This work shows that poly(A) site choice is affected in 45% or more of all genes in an Arabidopsis thaliana mutant that lacks a core polyadenylation factor subunit and that a novel poly(A) signal exists that can function in the absence of the affected protein. These results provide new insight into mechanisms of alternative polyadenylation in plants.
Abstract
The Arabidopsis thaliana ortholog of the 30-kD subunit of the mammalian Cleavage and Polyadenylation Specificity Factor (CPSF30) has been implicated in the responses of plants to oxidative stress, suggesting a role for alternative polyadenylation. To better understand this, poly(A) site choice was studied in a mutant (oxt6) deficient in CPSF30 expression using a genome-scale approach. The results indicate that poly(A) site choice in a large majority of Arabidopsis genes is altered in the oxt6 mutant. A number of poly(A) sites were identified that are seen only in the wild type or oxt6 mutant. Interestingly, putative polyadenylation signals associated with sites that are seen only in the oxt6 mutant are decidedly different from the canonical plant polyadenylation signal, lacking the characteristic A-rich near-upstream element (where AAUAAA can be found); this suggests that CPSF30 functions in the handling of the near-upstream element. The sets of genes that possess sites seen only in the wild type or mutant were enriched for those involved in stress and defense responses, a result consistent with the properties of the oxt6 mutant. Taken together, these studies provide new insights into the mechanisms and consequences of CPSF30-mediated alternative polyadenylation.
INTRODUCTION
The polyadenylation of mRNAs in the nucleus is a key step in eukaryotic gene expression, as it results in the addition of a poly(A) tail that is integrally involved in various aspects of mRNA functionality (Edmonds, 2002; Lemay et al., 2010). The processes that result in polyadenylation are closely associated with other steps in mRNA biogenesis, including transcription initiation, elongation, and termination as well as transport of the mRNA from nucleus to the cytoplasm (Bentley, 2002,Bentley, 2005; Buratowski, 2005; Kim et al., 2010; Lemay et al., 2010; Mapendano et al., 2010). Beyond the links with mRNA synthesis, transport, and function, polyadenylation also affects gene expression by determining the coding and regulatory potential of an mRNA. Especially with genes whose mRNAs may be polyadenylated at more than one position within the primary transcript, poly(A) site choice has the potential to contribute to regulation and ultimately to gene function (Lutz and Moreira, 2011; Xing and Li, 2011).
In plants, mRNA polyadenylation is mediated by a complex that consists of subunits that are, for the most part, evolutionarily conserved (Belostotsky and Rose, 2005; Hunt, 2008). Of these subunits, one of the more enigmatic is the 30-kD subunit of the Cleavage and Polyadenylation Stimulatory Factor (termed in this report as CPSF30). The Arabidopsis thaliana CPSF30 possesses three characteristic CCCH zinc finger motifs; these correspond to three of the five motifs found in the mammalian CPSF30 and its yeast counterpart, YTH1p (Delaney et al., 2006; Addepalli and Hunt, 2007). Together, these three motifs constitute the most highly conserved parts of the protein. Like its mammalian and yeast counterparts, the Arabidopsis CPSF30 is an RNA binding protein (Delaney et al., 2006; Addepalli and Hunt, 2007). The RNA binding activity of the Arabidopsis CPSF30 is largely determined by the N-terminal zinc finger motif (Addepalli and Hunt, 2007). Arabidopsis CPSF30 is also an endonuclease, the action of which leaves a 3′-OH group that is a suitable substrate for poly(A) polymerase (Addepalli and Hunt, 2007). The endonuclease activity is attributable to the C-terminal zinc finger motif. Beyond these biochemical activities, the Arabidopsis CPSF30 sits at the center of a hub of protein–protein interactions involving other CPSF subunits as well as other components of the polyadenylation complex (Hunt et al., 2008; Zhao et al., 2009).
Mutant plants deficient in CPSF30 expression are more tolerant than the wild type to oxidative stresses (Zhang et al., 2008), suggesting that the protein has a regulatory role in gene expression. Consistent with this suggestion, the biochemical activities of the Arabidopsis CPSF30, RNA binding and endonuclease activity, are affected in vitro by calmodulin and sulfhydryl reagents, respectively (Delaney et al., 2006; Addepalli and Hunt, 2008). In addition, one of the three zinc finger motifs of the protein is engaged in a dithiothreitol-sensitive disulfide bond (Addepalli et al., 2010). These properties are suggestive of communication of the protein with calcium and redox cellular signaling pathways and provide conceptual links between these signaling pathways and alternative poly(A) site choice (the expected outcome of alteration of the activities of CPSF30 in the cell).
These studies reveal CPSF30 to be a possible mediator of regulated alternative polyadenylation in Arabidopsis, but they leave other questions unanswered. Among these are the nature of the role(s) of CPSF30 in the polyadenylation reaction and the scope of possible CPSF30-mediated alternative poly(A) site choice. To address these questions, a high-throughput sequencing approach (Wu et al., 2011) has been adapted to the study of poly(A) site choice in an Arabidopsis mutant (oxt6) deficient in CPSF30 (Delaney et al., 2006; Zhang et al., 2008). The results reveal a large alteration of poly(A) site choice in the mutant, but a more modest alteration of protein-coding capacity of mRNAs. In addition, the results suggest that CPSF30 is important for the functioning of one of the three cis-elements that together constitute canonical plant polyadenylation signals (Hunt, 2008; Xing and Li, 2011). Thus, these studies reveal the existence of a new class of plant poly(A) signal that apparently lacks one of the three cis-elements that constitutes the canonical signal. Finally, they argue against a hypothesis arising from previous studies (Addepalli and Hunt, 2007) that proposed a role for CPSF30 in the pre-mRNA cleavage reaction that precedes the addition of the poly(A) tail.
RESULTS
Genome-Wide Characterization of Poly(A) Site Distribution in the oxt6 Mutant
To study the functioning of CPSF30 in poly(A) site choice in vivo, the genome-wide distribution of poly(A) sites was studied in an Arabidopsis mutant deficient in CPSF30 expression, oxt6 (Delaney et al., 2006; Zhang et al., 2008), using the high-throughput DNA sequencing approach described by Wu et al. (2011). Briefly, cDNA was produced using an anchored oligo(dT) primer that had at its 5′ end a sequence compatible with Illumina DNA sequencing protocols and a template-switching oligonucleotide intended to capitalize on the propensity of Moloney Murine Leukemia Virus-derived reverse transcriptases to add short oligo-dC tracts to the 3′ ends of first-strand cDNAs (Zhu et al., 2001). The resulting cDNAs were converted to double-stranded form, digested with one of two restriction enzymes, and attached to linkers containing Illumina-compatible sequences. These poly(A) tags (termed herein as PATs) were amplified, purified, and sequenced using the Illumina high-throughput DNA sequencing platform. The sequencing output is summarized in Supplemental Figure 1 online; six data sets representing three biological replicates for the wild type and mutant were obtained, consisting of between 35,000 and 4,110,000 individual mapped PATs. This range in data set sizes reflects differences in total sequence output, different levels of multiplexing (that reduced the number of sample-specific tags), and the quality of the samples and sequencing outputs. The resulting tag sequences were processed and analyzed in a number of ways as described below.
Given that CPSF30 is a known polyadenylation factor subunit that may be involved in transcriptional pausing and termination (Nag et al., 2007), it is possible that the oxt6 mutant possesses significantly more polyadenylation outside of 3′-untranslated regions (UTRs) as a consequence of impaired termination and subsequent production of read-through RNAs. To test this possibility, the genomic distributions of PATs were determined. In both the wild type and mutant, more than 90% of PATs fell within 3′-UTRs (Table 1), as expected for the locations of poly(A) sites. Recently, it was reported that PATs that map to protein-coding regions may be artifactual due to internal priming by reverse transcriptase when using an oligo(dT)-based primer (Sherstnev et al., 2012). Even taking this into consideration (and thus removing coding sequence [CDS]-localized PATs from the calculations), the fraction of PATs that mapped to 3′-UTRs was greater than 90%. In addition, regardless of whether CDS-localized PATs were included or excluded from consideration, the relative proportions of PATs in intergenic regions or promoters were lower in the mutant than the wild type; these locations are the expected places for PATs arising from read-through transcription. Taken together, these data suggest that there is not a large increase in polyadenylation outside of annotated 3′-UTRs in the oxt6 mutant.
Table 1. Genomic Distribution of PAT in the Wild Type and oxt6 Mutant.
| The Wild Type |
oxt6 |
|||
|---|---|---|---|---|
| Regiona | PAT No.b | PAT (%)c | PAT No. | PAT (%) |
| 3′-UTR | 3011277 | 92.50 | 6148434 | 95.60 |
| Intergenic | 93661 | 2.88 | 79802 | 1.24 |
| Promoter | 46769 | 1.44 | 51366 | 0.80 |
| CDS | 14339 | 0.44 | 71745 | 1.12 |
| Intron | 14072 | 0.43 | 13734 | 0.21 |
| 5′-UTR | 10727 | 0.33 | 3722 | 0.06 |
| Exon | 4401 | 0.14 | 7046 | 0.11 |
| Pseudogenic exon | 1740 | 0.05 | 1638 | 0.03 |
| AMB | 55694 | 1.71 | 51381 | 0.80 |
Genomic region as defined in the TAIR9 database. As explained by Wu et al. (2011), the 3′-UTRs were extended by 120 nucleotides. The default length for the promoter is 2000 nucleotides. If a given intergenic region between two divergently transcribed genes is shorter than 2000 nucleotides, then the promoter is the intergenic region. AMB, regions outside of the longest annotation unit that could not be unambiguously assigned (for example, for closely spaced genes, these could be either promoter or intergenic, depending on the context being used to define the region).
Total number of curated poly(A) site tags that map to the respective genomic regions.
An additional possible consequence of an alteration of polyadenylation efficiency (as might be expected in the oxt6 mutant) is the generation of increased numbers of antisense transcripts due to read-through into adjacent, convergently transcribed genes. To examine this possibility, the genomic distributions of PATs that correspond to antisense transcripts were tabulated (Table 2). About 25% of all PATs in this study had an antisense orientation with respect to an annotated Arabidopsis gene. The vast majority of antisense-oriented PATs were associated with overlapping transcription units and were thus at once “sense” with respect to some genes and “antisense” with respect to others; this reflects the compact nature of the Arabidopsis genome. Of the antisense PATs that might be more associated with read-through transcription (those that fall in the nearby and orphan classes in Table 2), there were fewer in the oxt6 mutant than the in the wild type. This is the opposite of what would be expected, were there to be an increase in read-through transcription. This result corroborates those shown in Table 1 and argues against a significant increase in read-through transcription in the oxt6 mutant.
Table 2. Genomic Distribution of Antisense-Oriented PATs in the Wild Type and oxt6 Mutant.
| The Wild Type |
oxt6 |
|||||
|---|---|---|---|---|---|---|
| Classa | PAT No.b | aPAT (%)c | tPAT (%)d | PAT No. | aPAT (%) | tPAT (%) |
| Overlapping | 1,208,188 | 94 | 24.00 | 2,669,472 | 97.0 | 25.00 |
| Nearby | 53,652 | 4.1 | 0.70 | 51,972 | 1.9 | 0.31 |
| Orphan | 23,943 | 1.9 | 0.80 | 22,574 | 1.1 | 0.35 |
Classification of the gene or transcription unit associated with an antisense PAC. Overlapping, antisense-oriented PACs associated with overlapping, convergently transcribed genes. Nearby, antisense PACs that lie downstream from nearby, convergently transcribed genes. Orphan, antisense PACs that cannot be associated with identifiable transcription units.
Total number of curated antisense-oriented poly(A) site tags that map to the respective class.
To further explore the consequences of the oxt6 mutation, PATs and poly(A) site clusters (PACs; defined by groups of individual sites that fall within 24 nucleotides of each other; Wu et al., 2011) that were seen only in the wild type or the mutant were identified and tabulated. There were some 33,000 wild-type-specific PACs, ∼17,000 oxt6- specific PACs, and 21,000 PACs that were seen in both the wild type and mutant (wt, oxt6, and common, respectively, in Table 3). The large majority (97%) of all PATs mapped to the common PACs. Moreover, more than 93% of all PATs were situated largely within PACs located in 3′-UTRs (Table 3). These results indicate that there is not a large-scale shift in the oxt6 mutant of poly(A) site usage away from annotated 3′-UTRs. Nonetheless, a number of poly(A) sites that are specific for either the wild type or mutant can be seen. Compared with the genomic distributions of common PACs, the wild-type- and oxt6-specific PACs were more likely to be situated in regions apart from 3′-UTRs; thus, ∼54% of the wild-type-specific PACs and 65% of the oxt6-specific PACs were found in regions other than 3′-UTRs. Only 22% of the common PACs were similarly situated. Removal of PATs and PACs that map to protein-coding regions (CDS in Table 3) from these assessments did not substantially change the outcomes; thus, 50% of the wild-type-specific PACs and 57% of the oxt6-specific PACs were found in regions other than 3′-UTRs. By contrast, 18% of the common PACs fell outside of annotated 3′-UTRs, and more than 90% of all PATs defined common PACs that fell within annotated 3′-UTRs.
Table 3. Poly(A) Sites Seen Only in the Wild Type or oxt6 Mutant.
| PATa |
PACb |
|||||
|---|---|---|---|---|---|---|
| Regionc | The Wild Type | oxt6 | Common | The Wild Type | oxt6 | Common |
| 3′-UTR | 79,717 | 37,765 | 9,047,220 | 15,378 | 5,909 | 16,543 |
| Intergenic | 48,614 | 25,344 | 99,478 | 7,902 | 4,151 | 1,596 |
| Promoter | 20,090 | 16,304 | 61,768 | 3,449 | 1,853 | 819 |
| CDS | 5,375 | 20,793 | 53,530 | 2,818 | 3,066 | 964 |
| Intron | 6,670 | 4,625 | 16,496 | 2,195 | 1,037 | 412 |
| 5′-UTR | 1,488 | 1,916 | 11,045 | 201 | 100 | 54 |
| Exon | 1,421 | 1,209 | 8,815 | 286 | 112 | 94 |
| Pseudogenic exon | 285 | 69 | 3,024 | 77 | 26 | 29 |
| AMB | 4,206 | 2,489 | 101,792 | 1,018 | 464 | 763 |
| Totals | 167,866 | 110,514 | 9,403,168 | 33,324 | 16,718 | 21,274 |
Seventy-nine percent of the genes with at least one wild-type- or oxt6-specific site possessed multiple poly(A) sites (apart from sites situated in protein coding regions; see Supplemental Data Set 1 online). Of these genes, 12% had a wild-type-specific major PAC, 2% had an oxt6-specific major PAC, and 49% had a common major PAC (a major PAC is defined as a PAC with the maximum number of PATs and at least five PATs). The remainder had no clearly identifiable major PAC. For those genes that possessed multiple sites, at least one of which was a wild-type-specific site, and had an identifiable major PAC, 56% had wild-type-specific sites that were upstream of the major PAC, 59% had wild-type-specific sites that were downstream of the major PAC, 18% had both upstream and downstream wild-type-specific sites, and 24% had a wild-type-specific major PAC. For those genes that possessed multiple sites, at least one of which was an oxt6-specific PAC, 69% had oxt6-specific sites upstream from the major PAC, 37% had oxt6-specific sites downstream from the major PAC, 10% had both upstream and downstream oxt6-specific PACs, and 7% had an oxt6-specific major PAC. (Note that in these tabulations some genes can fit into more than one class; therefore, the total of percentages can exceed 100.) From these observations, it is apparent that wild-type-specific poly(A) sites may lie with somewhat equal likelihood upstream or downstream from the major PAC in a gene, while oxt6-specific sites are more likely to lie upstream of the major PAC in a gene.
Previously, more than 350 Arabidopsis genes were identified by microarray experiments whose expression changed by more than twofold in the oxt6 mutant compared with its wild-type counterpart (Zhang et al., 2008). Of these, 69% possessed multiple PACs, of which at least one was a wild-type- or oxt6-specific site based on the PAT data presented herein. Of the genes with multiple PACs, 16% had a wild-type-specific major PAC, 1.3% had an oxt6-specific major PAC, and 67% had a common major PAC. For those genes that possessed multiple sites, at least one of which was a wild-type-specific site, 48% had wild-type-specific sites that were upstream of the major PAC, 49% had wild-type-specific sites that were downstream of the major PAC, and 16% had both upstream and downstream wild-type-specific sites. For those genes that possessed multiple sites, at least one of which was an oxt6-specific PACs, 53% had oxt6-specific sites upstream from the major PAC, 30% had oxt6-specific sites downstream from the major PAC, and 2.6% had both upstream and downstream oxt6-specific PACs. These trends are similar to those seen with the complete set of Arabidopsis genes represented in the analysis summarized in the preceding paragraph.
On a global basis, wild-type- and oxt6-specific PACs are defined by a small fraction (roughly 1.7%) of all PATs (Tables 1 and 3). This global trend is also seen, for the most part, on a gene-by-gene basis. Thus, for most genes, the number of PATs that define sites seen only in the wild type (CPSF30-dependent sites) was a small fraction of all of the PATs that mapped to the respective gene, <0.18 for 75% of all genes with at least one wild-type-specific PAC (see Supplemental Figure 2 online). The fraction of PATs that define sites seen only in the oxt6 mutant was even lower, <0.06 for 75% of all genes with at least one oxt6-specific PAC (see Supplemental Figure 2 online). However, there is a great deal of variability, with several individual genes possessing values far greater than the norm.
While wild-type- and oxt6-specific PACs are represented by a small percentage of the PATs (Table 3; see Supplemental Data Set 1 online), changes in the usage of these sites are likely to have consequences in terms of mRNA and gene function; this follows from the observation that more than 50% of these sites fall outside of annotated 3′-UTRs and thus are expected to have dramatic effects on features such as exonic contents of mRNAs or the susceptibility of affected mRNAs to quality control or surveillance mechanisms. To explore this, the nature of genes possessing sites unique to the wild type or oxt6 mutant that fell within introns and 5′-UTRs was studied. As indicated in Supplemental Table 1 online, genes that encode proteins involved in defense responses are enriched in the set of genes that possess intronic or 5′-UTR–situated poly(A) sites that are seen only in the wild type. Moreover, this set of genes is more likely to encode receptors or possess properties (Leu-rich repeat, Toll/Interleukin-1 receptor, and NB-ARC [nucleotide-binding adaptor shared by APAF-1, R proteins, and CED-4] core domains) associated with receptor functions (see Supplemental Table 1 online). Intriguingly, genes associated with chloroplast and transporter functions are particularly enriched in the set of genes with oxt6-specific intronic or 5′-UTR–situated PACs (see Supplemental Table 1 online). These results indicate that the distribution of CPSF30-dependent poly(A) sites in the Arabidopsis genome is decidedly nonrandom, at least as it pertains to the association with functional classes of genes and proteins.
Genome-Wide Differences in Poly(A) Site Choice in Poly(A) Sites That Are Situated within 3′-UTRs
The data presented in Table 3 suggest that poly(A) site choice is affected in the oxt6 mutant, but perhaps not to the extent expected for a mutant that is lacking a core CPSF subunit. However, it is possible that there are more subtle shifts in poly(A) site choice in the mutant that would be missed in a binary analysis that focuses on sites present exclusively in the wild type or mutant. Accordingly, a more nuanced analysis of poly(A) site choice was conducted. This consisted of characterizing sites that map to extended 3′-UTRs using an assay designed to provide a quantitative assessment of shifts in poly(A) site choice on a gene-by-gene basis. The focus on 3′-UTR–situated sites reflects the fact that the overwhelming majority of PATs map to 3′-UTRs (Table 1); the use of a 3′-UTR database that was extended by 500 nucleotides was intended to capture downstream alternative polyadenylation events associated with read-through transcription, as might be expected in a mutant lacking CPSF30 (Nag et al., 2007).
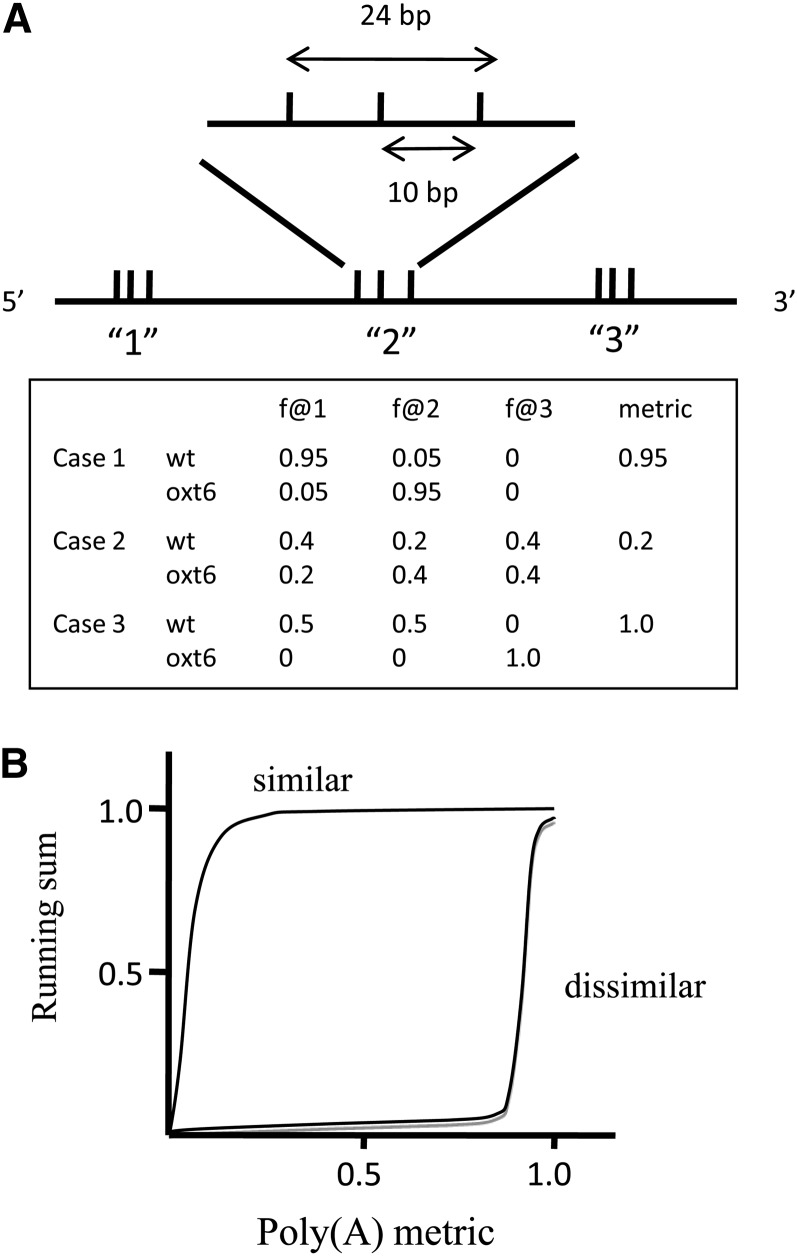
The assay developed for these studies is described in Methods and illustrated in Figure 1A. In this assay, a value is assigned to each gene that reflects differences in poly(A) site choice between two tag data sets. Thus, for a reference that showed identical distributions of tags in two different samples, the value of this metric would be 0. For another reference that showed completely different poly(A) sites in the two samples, the value would be 1. The resulting set of values was grouped in increments of 0.05 and the running sums of numbers of genes whose values fall within a given increment plotted as described in Methods and shown in Figure 1B. As displayed, data sets that are similar yield curves shifted to the left, while data sets that are very different yield curves shifted to the right.
Figure 1.
Strategy for Assessing Poly(A) Site Choice.
(A) Illustration of the clustering approach used to group closely situated poly(A) sites and of hypothetical results that may be used to generate the metric values for further analysis. The hypothetical reference sequence is at the bottom, bounded by 5′ and 3′. This reference has two clusters of poly(A) sites that are defined by the existence of sequence tags that end at the indicated positions (vertical tics). One such cluster is expanded at the top. Clustering of poly(A) sites is constrained such that the maximum distance between distinct sites is set at 10 nucleotides and the maximum span of a single cluster set to be 24 nucleotides. The table beneath illustrates three cases for illustrative purposes. In the three cases, the fraction of all tags that map to one of the two clusters in the reference sequence is calculated. From this, the absolute values of the differences between the two data sets (here, the wild type [wt] and mutant oxt6) is calculated and summed and the result divided by two to yield the value for the metric.
(B) Illustration of the two extreme hypothetical outcomes of the assay. For this, the set of metrics for a data set are divided into 20 steps of 0.05 [the values of the poly(A) metric] and the numbers of genes whose metrics fall into one of these 20 steps counted. The running sum (normalized so that the final value is 1.0) is then calculated and plotted as shown. The plots expected if two data sets are largely similar and largely dissimilar are shown. These curves represent the probable extremes, between which will fall the results obtained from actual data.
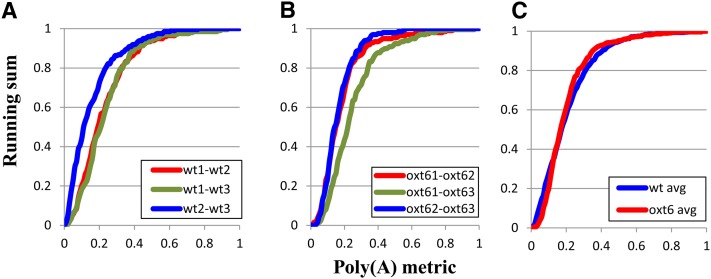
Given the complexity of the PAT protocol, a degree of inherent variability between different PAT data sets is expected. This variability would be manifest as curves that deviated significantly from the steep line expected if almost all reference sequences yielded identical results (the left-most extreme in Figure 1B). To gauge the extent of this variability, comparisons of the different members of the triplicate data sets for the wild-type and oxt6 mutant plants were conducted and plotted. As shown in Figure 2A, the plots obtained from the three wild-type comparisons were very similar. This was true even though the numbers of reference sequences that contributed to each plot varied by more than a factor of 10 (between 290 and 4600 genes). (These ranges reflect differences in the sizes of the various sequence data sets, and the fact that the comparison requires that a given reference sequence be represented by at least 15 individual tags in both data sets.) Similar results were obtained for the comparisons of the three oxt6 PAT data sets (Figure 2B). Either collectively (see Supplemental Figure 3 online) or when averaged (Figure 2C), the plots for the wild type and oxt6 comparisons were indistinguishable. These results provide a baseline estimate of the variability inherent in the genome-wide poly(A) site choice assay, and they show that the method used is highly reproducible and allows for comparisons of data sets that are very different in size.
Figure 2.
Plots of the Pairwise Comparisons of the Three Wild-Type (wt1, wt2, and wt3) and Three oxt6 (oxt61, oxt62, and oxt63) Data Sets.
(A) Results of the three wild type–wild type (wt) comparisons (wt1-wt2, wt1-wt3, and wt2-wt3) denote the three pairwise comparisons that were made. These datasets are derived from the sequencing samples as described in Methods.
(B) Results of the three oxt6-oxt6 comparisons (oxt61-oxt62, oxt61-oxt63, and oxt62-oxt63) denote the three pairwise comparisons that were made. These data sets are derived from the sequencing samples described in Methods.
(C) Plots of the averages (avg) of the three respective individual comparisons.
[See online article for color version of this figure.]
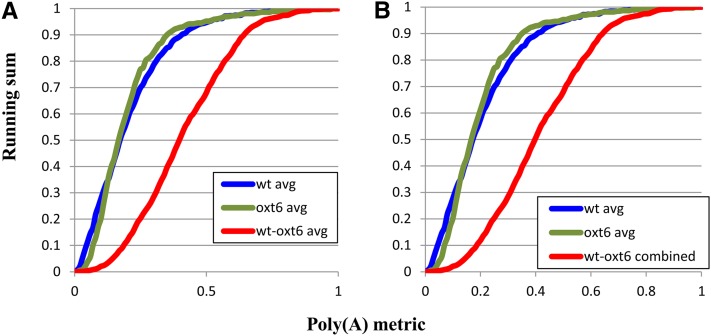
To assess the effects of the oxt6 mutation on poly(A) site choice genome-wide, the wild-type and mutant data sets were compared with each other in a systematic pairwise fashion. Two approaches were taken. For one, all nine possible pairwise comparisons were made and the results of the individual comparisons plotted (see Supplemental Figure 4 online). The results of these nine pairwise analyses were also averaged and plotted (Figure 3A). In these two instances, the curves shown in Figure 2C were included in the graphs so as to identify possible trends or differences that exceed the variability inherent in the technique. The individual plots (see Supplemental Figure 4 online) showed considerably more variation than did the plots shown in Figures 2A and 2B, but in all cases the curves deviated substantially from the wild type–wild type or oxt6-oxt6 curves. The plot of the averaged values (Figure 3A) readily showed this deviation.
Figure 3.
Plots of the Pairwise Comparisons of the Wild-Type Data Sets.
The curves showing the averages of the wild type–wild type (wt) and oxt6-oxt6 comparisons from Supplemental Figure 3 online and Figure 2C are provided for comparison’s sake.
(A) Plot of the average of the nine comparisons shown in Supplemental Figure 4 online.
(B) Plot of the results of a comparison of mappings using the combined tag data of the three sequencing runs. For this, the pooled collection of wild-type tags (e.g., wt1 + wt2 + wt3) were mapped to the extended 3′-UTR database, as was the pooled collection of oxt6 tags. avg, average.
[See online article for color version of this figure.]
The pairwise comparisons of the mappings of individual sequence collections yield between 300 and 3000 genes. Since there were no discernible differences in the three wild-type sequence data sets or in the three oxt6 sequence data sets, as indicated by the high degree of agreement in the curves shown in Figures 2A and 2B, the wild-type and oxt6 tag collections were pooled and these combined sequences mapped onto the extended 3′-UTR reference sequences. The mapping results were then used to perform a comparison, and the results plotted as shown in Figure 3B. These results corroborate those presented in Figure 3A and reveal a significant and extensive degree of variation in poly(A) site choice in the oxt6 mutant compared with the wild type. Inspection of the curves for the wild type–oxt6 comparisons (Figures 3A and 3B; see Supplemental Figure 4 online) suggests that as many as 90% of all genes are affected by the oxt6 mutation; this is apparent by noting the point at which the wild type–oxt6 curve begins to deviate from the control curves (near the y axis value of 0.1 in Figure 3; see Supplemental Figure 4 online).
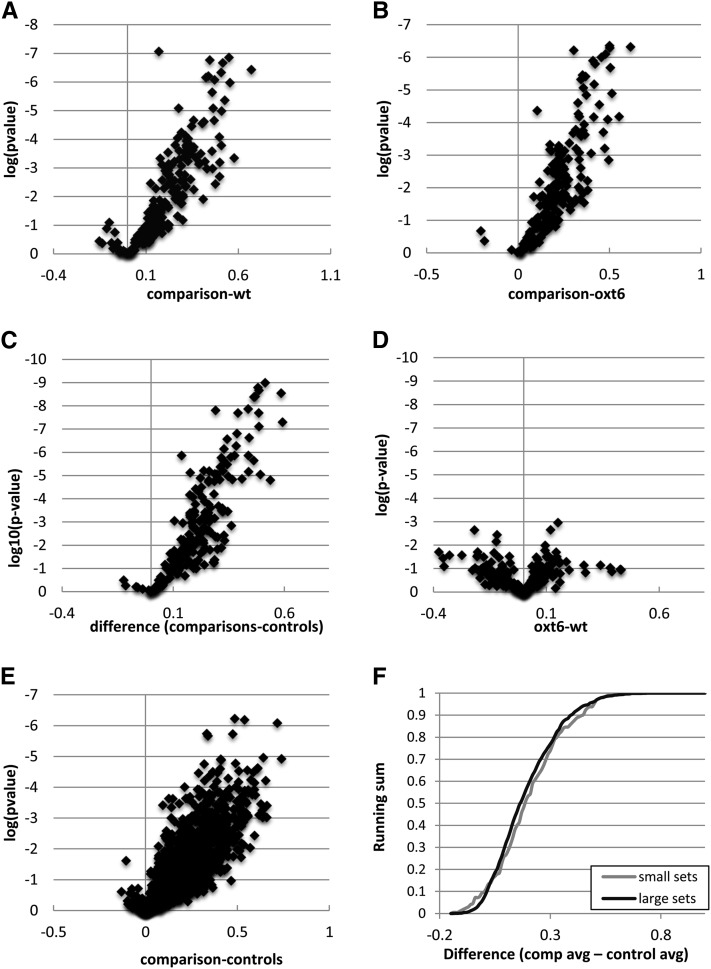
To supplement the plots shown in Figures 2 and 3, additional gene-by-gene analyses were performed. For each gene, the differences between the average metric (as defined in Figure 1) for the pairwise comparisons of “like” samples (e.g., the wt1-wt2 comparison, the wt1-wt3 comparison, etc.) and the average metric obtained for the different wild type–oxt6 comparisons (e.g., wt1-oxt61, wt1-oxt62, etc.) were calculated. In addition, the P value (calculated using a Student’s t test) of the test of the hypothesis that the two averages are the same was determined. Using these values, volcano plots as shown in Figure 4 were generated. One such plot involved 196 genes; these are all of the genes represented in all six data sets. (That there are relatively few such genes reflects the fact that one of the wild-type and one of the oxt6 data sets are ∼10% of the sizes of the other data sets [see Supplemental Figure 1 online]. Thus, there were a relatively small number of genes with the number of PATs [at least 15] required by the analysis that were also shared in all of the comparisons.) When the between-sample biological replicates (e.g., the nine wild type–oxt6 comparisons) were compared with the three wild type–wild type comparisons, the results shown in Figure 4A were obtained. Similar results were obtained when the between-sample replicates were compared with the three oxt6-oxt6 comparisons (Figure 4B). Moreover, when the nine between-sample comparisons were compared with the six within-sample comparisons, the latter taken as a single control, the results shown in Figure 4C were obtained. By contrast, when the differences between the three wild type–wild type comparisons and the three oxt6-oxt6 comparisons were similarly plotted, the results shown in Figure 4D were obtained. These results show two things: First, there is little or no significant difference between the within-sample comparisons for these 196 genes (Figure 4D); second, for most genes for which the difference in poly(A) metric exceeds 0.2, the differences are significant at the P < 0.05 level or below.
Figure 4.
Gene-by-Gene Analysis of Poly(A) Site Choice in the Wild Type and oxt6 Mutant.
(A) to (E) Two parameters were plotted. One was the difference, for each gene, of the average poly(A) metric (as described in Figure 1) obtained in different pairwise comparisons. The other was the log(10) of the P value derived from a two-tailed Student’s t test that tests the hypothesis that the means of the differences in the within-sample (e.g., wild type–wild type and oxt6-oxt6 comparisons) and between-sample (e.g., wild type–oxt6 comparisons) are the same. A flowchart is given in Supplemental Figure 6 online that elaborates on these calculations. The y axes in the plots are inverted, such that lower P values are plotted in increasing fashion. (A) to (D) show the results obtained with a small gene set, as described in the text. (E) shows the results obtained with the large gene set.
(A) A plot of the differences between the wild type (wt)–oxt6 comparison (“comparison” in all panels of this figure) and the wild type–wild type comparisons.
(B) A plot of the differences between the wild type–oxt6 comparison (“comparison” in all panels of this figure) and the oxt6-oxt6 comparisons (oxt6) as a function of the P value derived from the described Student’s t test.
(C) A plot of the differences between the wild type–oxt6 comparison (“comparison” in all panels of this figure) and the means of the control comparisons (wild type–wild type and oxt6-oxt6).
(D) A plot of the differences between the control comparisons (wild type–wild type and oxt6-oxt6).
(E) A plot of the differences between the wild type–oxt6 comparison (“comparison” in all panels of this figure) and the means of the control comparisons (wild type–wild type and oxt6-oxt6).
(F) Plot of the running sum of genes that possess increasing differences in the poly(A) metric; plots for the small (n = 196) and large (n = 1025) gene sets are shown.
To assess whether these trends are also seen in a larger number of genes, a similar analysis was done for comparisons of the larger tag data sets. This permitted only a single within-sample comparison for the wild-type and oxt6 data sets, but four wild type–oxt6 comparisons could be done; in these comparisons, 1025 genes could be assessed. A similar plot of the between-sample and within-sample data (the latter consisting of the average of the single wild type–wild type and oxt6-oxt6, analogous to that shown in Figure 4C) yielded the results shown in Figure 4E. While the statistical power of the analysis was more limited, owing to the smaller number of replicates, the same trend was seen in Figure 4E as in Figures 4A to 4C; thus, for the majority of genes with a metric difference of 0.2 or greater, the difference was significant at the P < 0.1 level or below.
These plots establish a baseline of sorts, in that they show that a difference in the poly(A) metric in the mutant and wild type of 0.2 or greater is indicative of a statistically significant change in poly(A) site choice in the mutant. This allows an estimation of the fraction of genes whose poly(A) site profiles change significantly in the mutant, by plotting the cumulative fraction of genes for which the metric differences fall between different windows (in increments of 0.05, much as is illustrated in Figure 1B and implemented in Figures 2 and 3). This was done for the 196 and 1025 gene sets analyzed in Figures 4A to 4E. The results (Figure 4F) show that there is no discernible difference between the small and large gene sets and provide a justification for an extrapolation of the conclusions drawn from these limited samples to most Arabidopsis genes. Based on the plots shown in Figure 4F, poly(A) site choice in at least 45% of all Arabidopsis genes is altered in the oxt6 mutant. This value and that derived from consideration of the plots shown in Figure 3 provide a range (45 to 90%) of the fraction of Arabidopsis genes affected by the oxt6 mutation.
Several individual reference sequences were further studied to provide additional corroboration of these results. On the basis of PAT abundance, most of the variability in poly(A) site choice was localized to 3′-UTRs and results in polymorphisms that cannot easily (if at all) be resolved by RNA gel blotting; this is compounded by the extensive occurrence, in Arabidopsis, of multiple mRNA isoforms arising from alternative promoter usage and splicing. Accordingly, 3′-rapid amplification of cDNA ends (RACE) was used for further corroboration. Four of the selected sequences are derived from genes that were studied previously using 3′-RACE (Zhang et al., 2008); the results from this previous study, plus four other sequences chosen for additional 3′-RACE corroboration, were used for the analysis shown in Supplemental Figure 5 online. For all eight genes, the 3′-RACE results show substantial differences in poly(A) site choice between the wild type and mutant. For five of these (At1g64230, At1g10410, At3g09390, At5g36910, and At5g38410), there was a relatively good correspondence in the PAT and 3′-RACE results between relative usages of sites. For two genes (At1g12390 and At2g17710), several sites not seen in the PAT data were apparent; however, in these genes, there were differences between the wild type and oxt6. For one of these genes (At1g20430), the wild-type PAT and 3′-RACE data were consistent, but the oxt6 PAT and 3′-RACE data were not. However, even in this case, there were differences between the wild type and mutant in the 3′-RACE results. While it has not been explored in detail, the discrepancies between the 3′-RACE and PAT data probably reflect the very limited sizes of the 3′-RACE data sets and possible biases due to additional manipulations associated with the 3′-RACE protocol that are absent from the PAT preparation.
Analysis of CPSF30-Dependent and -Independent Polyadenylation Signals
The fact that three classes of poly(A) sites, defined by their occurrence in either or both the wild type or oxt6 mutant, can be identified suggests that there may be different poly(A) signals in these different sets of sites. To study this possibility, a characterization of the single-nucleotide base compositions surrounding poly(A) sites was conducted; such characterizations are useful in identifying probable polyadenylation-related cis-elements (Graber et al., 1999; Loke et al., 2005). For this analysis, poly(A) sites were grouped into three sets: those seen only in the wild type (CPSF30-dependent), those seen only in the oxt6 mutant (CPSF30-independent), and those seen in both the wild type and mutant (common). Each set was subdivided according to the genomic position (e.g., falling within the extended 3′-UTR, introns, protein-coding regions, or intergenic regions). This latter subdivision was performed because previous genome-wide poly(A) site studies indicated differences in nucleotide compositions between sites located in different genomic positions (Wu et al., 2011).
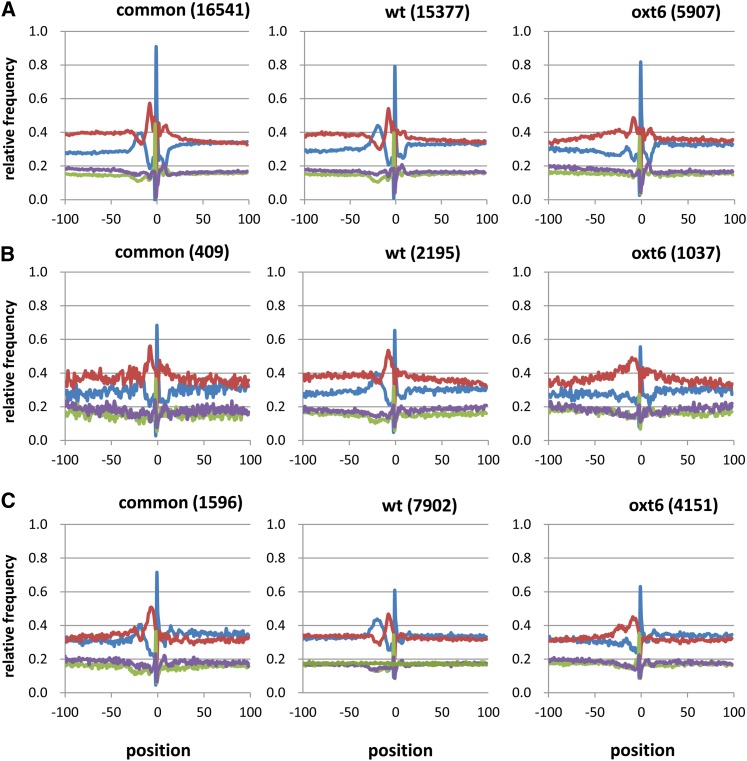
Plant poly(A) sites are flanked by regions with distinctive nucleotide composition preferences (Loke et al., 2005); these are illustrated with the “common” profiles shown in Figure 5A and include a generally high U composition within 100 nucleotides upstream of the poly(A) site, an A-rich peak centered around 20 nucleotides upstream from the poly(A) site, elevated U content immediately surrounding the poly(A) site, and a strong preference for the dinucleotide YA at the poly(A) site itself. As shown in Figure 5A, CPSF30-dependent poly(A) sites that lie within 3′-UTRs exhibit a similar pattern of nucleotide compositional trends. By contrast, CPSF30-independent sites lacked the distinctive A-rich region around −20 (Figure 5A); instead, there is a marked increase in U content, along with more subtle increases in C and G content. Similar trends are seen in poly(A) sites that fall within introns (Figure 5B) and intergenic regions (Figure 5C); in the latter cases, the common and CPSF30-dependent sites have the characteristic A-rich region around −20, while the CPSF30-independent sites lack this characteristic. Interestingly, the distinctive base compositions that correspond to the far-upstream element (FUE; upstream of −50) and cleavage element (CE) regions (−5 to +10) are indistinguishable in the three sets of sites. [For reasons mentioned above and in Sherstnev et al., (2012), poly(A) sites that fall within protein-coding regions were not included in this analysis.]
Figure 5.
Position-by-position analysis of average base composition of the regions surrounding PACs.
(A) PACs that fall within extended 3′-UTRs.
(B) PACs that fall within introns.
(C) PACs that fall in intergenic regions. In all cases, plots were generated as described previously (Loke et al., 2005). “common” - poly(A) sites seen in both the wild-type and mutant; “wt” - sites seen only in the wild-type; “oxt6” - sites seen only in the oxt6 mutant. The numbers in the parentheses are the number of sequences used for the plots. Blue traces - A; red traces - U; green traces - C; purple traces - G.
DISCUSSION
Insight into the Nature of Plant Polyadenylation Signals
In the current model for a canonical plant polyadenylation signal, three discreet cis-elements collaborate to effect efficient mRNA 3′ end formation (Graber et al., 1999; Loke et al., 2005; Shen et al., 2008; Xing et al., 2010). All of these elements are somewhat degenerate in terms of sequence composition; thus, the FUE consists of an extended U+G-rich region situated more than 50 nucleotides upstream from the poly(A) site, the near-upstream element (NUE) is an A-rich region of 6 to 10 nucleotides situated 10 to 30 nucleotides upstream from the poly(A) site, and the CE is a U-rich region centered around the poly(A) site that itself is typically a YA dinucleotide. While mutational analysis has shown that each element contributes to a high-efficiency signal (Mogen et al., 1990,Mogen et al., 1992; MacDonald et al., 1991; Sanfacon et al., 1991; Rothnie et al., 1994; Li and Hunt, 1995), most aspects of the functioning of these elements have not been defined. The results presented in this article indicate that one polyadenylation factor subunit, CPSF30, plays a role of the functioning of the NUE; this is based on the observations that poly(A) sites that are used only in the wild type, and not the oxt6 mutant, possess the characteristic A-rich NUE signature, while sites used only in the oxt6 mutant lack this signature (Figure 5). These latter sites define a new class of plant polyadenylation signal, one that lacks the A-rich NUE.
While there is a correlation between the presence of the A-rich NUE and dependence on CPSF30 for many sites, many other poly(A) sites are used in both the wild type and mutant, and these sites possess the A-rich NUE (“common” in Figure 5). Therefore, many NUE-containing sites are able to function in the absence of CPSF30. There are several possible explanations for this result. There may be two functional classes of poly(A) signal in Arabidopsis, defined by the relationship between the respective NUE and CPSF30; some NUEs may require CPSF30 for function, while others may not. Alternatively, all NUEs may require CPSF30, but some poly(A) signals may have stronger FUEs and/or CEs that can compensate for the loss of NUE function in the oxt6 mutant, while others would not. It may also be that aspects of both of these scenarios are correct; thus, there may be CPSF30-dependent and CPSF30-independent NUEs as well as various combinations of strong and weak FUEs and CEs. While perhaps not conclusive, the results presented in Figure 3 are more supportive of the second and third of these possibilities; a strict dualistic NUE model would predict that a significant fraction of genes should have a poly(A) metric that is indistinguishable from the within-sample variability, a prediction that is not borne out. Models that incorporate combinations of strong and weak FUEs and CEs may also explain the observation that the bulk of all mapped PATs define sites that are common to the wild type and oxt6 mutant (Table 3; see Supplemental Figure 2 online); these sites would be expected to have strong elements and thus be better disposed to function in the absence of CPSF30.
The Roles of CPSF30 in mRNA 3′ End Processing in Plants
While CPSF30 is a core subunit of CPSF (Jenny et al., 1996; Shi et al., 2009), and its yeast ortholog YTH1p is a core subunit of the cleavage and polyadenylation factor (CPF) (Preker et al., 1997; Ohnacker et al., 2000; Nedea et al., 2003), the precise roles of these proteins in the polyadenylation reaction remains unclear. The RNA binding properties of the proteins and direct RNA binding assays are consistent with an involvement in binding at or near the actual processing site of the pre-mRNA (Barabino et al., 1997,Barabino et al., 2000). The Drosophila melanogaster and Arabidopsis proteins possess endonucleolytic activity (Bai and Tolias, 1996; Addepalli and Hunt, 2007), suggestive of a role in the processing reaction and thus consistent with an association with the processing site. However, such a role is not easy to reconcile with the probable functioning of CPSF73 as a processing endonuclease (Ryan et al., 2004; Mandel et al., 2006). YTH1p is the subunit of CPF to which FIP1p binds (Barabino et al., 2000; Helmling et al., 2001; Tacahashi et al., 2003); thus, CPSF30 may be the link between CPF-associated RNA and poly(A) polymerase (through Fip1).
The mechanistic link between CPSF30 and NUE is not clear; as stated in the preceding paragraph, the in vitro RNA binding properties of the animal and yeast CPSF30 orthologs are not consistent with an association with the NUE, but rather with either the FUE or CE. Moreover, the results of RNA binding assays of the Arabidopsis protein are not consistent with a direct interaction between CPSF30 and the NUE (Addepalli and Hunt, 2007). It may be that CPSF30 acts as a bridge between the FUE or CE and the factor (probably CPSF, via CPSF160; Murthy and Manley, 1995) that binds the NUE; this bridging function might serve to enhance or stabilize a larger network of RNA–protein and protein–protein interactions associated with the recognition of the three cis-elements. For sites that function in both the wild type and mutant, the larger network of interactions would still be able to form in the absence of CPSF30 or the NUE such that the actual positions of poly(A) sites associated with each FUE-CE combination would be unaffected, but the relative efficiencies of sites would be altered.
Previously, it was reported that the Arabidopsis CPSF30 was an endonuclease that left a 3′-OH group suited for subsequent polyadenylation (Addepalli and Hunt, 2007). This characteristic is consistent with the suggestion that the plant CPSF30 is the endonuclease that processes the pre-mRNA prior to subsequent poly(A) addition. However, if the Arabidopsis CPSF30 is indeed a processing nuclease, then the results described in this article indicate that it cannot be the sole nuclease in the polyadenylation complex. This follows from two observations. First, 53% of all of the poly(A) sites seen collectively in the wild type and oxt6 mutant can function as poly(A) sites in the oxt6 mutant (the sum of the total PACs in the “oxt6” and “common” categories in Table 3) and thus can be processed in the absence of CPSF30. Were CPSF30 to be the sole processing endonuclease, these classes of sites would not exist.
Second, it is possible that the plant polyadenylation complex includes more than one processing endonuclease, one of which is CPSF30. In this case, polyadenylation might be rescued in the oxt6 mutant by other processing nucleases (such as CPSF73; Ryan et al., 2004; Mandel et al., 2006). However, ∼30% of all poly(A) sites are used in both the wild type and mutant (the “common” class in Table 3); with these sites, the actual positions of cleavage and polyadenylation are not affected by the presence or absence of CPSF30. It seems unlikely that alternative enzymes could assume the exact same position in the processing complex as CPSF30, such that poly(A) site position is unaffected by the removal of CPSF30. Thus, for the “common” sites in Table 3, the results suggest that CPSF30 is not the processing endonuclease. It remains possible that CPSF30 is the processing endonuclease responsible for handling of the wild-type class of PACs summarized in Table 3. However, the simplest explanation for all of the results is that CPSF30 is in fact not the processing enzyme for polyadenylation in plants.
These considerations raise the possibility of other roles for the endonuclease activity of CPSF30 apart from one in processing of the pre-mRNA prior to poly(A) addition. Such roles might be associated with other interactions or localizations of CPSF30. For example, the Arabidopsis CPSF30 accumulates in cytoplasmic locales that also possess Dcp2 (Rao et al., 2009); the nuclease activity of CPSF30 might be important for the functioning of CPSF30 in these locations, perhaps as a factor in RNA storage, transport, or degradation in the cytoplasm.
Implications for CPSF30-Mediated Alternative Polyadenylation
A defining characteristic of the Arabidopsis CPSF30 protein is that it is a calmodulin binding protein and that RNA binding by CPSF30 in vitro is inhibited by calmodulin in a calcium-dependent fashion (Delaney et al., 2006). The Arabidopsis CPSF30 is also inhibited by sulfhydryl reagents (Addepalli and Hunt, 2008), presumably through disruption of a disulfide linkage that involves two of the Cys residues in the third zinc finger motif of the protein (Addepalli et al., 2010). These properties suggest that calcium- and redox-mediated cellular signaling may inactivate CPSF30, thereby leading to numerous changes in poly(A) site choice. The results presented in this article provide insight into the possible scope of such changes. Thus, it is likely that many (between 45 and 90%, as explained above) genes in cells subjected to stimuli that activate redox- or calmodulin-mediated signaling pathways would be affected in poly(A) site choice. This global remodeling would have the potential to alter the regulatory contents of numerous 3′-UTRs, depending on the respective locations of poly(A) sites and regulatory elements such as microRNA binding sites.
In addition, as indicated in Table 3, a number of sites located outside of 3′-UTRs would be affected by inhibition of CPSF30, with wild-type-specific sites being lost and oxt6-specific sites activated. In such cases, inhibition of CPSF30 would affect the production of truncated mRNAs with altered or no function. Some interesting trends are apparent in the Gene Ontology analysis of such genes (see Supplemental Table 1 online). Thus, a disproportionate number of genes that encode defense receptor-like proteins possess poly(A) sites outside of the 3′-UTR that are not seen in the oxt6 mutant; this implies that conditions that might be expected to inhibit CPSF30 activity (such as challenge with pathogens or other treatments that would increase reactive oxygen species in the cell) would reduce the production of nonproductive transcripts encoded by genes that give rise to defense receptors. In addition, genes associated with plastid and transporter functions are more likely to possess poly(A) sites that are used only in the oxt6 mutant (see Supplemental Table 1 online). While the significance of this observation remains to be determined, it may be that CPSF30-mediated alternative polyadenylation plays role in redirecting the transcriptional output of a cell away from plastid functionality in times of stress and in fine-tuning the profile of transporter activities under these conditions.
Of course, many questions are raised by these results. For example, while it is clear that the potential scope of CPSF30-mediated alternative poly(A) site choice is considerable, it is also true that the sum total of all PATs that define CPSF30-dependent and -independent sites is rather low (Table 3; see Supplemental Figure 2 online). Thus, for most genes, PATs that define strain-specific poly(A) sites are a minor component of the total set of PATs for any given gene. This may mean that strain-specific sites do not much affect overall levels of expression of genes that possess them. Alternatively, mRNAs ending at these sites may be relatively unstable, thus representing a disproportionately large component of the transcriptional output of the respective gene. Also, truncated mRNAs may encode polypeptides that are particularly toxic or have potent regulatory activities; small quantities of mRNAs that encode such polypeptides could have effects that are out of proportion to the their steady state levels. These and other unanticipated questions and outcomes are matters that will be resolved by future research
METHODS
PAT Preparation and Data Analysis
PATs were prepared from three different plant samples for the wild type and oxt6 mutant; the wild type and mutant have been described elsewhere (Delaney et al., 2006; Zhang et al., 2008). The procedures for growing Arabidopsis thaliana plants, isolating RNA, preparing and sequencing PATs, and data analysis have been described in detail elsewhere (Wu et al., 2011); data summaries for this study are provided in Supplemental Figure 1 and Supplemental Data Sets 1 to 6 online. Sample accessions for the six sequence data sets are as follows: wt1, SRS282313; wt2, SRS282312; wt3, SRS282318; oxt61, SRS282313; oxt62, SRS282318; and oxt63, SRS282315. Note that some of the sequencing data sets had more than one sequencing sample.
To compare poly(A) site choice genome wide (Figures 1 to 4), the TAIR10 3′-UTR database (www.Arabidopsis.org) was modified so that each entry possessed an additional 500 nucleotides of sequence downstream from the end of the annotated unit; this was done to account for the possibility that sites in the oxt6 mutant might be unusually distant from the annotated unit, perhaps due to very inefficient polyadenylation or transcription termination. Curated tag sequences were mapped to the reverse complement of the modified 3′-UTR sequence file using CLC Genomics Workbench (CLC Bio) and sam files generated for each mapping. These sam files were analyzed using a Java program that generates a value, for a given gene, that reflects differences in relative poly(A) site choice in different tag datasets. To generate this value, individual poly(A) sites were grouped into clusters, with the maximum cluster size set at 24 nucleotides and the maximum distance between individual sites in a cluster set at 10 nucleotides. Subsequently, this relative proportion of tags that defined each cluster in the 3′-UTR was determined, the position-by-position difference between the two sets being compared was calculated, and the sum of the absolute values for all of the PACs in the reference calculated. This sum was divided by 2 (to provide a 0 to 1 scale) and assigned to the respective reference sequence. The resulting set of values was analyzed in two ways. The numbers of reference sequences that possessed values that increased in increments of 0.05 were determined, these values were normalized to the total number of reference sequences, and the running sum of the outputs was calculated and plotted. In addition, the averages for genes present in all replicates or in the four larger data sets (wt1, wt3, oxt62, and oxt63) were determined and assessed using a two-tailed Student’s t test. Two values, the differences in average poly(A) metric obtained from within-sample (e.g., wt1-wt2, oxt61-oxt62, etc.) and between-sample (wt1-oxt61, etc.) comparisons and the negative logarithms of the results of the t tests analyzing these comparisons, were used to generate so-called volcano plots as well as a plot of the running sum of the differences in poly(A) metrics (Figure 4). (The approach for generating the plots shown in Figure 4 is summarized in the flowchart in Supplemental Figure 6 online.)
3′-RACE Confirmation of Poly(A) Sites
First-strand cDNA synthesis was performed with an oligo(dT)V primer (see Supplemental Table 2 online) and SuperScript III reverse transcriptase (Invitrogen), using total RNA (isolated as described in Wu et al., 2011) as the template. Two rounds of PCR were performed; for each round, the same 3′ primer (“3′-adaptor” in Supplemental Table 2 online) was used. For the gene-specific primers, the one more distal to the 3′ end was used in the first round and the more proximal nested primer in the second round. PCR products were then cloned into pGEM-T Easy (Promega), and 20 random clones of each reaction were sequenced (Functional Biosciences).
Accession Numbers
The short DNA sequences generated in this study have been deposited in the National Center for Biotechnology Information Sequence Read Archive under accession number SRA048565. Accessions for the six sequence data sets are as follows: wt1, SRS282313; wt2, SRS282312; wt3, SRS282318; oxt61, SRS282313; oxt62, SRS282318; and oxt63, SRS282315.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Summary of PAT Mapping and Poly(A) Site Curation.
Supplemental Figure 2. Box Plots Showing the Fraction of All PATs That Are CPSF30 Dependent or CPSF30 Independent.
Supplemental Figure 3. Plots of the within-Genotype Pairwise Comparisons of the Wild Type and Data Sets.
Supplemental Figure 4. Plots of the between-Genotype Pairwise Comparisons of the Wild Type and oxt6 Data Sets.
Supplemental Figure 5. Comparison of Poly(A) Site Distributions as Determined by High-Throughput Sequencing (PAT) and 3′-RACE.
Supplemental Figure 6. Flowchart and Examples of the Calculations Used in Figure 4.
Supplemental Table 1. Functional Classes Overrepresented in the Set of Genes Possessing Poly(A) Sites Located outside of 3′-UTRs and Coding Regions.
Supplemental Table 2. DNA Primers Used for Confirmation.
Supplemental Data Set 1. Sense-Oriented PACs Identified in This Study.
Supplemental Data Set 2. Antisense-Oriented PACs Identified in This Study.
Supplemental Data Set 3. Genes Used in the DAVID Functional Classification Analysis.
Supplemental Data Set 4. Raw Outputs of the Analysis Presented in Supplemental Figure 3.
Supplemental Data Set 5. Poly(A) Metric Values Obtained for Each of the 196 Genes Shared by All of the 15 Pairwise Comparisons Presented in Supplemental Data Set 4.
Supplemental Data Set 6. Poly(A) Metric Values Obtained for Each of the 1025 Genes Shared by the Six Pairwise Comparisons of the Four Largest Data Sets (wt2, wt3, oxt62, and oxt63).
Acknowledgments
We thank Carol Von Lanken for technical assistance. This work was supported by U.S. National Science Foundation (IOS-0817818 to A.G.H. and Q.Q.L.), the U.S. National Institutes of Health (1R15GM094732-01 A1 to Q.Q.L.), the University of Kentucky Executive Vice President for Research, and Miami University Office for the Advancement of Research and Scholarship, and IT Service (the use of Redhawk Cluster). This project was also funded by funds from the National Natural Science Foundation of China (61174161 to G.J.), Research Funds for the Doctoral Programs of Higher Education of China (20090121110022), and the Fundamental Research Funds for the Key Universities of China, Xiamen University (2011121047, 201112G018, and CXB2011035).
AUTHOR CONTRIBUTIONS
P.E.T., X.W., M.L., B.G., and A.G.H. designed and performed most of the research. P.E.T. and X.W. developed the analytical and computational tools that are unique to this study. P.E.T., X.W., M.L., G.J., Q.Q.L., and A.G.H. participated in the data analysis. A.G.H., X.W., and Q.Q.L. wrote, edited, and revised the article.
Glossary
- UTR
untranslated region
- PAT
poly(A) tag
- CDS
coding sequence
- PAC
poly(A) site cluster
- RACE
rapid amplification of cDNA ends
- FUE
far-upstream element
- NUE
near-upstream element
- CE
cleavage element
- CPF
cleavage and polyadenylation factor
References
- Addepalli B., Hunt A.G. (2007). A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-kDa subunit of cleavage and polyadenylation specificity factor. Nucleic Acids Res. 35: 4453–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addepalli B., Hunt A.G. (2008). Redox and heavy metal effects on the biochemical activities of an Arabidopsis polyadenylation factor subunit. Arch. Biochem. Biophys. 473: 88–95 [DOI] [PubMed] [Google Scholar]
- Addepalli B., Limbach P.A., Hunt A.G. (2010). A disulfide linkage in a CCCH zinc finger motif of an Arabidopsis CPSF30 ortholog. FEBS Lett. 584: 4408–4412 [DOI] [PubMed] [Google Scholar]
- Bai C., Tolias P.P. (1996). Cleavage of RNA hairpins mediated by a developmentally regulated CCCH zinc finger protein. Mol. Cell. Biol. 16: 6661–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino S.M., Hübner W., Jenny A., Minvielle-Sebastia L., Keller W. (1997). The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 11: 1703–1716 [DOI] [PubMed] [Google Scholar]
- Barabino S.M., Ohnacker M., Keller W. (2000). Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J. 19: 3778–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belostotsky D.A., Rose A.B. (2005). Plant gene expression in the age of systems biology: Integrating transcriptional and post-transcriptional events. Trends Plant Sci. 10: 347–353 [DOI] [PubMed] [Google Scholar]
- Bentley D. (2002). The mRNA assembly line: Transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14: 336–342 [DOI] [PubMed] [Google Scholar]
- Bentley D.L. (2005). Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17: 251–256 [DOI] [PubMed] [Google Scholar]
- Buratowski S. (2005). Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 17: 257–261 [DOI] [PubMed] [Google Scholar]
- Delaney K.J., Xu R., Zhang J., Li Q.Q., Yun K.Y., Falcone D.L., Hunt A.G. (2006). Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol. 140: 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds M. (2002). A history of poly A sequences: From formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 71: 285–389 [DOI] [PubMed] [Google Scholar]
- Graber J.H., Cantor C.R., Mohr S.C., Smith T.F. (1999). In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. USA 96: 14055–14060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmling S., Zhelkovsky A., Moore C.L. (2001). Fip1 regulates the activity of Poly(A) polymerase through multiple interactions. Mol. Cell. Biol. 21: 2026–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A.G. (2008). Messenger RNA 3′ end formation in plants. Curr. Top. Microbiol. Immunol. 326: 151–177 [DOI] [PubMed] [Google Scholar]
- Hunt A.G., et al. (2008). Arabidopsis mRNA polyadenylation machinery: Comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genomics 9: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A., Minvielle-Sebastia L., Preker P.J., Keller W. (1996). Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science 274: 1514–1517 [DOI] [PubMed] [Google Scholar]
- Kim H., Erickson B., Luo W., Seward D., Graber J.H., Pollock D.D., Megee P.C., Bentley D.L. (2010). Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 17: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J.F., Lemieux C., St-André O., Bachand F. (2010). Crossing the borders: poly(A)-binding proteins working on both sides of the fence. RNA Biol. 7: 291–295 [DOI] [PubMed] [Google Scholar]
- Li Q., Hunt A.G. (1995). A near-upstream element in a plant polyadenylation signal consists of more than six nucleotides. Plant Mol. Biol. 28: 927–934 [DOI] [PubMed] [Google Scholar]
- Loke J.C., Stahlberg E.A., Strenski D.G., Haas B.J., Wood P.C., Li Q.Q. (2005). Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 138: 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C.S., Moreira A. (2011). Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip. Rev. RNA 2: 22–31 [DOI] [PubMed] [Google Scholar]
- MacDonald M.H., Mogen B.D., Hunt A.G. (1991). Characterization of the polyadenylation signal from the T-DNA-encoded octopine synthase gene. Nucleic Acids Res. 19: 5575–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel C.R., Kaneko S., Zhang H., Gebauer D., Vethantham V., Manley J.L., Tong L. (2006). Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapendano C.K., Lykke-Andersen S., Kjems J., Bertrand E., Jensen T.H. (2010). Crosstalk between mRNA 3′ end processing and transcription initiation. Mol. Cell 40: 410–422 [DOI] [PubMed] [Google Scholar]
- Mogen B.D., MacDonald M.H., Graybosch R., Hunt A.G. (1990). Upstream sequences other than AAUAAA are required for efficient messenger RNA 3′-end formation in plants. Plant Cell 2: 1261–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogen B.D., MacDonald M.H., Leggewie G., Hunt A.G. (1992). Several distinct types of sequence elements are required for efficient mRNA 3′ end formation in a pea rbcS gene. Mol. Cell. Biol. 12: 5406–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy K.G., Manley J.L. (1995). The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 9: 2672–2683 [DOI] [PubMed] [Google Scholar]
- Nag A., Narsinh K., Martinson H.G. (2007). The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat. Struct. Mol. Biol. 14: 662–669 [DOI] [PubMed] [Google Scholar]
- Nedea E., He X., Kim M., Pootoolal J., Zhong G., Canadien V., Hughes T., Buratowski S., Moore C.L., Greenblatt J. (2003). Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 278: 33000–33010 [DOI] [PubMed] [Google Scholar]
- Ohnacker M., Barabino S.M., Preker P.J., Keller W. (2000). The WD-repeat protein pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J. 19: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P.J., Ohnacker M., Minvielle-Sebastia L., Keller W. (1997). A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 16: 4727–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S., Dinkins R.D., Hunt A.G. (2009). Distinctive interactions of the Arabidopsis homolog of the 30 kD subunit of the cleavage and polyadenylation specificity factor (AtCPSF30) with other polyadenylation factor subunits. BMC Cell Biol. 10: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothnie H.M., Reid J., Hohn T. (1994). The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3′-end formation in plants. EMBO J. 13: 2200–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K., Calvo O., Manley J.L. (2004). Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon H., Brodmann P., Hohn T. (1991). A dissection of the cauliflower mosaic virus polyadenylation signal. Genes Dev. 5: 141–149 [DOI] [PubMed] [Google Scholar]
- Shen Y., Ji G., Haas B.J., Wu X., Zheng J., Reese G.J., Li Q.Q. (2008). Genome level analysis of rice mRNA 3′-end processing signals and alternative polyadenylation. Nucleic Acids Res. 36: 3150–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherstnev A., Duc C., Cole C., Zacharaki V., Hornyik C., Ozsolak F., Milos P.M., Barton G.J., Simpson G.G. (2012). Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat. Struct. Mol. Biol. 19: 845–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Chan S., Martinez-Santibañez G. (2009). An up-close look at the pre-mRNA 3′-end processing complex. RNA Biol. 6: 522–525 [DOI] [PubMed] [Google Scholar]
- Tacahashi Y., Helmling S., Moore C.L. (2003). Functional dissection of the zinc finger and flanking domains of the Yth1 cleavage/polyadenylation factor. Nucleic Acids Res. 31: 1744–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Liu M., Downie B., Liang C., Ji G., Li Q.Q., Hunt A.G. (2011). Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA 108: 12533–12538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing A., Moon B.P., Mills K.M., Falco S.C., Li Z. (2010). Revealing frequent alternative polyadenylation and widespread low-level transcription read-through of novel plant transcription terminators. Plant Biotechnol. J. 8: 772–782 [DOI] [PubMed] [Google Scholar]
- Xing D., Li Q.Q. (2011). Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip. Rev. RNA 2: 445–458 [DOI] [PubMed] [Google Scholar]
- Zhang J., Addepalli B., Yun K.Y., Hunt A.G., Xu R., Rao S., Li Q.Q., Falcone D.L. (2008). A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS ONE 3: e2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Xing D., Li Q.Q. (2009). Unique features of plant cleavage and polyadenylation specificity factor revealed by proteomic studies. Plant Physiol. 151: 1546–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.Y., Machleder E.M., Chenchik A., Li R., Siebert P.D. (2001). Reverse transcriptase template switching: A SMART approach for full-length cDNA library construction. Biotechniques 30: 892–897 [DOI] [PubMed] [Google Scholar]