability and DNA repair in plants. Both the development of novel assays and the importance of applying appropriate assays to specific analyses of specific questions are of unquestioned value and importance. The application of other assays for genome instability or the use of techniques, such as laser-assisted microdissection, suggested by Ülker et al. (2012) will certainly help to complement our understanding of mechanisms that are required for genome stability in plants. As is the case with most techniques, in the long run, SHR lines will become obsolete and will be replaced by superior assays. Because of the rapid development of new techniques and the improvement of existing techniques, molecular biology is evolving rapidly. The strength of the SHR lines is that one can easily measure the stability of a unique repeated sequence in a single locus in millions of cells, in some cases even in living plants. Using modern genome sequencing techniques, it might be more convenient in the long run to look at variation of all repeated sequences in the genomes of a few representative cells during plant growth. An alternative would be to define a limited number of natural repeats and test their respective stability in DNA extracted from whole plants over the cell population by single molecule PCR. Such approaches do not rely on gene expression or the integration of transgenes; therefore, not only could artifacts caused by the assay system be excluded beforehand, but also results might be obtained more quickly.
Accession Numbers
The following sequences have been deposited in GenBank: DGU.US-1 HR reporter construct T-DNA, complete sequence (JX475904); DU.GUS-8 DU.GUS HR reporter construct T-DNA, complete sequence (JX475905); IU.GUS HR reporter construct T-DNA, complete sequence (JX475903).
Acknowledgments
We apologize that most publications dealing with an in planta SHR system could not be cited because of space limitation. We thank Dacheng Tian, Stefanie Dukowic-Schulze, and Frank Hartung for sharing unpublished information on insertion sites of SHR lines and Seiichi Toki for information on the use of rice reporters for SHR. We also thank Charles White for critical reading of the article, David Schürmann for supplying Figure 1, and Alexander Knoll and Frieder Fauser for sequence processing. The SHR lines and specific information about them are available on request to H.P.
AUTHOR CONTRIBUTIONS
Both authors contributed to writing the article.
References
- Abe K., Osakabe K., Nakayama S., Endo M., Tagiri A., Todoriki S., Ichikawa H., Toki S. (2005). Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol. 139: 896–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A., Blevins T., Yao Y., Golubov A., Bilichak A., Ilnytskyy Y., Hollunder J., Meins F., Jr., Kovalchuk I. (2010). Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS ONE 5: e9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurazzi M., Ray A., Viret J.F., Perera R., Wang X.H., Lloyd A.M., Signer E.R. (1996). Enhancement of somatic intrachromosomal homologous recombination in Arabidopsis by the HO endonuclease. Plant Cell 8: 2057–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L., Hunter B., Sheikh M., Jauvion V., Gasciolli V., Vaucheret H., Matzke M., Furner I. (2008). Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends Plant Sci. 13: 4–6 [DOI] [PubMed] [Google Scholar]
- Endo M., Ishikawa Y., Osakabe K., Nakayama S., Kaya H., Araki T., Shibahara K., Abe K., Ichikawa H., Valentine L., Hohn B., Toki S. (2006). Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 25: 5579–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F., Roth N., Pacher M., Ilg G., Sánchez-Fernández R., Biesgen C., Puchta H. (2012). In planta gene targeting. Proc. Natl. Acad. Sci. USA 109: 7535–7540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch O., Benvenuto G., Bowler C., Molinier J., Hohn B. (2004). The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol. Cell 16: 479–485 [DOI] [PubMed] [Google Scholar]
- Gherbi H., Gallego M.E., Jalut N., Lucht J.M., Hohn B., White C.I. (2001). Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2: 287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Knoll A., Wurz-Wildersinn R., Puchta H. (2008). Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 4: e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Puchta H. (2007). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18836–18841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilnytskyy Y., Boyko A., Kovalchuk I. (2004). Luciferase-based transgenic recombination assay is more sensitive than beta-glucoronidase-based. Mutat. Res. 559: 189–197 [DOI] [PubMed] [Google Scholar]
- Kilby N.J., Leyser H.M., Furner I.J. (1992). Promoter methylation and progressive transgene inactivation in Arabidopsis. Plant Mol. Biol. 20: 103–112 [DOI] [PubMed] [Google Scholar]
- Kinoshita Y., Saze H., Kinoshita T., Miura A., Soppe W.J., Koornneef M., Kakutani T. (2007). Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 49: 38–45 [DOI] [PubMed] [Google Scholar]
- Kirik A., Pecinka A., Wendeler E., Reiss B. (2006). The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18: 2431–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A., Higgins J.D., Seeliger K., Reha S.J., Dangel N.J., Bauknecht M., Schröpfer S., Franklin F.C., Puchta H. (2012). The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 24: 1448–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A., Puchta H. (2011). The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J. Exp. Bot. 62: 1565–1579 [DOI] [PubMed] [Google Scholar]
- Kovalchuk I., Kovalchuk O., Arkhipov A., Hohn B. (1998). Transgenic plants are sensitive bioindicators of nuclear pollution caused by the Chernobyl accident. Nat. Biotechnol. 16: 1054–1059 [DOI] [PubMed] [Google Scholar]
- Kovalchuk I., Kovalchuk O., Kalck V., Boyko V., Filkowski J., Heinlein M., Hohn B. (2003). Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 423: 760–762 [DOI] [PubMed] [Google Scholar]
- Lebel E.G., Masson J., Bogucki A., Paszkowski J. (1993). Stress-induced intrachromosomal recombination in plant somatic cells. Proc. Natl. Acad. Sci. USA 90: 422–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Santerre-Ayotte S., Boivin E.B., Jean M., Belzile F. (2004). A novel reporter for intrachromosomal homoeologous recombination in Arabidopsis thaliana. Plant J. 40: 1007–1015 [DOI] [PubMed] [Google Scholar]
- Liang L., Flury S., Kalck V., Hohn B., Molinier J. (2006). CENTRIN2 interacts with the Arabidopsis homolog of the human XPC protein (AtRAD4) and contributes to efficient synthesis-dependent repair of bulky DNA lesions. Plant Mol. Biol. 61: 345–356 [DOI] [PubMed] [Google Scholar]
- Lucht J.M., Mauch-Mani B., Steiner H.Y., Metraux J.P., Ryals J., Hohn B. (2002). Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat. Genet. 30: 311–314 [DOI] [PubMed] [Google Scholar]
- Mannuss A., Dukowic-Schulze S., Suer S., Hartung F., Pacher M., Puchta H. (2010). RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell 22: 3318–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Ries G., Bonhoeffer S., Hohn B. (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Ries G., Zipfel C., Hohn B. (2006). Transgeneration memory of stress in plants. Nature 442: 1046–1049 [DOI] [PubMed] [Google Scholar]
- Opperman R., Emmanuel E., Levy A.A. (2004). The effect of sequence divergence on recombination between direct repeats in Arabidopsis. Genetics 168: 2207–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orel N., Kyryk A., Puchta H. (2003). Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. Plant J. 35: 604–612 [DOI] [PubMed] [Google Scholar]
- Pâques F., Haber J.E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A., Rosa M., Schikora A., Berlinger M., Hirt H., Luschnig C., Mittelsten Scheid O. (2009). Transgenerational stress memory is not a general response in Arabidopsis. PLoS ONE 4: e5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhans A., Schlüpmann H., Basse C., Paszkowski J. (1990). Intrachromosomal recombination in plants. EMBO J. 9: 3437–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H. (2005). The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 56: 1–14 [DOI] [PubMed] [Google Scholar]
- Puchta H., Dujon B., Hohn B. (1996). Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. USA 93: 5055–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H., Swoboda P., Hohn B. (1995). Induction of intrachromosomal homologous recombination in whole plants. Plant J. 7: 203–210 [Google Scholar]
- Qin X.F., Holuigue L., Horvath D.M., Chua N.H. (1994). Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell 6: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries G., Heller W., Puchta H., Sandermann H., Seidlitz H.K., Hohn B. (2000). Elevated UV-B radiation reduces genome stability in plants. Nature 406: 98–101 [DOI] [PubMed] [Google Scholar]
- Roth N., Klimesch J., Dukowic-Schulze S., Pacher M., Mannuss A., Puchta H. (October 1, 2012). The requirement for recombination factors differs considerably between different pathways of homologous double-strand break repair in somatic plant cells. Plant J. http://dx.doi.org/10.1111/j.1365-313X.2012.05119.x [DOI] [PubMed] [Google Scholar]
- Schuermann D., Fritsch O., Lucht J.M., Hohn B. (2009). Replication stress leads to genome instabilities in Arabidopsis DNA polymerase delta mutants. Plant Cell 21: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann D., Molinier J., Fritsch O., Hohn B. (2005). The dual nature of homologous recombination in plants. Trends Genet. 21: 172–181 [DOI] [PubMed] [Google Scholar]
- Seeliger K., Dukowic-Schulze S., Wurz-Wildersinn R., Pacher M., Puchta H. (2012). BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana. New Phytol. 193: 364–375 [DOI] [PubMed] [Google Scholar]
- Shalev G., Levy A.A. (1997). The maize transposable element Ac induces recombination between the donor site and an homologous ectopic sequence. Genetics 146: 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert R., Puchta H. (2002). Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell 14: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Zhang Y., Yang S., Chen J.Q., Hohn B., Tian D. (2008). Insertion DNA promotes ectopic recombination during meiosis in Arabidopsis. Mol. Biol. Evol. 25: 2079–2083 [DOI] [PubMed] [Google Scholar]
- Swoboda P., Gal S., Hohn B., Puchta H. (1994). Intrachromosomal homologous recombination in whole plants. EMBO J. 13: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland B., Hohn B., Puchta H. (1994). Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus. Proc. Natl. Acad. Sci. USA 91: 8000–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B., Rahman M., Hommelsheim C.M., Berson T., Thomas S., Chandrasekar B., Olcay A.C., Berendzen K.W., Frantzeskakis L. (2012). Re-evaluation of the reliability and usefulness of the somatic homologous recombination reporter lines. Plant Cell 24: 4314–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., White C.I., Rendtlew Danielsen J.M., Yang Y.G., Qi Y. (2012). A role for small RNAs in DNA double-strand break repair. Cell 149: 101–112 [DOI] [PubMed] [Google Scholar]
- Xiao Y.L., Peterson T. (2000). Intrachromosomal homologous recombination in Arabidopsis induced by a maize transposon. Mol. Gen. Genet. 263: 22–29 [DOI] [PubMed] [Google Scholar]