This work identifies 6480 long intergenic noncoding RNAs in Arabidopsis, many of which show organ-specific and stress-responsive expression. The biogenesis of a group of long intergenic noncoding RNAs is coregulated by the RNA processing proteins SERRATE, CBP20, and CBP80.
Abstract
Long intergenic noncoding RNAs (lincRNAs) transcribed from intergenic regions of yeast and animal genomes play important roles in key biological processes. Yet, plant lincRNAs remain poorly characterized and how lincRNA biogenesis is regulated is unclear. Using a reproducibility-based bioinformatics strategy to analyze 200 Arabidopsis thaliana transcriptome data sets, we identified 13,230 intergenic transcripts of which 6480 can be classified as lincRNAs. Expression of 2708 lincRNAs was detected by RNA sequencing experiments. Transcriptome profiling by custom microarrays revealed that the majority of these lincRNAs are expressed at a level between those of mRNAs and precursors of miRNAs. A subset of lincRNA genes shows organ-specific expression, whereas others are responsive to biotic and/or abiotic stresses. Further analysis of transcriptome data in 11 mutants uncovered SERRATE, CAP BINDING PROTEIN20 (CBP20), and CBP80 as regulators of lincRNA expression and biogenesis. RT-PCR experiments confirmed these three proteins are also needed for splicing of a small group of intron-containing lincRNAs.
INTRODUCTION
Recent advances in DNA sequencing technology and transcriptome analysis have challenged the traditional view that protein coding genes are the only effectors of gene function. Noncoding RNAs (ncRNAs) have emerged as major products of the eukaryotic transcriptome with regulatory importance (Laporte et al., 2007; Rymarquis et al., 2008; Guttman et al., 2009; Fabbri and Calin, 2010; Zhang et al., 2010). Based on their characteristics, which are distinct from those of housekeeping ncRNAs, including rRNAs, tRNAs, and small nucleolar RNAs, ncRNAs can be classified as (1) small RNAs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs); (2) natural antisense transcripts (NATs); (3) long intronic noncoding RNAs; and (4) long intergenic noncoding RNAs (lincRNAs). Genome-wide computational analysis has largely been performed on small RNAs owing to their ease of cloning. Previous genome-wide analyses have identified more than 2000 cis- and trans-NATs in Arabidopsis (Wang et al., 2005; Wang et al., 2006), but these are mainly mRNAs. Some lincRNAs, such as INDUCED BY PHOSPHATE STARVATION1 (IPS1) and AT4, are known to function as target mimics of miRNAs (Shin et al., 2006; Franco-Zorrilla et al., 2007). COLDAIR, a long intronic ncRNA, has recently been implicated in the epigenetic repression of FLC during vernalization (Heo and Sung, 2011).
lincRNAs have been described in yeast as well as higher eukaryotes (Bumgarner et al., 2009; Khalil et al., 2009; Ulitsky et al., 2011), and genome-wide analysis has uncovered more than 8000 lincRNA genes in the human genome (Khalil et al., 2009; Chen and Carmichael, 2010; Cabili et al., 2011). Mammalian lincRNAs are suggested to be transcribed by RNA polymerase II and processed by both 5′-capping and 3′ poly(A) addition (Guttman et al., 2009), and many contain introns (Managadze et al., 2011; Ulitsky et al., 2011). lincRNAs are expressed in a tissue-specific manner, and many lincRNA genes are regulated by stress (Dinger et al., 2008; Cabili et al., 2011). Moreover, ∼20% of the 3300 lincRNAs in human cells are associated with polycomb repressor complex 2 (Hekimoglu and Ringrose, 2009). Emerging evidence supports the view that lincRNAs play important roles in many fundamental biological processes (Hekimoglu and Ringrose, 2009; Chen and Carmichael, 2010; Tsai et al., 2010; K.C. Wang et al., 2011; Cabianca et al., 2012). Consistent with this view, knockdown of a group of lincRNAs in mouse embryonic stem cells disrupted pluripotency and/or altered expression levels of differentiation markers (Guttman et al., 2011). In addition, genetic mutations of human lincRNAs have been associated with diseases and pathophysiological conditions (Gupta et al., 2010; Hu et al., 2011; Zhu et al., 2011; Cabianca et al., 2012).
In plants, systematic searches for ncRNAs have been conducted in Arabidopsis thaliana (MacIntosh et al., 2001; Marker et al., 2002; Rymarquis et al., 2008; Song et al., 2009; Jouannet and Crespi, 2011) and Medicago truncatula (Wen et al., 2007). However, lincRNAs have not yet been identified and investigated on a genome scale. Genome-wide bioinformatics analysis based on full-length cDNA databases identified 76 Arabidopsis non-protein-coding RNAs; 14 of these RNAs are NATs and six are associated with small RNAs (Hirsch et al., 2006; Ben Amor et al., 2009). The Arabidopsis Information Resource (TAIR) version 9 annotated 350 transcripts as “the other RNAs,” many of which are transcribed from intergenic regions. Like the group of 76 non-protein-coding RNAs, the 350 “other RNAs” comprise NATs, small RNA-related transcripts, and potential lincRNAs, as well as some transcripts of high protein-coding potential. Another analysis of tiling arrays uncovered a large number of transcripts derived from intergenic regions of the Arabidopsis genome (Matsui et al., 2008). With increasing evidence implicating important biological roles of lincRNAs in animal cells (Barsotti and Prives, 2010; Qureshi et al., 2010), a comprehensive genome-wide analysis of plant lincRNA is warranted.
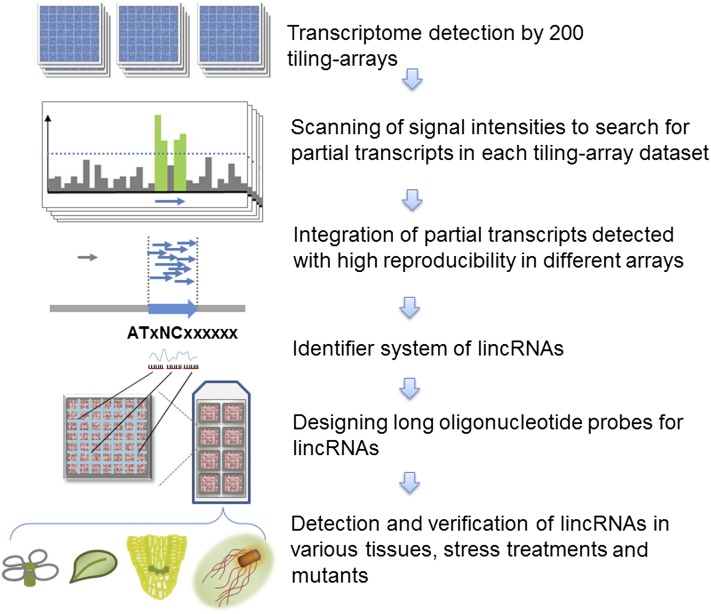
Here, we performed an integrative analysis of 200 Arabidopsis tiling array data sets using a specifically designed bioinformatics strategy and identified a total of 13,230 intergenic transcription units (TUs). Among these, 6480 TUs encoded transcripts unrelated to repeat sequences, and these transcripts were identified as lincRNAs. Moreover, we sequenced RNAs derived from four different organ samples by RNA sequencing (RNA-seq) and identified 2708 intergenic TUs encoding lincRNAs. To further validate and investigate the newly identified lincRNAs, we used a custom long-oligonucleotide expression array (ATH lincRNA v1 array) to profile lincRNA expression in various organs of wild-type plants, wild-type samples in response to environmental treatments, and Arabidopsis mutant plants (Figure 1). We also profiled expression changes of lincRNAs by reanalysis of tiling array data sets in 11 Arabidopsis mutants. Finally, we found a subgroup of lincRNAs was coregulated by CAP BINDING PROTEIN20 (CBP20), CBP80, and SERRATE (SE).
Figure 1.
Flow Chart of RepTAS.
[See online article for color version of this figure.]
RESULTS
Previously Characterized lincRNAs in Arabidopsis
By analysis of Arabidopsis EST and tiling array data sets, several groups have identified thousands of ncRNAs (MacIntosh et al., 2001; Marker et al., 2002; Rymarquis et al., 2008; Song et al., 2009; Jouannet and Crespi, 2011). However, the majority of the reported ncRNAs are NATs (Matsui et al., 2008; Okamoto et al., 2010), and it is unclear how many of these are indeed transcribed from intergenic regions. Therefore, as a first step, we reanalyzed these ncRNAs in an attempt to identify bona fide lincRNAs. In general, all intergenic transcripts could be considered as potential lincRNAs; however, some of them may also be related to other types of transcripts, such as truncated mRNAs, by-products of protein-coding genes, expressed repeats, or other ncRNAs, all of which are functionally distinct from lincRNAs (Zhang et al., 2010). Such transcripts may confound the analysis of bona fide lincRNAs. Therefore, to facilitate further investigation of lincRNAs, we used the following criteria to provide a strict definition for lincRNAs: (1) The transcript length must by ≥200 nucleotides; (2) the transcript must contain no open reading frame (ORF) encoding >100 amino acids; (3) TUs encoding lincRNAs must be located at least 500 bp away from any known protein-coding genes and genes for housekeeping ncRNAs; and (4) the TUs must not encode any transposable elements (TEs) and must not overlap with those encoding NATs. Other than the lincRNA genes, the remaining intergenic TUs were classified into the following groups (see Methods): TUs for NATs; TUs overlapping with TEs and/or repeats, called repeat-containing transcription units (RCTUs); gene-associated transcription units (GATUs); TUs encoding transcripts with long ORFs suggesting novel protein-coding genes, also named transcription units of unknown coding potential (TUCPs) (Cabili et al., 2011); and other intergenic transcription units (OITUs).
The TAIR9 version of the Arabidopsis genome annotated 350 transcripts as “other RNAs.” Applying our criteria for lincRNAs, we found only 36 transcripts could be considered as lincRNAs. Supplemental Data Set 1 online lists the reclassification details of the 350 “other RNAs.” We also searched for lincRNAs by reanalysis of 1,045,472 cDNA and/or EST sequences and identified 36 new lincRNAs (see Supplemental Data Set 2 online). Moreover, two studies of tiling array analysis based on the ARTADE algorithm provided transcriptional evidence for 6105 and 7719 ncRNAs in seed and seedling samples, respectively (Toyoda and Shinozaki, 2005; Matsui et al., 2008; Okamoto et al., 2010). After having updated their genomic loci into TAIR9, we found the majority of these ncRNAs were NATs and/or repeats, and only 32 and 61 ncRNAs in seed and seedling samples, respectively, could be considered as lincRNAs (see Supplemental Data Sets 3 and 4 online).
The reanalysis and reclassification of previously reported ncRNAs confirmed the existence of lincRNAs in Arabidopsis. However, the total number of lincRNAs (<200) was still far below the number of reported lincRNAs (550 to ∼8000) in animals (Guttman et al., 2009, 2010; Khalil et al., 2009; Cabili et al., 2011; Ulitsky et al., 2011; Pauli et al., 2012). Assuming that a large number of lincRNAs may still remain undiscovered in Arabidopsis, we developed a bioinformatics strategy to identify lincRNAs on a genome-wide basis.
A New Approach for lincRNAs Identification Based on Tiling Array Analysis
During the past decade, hundreds of tiling array data sets have been deposited in public databases. In principle, it should be possible to use hybridization data sets derived from tiling arrays to identify any lincRNAs transcribed from intergenic regions. However, the application of this approach for lincRNA analysis is complicated by the low signal-to-noise ratio, thereby generating unacceptable levels of false positives and false negatives. For example, many lincRNAs are expressed at low levels and their expression becomes detectable only in a few tissues, mutants, and/or in plants subjected to certain treatments (Matsui et al., 2008). To avoid the inclusion of false negatives, we analyzed as many high-quality tiling array data sets as were available, and to avoid the inclusion of false positives, we required reproducible detection of each putative lincRNA in at least three data sets. We argued that systemic noise would appear in a random fashion, whereas bona fide lincRNAs should be reproducibly detected in a certain number of tiling array data sets (see Methods). We referred to this strategy as reproducibility-based tiling array analysis strategy (RepTAS).
We analyzed 200 tiling array data sets comprising 100 ATH 1.0F arrays and 100 ATH 1.0R arrays (see Supplemental Data Set 5 online). These data sets were derived from tiling array experiments of 14 Arabidopsis mutants, 18 different stress treatments, and six different organs/tissues. All arrays were hybridized with labeled RNAs based on poly(A) selection. Using RepTAS, we identified 13,230 TUs in intergenic regions based on TAIR9 genome annotation (see Supplemental Data Set 6 online). Applying our criteria for bona fide lincRNA genes, we found 6728 RCTUs, 22 TUCPs, and 6480 TUs encoding lincRNAs (see Supplemental Data Set 6 online). We also found that the frequency distribution of the relative repeat-overlapping length of RCTUs displayed two peaks (see Supplemental Figure 1 and Supplemental Data Set 7 online), suggesting two independent Gaussian distributions. More than 49% RCTUs showed complete overlapping sequences with those of TEs or repeats (see Supplemental Figure 1 online, right peak). It is possible that some of the RCTU-encoding transcripts with partial sequence overlap with repeat sequences may still function as lincRNAs. To be conservative, we focused on only the 6480 strictly defined lincRNAs for further analysis.
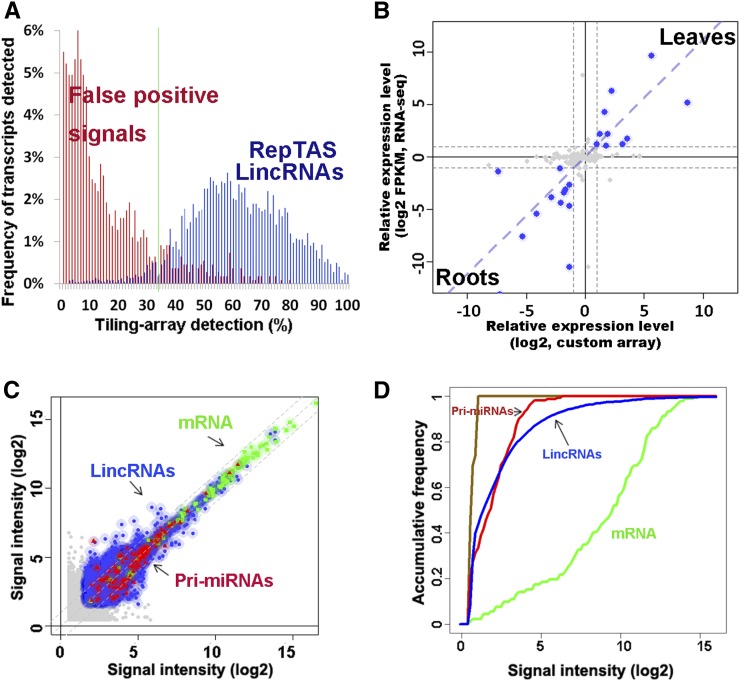
The lincRNAs were ∼200 to 1000 nucleotides in length with the majority having a length centering around 200 to 300 nucleotides (see Supplemental Figure 2 online). Figure 2A shows >95% of the lincRNAs were reproducibly detected on >35% of the tiling arrays. By contrast, 85- to 140-nucleotide signal peaks or partial transcripts that were not reproducibly detected may constitute false positive detection by tiling array probes or background noises. Probes on ATH 1.0F arrays and 100 ATH 1.0R arrays were selected to avoid cross-hybridization due to partial sequence similarity between transcripts (Naouar et al., 2009). However, signals detected by tiling array may still potentially arise from cross-hybridization (Müller et al., 2012). To address this issue, we used BLAST to compare each sequence of the 6480 lincRNAs against sequences of the remaining 6479 lincRNAs and of the mRNAs annotated by TAIR9. We found only 365 lincRNAs (∼6%) shared more than 100 nucleotides of homologous sequences with other transcripts. By analysis of tiling array data sets (Matsui et al., 2008), we compared the 365 lincRNAs and those transcripts with homologous sequences and found no significant correlation in their expression levels (Pearson correlation coefficient [PCC] = 0.11, P value > 0.26; see Supplemental Figure 3 online). These results strongly suggest that the lincRNAs detected by the RepTAS constitute a distinct group rather than a result of cross-hybridization signals.
Figure 2.
Detection of Arabidopsis lincRNAs by RepTAS and ATH lincRNA v1 Arrays.
(A) Distribution in 200 tiling arrays of lincRNAs detected by RepTAS. Blue bars show predicted lincRNAs, whereas red bars show 85 to ∼140-nucleotide signal peaks or partial transcripts detected by only two neighboring positive probes. Such partial transcripts were considered to be false positives. See Methods for details.
(B) Changes of lincRNA expression levels in roots and leaves detected by custom array and RNA-seq. lincRNAs with more than twofold change in expression level are represented by blue solid circles. Gray dashed lines show a twofold change in expression level. FPKM, fragments per kilobase of exon per million fragments mapped (Cabili et al., 2011).
(C) Expression levels of lincRNAs, pri-miRNAs, and mRNAs in two independent flower samples detected by ATH lincRNA v1 arrays. The x axis and y axis give log2 values of signal intensity detected in two biological replicates. Blue solid circles, lincRNAs; green squares, mRNAs; red triangles, pri-miRNAs; and gray solid circles, lincRNAs with signal intensities below the background value. Shadow shows the range of twofold change in signal intensity of each transcript.
(D) Accumulative distribution of lincRNA, pri-miRNA, and mRNA expression levels detected by ATH lincRNA v1 arrays. Green, mRNAs; red, lincRNAs; blue, pri-miRNAs; and brown, negative control probes. Average signal values of two independent flower samples are given.
Transcriptome Detection of lincRNAs by Custom Array
To verify expression of the identified lincRNAs and to facilitate data analysis, we designed a custom 60-mer long-oligonucleotide expression array. A similar approach was used to detect lincRNAs in mouse and human where the authors designed probes for 350 randomly selected intergenic regions (Guttman et al., 2009). Applying this strategy with some modifications, we selected a group of representative lincRNAs and other intergenic transcripts based on RepTAS (see Methods). The final array, named the ATH lincRNA v1 array, included probes for 3718 lincRNAs, 833 RCTU-derived transcripts, 45 GATU-derived transcripts, 173 precursors of miRNAs (pri-miRNAs), and 90 well-characterized mRNAs (see Supplemental Data Set 8 online). The array design and data were submitted to the Gene Expression Omnibus (GEO) database under accession numbers GPL13750 and GPL13751.
Nine ATH lincRNA v1 arrays (hereafter referred to as custom arrays) were hybridized with RNAs from Arabidopsis flowers, leaves, and roots, each with three biological replicates. Quality control analysis of the custom arrays showed the signal intensities of the spike-in probes were highly correlated with their RNA concentrations (see Supplemental Figure 4 online; PCC > 0.99). We used two criteria to identify detectable lincRNAs in each sample: (1) the signal intensity of lincRNA must be higher than the average signal intensity of the first two spike-in probes, which corresponded to an RNA concentration of ∼0.0022 fg/μL (see Supplemental Figure 4 online); (2) P values of the Mann-Whitney U test between signal intensities of lincRNAs and those of negative control probes must be lower than 0.001. Based on the cutoff values, 60 to ∼80% lincRNAs could be detected in each organ, and >92% lincRNAs were detectable in at least two custom arrays (see Supplemental Figure 5 online).
Identification and Verification of lincRNA by RNA Sequencing
Other than tiling arrays and microarrays, high-throughput RNA sequencing (RNA-seq) has been used for transcriptome profiling and lincRNA identification (Cabili et al., 2011; Ulitsky et al., 2011; Young et al., 2012). To verify and identify lincRNAs, we subjected four RNA libraries derived from roots, leaves, flowers, and siliques to RNA-seq. Each RNA library yielded 223 to 250 million 101-bp single-end sequences (235 million on average). The total number of sequencing reads approaching 1 billion was comparable to or even higher than those reported by several RNA-seq studies in other species (Guttman et al., 2009, 2010; Khalil et al., 2009; Ulitsky et al., 2011; Pauli et al., 2012). We aligned RNA sequences to TAIR9 using Tophat and SAMtools (Trapnell et al., 2009). We found that 4796 (36%) of 13,230 intergenic TUs, including 2708 (42%) of 6480 lincRNA genes, have RNA-seq sequences (see Supplemental Data Set 9 online). The mapped sequences were then assembled into transcripts using Cufflinks and Cuffcompare (Trapnell et al., 2009; Cabili et al., 2011; Trapnell et al., 2012), yielding 30,199 to ∼30,650 transcripts in each organ (see Supplemental Data Set 10 online). Of these, 29,194 to ∼29,895 transcripts (∼97%) mapped to the genomic regions of annotated Arabidopsis genes (see Supplemental Data Set 10 online). The remaining transcripts derived from intergenic regions were merged into 1340 transcripts using Cuffcompare (Cabili et al., 2011; Trapnell et al., 2012). Applying our criteria for lincRNAs, these intergenic transcripts could be classified into transcripts encoded by 678 RCTUs, 370 GATUs, seven TUCPs, seven OITUs, and 278 lincRNA genes (see Supplemental Data Set 11 online). The number of intergenic transcripts we identified by RNA-seq was much higher than that obtained by reclassification of previously reported ncRNAs (Table 1). Analysis of lincRNA abundance obtained from the two different platforms showed that the fragments per kilobase of exon per million fragments mapped were highly correlated with the signal intensities detected by custom arrays (Figure 2B; PCC = 0.79 and P value < 1.1e−05). In addition, 15 lincRNAs detected by custom arrays and/or RNA-seq were verified using quantitative RT-PCR (qRT-PCR; see Supplemental Data Set 12 online). Taken together, our results indicate the high reproducibility and reliability of transcriptome analysis profiled by the two platforms and also provide evidence that many lincRNAs are bona fide transcripts.
Table 1. Summary of Intergenic TUs Identified by Various Approaches.
| TU Type | Other RNAs | EST | Tiling Array Analysis (Seedlings) | Tiling Array Analysis (Seeds) | RepTAS | RNA-seq |
|---|---|---|---|---|---|---|
| lincRNAs | 36 | 36 | 32 | 61 | 6,480 | 278 |
| GATUs | 52 | 69 | 369 | 434 | * | 370 |
| TUCPs | 5 | 0 | 0 | 0 | 22 | 7 |
| RCTUs | 55 | 83 | 172 | 237 | 6,728 | 678 |
| OITUs | 1 | 8 | 1 | 6 | * | 7 |
| Total intergenic TUs | 149 | 196 | 574 | 738 | 13,230 | 1,340 |
*In our RepTAS analysis (Methods), GATUs and OITUs could not meet the identification criteria and therefore these TUs were filtered out.
Design of an Identifier System of Arabidopsis lincRNAs
Compared with lincRNAs previously identified by multiple platforms, 14 of the 36 lincRNAs derived from analysis of the EST data set (see Supplemental Data Set 13A online), 26 of the 32 lincRNAs and 39 of the 60 lincRNAs from two different tilling array data sets (see Supplemental Data Sets 13B and 13C online), and 167 of the 278 lincRNAs from our RNA-seq data sets were also detected by the RepTAS (see Supplemental Data Set 13D online). These results show that most previously identified lincRNAs (collectively 60%) can also be recovered by our new bioinformatics strategy. Moreover, >98% lincRNAs identified by RepTAS and RNA-seq in this study are novel transcripts in Arabidopsis (see Supplemental Data Set 13 online).
To provide an integrated and unified list for Arabidopsis lincRNAs, we developed an identifier system to annotate all the lincRNAs. For convenience, we used the nomenclature “ATxNCxxxxxx,” which is similar to the current TAIR identifier “ATxGxxxxxx,” except with the change of G to NC denoting ncRNA.
About 41% of the lincRNAs in human and mouse contain introns (Managadze et al., 2011). Among the 36 Arabidopsis lincRNAs annotated in TAIR9, 18 lincRNAs have introns (see Supplemental Data Set 14A online). Using SplicePort with Arabidopsis-specific parameters to scan for possible intron-exon splicing sites on lincRNAs (Dogan et al., 2007), we predicted full intron features with both conserved splicing donor and acceptor motifs on 595 lincRNAs and partial intron features on 1295 lincRNAs (see Supplemental Data Set 14B online).
Many different types of noncoding transcripts have been reported to be associated with promoter regions or to be derived from sequences close to the transcription start site of genes (Davis and Ares, 2006; Parkhomchuk et al., 2009; Zhao et al., 2010). Theoretically, TUs for some of these transcripts may also appear in the intergenic regions. Considering their genomic relationship with protein-coding genes, the promoter-associated TUs are preferentially located at the terminal portions of the intergenic region or more randomly positioned when the intergenic regions where they are located are short. Using TAIR9, we found that the length of lincRNA-encoding intergenic regions was comparatively longer than that of the average intergenic region of the whole genome (see Supplemental Figure 6A online), and TUs encoding lincRNA were preferentially located in the central part of intergenic regions (see Supplemental Figure 6B online). These results suggest that the identified lincRNAs are distinct from promoter-associated transcripts.
After aligning Arabidopsis lincRNA sequences with the genomic sequences of six other plant species, we found <2% lincRNAs displayed significant evolutionary conservation (see Supplemental Figure 7 online). Twenty-nine (∼5%) of the 550 zebra fish lincRNAs and 4.1 to 5.5% of the 3122 mouse lincRNAs were evolutionally conserved as measured by alignment of primary sequences, suggesting a rapid sequence evolution of lincRNAs (Ponjavic et al., 2007; Marques and Ponting, 2009; Ulitsky et al., 2011). Other studies, which used more comprehensive evolutionary analytical methods, reported a higher conservation rate (∼11%) of lincRNAs in mouse (Guttman et al., 2009; Chodroff et al., 2010). The low level of evolutionary conservation of Arabidopsis lincRNA in primary sequence is consistent with that previously reported in mammals.
The Majority of lincRNAs Are Expressed at Levels between Those of mRNAs and pri-miRNAs
We found that lincRNAs were expressed at levels higher than those of pri-miRNAs but lower than those of mRNAs as shown by scatterplots (Figure 2C), accumulative frequency of detection (Figure 2D), and distribution of expression levels in different organs (see Supplemental Figure 8 online). To confirm this finding, we analyzed nine tiling array data sets obtained from different experiments performed by various groups (Laubinger et al., 2008, 2010; Zeller et al., 2009). Again, similar expression patterns appeared in all nine independent experiments that used various tissues and different stress treatments (see Supplemental Figure 9 online). We concluded that a general feature of Arabidopsis lincRNAs is that the majority of them are expressed ∼30- to ∼60-fold lower than mRNA levels (see Supplemental Figures 8D to 8F online). Similar expression patterns were observed with mammalian lincRNAs (Khalil et al., 2009). These results suggest that lincRNAs may differ from mRNAs in their biogenesis, processing, and/or stability. Moreover, the relatively low expression level explains why only a few hundred lincRNAs have been identified based on cDNA and EST libraries.
lincRNAs Display Organ Preferential Expression
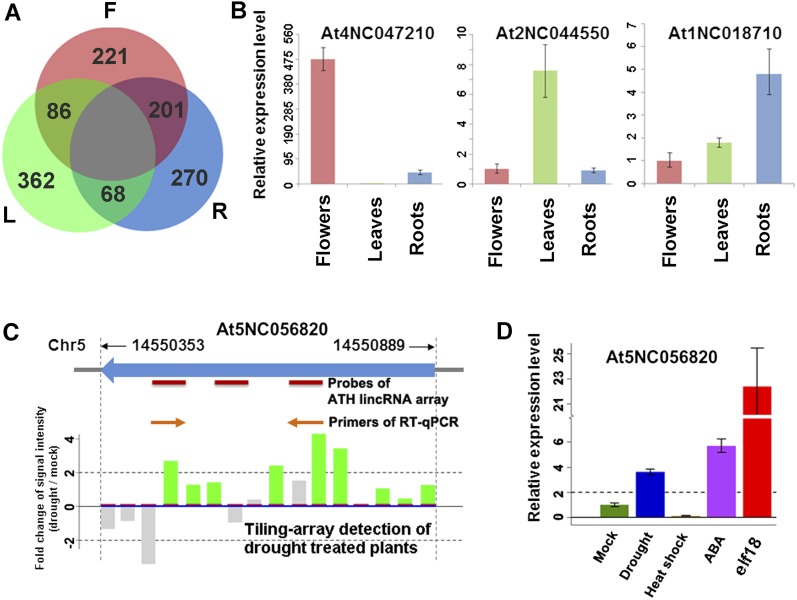
To identify lincRNAs with possible organ preferential expression, we analyzed the nine custom array data sets of flowers, leaves, and roots. Measuring differential expression of lincRNAs by empirical Bayes analysis of variance (P value of eBays ANOVA < 0.01) with a cutoff of twofold change of signal intensity (Smyth, 2004), we identified 1208 (32%) organ preferential lincRNAs from a total of 3718 lincRNAs represented on the customs arrays (see Supplemental Data Set 8B online). This group included 212 lincRNAs preferentially expressed in flowers, 362 in leaves, and 272 in roots (Figure 3A); note that a group of lincRNAs was preferentially expressed in two organs compared with the third organ (Figure 3A; see Supplemental Data Set 8B online). The expression changes of lincRNAs were reproducibly detected in three biological replicates (see Supplemental Figure 10 online). We also profiled lincRNA expression using RNA-seq and found 79 organ preferential lincRNAs from 278 assembled lincRNAs (see Supplemental Data Set 11A online). Finally, the organ preferential expression patterns of 15 representative lincRNAs were validated by qRT-PCR (see Supplemental Data Set 12 online). Figure 3B shows three typical examples of differentially expressed lincRNAs.
Figure 3.
Expression Profiles of lincRNAs in Different Arabidopsis Plant Organs and in Response to Biotic and Abiotic Stresses.
(A) A Venn diagram showing preferential expression of lincRNAs in different organs. F, flowers; L, leaves; and R, roots.
(B) Organ preferential expression of three selected lincRNAs. Relative expression levels of lincRNAs were measured by qRT-PCR. Other examples are shown in Supplemental Data Set 12 online.
(C) and (D) Detection and experimental verification of At5NC056820, a predicted lincRNA. Expression levels are given with sd bars (n = 3). Note that At5NC056820 was highly induced by elf18 and moderately induced by ABA and drought treatment.
[See online article for color version of this figure.]
Stress-Responsive lincRNAs
We analyzed the expression profile of lincRNAs in published tiling array data sets derived from stress treatments (Matsui et al., 2008) and detected 1832 lincRNAs that were significantly altered after 2 h and/or 10 h of drought, cold, high-salt, and/or abscisic acid (ABA) treatments (see Supplemental Figure 11 and Supplemental Data Set 15 online). We also investigated the expression of four representative lincRNAs by qRT-PCR (see Supplemental Figure 12 online). All of the four lincRNAs showed similar induction pattern during drought stress or ABA treatment. When this analysis was extended to several additional treatments, we found that heat shock did not elevate levels of these lincRNAs. However, treatment by elf18 (EF-Tu), which triggers pathogen-associated molecular pattern responses (Kunze et al., 2004), increased At5NC056820 expression level by 22-fold compared with the control level (Figures 3D and 3E).
The Majority of lincRNAs Are Not Associated with Small RNAs
To explore whether some lincRNAs may act as precursors of miRNAs and/or siRNAs, we aligned our published collection of small RNA sequences to lincRNAs. The small RNA data sets were derived from wild-type (Columbia-0 [Col-0]), AGO1, and AGO4 immunoprecipitation samples in flowers, leaves, roots, and seedlings (H. Wang et al., 2011). Only 163 (2.5%) out of 6516 lincRNAs (6480 identified by RepTAS and 36 by reanalysis of TAIR9 other RNAs) had related small RNAs (see Supplemental Data Set 16 online). In this group, 24 lincRNAs mainly generated 19- to ∼22-nucleotide small RNAs that were more likely to be associated with AGO1 and 129 produced 24-nucleotide small RNAs that were mainly associated with AGO4. Our results suggest that the majority of lincRNAs are processed by small RNA–independent pathways.
SE, CBP20, and CBP80 Regulate lincRNA Biogenesis
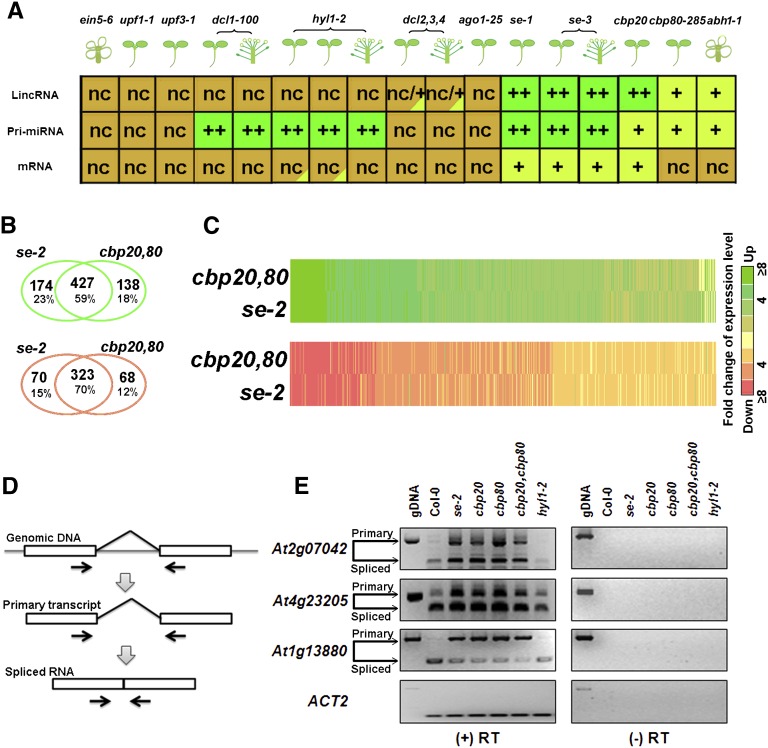
Accumulative frequency analysis has been widely used to compare expression levels or evolutionary conservation of different transcript categories (Guttman et al., 2009, 2010). This approach compares global changes in expression levels between transcript groups. To investigate possible regulators involved in lincRNA biogenesis and processing, we applied this approach to analyze lincRNA expression levels as well as those of pri-miRNAs and mRNAs in tiling array data sets derived from three different organs and 11 mutant samples. These mutants were se-1, se-3, abh1-1/cbp80-285, cbp20-1, upf1-1, upf3-1, dcl1-100, dcl2,3,4, ago1-25, hyl1-2, and ein5-6 (Gregory et al., 2008; Laubinger et al., 2008, 2010; Kurihara et al., 2009a, 2009b). Supplemental Figures 13 and 14 online compare accumulative frequency distributions of expression levels of lincRNAs, pri-miRNAs, and mRNAs between the wild type and each of the mutants. In all cases, lincRNA expression levels were lower than those of mRNAs but slightly higher than those of pri-miRNAs. Figure 4A summarizes expression patterns of the three transcript categories in these samples.
Figure 4.
Global Changes of Expression Levels of Three Transcript Categories in 11 Arabidopsis Mutants.
(A) Global changes of transcript levels in 11 mutants compared with the wild type. Plant organs are shown in cartoon formats. Dark-green “++” shows highly upregulated transcripts compared with the wild type. Light-green “+” shows slightly upregulated transcripts compared with the wild type. Red “-” shows downregulated transcripts compared with the wild type. Brown “nc” indicates no change.
(B) A Venn diagram of upregulated (green) and downregulated (red) lincRNAs in se-2 and cbp20/80 double mutant.
(C) Heat maps of 734 lincRNAs in se-2 and the cbp20/80 double mutant. Data in (B) were used for this analysis.
(D) Detection of lincRNA splicing using RT-PCR. Primers of PCR/RT-PCR are shown by arrows.
(E) Two intron retention events in two lincRNAs (AT2G07042 and AT4G23205) detected by RT-PCR in se, cbp20, cbp80, and the cbp20/80 double mutant. We used hyl1-2 as a negative control of splicing regulated by SE, CBP20, and CBP80. The AT1G13880 is an mRNA previously shown to be regulated by SE, CBP20, and CBP80 (Laubinger et al., 2008); this served as a positive control. RT-PCR products were verified by sequencing. gDNA, genomic DNA.
Among the mutants, ein5-6 (xrn4) is defective in a 5′-3′ exonuclease (Gregory et al., 2008), whereas ufp1-1 and upf3-1 are deficient in non-sense-mediated decay of transcripts (Kurihara et al., 2009b). As expected, these three mutants did not show any global expression level changes in all the three transcript categories (Figure 4A; see Supplemental Figure 13 online). The NMD mutants, upf1-1 and upf3-1, are known to accumulate aberrant transcripts (Kurihara et al., 2012), which must be clearly distinct from lincRNAs since the latter did not accumulate in these two mutants (Figure 4A; see Supplemental Figure 13 online).
We next analyzed the tiling array data sets related to dcl1-100, dcl2,3,4 triple mutant, hyl1-2, and ago1-25. Pri-miRNA expression levels were higher in dcl1-100 and hyl1-2 compared with the wild type, but no changes were seen in dcl,2,3,4 and ago1-25 (Figure 4A; see Supplemental Figure 13 online). These results are consistent with the role of DCL1 and HYL1 but not DCL2, 3, and 4 in pri-miRNA processing (Laubinger et al., 2010). A deficiency of DCL1 and HYL1 would lead to an accumulation of miRNA precursors (Laubinger et al., 2010). However, dcl1-100, hyl1-2, and ago1-25 did not show any global changes in lincRNA expression levels, suggesting that DCL1, HYL1, and AGO1 do not play a prominent role in lincRNA biogenesis and processing (Figures 4C and 4F; see Supplemental Figure 13 online).
SE is known to have multiple functions. This protein cooperates with HYL1 and DCL1 to process pri-miRNA (Yang et al., 2006), and along with ABH1/CBP80 and CBP20, it is required for mRNA and pri-miRNA splicing (Laubinger et al., 2008). The SE-mediated splicing event is believed to prevent generation of related siRNAs to trigger posttranscriptional gene silencing (Christie and Carroll, 2011; Christie et al., 2011). In addition, SE may also act as a transcription mediator (Voisin et al., 2009). By analysis of public tiling array data sets, we found that se-1, se-3, cbp20-1, and cbp80-285 indeed accumulated higher levels of pri-miRNAs compared with the wild type (see Supplemental Figure 14 online), confirming previous results (Laubinger et al., 2008). Moreover, we found a global increase of lincRNA expression levels in these four mutants (see Supplemental Figure 14 online) with the following order of severity: cbp20, se-3, se-1, and cbp80. The expression levels of 189 lincRNAs were upregulated (P value of Mann-Whitney U test < 0.05) in at least one of the three mutants (see Supplemental Figure 15 online). Note that around 50% of these lincRNAs showed elevated expression levels in all the three mutants (see Supplemental Figure 15 online). This result suggests that a group of lincRNAs are coregulated by SE, CBP20, and CBP80.
To further investigate lincRNA regulation by SE, CBP20, and CBP80, we used our custom arrays to detect lincRNA expression in se-2 and cbp20,80 double mutants, which have more severe phenotypes (Grigg et al., 2005). We found the expression levels of 750 lincRNAs (20%) out of the 3718 lincRNAs with probes on custom array were significantly changed in the two mutants (Figures 4B and 4C; P value of eBays ANOVA < 0.05 and fold change of signal intensity ≥ 2). This group included 427 co-upregulated and 323 co-downregulated lincRNAs (see Supplemental Data Set 17 online). Supplemental Data Set 18 online shows qRT-PCR verification of expression levels of 10 lincRNAs in the wild type, se-2, cbp20-1, cbp80-285, and cbp20,80 as well as hyl1-2 as a negative control. We also found a larger number and proportion of lincRNAs coregulated by SE and CBPs compared with those obtained from analysis of previous tiling array data. This difference may be attributed to the different severity of the mutant alleles used and the increased sensitivity of our custom arrays. Together, our results provide evidence that a group of lincRNAs are coregulated by SE, CBP20, and CBP80.
In a genome-wide study, Laubinger et al. (2008) detected 140 intron retention events (0.5%) out of 30,615 introns of pri-miRNAs and mRNAs in se, cbp20, and cbp80. Of the 36 lincRNAs annotated in TAIR9 as “other RNAs,” four were co-upregulated by SE and CBPs and at least three lincRNAs contained introns (see Supplemental Data Set 14C online). If SE, CBP20, and CBP80 indeed regulate lincRNA splicing, we would expect accumulation of unspliced lincRNAs among mutants deficient in one of these three proteins (Figure 4D). RT-PCR experiments detected two intron retention events in two annotated lincRNA in se-2, cbp20-1, cbp80-285, and cbp20,80 double mutants (Figure 4E). As a negative control, the intron retention events were not detected in the hyl1-2 mutant. These results confirm that SE, CBP20, and CBP80 indeed regulate intron splicing of some lincRNAs, similar to their regulation of mRNAs and pri-miRNAs (Laubinger et al., 2008).
Summary of Expression Evidence of lincRNAs
Here, we used RepTAS to predict 6480 lincRNAs in the Arabidopsis genome. Because transcripts of 2708 (∼42% out of 6480) lincRNAs were directly detected by RNA-seq (see Supplemental Data Set 19 online), this lincRNA group has the most solid experimental support. Of the remaining 3772 lincRNAs, expression levels of 1629 (25% out of 6480) lincRNAs showed significant expression changes in different organs under various stress treatments and/or in se and cbp20/80 mutants (see Supplemental Data Set 19 online). The expression specificity of these lincRNAs argues that they are not products of random transcription noise and suggest they have biological functions. The remaining 2143 lincRNAs, which were detected as hybridization signals on tiling arrays, could be considered as putative lincRNAs.
DISCUSSION
Multiple Technical Platforms Confirmed lincRNAs as Bona Fide Transcripts
Reproducible detection of lincRNAs in tiling arrays, custom arrays, and RNA-seq as well as by qRT-PCR provides strong evidence that they are bona fide transcripts rather than random products of transcriptional noise. Significant changes observed with many lincRNAs in different organs or during stress treatments suggest they are dynamically regulated. Moreover, specifically expressed lincRNAs are likely functional in development as well as in various stress responses. Further work will be directed toward addressing functions of selected lincRNAs using molecular techniques.
RepTAS Is an Effective Strategy to Identify lincRNAs
Fourteen out of the 36 lincRNAs identified by analysis of EST data sets were detected by RepTAS and included in our list (see Supplemental Data Set 13A online). Of the 32 and 60 lincRNAs identified by two different tiling array experiments (Lister et al., 2008; Matsui et al., 2008; Okamoto et al., 2010), 26 and 39, respectively, were detected by our analysis (see Supplemental Data Sets 13B and 13C online). This result confirmed the reliability of our bioinformatics approach for lincRNA identification. On the other hand, two reasons may account for the observation that some previously identified lincRNAs escaped detection by RepTAS. (1) The expression level of some lincRNAs may be too low to be reproducibly detected; and (2) some lincRNAs may be only expressed under certain specific conditions or in some mutants. These lincRNAs may be scored as false positive signals in our RepTAS analysis.
Another platform for genome-wide lincRNA identification is RNA-seq. Here, we collected an Arabidopsis RNA-seq data set of ∼1 billion sequences and identified 2708 lincRNAs. In zebra fish, mouse, and human, 567, 1457, and 8195 intergenic transcripts were identified from RNA-seq data sets consisting of ∼135, ∼152 million and ∼4 billion sequences, respectively (Guttman et al., 2010; Cabili et al., 2011; Ulitsky et al., 2011). The number of lincRNAs identified by RNA-seq largely depends on the depth of sequencing and the variety of samples used (Cabili et al., 2011). The number of identified lincRNAs in this study is comparable with the number of lincRNAs reported in human (Cabili et al., 2011). Our study provides an alternative and robust strategy for lincRNA identification and uncovers thousands of lincRNAs in Arabidopsis for future functional exploration.
Biogenesis and Regulation of lincRNA
Although thousands of lincRNAs have been identified in yeast and mammals, little is known about proteins that can specifically regulate lincRNA transcription, processing, and maturation. Here, we identified SE, CBP20, and CBP80 as major regulators of Arabidopsis lincRNA biogenesis, but the precise mechanism remains to be elucidated. CBP80 and CBP20 are conserved in all eukaryotes and mice and encode a protein called ARS2 that is homologous to the Arabidopsis SE (Gruber et al., 2009) Moreover, like SE, ARS2 also forms a complex with CBP80 (Gruber et al., 2009). In view of our results, it would not be surprising if lincRNA expression in these organisms is also regulated by ARS2, CBP20, and CBP80.
Although a large number of lincRNAs were upregulated in se and cbp20,80 mutants, in Arabidopsis, only 96 lincRNAs carried introns or predicted introns (see Supplemental Data Sets 14 and 17 online). This observation suggests that many non-intron-carrying lincRNAs can be also regulated by SE and CBPs. In addition, 415 lincRNAs were downregulated in se and cbp20,80 mutants, suggesting SE and/or CBPs may function as positive regulators of these lincRNAs.
In summary, we identified 6480 Arabidopsis lincRNAs by a bioinformatics approach and directly profiled 3718 lincRNAs by arrays and obtained RNA-seq evidence for 2708 lincRNAs. Our current data set provides a solid and excellent platform for future exploration of Arabidopsis lincRNA regulation and function. In mice, more than 660 lincRNAs are physically associated with polycomb repressive complexes (Khalil et al., 2009), and lincRNAs are known to interact with chromatin proteins to positively or negatively regulate expression of neighboring genes (K.C. Wang et al., 2011). The cis-function model may also operate in plants as has been recently reported with an intronic ncRNA named COLDAIR (Heo and Sung, 2011). Our genome analysis uncovered a number of lincRNAs specifically expressed under certain treatments. We believe our high-throughput lincRNA detection platform coupled with the availability of a large number of well-characterized Arabidopsis mutants will greatly facilitate the elucidation of regulatory functions of lincRNAs, and the ensuing results should provide mechanistic insights relevant to other eukaryotes.
METHODS
Plant Materials and Stress Treatment
Plants of Arabidopsis thaliana were grown in a greenhouse under long-day conditions (22°C, 16/8 h photoperiod cycles). Flowers buds and some very young siliques were separately collected from 4-week-old wild-type (Col-0) plants. Root and leaf samples were collected from 30-d-old wild-type plants (Col-0) grown hydroponically in full-strength MGRL medium under long-day conditions (Fujiwara et al., 1992). Seedling samples were collected from 2-week-old wild-type (Col-0), se-2, hyl1-1, cbp20-1, cbp80-285, and cbp20,80 plants grown on solid Murashige and Skoog (MS) medium under long-day conditions. For chemical treatments, 2-week-old seedlings were incubated in liquid MS medium containing 10 µM ABA (Sigma-Aldrich) or 1 µM elf18 (synthesized by the Proteomics Center, Rockefeller University) solutions at 22°C for 3 h under continuous light (Kunze et al., 2004). Abiotic stresses were applied to 2-week-old seedlings either by drying on Whatman 3MM papers (dehydration treatment) or incubating on solid MS medium at 37°C (heat treatment) for 3 h under continuous light. After each treatment, seedlings were harvested and frozen immediately in liquid nitrogen until use. Total RNA was extracted and treated with DNase I using RNeasy plant mini kit (Qiagen).
Summary of EST, cDNA, and Reported ncRNA Data Sets
TAIR9 annotation files (Swarbreck et al., 2008) were downloaded from the FTP server of TAIR (ftp://ftp.Arabidopsis.org/), and the sequences and genomic loci of 350 “other RNAs” were parsed from the gff-formatted file. The National Center for Biotechnology Information UniGene database build 73 contains 1,045,472 Arabidopsis EST or cDNA sequences. These EST sequences have been clustered by UniGene into 30,595 nonredundant gene-oriented clusters, also known as UniGene clusters. We downloaded the UniGene clusters and aligned them against the Arabidopsis genome sequence (version TAIR9) by BLAT with a cutoff of match ratio >95% (Kent, 2002). For each sequence, we searched for its best and unique match in the genome, and 28,235 UniGene clusters were selected for further analysis. Small RNA data sets were analyzed as described previously (H. Wang et al., 2011).
Transcriptome Analysis of Data Sets from Tiling Array Experiments
We collected 200 tiling array data sets from the GEO database (see Supplemental Data Set 5 online). Since the probe sequences of Affymetrix GeneChip Arabidopsis tiling array set (1.0F and 1.0R) were originally based on TIGR 5 (Zhang et al., 2006), we aligned these sequences against the Arabidopsis genome (TAIR9) by BLASTn. For further analysis, we selected only probes with sequences having perfect and unique sequence matches to those of the Arabidopsis genome (TAIR9). For each tiling array data set, the sum of the median signal intensity and the standard deviation of signal intensities derived from all perfect matched probes were taken as the background signal intensity. All probes with signal intensities higher than the background signal intensity were considered to be positive. The following criteria were used to scan signal peaks (or partial transcripts) on chromosomes: (1) The peak should cover a region of at least 200-bp long; (2) the peak should be detected by at least three positive probes; (3) there should be signal detected by at least one positive probe in every 120 bp of the peak region; (4) no positive signals should be detected on either the 5′ or the 3′ flanking 200-nucleotide regions of the peak; and (5) P values of Mann-Whitney U test between signal intensities detected by probes corresponding to the peak region and those detected by the negative probe group should be lower than 0.001. If partial transcripts identified from at least three different tiling arrays shared an overlapping genomic locus, we merged them into a TU and the reproducibility of TU detection in 200 tiling array data sets was determined. In addition, for quality control analysis, we also defined those signal peaks and reproducibility detected by only two neighboring positive probes as false positives.
Analysis of tiling array data by Matsui et al. (2008) and Okamoto et al. (2010) identified 7719 and 6105 ncRNAs expressed in seedlings and seeds, respectively. Because the genomic loci of these ncRNAs were based on the Arabidopsis genome versions TAIR7 and TIGR5, as the first step, we parsed these genomic sequences from their loci and aligned the ncRNA sequences to TAIR9 using BLAT with a cutoff of match ratio >95% (Kent, 2002). Only ncRNAs with the best and unique matches in TAIR9 were selected for further analysis.
We also profiled transcriptomes of plants subjected to abiotic stresses and of several mutants by reanalysis of the tiling array data sets deposited in the GEO database. CEL files of tiling array data sets in each experiment were separately normalized by the Quantile method using R (Bolstad et al., 2003). Expression levels of lincRNAs, pri-miRNAs, and mRNAs were calculated by Tukey’s Median Polish procedure (Li et al., 2008) using signal intensities derived from positive probes (n ≥ 3).
Transcriptome Detection by RNA-seq
We sequenced polyadenylated RNA libraries derived from flower, leaf, root, and silique samples using Illumina HiSequation 2000 with 101-cycle single-end sequencing protocol. Each sample was sequenced in a single lane. Fastaq-formatted data sets were uploaded to the GEO database under accession number GSE38612. Sequences were aligned to TAIR9 using TopHat version V1.3.0 (Trapnell et al., 2009). The mapped sequences of each sample were assembled by Cufflinks version 1.3.0 with TAIR9 annotation as the reference (Trapnell et al., 2012). Fragments per kilobase of exon per million fragments mapped of assembled transcripts were calculated using Cuffcompare (Trapnell et al., 2012).
Criteria and Five-Step Analysis for lincRNA Identification
We used the following criteria and five-step analysis to identify lincRNAs (see Supplemental Figure 16 online). (1) The genomic loci of TUs identified by reclassification of various data sets were compared with those of TAIR9 annotated genes. TUs located on the same DNA strand but overlapped with annotated genes were defined as “TUs overlapped with annotated genes,” whereas TUs located on the antisense DNA strand and complementary to annotated genes were referred to as TUs encoding NATs. The remaining TUs were considered as “intergenic TUs” for further analysis. (2) By comparing genomic positions of intergenic TUs with those of TEs, we found a group of TUs that overlapped with TEs, and these were defined as RCTUs. (3) For the remaining TUs, we searched for their proximity to neighboring annotated genes. TUs with annotated genes located within their 500-bp flanking regions were classified as GATUs. (4) The other TUs were scanned for their protein-coding potential applying GenScan to predict ORFs with Arabidopsis specific parameters (Burge and Karlin, 1997). Those that encode more than 100 amino acids were defined as TUCPs. (5) Finally, intergenic TUs encoding transcripts longer than 200 nucleotides were defined as TUs for lincRNAs, whereas those encoding shorter transcripts were considered as OITUs.
Design of ATH lincRNA v1 Array
The ATH lincRNA v1 array was an Agilent 8 × 15 formatted array with 60-mer oligonucleotides probes that were designed using Agilent Earray with the following steps. (1) We used base composition methodology to scan probe candidates complementary to target sequences. (2) We applied the Arabidopsis-specific parameters provided by Agilent Earray to select for probe candidates. (3) Probe candidates with unique locations on TAIR9 were obtained, and among these, three probes with the best position distribution on each target transcript were selected. These three steps produced a total of 15,208 probes for transcript detection. In addition, the control probes (536) provided by Agilent were included, giving an ATH lincRNA v1 array with 15,744 probes. The array design information and data were submitted to the GEO database under accession numbers GPL13750 and GPL13751.
Transcriptome Detection by Custom Array
We performed RNA labeling, hybridization, and scanning according to the protocols of Agilent. Cyanine-3 (Cy3)–labeled cRNA was prepared from 0.5 µg RNA using the One-Color Low RNA Input Linear Amplification PLUS kit (Agilent). Cy3-labeled cRNA (1.5 µg) was fragmented at 60°C for 30 min in 250 μL containing 1× Agilent fragmentation buffer and 2× Agilent blocking agent. On completion of the reaction, 250 μL of 2× Agilent hybridization buffer was added to the mixture and hybridized to ATH lincRNA v1 array for 17 h at 65°C in a rotating Agilent hybridization oven. After hybridization, microarrays were washed 1 min at room temperature with GE Wash Buffer 1 (Agilent) and 1 min with 37°C GE Wash buffer 2 (Agilent) and then dried immediately by brief centrifugation. Slides were scanned immediately after washing on an Agilent DNA microarray scanner using one color scan setting for 8 × 15k array slides
Raw signals from ATH lincRNA v1 arrays scanned by Agilent Feature Extraction Software were normalized using GeneSpring by the Quantile method (Bolstad et al., 2003). Quality control analysis was performed using default parameters of GeneSpring. The log2 values of normalized signal intensities were analyzed using R. Differential expression patterns of transcripts were measured by eBays ANOVA using R with the limma package (Smyth, 2004). If a transcript was significantly detected by multiple probes, we selected the probe with the lowest eBays ANOVA P value for further analysis. We uploaded the raw data and normalized signal intensities of three organs from wild-type plants and se-2 and cbp20,80 samples to the GEO database under accession numbers GSE30394 and GSE35963.
qRT-PCR
A total of 1 to 2 μg of RNA previously treated with DNase I (RNeasy plant mini kit) was reverse transcribed using SuperScript III (Invitrogen) and oligo(dT) primer. cDNA was analyzed by quantitative PCR using SYBR Green Jump-Start Taq ReadyMix (Sigma-Aldrich) and the Applied Biosystems 7900HT real-time PCR system. All qRT-PCR reactions were performed in triplicates for each cDNA sample with an annealing temperature of 60°C and a total of 40 cycles of amplification. Expression levels were quantified relative to that of the housekeeping gene ACTIN2. The comparative cycle threshold method was used to quantify relative expression levels of target transcripts. Primer sequences are presented in Supplemental Figure 17 online.
Accession Numbers
The design information of ATH lincRNA v1 array and hybridization data sets are available in the GEO database under accession numbers GPL13750, GPL13751, GSE30394, and GSE35963. RNA-seq data sets are available in the Sequence Read Archive database under accession number SRP013631 and in the GEO database under accession number GSE38612. Supplemental data sets have been uploaded to datadryad.org under accession number doi:10.5061/dryad.n40hc.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Distribution of Relative Lengths of Overlapping Regions between RCTUs and Repeats.
Supplemental Figure 2. Length Distribution of lincRNAs.
Supplemental Figure 3. No Significant Correlation of Expression Levels between lincRNAs and Transcripts with Partial Homologous Sequence.
Supplemental Figure 4. Quality Control Analysis of ATH lincRNA v1 Arrays and Expression Profiling of lincRNAs.
Supplemental Figure 5. Relative Number of lincRNAs Detected in Three Different Organs.
Supplemental Figure 6. Relative Position of lincRNA in Intergenic Region.
Supplemental Figure 7. Evolutionary Conservation of lincRNAs.
Supplemental Figure 8. Comparison of lincRNA Expression Level with Those of pri-miRNAs and mRNAs.
Supplemental Figure 9. Accumulative Frequency of lincRNA Expression Level Detected in Tiling Array Data Sets.
Supplemental Figure 10. Detection of lincRNAs with Organ-Preferential Expression in Three Different Biological Replicates.
Supplemental Figure 11. Heat Maps of lincRNAs Induced by Stresses.
Supplemental Figure 12. Verification of Biotic and Abiotic Stress-Responsive lincRNAs by qRT-PCR.
Supplemental Figure 13. Accumulative Frequency of Expression Levels of pri-miRNAs, lincRNAs, and mRNAs in Different Mutants.
Supplemental Figure 14. Accumulative Frequency of Expression Levels of pri-miRNAs, lincRNAs, and mRNAs in se, cbp20, and cbp80 Mutants.
Supplemental Figure 15. Changes in Expression Levels of Upregulated lincRNAs in se-1, cbp20-1, and cbp80-285.
Supplemental Figure 16. Five-Step Analysis for Identification of TUs for lincRNA from Intergenic TUs.
Supplemental Figure 17. Primer Sequences for RT-PCR.
Supplemental Data Set 1. Classification of TAIR9 Other RNAs.
Supplemental Data Set 2. Classification of the TUs Identified by ESTs.
Supplemental Data Set 3. Classification of the TUs Previously Reported by Matsui et al. (2008).
Supplemental Data Set 4. Classification of the TUs Previously Reported by Okamoto et al. (2010).
Supplemental Data Set 5. Tiling Array Data Sets We Used for Analysis.
Supplemental Data Set 6. Classification of the TUs Identified by RepTAS.
Supplemental Data Set 7. RCTUs Identified by RepTAS.
Supplemental Data Set 8. Design of ATH LincRNA v1 Array.
Supplemental Data Set 9. lincRNAs Identified by Both RepTAS and RNA-seq.
Supplemental Data Set 10. Summary of Assembled Transcripts by RNA-seq.
Supplemental Data Set 11. Classification of the TUs Identified by RNA-seq.
Supplemental Data Set 12. Verification of Organ-Preferentially Expressed lincRNA by qRT-PCR.
Supplemental Data Set 13. Comparison of lincRNAs Identified by Multiple Platforms.
Supplemental Data Set 14. Predicted Introns of lincRNAs.
Supplemental Data Set 15. Stress-Responsive lincRNAs.
Supplemental Data Set 16. lincRNAs Associated with smRNAs.
Supplemental Data Set 17. lincRNAs Regulated by SERRATE and CBPs.
Supplemental Data Set 18. Verification of SE and CBPs Regulated lincRNAs by qRT-PCR.
Supplemental Data Set 19. Summary of Expression Evidence of lincRNA.
Acknowledgments
We thank Connie Zhao and Bin Zhang (Genomics Resource Center, Rockefeller University) for their technical support and members of the Chua lab for discussion. This work was supported in part by National Institutes of Health GM44640 and in part by the Cooperative Research Program for Agriculture Science and Technology Development (Project PJ906910), Rural Development Administration, Republic of Korea. C.A.-H. was supported by a Consejo Nacional de Ciencia y Tecnologia postdoctoral fellowship. L.B. was supported by a postdoctoral fellowship from the Spanish Ministerio de Educación (AGR-2009-0486).
AUTHOR CONTRIBUTIONS
J.L., J.X., H.W., and N.-H.C. designed experiments. J.L. and H.W. collected data and preformed bioinformatic and statistical analysis. J.L. and J.X. designed ATH lincRNA v1 arrays. S.D. performed RNA-seq experiment. J.X. and C.J. performed array hybridizations. C.J., L.B., and C.A.-H. conducted qRT-PCR verifications. J.L., J.X., and N.-H.C. wrote the article, which was reviewed and approved by all authors.
Glossary
- pri-miRNA
to be defined
- ncRNA
noncoding RNA
- miRNA
microRNA
- siRNA
small interfering RNA
- NAT
natural antisense transcript
- lincRNA
long intergenic noncoding RNA
- TAIR
The Arabidopsis Information Resource
- TU
transcription unit
- ORF
open reading frame
- TE
transposable element
- RCTU
repeat-containing transcription unit
- GATU
gene-associated transcription unit
- TUCP
transcription units of unknown coding potential
- OITU
other intergenic transcription unit
- RepTAS
reproducibility-based tiling array analysis strategy
- PCC
Pearson correlation coefficient
- GEO
Gene Expression Omnibus
- RNA-seq
RNA sequencing
- qRT-PCR
quantitative RT-PCR
- eBays ANOVA
empirical Bayes analysis of variance
- ABA
abscisic acid
- Col-0
Columbia-0
- MS
Murashige and Skoog
References
- Barsotti A.M., Prives C. (2010). Noncoding RNAs: The missing “linc” in p53-mediated repression. Cell 142: 358–360 [DOI] [PubMed] [Google Scholar]
- Ben Amor B., et al. (2009). Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 19: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Bumgarner S.L., Dowell R.D., Grisafi P., Gifford D.K., Fink G.R. (2009). Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. USA 106: 18321–18326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C., Karlin S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Cabianca D.S., Casa V., Bodega B., Xynos A., Ginelli E., Tanaka Y., Gabellini D. (2012). A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.L., Carmichael G.G. (2010). Decoding the function of nuclear long non-coding RNAs. Curr. Opin. Cell Biol. 22: 357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodroff R.A., Goodstadt L., Sirey T.M., Oliver P.L., Davies K.E., Green E.D., Molnár Z., Ponting C.P. (2010). Long noncoding RNA genes: Conservation of sequence and brain expression among diverse amniotes. Genome Biol. 11: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie M., Carroll B.J. (2011). SERRATE is required for intron suppression of RNA silencing in Arabidopsis. Plant Signal. Behav. 6: 2035–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie M., Croft L.J., Carroll B.J. (2011). Intron splicing suppresses RNA silencing in Arabidopsis. Plant J. 68: 159–167 [DOI] [PubMed] [Google Scholar]
- Davis C.A., Ares M., Jr (2006). Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103: 3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger M.E., et al. (2008). Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 18: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan R.I., Getoor L., Wilbur W.J., Mount S.M. (2007). SplicePort–An interactive splice-site analysis tool. Nucleic Acids Res. 35: W285– W291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M., Calin G.A. (2010). Beyond genomics: Interpreting the 93% of the human genome that does not encode proteins. Curr. Opin. Drug Discov. Devel. 13: 350–358 [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Hirai M.Y., Chino M., Komeda Y., Naito S. (1992). Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 99: 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory B.D., O’Malley R.C., Lister R., Urich M.A., Tonti-Filippini J., Chen H., Millar A.H., Ecker J.R. (2008). A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Grigg S.P., Canales C., Hay A., Tsiantis M. (2005). SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Gruber J.J., et al. (2009). Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell 138: 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.A., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., et al. (2011). lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., et al. (2010). Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 28: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekimoglu B., Ringrose L. (2009). Non-coding RNAs in polycomb/trithorax regulation. RNA Biol. 6: 129–137 [DOI] [PubMed] [Google Scholar]
- Heo J.B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hirsch J., Lefort V., Vankersschaver M., Boualem A., Lucas A., Thermes C., d’Aubenton-Carafa Y., Crespi M. (2006). Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol. 140: 1192–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Yuan B., Flygare J., Lodish H.F. (2011). Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 25: 2573–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouannet V., Crespi M. (2011). Long nonprotein-coding RNAs in plants. Prog. Mol. Subcell. Biol. 51: 179–200 [DOI] [PubMed] [Google Scholar]
- Kent W.J. (2002). BLAT—The BLAST-like alignment tool. Genome Res. 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A.M., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Kaminuma E., Matsui A., Kawashima M., Tanaka M., Morosawa T., Ishida J., Mochizuki Y., Shinozaki K., Toyoda T., Seki M. (2009a). Transcriptome analyses revealed diverse expression changes in ago1 and hyl1 Arabidopsis mutants. Plant Cell Physiol. 50: 1715–1720 [DOI] [PubMed] [Google Scholar]
- Kurihara Y., et al. (2009b). Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 2453–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Schmitz R.J., Nery J.R., Schultz M.D., Okubo-Kurihara E., Morosawa T., Tanaka M., Toyoda T., Seki M., Ecker J.R. (2012). Surveillance of 3′ noncoding transcripts requires FIERY1 and XRN3 in Arabidopsis. G3 (Bethesda) 2: 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte P., Merchan F., Amor B.B., Wirth S., Crespi M. (2007). Riboregulators in plant development. Biochem. Soc. Trans. 35: 1638–1642 [DOI] [PubMed] [Google Scholar]
- Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Rätsch G., Weigel D. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 8795–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Zeller G., Henz S.R., Buechel S., Sachsenberg T., Wang J.W., Rätsch G., Weigel D. (2010). Global effects of the small RNA biogenesis machinery on the Arabidopsis thaliana transcriptome. Proc. Natl. Acad. Sci. USA 107: 17466–17473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., He H., Zhang J., Wang X., Bai S., Stolc V., Tongprasit W., Young N.D., Yu O., Deng X.W. (2008). Transcriptional analysis of highly syntenic regions between Medicago truncatula and Glycine max using tiling microarrays. Genome Biol. 9: R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., O’Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh G.C., Wilkerson C., Green P.J. (2001). Identification and analysis of Arabidopsis expressed sequence tags characteristic of non-coding RNAs. Plant Physiol. 127: 765–776 [PMC free article] [PubMed] [Google Scholar]
- Managadze D., Rogozin I.B., Chernikova D., Shabalina S.A., Koonin E.V. (2011). Negative correlation between expression level and evolutionary rate of long intergenic noncoding RNAs. Genome Biol. Evol. 3: 1390–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marker C., Zemann A., Terhörst T., Kiefmann M., Kastenmayer J.P., Green P., Bachellerie J.P., Brosius J., Hüttenhofer A. (2002). Experimental RNomics: Identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol. 12: 2002–2013 [DOI] [PubMed] [Google Scholar]
- Marques A.C., Ponting C.P. (2009). Catalogues of mammalian long noncoding RNAs: Modest conservation and incompleteness. Genome Biol. 10: R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A., et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49: 1135–1149 [DOI] [PubMed] [Google Scholar]
- Müller M., Patrignani A., Rehrauer H., Gruissem W., Hennig L. (2012). Evaluation of alternative RNA labeling protocols for transcript profiling with Arabidopsis AGRONOMICS1 tiling arrays. Plant Methods 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naouar N., Vandepoele K., Lammens T., Casneuf T., Zeller G., van Hummelen P., Weigel D., Rätsch G., Inzé D., Kuiper M., De Veylder L., Vuylsteke M. (2009). Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J. 57: 184–194 [DOI] [PubMed] [Google Scholar]
- Okamoto M., et al. (2010). Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. Plant J. 62: 39–51 [DOI] [PubMed] [Google Scholar]
- Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., Krobitsch S., Lehrach H., Soldatov A. (2009). Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A., Valen E., Lin M.F., Garber M., Vastenhouw N.L., Levin J.Z., Fan L., Sandelin A., Rinn J.L., Regev A., Schier A.F. (2012). Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 22: 577–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J., Ponting C.P., Lunter G. (2007). Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 17: 556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi I.A., Mattick J.S., Mehler M.F. (2010). Long non-coding RNAs in nervous system function and disease. Brain Res. 1338: 20–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymarquis L.A., Kastenmayer J.P., Hüttenhofer A.G., Green P.J. (2008). Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci. 13: 329–334 [DOI] [PubMed] [Google Scholar]
- Shin H., Shin H.S., Chen R., Harrison M.J. (2006). Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J. 45: 712–726 [DOI] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3. [DOI] [PubMed]
- Song D., Yang Y., Yu B., Zheng B., Deng Z., Lu B.L., Chen X., Jiang T. (2009). Computational prediction of novel non-coding RNAs in Arabidopsis thaliana. BMC Bioinformatics 10(Suppl 1): S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D., et al. (2008). The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 36(Database issue): D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda T., Shinozaki K. (2005). Tiling array-driven elucidation of transcriptional structures based on maximum-likelihood and Markov models. Plant J. 43: 611–621 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. (2011). Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin D., Nawrath C., Kurdyukov S., Franke R.B., Reina-Pinto J.J., Efremova N., Will I., Schreiber L., Yephremov A. (2009). Dissection of the complex phenotype in cuticular mutants of Arabidopsis reveals a role of SERRATE as a mediator. PLoS Genet. 5: e1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Chua N.H., Wang X.J. (2006). Prediction of trans-antisense transcripts in Arabidopsis thaliana. Genome Biol. 7: R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang X., Liu J., Kiba T., Woo J., Ojo T., Hafner M., Tuschl T., Chua N.H., Wang X.J. (2011). Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J. 67: 292–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.C., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.J., Gaasterland T., Chua N.H. (2005). Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 6: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Parker B.J., Weiller G.F. (2007). In silico identification and characterization of mRNA-like noncoding transcripts in Medicago truncatula. In Silico Biol. (Gedrukt) 7: 485–505 [PubMed] [Google Scholar]
- Yang L., Liu Z., Lu F., Dong A., Huang H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47: 841–850 [DOI] [PubMed] [Google Scholar]
- Young R.S., Marques A.C., Tibbit C., Haerty W., Bassett A.R., Liu J.L., Ponting C.P. (2012). Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol. Evol. 4: 427–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller G., Henz S.R., Widmer C.K., Sachsenberg T., Rätsch G., Weigel D., Laubinger S. (2009). Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J. 58: 1068–1082 [DOI] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Ecker J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu J., Jia C., Li T., Wu R., Wang J., Chen Y., Zou X., Chen R., Wang X.J., Zhu D. (2010). Systematic identification and evolutionary features of rhesus monkey small nucleolar RNAs. BMC Genomics 11: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T. (2010). Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Yu M., Li Z., Kong C., Bi J., Li J., Gao Z. (2011). ncRAN, a newly identified long noncoding RNA, enhances human bladder tumor growth, invasion, and survival. Urology 77: 510.e1-5. [DOI] [PubMed]