The authors developed a strategy to identify putative transposon insertions that are present in a resequenced soybean population but absent in the soybean reference genome. Comparison of the distribution patterns of these elements and the patterns in the reference genome revealed evolutionary processes that reshaped the landscape of transposons after their integration in the host genome.
Abstract
Preferential accumulation of transposable elements (TEs), particularly long terminal repeat retrotransposons (LTR-RTs), in recombination-suppressed pericentromeric regions seems to be a general pattern of TE distribution in flowering plants. However, whether such a pattern was formed primarily by preferential TE insertions into pericentromeric regions or by selection against TE insertions into euchromatin remains obscure. We recently investigated TE insertions in 31 resequenced wild and cultivated soybean (Glycine max) genomes and detected 34,154 unique nonreference TE insertions mappable to the reference genome. Our data revealed consistent distribution patterns of the nonreference LTR-RT insertions and those present in the reference genome, whereas the distribution patterns of the nonreference DNA TE insertions and the accumulated ones were significantly different. The densities of the nonreference LTR-RT insertions were found to negatively correlate with the rates of local genetic recombination, but no significant correlation between the densities of nonreference DNA TE insertions and the rates of local genetic recombination was detected. These observations suggest that distinct insertional preferences were primary factors that resulted in different levels of effectiveness of purifying selection, perhaps as an effect of local genomic features, such as recombination rates and gene densities that reshaped the distribution patterns of LTR-RTs and DNA TEs in soybean.
INTRODUCTION
Transposable elements (TEs) are ubiquitous DNA components of all eukaryotic genomes so far investigated. Based on molecular mechanisms responsible for their transposition, TEs are traditionally categorized into two major classes: retrotransposons and DNA transposons. Retrotransposons (RTs) can be divided into long terminal repeat (LTR) RTs and the non-LTR-RTs, such as long interspersed nuclear elements and short interspersed nuclear elements, whereas DNA transposons can be divided into at least 10 superfamilies (Wicker et al., 2007). Elements within a superfamily are generally grouped into different families on the basis of their sequence similarity. The abundance of individual superfamilies and families of TEs varies among species. For example, non-LTR-RTs are most abundant in vertebrates, while LTR-RTs make up the largest fraction of repetitive DNA in plants (Kumar and Bennetzen, 1999). Comparative analyses of plant TEs revealed that the scales and time frames over which they proliferated, persisted, and were purged vary considerably across families, lineages, and species (Wicker et al., 2003; Ma and Bennetzen, 2004; Piegu et al., 2006; Du et al., 2010b). Such variations are largely responsible for difference in genome size among species, subspecies, and even different individuals of a same species (Hirochika et al., 1996; Ma et al., 2004; Wang and Dooner, 2006; Huang and Dooner, 2008).
Regardless of the sizes of their host genomes, TEs are found to be enriched in the pericentromeric regions of many plant genomes (Arabidopsis Genome Initiative, 2000; International Rice Genome Sequencing Project, 2005; Paterson et al., 2009; Schnable et al., 2009; Schmutz et al., 2010). In contrast with the chromosomal arms, pericentromeric regions generally show suppressed genetic recombination (GR) and have low gene densities; thus, negative associations of TE contents with both GR rates and gene densities are often observed between the two distinct chromatin environments. If TE insertions more frequently cause deleterious mutations in gene-rich chromosomal arms than in gene-poor pericentromeric regions of the host genome, TEs should be preferentially accumulated in the latter regions, where natural selection is inefficient or less efficient (Gaut et al., 2007). Alternatively, the biased accumulation of TEs in pericentromeric regions could result from preferential TE insertions in those regions.
In attempts to minimize potential centromere effects, recombination-suppressed pericentromeric regions were generally excluded or analyzed separately in previous investigations of correlation between local GR rates and genomic features in plants (Zhang and Gaut, 2003; Rizzon et al., 2006; Tian et al., 2009). When only chromosomal arms were analyzed, some puzzling results from different species were often obtained. For example, the GR rates were found to be positively correlated with gene densities and negatively correlated with TE contents in rice (Oryza sativa) (Tian et al., 2009), whereas such correlations were not detected in Arabidopsis thaliana (Wright et al., 2003). In some cases, opposite correlations between the GR rates and genomic features were even observed (Duret et al., 2000; Rizzon et al., 2002; Tian et al., 2009), suggesting that the nature and relative strengths of the forces acting on TE accumulation may vary across organisms.
Nevertheless, the GR rates and genomic features were often compared on different time scales in previous studies. For example, the local GR rates (centimorgan/Mb) along chromosomes were generally estimated by integrating physical and genetic maps, with the latter routinely constructed using a mapping population derived from two varieties capable of intercrossing (Gaut et al., 2007). By contrast, the observed distribution patterns of TEs were the outcomes of millions of years of coevolution of TEs and their host genomes and were largely determined by the competing activities of TE amplification and the generation of small deletions (Devos et al., 2002; Ma et al., 2004; Bennetzen et al., 2005). Given that the local GR rates and genomic features vary over evolutionary times, their potential correlations would be more accurately assessed if comparisons can be made on a similar time scale.
Sequencing of the 1.1-gigabase genome of the paleopolyploid soybean (Glycine max) (Schmutz et al., 2010), one of the most economically important legumes crop domesticated from its wild progenitor species Glycine soja ∼5000 million years ago (Carter et al., 2004), revealed a few striking genomic features in comparison with the 0.4-gigabase rice genome: (1) Approximately 57% of the soybean genome sequence occurred in recombination-suppressed pericentromeric regions (Schmutz et al., 2010) versus only ∼12% in the pericentromeric regions of rice (International Rice Genome Sequencing Project, 2005); (2) the proportions of LTR-RTs in the pericentromeric regions and chromosomal arms are 63 and 11% in soybean (Du et al., 2010b) versus 39 and 17% in rice (Tian et al., 2009); (3) the proportions of DNA transposons in the pericentromeric regions and chromosomal arms are 21.5 and 8.9% in soybean (Du et al., 2010b) versus 7.5 and 13.8% in rice (Tian et al., 2009). It remains unclear why the patterns of TE distributions between the two genomes are so different.
Theoretically, whether a pattern of TE distribution primarily results from preferential insertion or selection against genic insertions could be determined by comparisons of de novo TE insertions with accumulated ones (Gaut et al., 2007), given that de novo insertions such as those activated and amplified in plant tissue cultures have undergone no or little selection pressure (Naito et al., 2009). However, because very few families of active TEs were identified in plants, such analyses were limited to a few active families (e.g., the LTR-RT family Tos17 [Miyao et al., 2003] and the miniature inverted-repeat transposable element family mPing [Naito et al., 2009] in rice). Alternatively, potential interplay among the GR rates, preferential TE insertions, and selection could be deduced by comparison of relatively young insertions with accumulated ones.
PCR-based repeat-junction markers have been used to investigate haplotype variations of TE insertions (Ma and Bennetzen, 2004; Devos et al., 2005; Luce et al., 2006; You et al., 2010), but this method is only suitable for analyses of a limited numbers of TE families. Sequencing and recent resequencing of many plant genomes have provided unprecedented opportunities to investigate genome-wide TE insertions within a recent evolutionary time frame and at a population level. Theoretically, the probability that two LTR-RTs share an identical 5-bp target site duplication in a genome is <0.1% (4−5) (Ma et al., 2004). If integrations of two copies of TEs belonging to a same family are random events, the chance that they share >20-bp flanking sequences in a same genome would be <4−20. Therefore, TE junction sequences have been used to identify TE insertion polymorphisms among individual genomes of the same species (Ma and Bennetzen, 2004; Devos et al., 2005; Tian et al., 2011). More recently, the next-generation sequencing (NGS) short reads have been used to characterize human RT insertion polymorphisms, yielding an unprecedented catalog of common and rare variants (e.g., nonreference TEs) due to insertional mutagenesis (Ewing and Kazazian, 2011).
In this study, a semiautomated bioinformatics pipeline was developed to identify and map nonreference TE insertions in 31 resequenced soybean genomes (Lam et al., 2010) using the Williams82 genome (Schmutz et al., 2010) as a reference. TE insertions present in the 31 accessions but absent in Williams82 and TE insertions unique to the G. max accessions but absent in the G. soja accessions were identified. Following genome-wide profiling of nonreference TE insertions among the 31 genomes, we compared the distribution patterns of the TE insertions present in G. max but absent in G. soja and the accumulated ones in the context of GR rates and gene densities in the reference genome. Our analyses revealed that, although both LTR-RTs and DNA TEs were preferentially accumulated in pericentromeric regions in contrast with chromosomal arms of the host genome, these two classes of TEs had distinct insertional preferences for the two distinct chromatin environments and thus had undergone different levels of purifying selections as a major force purging TE DNA from chromosomal arms.
RESULTS
Genome-Wide Identification of Nonreference TE Insertions in the 31 Soybean Genomes Using the Resequencing Short Reads
Following the methodology described by Ewing and Kazazian (2011), we developed a semiautomated bioinformatics pipeline strategy, as illustrated in Supplemental Figure 1 online, to identify TE insertions that are present in the 31 resequenced soybean genomes but absent in the Williams82 reference genome (Schmutz et al., 2010) using ∼1408 million 75-bp genome resequencing short reads (Lam et al., 2010). The pipeline runs in the following order: (1) Extract the 75-bp short reads and identify the ones each containing a TE-flanking sequence junction site from a resequenced genome; (2) remove the reads with TE junction sites that are shared by either assembled or unassembled Williams82 whole-genome sequence (WGS); (3) map the short reads retained from the last step to the reference genome based on the non-TE portions of TE junctions and remove unmappable nonreference TE insertions; (4) compare the TE junction sequences among the 31 resequenced genomes to identify nonredundant nonreference TE insertion candidates; and (5) manually inspect each of the nonredundant nonreference TE candidates mapped to a single site of the reference genome sequence. The strategy is described in more detail in Methods.
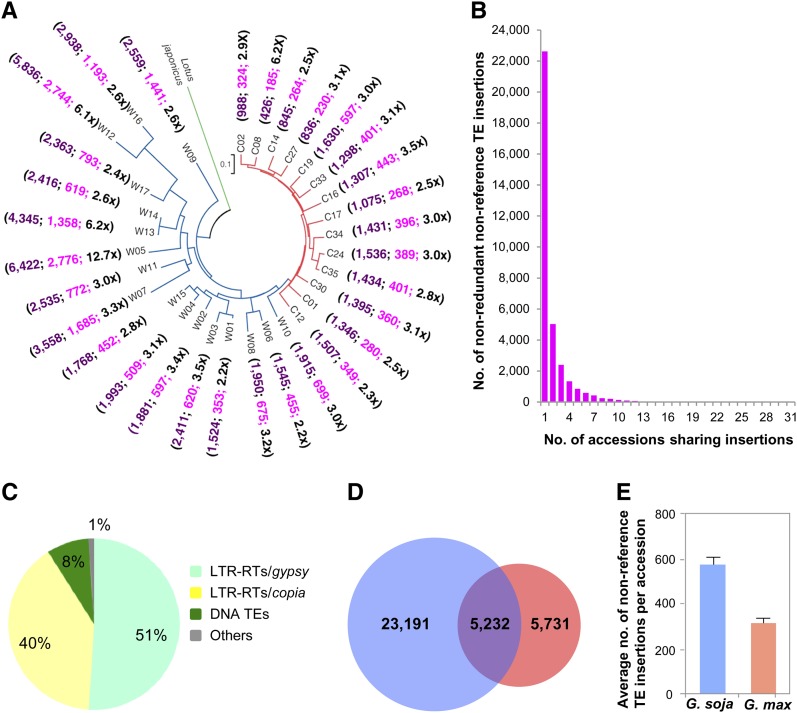
Using this approach, we identified a total of 34,154 nonredundant TE insertions in the 17 G. soja and 14 G. max accessions that were mapped to unique sites of the reference genome sequence. These insertions were absent in Williams82 and thus referred to as nonreference TE insertions (Figure 1A). These include 22,628 (66%) insertions detected only in single accessions (on average, 730 per accession), 5035 (15%) detected in two accessions, and 6491 (19%) detected in more than three accessions (Figure 1B). Approximately 91% of these 34,154 insertions are LTR-RTs (51% LTR/gypsy and 40% LTR/copia), 8% are DNA transposons, and 1% are other types of repeats, which were not further analyzed (Figure 1C). On average, 2100 nonreference insertions per accession, including nonredundant and redundant ones in the population, were identified (Figure 1A; see Supplemental Data Set 1 online).
Figure 1.
Nonreference TE Insertions Identified in the 31 Wild and Cultivated Soybean Genomes.
(A) Numbers of nonreference TE insertions identified in individual soybean accessions. Purple and pink numbers indicate unique insertions in each of the 31 accessions and insertions shared by two or multiple accessions with 2.2 to 12.7genome coverage of short reads (black numbers) The neighbor-joining phylogenetic relationship of the 31 accessions was adapted from Lam et al. (2010). The red, blue, and green lines in the phylogenetic tree indicate G. max, G. soja, and Lotus japonicus (outgroup) accessions, respectively.
(B) Number of nonredundant nonreference TE insertions present in one or shared by two or multiple accessions.
(C) Proportions of different categories of nonredundant nonreference TEs identified in the soybean population.
(D) Numbers of nonredundant nonreference TEs present in the wild soybean subpopulation (blue), the cultivated soybean subpopulation (orange), and shared by both subpopulations (overlap).
(E) Average numbers of unique nonreference TEs in each of the 31 genomes.
Error bars represent sd of uncertainty calculated based on data from 14 G. soja accessions and 17 G. max accessions.
The number of the nonreference TE insertions detected in individual accessions seemed to be associated with the sequencing depths of their genomes, but exceptions were also observed (Figure 1A). For example, 324 unique insertions were detected in the accession C02 with 2.9× genome coverage of short reads, whereas only 185 unique insertions were detected in the accession C08 with 6.2× genome coverage of short reads. Among the 31 accessions investigated, C08 is the only one developed in the US and was suggested to share higher similarity with Williams82 than any other accessions (Lam et al., 2010). This may partially explain why C08 has the lowest number of nonreference TE insertions. In spite of the low genome coverage of the 75-bp resequencing reads, with an average of 3.5× per accession, these reads from the 31 accessions together make up 108× genome coverage (Figure 1A). Therefore, if a nonreference TE insertion was detected in only a single accession, it is most likely that it was a relatively rare insertion in the whole population, although it may exist in more than one of the 31 accessions. Under this assumption, we deduce that the majority of the unique TE insertions detected in C02 and C08, as well as unique insertions detected in other accessions, reflect relatively young insertion events that have occurred during recent diversification of the 31 accessions.
Of the 34,154 nonreference TE insertions, 5731 were detected only in the G. max subpopulation, 23,191 were detected only in the G. soja subpopulation, and 5232 were detected in both subpopulations (Figure 1D). If few TE insertions were eliminated from Williams82 in the past a few thousand years, the time frame for the soybean domestication event, then it is reasonable to deduce that the majority of the unique insertions in the G. max subpopulation are likely to have occurred during recent soybean varietal differentiation. Overall, more insertions were detected in each of the G. soja accessions in comparison with the G. max accessions with similar genome coverage of short reads (Figures 1A, 1D, and 1E). This observation was expected given two facts: (1) the reference genome used for identification of nonreference TE insertions is from a G. max accession; and (2) the G. soja subpopulation has spanned a longer evolutionary time frame for divergence, within which more nonreference TEs would have been proliferated.
To evaluate the effectiveness of the pipeline for identification of the nonreference TE insertions, we used a PCR-based method (see details in Methods) to analyze a sample of nonreference TE insertions detected by this pipeline in the resequenced population. These include seven insertions predicted in multiple accessions by both end junctions of individual TE insertions, six insertions predicted in multiple accessions by single end junctions of individual TE insertions, and 18 insertions predicted in single accessions by both end junctions of individual TE insertions (see Supplemental Data Set 1 and Supplemental Table 1 online). As illustrated in Supplemental Figure 2 online, three primers (see Supplemental Table 2 online) were used to validate the presence or absence of one putative TE insertion site, two (i.e., P1 and P3) from the upstream and downstream of a particular TE insertion site, and the third from one of two termini of the TE. At these 31 nonredundant nonreference TE insertion sites determined based on the reference genome, a total of 135 insertions, including nonredundant and redundant ones, in the resequenced soybean population were predicted by the pipeline (see Supplemental Table 1 online). Of the 135 insertions, 126 (93%) were validated by PCR analysis (see Supplemental Table 1 online). Additionally, at these 31 insertion sites, we identified by PCR 150 redundant insertions in the population, which were not detected by the pipeline (see Supplemental Table 1 online). We thus estimate that ∼53% of the redundant non-TE insertions existing in the population were not predicted with the current depth of resequencing short reads solely by our bioinformatics pipeline. Nevertheless, all the 31 nonreference TE insertions in the population were predicted by the pipeline. By contrast, none of these 31 TE insertions were detected in Williams82 by PCR, indicating their absence in the reference genome.
Distribution of Accumulated TEs in the Context of GR Rates and Gene Densities in the Reference Genome
In an attempt to understand the potential preferences and patterns of nonreference TE insertions in the resequenced genomes, we first analyzed the distribution of accumulated TEs and genes in the reference genome and their relationships with local GR rates and gene densities. The distribution of TEs and genes, pooled in 1-Mb contiguous subregions along each of the 20 chromosomes (Figure 2C; see Supplemental Figure 3 online), were calculated based on previous analyses of the Williams82 reference genome (Du et al., 2010b; Schmutz et al., 2010). The local recombination rates (Figure 2D; see Supplemental Figure 3 online) were estimated by comparison of genetic and physical maps of soybean (Schmutz et al., 2010) using an R-based tool MareyMap (Rezvoy et al., 2007) (see Methods).
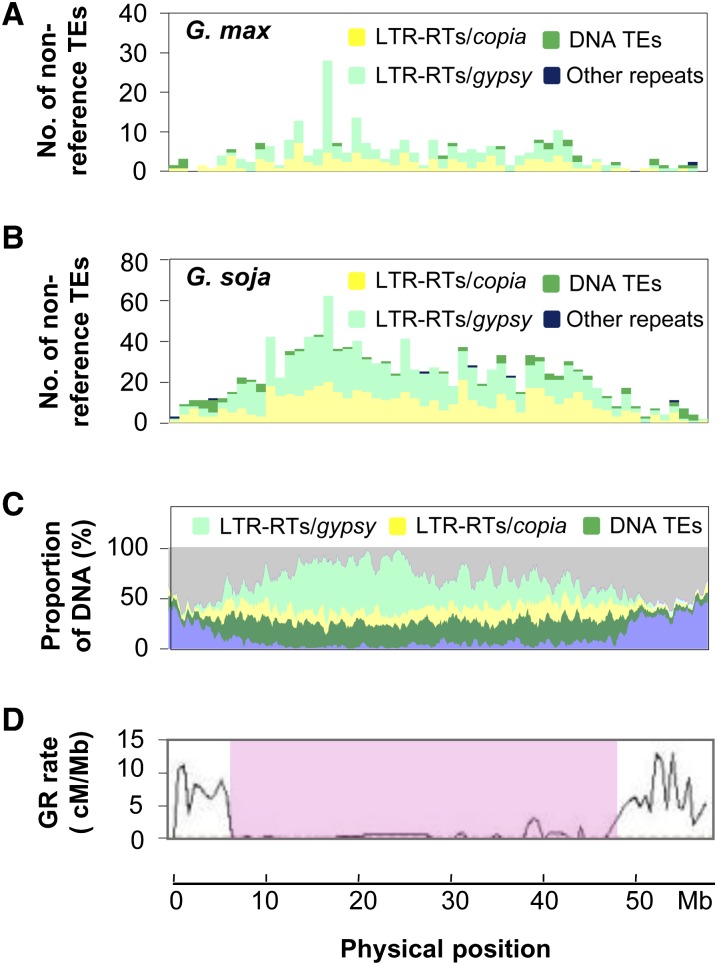
Figure 2.
Nonreference TE Insertion Sites and Genomic Features along the Soybean Chromosome 1.
(A) Distribution of nonreference TE insertions in the cultivated soybean accessions according to the reference genome.
(B) Distribution of nonreference TE insertions in the wild soybean accessions according to the reference genome.
(C) Distribution of accumulated TEs in the reference genome. The bottom blue curve represents gene densities.
(D) Variation of local GR rates. The pink highlighted area defines the pericentromeric region of the chromosome. cM, centimorgans.
Overall, our data reveal significant negative correlations of TE contents with both gene densities and local GR rates at the whole-genome level (Table 1). It has been reported that different classes or subclasses of TEs often exhibited distinct distribution patterns in host genomes (Gao et al., 2008). Hence, we analyzed gypsy LTR-RTs, copia LTR-RTs, and DNA TEs separately. As shown in Table 1, each of the three classes/subclasses of TEs showed the same relationships with gene contents and local GR rates as all TEs together did. Although the levels of significance were lower when recombination-suppressed pericentromeric regions (Schmutz et al., 2010) were excluded, the correlations of the compared parameters remain unchanged and significant. Together, these observations suggest that these two major DNA components (i.e., the accumulated TEs and genes) are organized along recombinational gradients in the soybean reference genome.
Table 1. Correlation of Accumulated TE Contents with Local GR Rates and Gene Densities in the Reference Genome.
| Whole Chromosomes |
Chromosomal Arms |
|||
|---|---|---|---|---|
| Features | ra | Pb | ra | Pb |
| TEs contents versus gene contents | −0.932 | <10−4 | −0.854 | <10−4 |
| TEs contents versus GR rates | −0.633 | <10−4 | −0.212 | <10−4 |
| LTR-RTs contents versus gene contents | −0.895 | <10−4 | −0.828 | <10−4 |
| LTR-RTs contents versus GR rates | −0.608 | <10−4 | −0.203 | <10−4 |
| LTR-RTs/copia contents versus gene contents | −0.811 | <10−4 | −0.817 | <10−4 |
| LTR-RTs/copia contents versus GR rates | −0.554 | <10−4 | −0.194 | 0.0002 |
| LTR-RTs/gypsy contents versus gene contents | −0.824 | <10−4 | −0.748 | <10−4 |
| LTR-RTs/gypsy contents versus GR rates | −0.559 | <10−4 | −0.189 | 0.0003 |
| DNA TEs contents versus gene contents | −0.801 | <10−4 | −0.818 | <10−4 |
| DNA TEs contents versus GR rates | −0.543 | <10−4 | −0.208 | <10−4 |
| Genes contents versus GR rates | 0.623 | <10−4 | 0.156 | 0.0031 |
Pearson correlation coefficient.
All P values calculated by 10,000 bootstrap resamplings.
Distribution of Nonreference TEs versus Accumulated TEs along Chromosomes
To evaluate the tendency of TE insertions, we performed comparative analyses among the distribution patterns of TEs inserted/accumulated within different evolutionary time frames. These analyses include (1) comparison between nonreference TEs in the G. max subpopulation and nonreference TEs in the G. soja subpopulation, (2) comparison between nonreference TEs in the G. max subpopulation and the accumulated TEs in the reference genome, and (3) comparison between nonreference TEs in the G. soja subpopulation and the accumulated TEs in the reference genome. The distribution of nonreference TE insertions and accumulated TE contents pooled in 1-Mb contiguous subregions along each of the 20 chromosomes, as described above, are illustrated in Figure 2 and Supplemental Figure 3 online. The analytical results are summarized in Table 2.
Table 2. Correlation of Nonreference TE Insertions in the Resequenced Genomes with Accumulated TEs in the Reference Genome.
| Whole Chromosomes |
Chromosomal Arms |
|||
|---|---|---|---|---|
| Features compareda | rb | Pc | rb | Pc |
| Densities of nonreference LTR-RTs: G. max versus G. soja | 0.778 | <10−4 | 0.669 | <10−4 |
| Densities of nonreference LTR-RTs/copia: G. max versus G. soja | 0.581 | <10−4 | 0.563 | <10−4 |
| Densities of nonreference LTR-RTs/gypsy: G. max versus G. soja | 0.754 | <10−4 | 0.526 | <10−4 |
| Densities of nonreference DNA TEs: G. max versus G. soja | 0.286 | <10−4 | 0.187 | <10−4 |
| Densities of nonreference LTR-RTs in G. max versus proportions of accumulated LTR-RTs | 0.707 | <10−4 | 0.497 | <10−4 |
| Densities of nonreference LTR-RTs/copia in G. max versus proportions of accumulated LTR-RTs/copia | 0.466 | <10−4 | 0.309 | <10−4 |
| Densities of nonreference LTR-RTs/gypsy in G. max versus proportions of accumulated LTR-RTs/gypsy | 0.630 | <10−4 | 0.468 | <10−4 |
| Densities of nonreference DNA TEs in G. max versus proportions of accumulated DNA TE DNA | −0.114 | 0.0004 | 0.021 | 0.6874 |
| Densities of nonreference LTR-RTs in G. soja versus proportions of accumulated LTR-RTs | 0.864 | <10−4 | 0.712 | <10−4 |
| Densities of nonreference LTR-RTs/copia in G. soja versus proportions of accumulated LTR-RTs/copia | 0.681 | <10−4 | 0.494 | <10−4 |
| Densities of nonreference LTR-RTs/gypsy in G. soja versus proportions of accumulated LTR-RTs/gypsy | 0.793 | <10−4 | 0.760 | <10−4 |
| Densities of nonreference DNA TEs in G. soja versus proportions of accumulated DNA TE DNA | −0.239 | <10−4 | −0.023 | 0.6553 |
For each class/subclass of the nonreference TEs, a positive correlation with regard to their chromosomal distributions was detected between the G. max and G. soja subpopulations regardless of whether pericentromeric regions are excluded in the analysis or not. For either the gypsy or copia subclasses, consistent positive correlations between the nonreference LTR-RT insertions and the proportions of accumulated LTR-RT DNA were observed, regardless of whether pericentromeric regions are excluded in the analysis or not. A negative correlation between the nonreference DNA TE insertions and the proportions of accumulated DNA TE contents was also observed at the whole-genome level, but such a correlation was not detected when pericentromeric regions were excluded. The relationships between the distributions of the nonreference TEs and accumulated TEs belonging to each of the three classes/subclasses described above remains unchanged, when the nonreference TEs present only in the G. max subpopulation and those present only in the G. soja subpopulation were separately compared with the accumulated TEs in the reference genome (Table 2). These results indicate that the distribution patterns of the accumulated LTR-RTs predict the distribution of new LTR-RT insertions, but the distribution patterns of DNA TEs vary considerably along evolutionary times.
Distribution of Nonreference TE Insertions in the Context of GR Rates and Gene Densities According to the Reference Genome
To understand whether the nonreference TE insertions are associated with local genomic features, we analyzed the correlations between the distribution of nonreference TE insertions and the distribution of genes and local GR rates using the same set of contiguous subregions along each of the 20 chromosomes of the reference genome described above. As shown in Table 3, negative associations of the nonreference LTR-RT (either gypsy, copia, or both subclasses) insertions with gene contents and local GR rates were detected, regardless of whether the recombination-suppressed pericentromeric regions were included or not. When the nonreference LTR-RT insertions in G. max and G. soja populations were analyzed separately, the correlation between the copia LTR-RT insertions in the G. max population and local GR rates was found to be insignificant. By contrast, DNA TEs exhibited distinct patterns of distribution in the context of gene densities and GR rates. At the whole-genome level, the nonreference DNA TE insertions in the G. max subpopulation, the G. soja subpopulation, or the whole population were found to be positively associated with gene contents and local GR rates. When the pericentromeric regions were excluded, negative correlations between the gene contents and nonreference DNA TE insertions were observed, but no association of local GR rates with the nonreference DNA TE insertions in either G. max, G. soja, or the whole population was detected.
Table 3. Correlation of Nonreference TE Insertions with Genomic Features of the Reference Genome.
| Whole Chromosomes |
Chromosomal Arms |
|||
|---|---|---|---|---|
| Features Compareda | rb | Pc | rb | Pc |
| Densities of nonreference LTR-RTs in G. max versus GR rate | −0.460 | <10−4 | −0.130 | 0.012 |
| Densities of nonreference LTR-RTs/copia in G. max versus GR rate | −0.391 | <10−4 | −0.074 | 0.1545 |
| Densities of nonreference LTR-RTs/gypsy in G. max versus GR rate | −0.408 | <10−4 | −0.152 | 0.0034 |
| Densities of nonreference DNA TEs in G. max versus GR rate | 0.109 | 0.0007 | −0.023 | 0.6553 |
| Densities of nonreference LTR-RTs in G. soja versus GR rate | −0.576 | <10−4 | −0.187 | 0.0003 |
| Densities of nonreference LTR-RTs/copia in G. soja versus GR rate | −0.568 | <10−4 | −0.181 | 0.0004 |
| Densities of nonreference LTR-RTs/gypsy in G. soja versus GR rate | −0.517 | <10−4 | −0.161 | 0.0019 |
| Densities of nonreference DNA TEs in G. soja versus GR rates | 0.197 | <10−4 | −0.046 | 0.373 |
| Densities of nonreference LTR-RTs in G. max versus densities of genes | −0.667 | <10−4 | −0.436 | <10−4 |
| Densities of nonreference LTR-RTs/copia in G. max versus densities of genes | −0.584 | <10−4 | −0.357 | <10−4 |
| Densities of nonreference LTR-RTs/gypsy in G. max versus densities of genes | −0.582 | <10−4 | −0.388 | <10−4 |
| Densities of nonreference DNA TEs in G. max versus densities of genes | 0.168 | <10−4 | −0.118 | 0.0234 |
| Densities of nonreference LTR-RTs in G. soja versus densities of genes | −0.831 | <10−4 | −0.614 | <10−4 |
| Densities of nonreference LTR-RTs/copia in G. soja versus densities of genes | −0.814 | <10−4 | −0.530 | <10−4 |
| Densities of nonreference LTR-RTs/gypsy in G. soja versus densities of genes | −0.750 | <10−4 | −0.594 | <10−4 |
| Densities of nonreference DNA TEs in G. soja versus densities of genes | 0.292 | <10−4 | −0.127 | 0.0141 |
Density refers to numbers of nonreference TE insertions per 1-Mb region along chromosomes according to the reference genome; densities of genes were estimated based on the reference genome.
Pearson correlation coefficient.
All P values calculated by 10,000 bootstrap resamplings.
Distribution of the Nonreference TEs versus the Accumulated TEs between Pericentromeric Regions and Chromosomal Arms
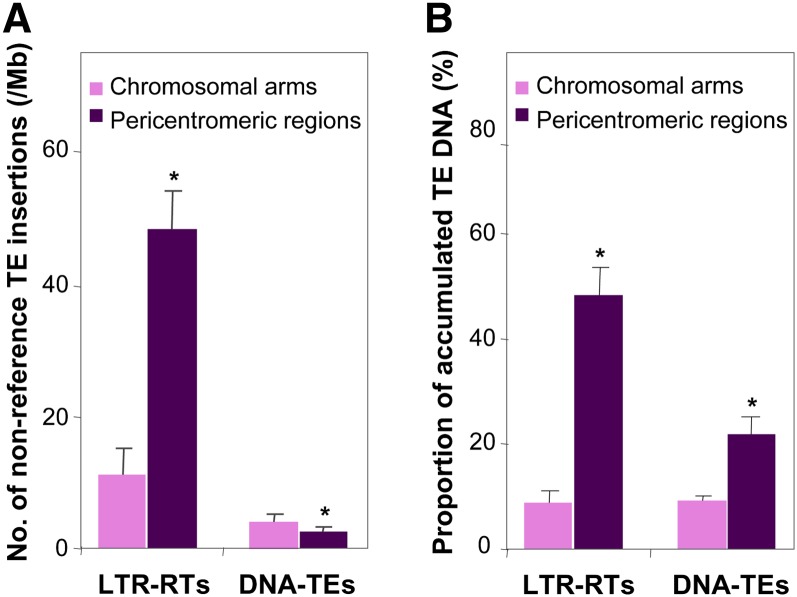
The soybean genome showed extremely contrasting genomic features with regard to the GR rates, gene densities, and TE distribution between the pericentromeric regions and chromosomal arms (Du et al., 2010b; Schmutz et al., 2010). As described in Table 1, the levels of correlations among the genomic features along chromosomes were generally reduced when pericentromeric regions were excluded, suggesting strong pericentromeric effects on shaping the genomic features between distinct chromatin environments. In an attempt to shed light on the nature and strength of such effects, in particular, on the biased accumulation of TEs in pericentromeric regions, we analyzed relative abundance of the 34,154 nonreference TE insertions versus accumulated TEs between pericentromeric regions and chromosomal arms.
We found that the densities of nonreference LTR-RTs (either gypsy or copia subclasses) detected in either the G. soja or the G. max subpopulations in pericentromeric regions were significantly higher than in chromosomal arms (Table 4), consistent with the distribution pattern of the accumulated LTR-RTs in these two types of chromatin environments in the reference genome (Du et al., 2012; Table 4, Figure 3). By contrast, the densities of the nonreference DNA TEs in both the G. max and G. soja subpopulations did not show significant differences between pericentromeric regions and chromosomal arms, although the accumulated DNA TEs were significantly enriched in pericentromeric regions compared with chromosomal arms of the reference genome (Du et al., 2012; Table 4, Figure 3). When the two subpopulations were combined as a single population, we detected significantly higher density of nonreference DNA TE insertions in chromosomal arms than in pericentromeric regions of the 20 chromosomes (Figure 3; see Supplemental Figure 4 online).
Table 4. Distribution of Nonreference TE Insertions between Chromosomal Arms and Pericentromeric Regions of the 20 Chromosomes.
| Densities of Nonreference TE Insertions or Proportions of TE DNA | Chromosomal Armsb | Pericentromeric Regionsb | P Valuea Pf>|t| |
|---|---|---|---|
| G. max subpopulation | |||
| No. of nonreference LTR-RT insertions per Mb | 6.06 ± 4.22 | 48.77 ± 38.61 | <10−4 |
| No. of nonreference LTR-R/copia insertions per Mb | 3.10 ± 1.92 | 20.17 ± 9.29 | <10−4 |
| No. of nonreference LTR-RT/gypsy insertions per Mb | 2.97 ± 2.08 | 28.60 ± 12.57 | <10−4 |
| No. of nonreference DNA TE insertions per Mb | 2.22 ± 1.31 | 1.94 ± 0.98 | 0.2181 |
| G. soja subpopulation | |||
| No. of nonreference LTR-RT insertions per Mb | 6.06 ± 4.22 | 48.77 ± 38.61 | <10−4 |
| No. of nonreference LTR-RT/copia insertions per Mb | 3.10 ± 1.92 | 20.17 ± 9.29 | <10−4 |
| No. of nonreference LTR-RT/gypsy insertions per Mb | 2.97 ± 2.08 | 28.60 ± 12.57 | <10−4 |
| No. of nonreference DNA TE insertions per Mb | 2.22 ± 1.31 | 1.94 ± 0.98 | 0.2181 |
| Reference genome | |||
| Proportions of LTR-RT DNA (%) | 8.70 ± 1.91 | 47.24 ± 5.60 | <10−4 |
| Proportions of LTR-RT/copia DNA (%) | 4.75 ± 0.88 | 13.11 ± 1.28 | <10−4 |
| Proportions of LTR-RT/gypsy DNA (%) | 3.95 ± 1.16 | 34.12 ± 4.89 | <10−4 |
| Proportions of DNA TE DNA (%) | 8.88 ± 1.21 | 21.54 ± 3.40 | <10−4 |
Student’s t test.
Mean ± sd.
Figure 3.
Comparison of Nonreference TEs and Accumulated TEs between Pericentromeric Regions and Chromosomal Arms.
Error bars represent sd of uncertainty calculated based on data from 20 chromosomes.
(A) Densities of nonreference LTR-RTs and DNA TEs.
(B) Proportions of accumulated LTR-RT and DNA TE DNA. Asterisks indicate significant difference at the level of P < 0.01.
[See online article for color version of this figure.]
Further comparisons between the pericentromeric regions and chromosomal arms of the 20 chromosomes revealed positive correlations between the nonreference LTR-RTs (either gypsy or copia subclasses) in either the G. max or G. soja subpopulations and the accumulated LTR-RT contents and negative correlations between the nonreference DNA TEs in either the G. max or G. soja subpopulations and the accumulated DNA TE contents (Table 5). By contrast, negative correlations of nonreference LTR-RTs (either gypsy or copia subclasses) in either the G. max or G. soja subpopulations with gene densities and positive correlations of nonreference DNA TEs with gene densities were detected (Table 5).
Table 5. Correlations of Nonreference TE Insertions with Accumulated TE DNA and Gene Densities in Chromosomal Arms and Pericentromeric Regions of the 20 Chromosomes.
|
G. max Subpopulation |
G. soja Subpopulation |
|||
|---|---|---|---|---|
| Features Compared | ra | Pb | ra | Pb |
| Densities of nonreference LTR-RT insertion versus proportions of accumulated LTR-RT DNA | 0.749 | <10−4 | 0.809 | <10−4 |
| Densities of nonreference LTR-RT/copia insertions versus proportions of accumulated LTR-RT/copia DNA | 0.788 | <10−4 | 0.841 | <10−4 |
| Densities of nonreference LTR-R/gypsy insertions versus proportions of accumulated LTR-RT/gypsy DNA | 0.732 | <10−4 | 0.802 | <10−4 |
| Densities. of nonreference DNA TE insertions versus proportions of accumulated DNA TE DNA | −0.043 | 0.791 | −0.110 | 0.500 |
| Densities of nonreference LTR-RT insertion versus densities of genes | −0.781 | <10−4 | −0.838 | <10−4 |
| Densities of nonreference LTR-RT/copia insertions versus densities of genes | −0.759 | <10−4 | −0.820 | <10−4 |
| Densities of nonreference LTR-RT/gypsy insertions versus densities of genes | −0.790 | <10−4 | −0.847 | <10−4 |
| Densities of nonreference DNA TE insertions versus densities of genes | 0.095 | 0.559 | 0.090 | 0.582 |
Pearson correlation coefficient.
All P values calculated by 10,000 bootstrap resamplings.
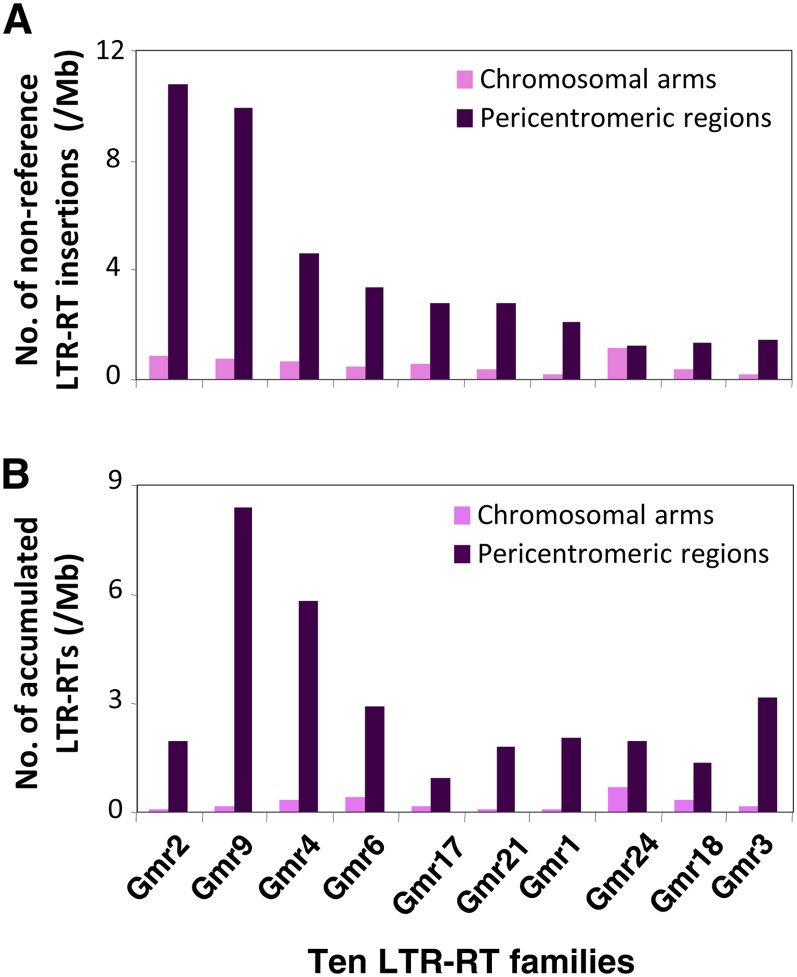
To further assess the dynamics of recent TE proliferation in the soybean genomes, we analyzed the abundance and distribution patterns of the 10 largest LTR-RT families accumulated in the reference genome versus corresponding nonreference TE insertions belonging to these same families identified in the resequenced population. Overall, large families of LTR-RTs in the reference genome also tended to correspond with a high number of nonredundant nonreference insertions in the resequenced population, but exceptions were also observed (Figure 4; see Supplemental Figure 5 online). For example, Gmr2 is the 8th largest family in the reference genome but has the largest number of nonreference insertions in the resequenced population, suggesting that the activities for TE proliferation vary among families within recent evolutionary time frames. All these 10 families showed higher densities of nonreference insertions in pericentromeric regions than in chromosomal arms, although the ratios of relative abundance of the nonreference insertions in the two distinct chromatin environments vary among families (Figure 4).
Figure 4.
Comparison of 10 Families of Nonreference and Accumulated LTR-RTs between Pericentromeric Regions and Chromosomal Arms.
(A) Densities of individual nonreference LTR-RTs.
(B) Densities of individual accumulated LTR-RTs.
[See online article for color version of this figure.]
Distinct Insertional Preferences of Nonreference TE Insertions: Pericentromeric Regions versus Chromosomal Arms
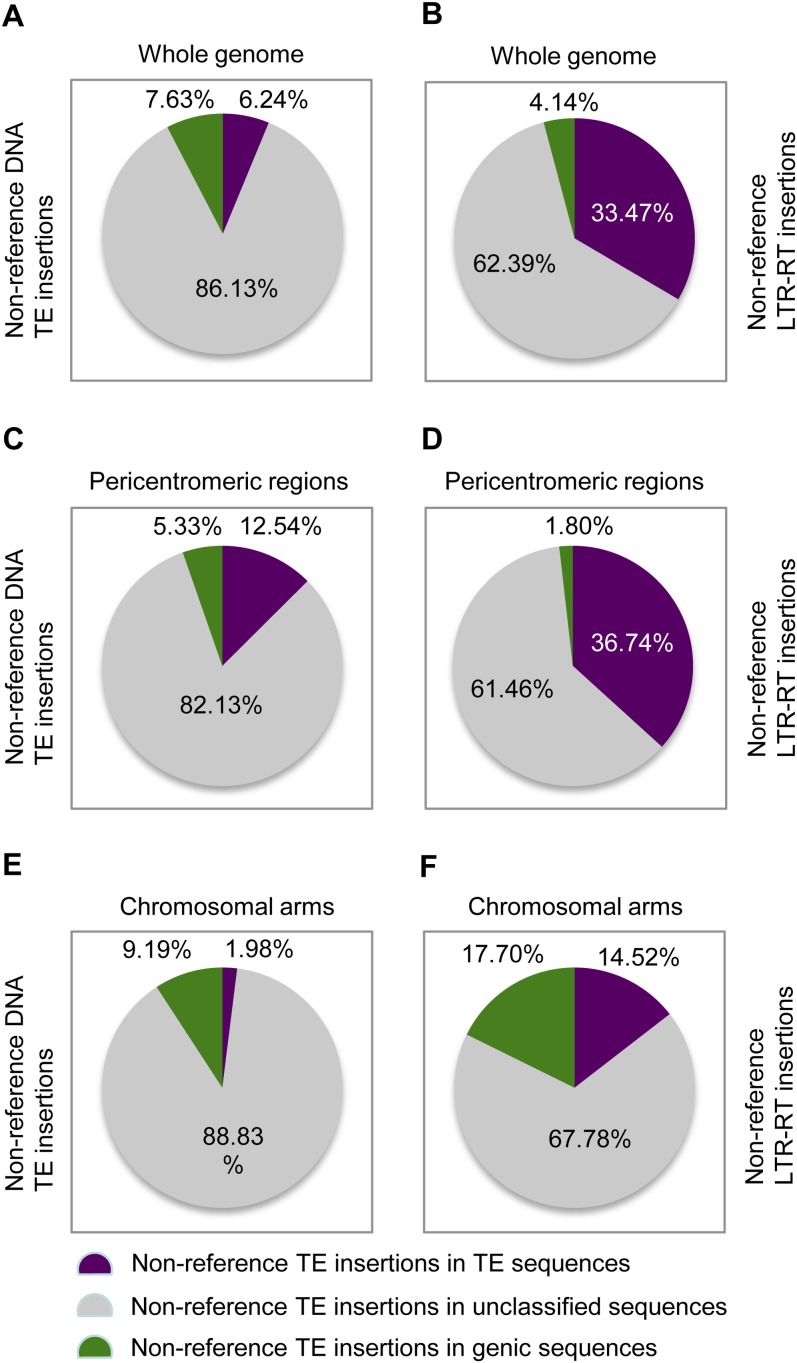
According to the sequence categories of their insertion sites, both the nonreference LTR-RTs and nonreference DNA TEs show different insertional preferences between pericentromeric regions and chromosomal arms (Figure 5; see Supplemental Data Set 1 online). A total of 36.74, 61.46, and 1.80% of nonreference LTR-RTs were found in repetitive sequences, unclassified intergenic sequences, and genic sequences, respectively, in pericentromeric regions (Figure 5D), versus 14.52, 67.78, and 17.70% of nonreference LTR-RT insertions in the three categories of sequences in chromosomal arms (Figure 5F). By contrast, 12.54, 82.13, and 5.33% of nonreference DNA TEs were found in repetitive sequences, unclassified intergenic sequences, and genic sequences, respectively, in pericentromeric regions (Figure 5C) versus 1.98, 88.83, and 9.19% of nonreference DNA TE insertions in the three types of sequences in chromosomal arms (Figure 5E). In addition, the proportions of each category of repetitive sequences flanking nonreference TE insertion sites and the proportions of each category of genic sequences flanking nonreference TE insertion sites also show different degrees of variations between pericentromeric regions and chromosomal arms (Tables 6 and 7).
Figure 5.
Proportions of Nonreference TE Insertions in Different Categories of Sequence Components According to the Reference Genome.
(A) Nonreference DNA-TE insertions in the whole genomes.
(B) Nonreference LTR-RTs insertions in the whole genomes.
(C) Nonreference DNA-TE insertions in pericentromeric regions.
(D) Nonreference LTR-RTs insertions in pericentromeric regions.
(E) Nonreference DNA-TE insertions in chromosomal arms.
(F) Nonreference LTR-RTs insertions in chromosomal arms.
[See online article for color version of this figure.]
Table 6. Categories of Repetitive Sequences Harboring Nonreference TE Insertions in the Resequenced Soybean Population.
| Categories of Repetitive Sequences Where Nonreference TE Inserted |
||||||||
|---|---|---|---|---|---|---|---|---|
|
LTR-RTs/copia |
LTR-RTs/gypsy |
DNA TEs |
Other Repeats |
|||||
| Types of Nonreference TEs | No. | % | No. | % | No. | % | No. | % |
| In the whole genome | ||||||||
| LTR-RTs/copia | 1217 | 28.80 | 2637 | 62.41 | 209 | 4.95 | 162 | 3.83 |
| LTR-RTs/gypsy | 1318 | 21.41 | 4189 | 68.05 | 202 | 3.28 | 447 | 7.26 |
| LTR-RTs | 2535 | 24.42 | 6826 | 65.75 | 411 | 3.96 | 609 | 5.87 |
| DNA TEs | 44 | 24.44 | 89 | 49.44 | 47 | 26.11 | 0 | 0.00 |
| LTR-RTs and DNA TEs | 2581 | 24.39 | 6924 | 65.44 | 463 | 4.38 | 613 | 5.79 |
| In chromosomal arms | ||||||||
| LTR-RTs/copia | 135 | 45.61 | 101 | 34.12 | 55 | 18.58 | 5 | 1.69 |
| LTR-RTs/gypsy | 159 | 43.32 | 150 | 40.87 | 53 | 14.44 | 5 | 1.36 |
| LTR-RTs | 294 | 44.34 | 251 | 37.86 | 108 | 16.29 | 10 | 1.51 |
| DNA TEs | 9 | 26.47 | 8 | 23.53 | 17 | 50.00 | 0 | 0 |
| LTR-RTs and DNA TEs | 303 | 43.22 | 261 | 37.23 | 126 | 17.97 | 11 | 1.57 |
| In pericentromeric regions | ||||||||
| LTR-RTs/copia | 1082 | 27.54 | 2536 | 64.55 | 154 | 3.92 | 157 | 4.00 |
| LTR-RTs/gypsy | 1159 | 20.02 | 4039 | 69.77 | 149 | 2.57 | 442 | 7.64 |
| LTR-RTs | 2241 | 23.06 | 6575 | 67.66 | 303 | 3.12 | 599 | 6.16 |
| DNA TEs | 35 | 23.97 | 81 | 55.48 | 30 | 20.55 | 0 | 0 |
| LTR-RTs and DNA TEs | 2278 | 23.06 | 6663 | 67.44 | 337 | 3.41 | 602 | 6.09 |
Table 7. Genic Sequences Harboring Nonreference TE Insertions in the Resequenced Soybean Population.
|
UTRs |
Exons |
Introns |
||||
|---|---|---|---|---|---|---|
| Types of Nonreference TEs | No. | % | No. | % | No. | % |
| In the whole genome | ||||||
| LTR-RTs/copia | 69 | 9.61 | 282 | 39.28 | 367 | 51.11 |
| LTR-RTs/gypsy | 37 | 6.55 | 211 | 37.35 | 317 | 56.11 |
| LTR-RTs | 106 | 8.26 | 493 | 38.43 | 684 | 53.31 |
| DNA TEs | 40 | 18.18 | 75 | 34.09 | 105 | 47.73 |
| LTR-RTs and DNA TEs | 152 | 9.54 | 573 | 35.95 | 869 | 54.52 |
| In chromosomal arms | ||||||
| LTR-RTs/copia | 45 | 10.04 | 172 | 38.39 | 231 | 51.56 |
| LTR-RTs/gypsy | 20 | 5.56 | 130 | 36.11 | 210 | 58.33 |
| LTR-RTs | 65 | 8.04 | 302 | 37.38 | 441 | 54.58 |
| DNA TEs | 30 | 18.99 | 45 | 28.48 | 83 | 52.53 |
| LTR-RTs and DNA TEs | 100 | 9.61 | 351 | 33.72 | 590 | 56.68 |
| In pericentromeric regions | ||||||
| LTR-RTs/copia | 24 | 8.89 | 110 | 40.74 | 136 | 50.37 |
| LTR-RTs/gypsy | 17 | 8.29 | 81 | 39.51 | 107 | 52.20 |
| LTR-RTs | 41 | 8.63 | 191 | 40.21 | 243 | 51.16 |
| DNA TEs | 10 | 16.13 | 30 | 48.39 | 22 | 35.48 |
| LTR-RTs and DNA TEs | 52 | 9.40 | 222 | 40.14 | 279 | 50.45 |
Distinct Insertional Preferences of Nonreference TEs: LTR-RTs versus DNA TEs
To compare the insertional preferences between nonreference LTR-RTs and DNA TEs, we analyzed the genomic sequences flanking the 34,154 nonreference TE insertion sites. Our data reveal that, of the 34,154 nonreference TE insertions predicted by our pipeline, 21,979 (64%) were found in unclassified sequences, 9905 (31%) inserted in TEs (29%) and other repeats (2%), and 1708 (5%) were found in genic sequences of the soybean genome (see Supplemental Data Set 1 online).
The nonreference LTR-RTs and DNA TEs show distinct insertional preferences according to the sequence categories of their insertion sites (Figure 5; see Supplemental Data Set 1 online). Of the 31,017 nonreference LTR-RT insertion sites, 10,381 (33.47%), 19,353 (62.39%), and 1283 (4.14%) were found within repetitive sequences, unclassified sequences, and genic sequences, respectively (Figure 5B). By contrast, 180 (6.24%), 2483 (86.13%), and 220 (7.63%) of the 2883 nonreference DNA TE insertion sites were found within repetitive sequences, unclassified sequences, and genic sequences, respectively (Figure 5A). We further compared categories of the repetitive sequences flanking the nonreference LTR-RT insertion sites and DNA TE insertion sites. A total of 90, 4, and 6% of LTR-RT insertion sites were found within LTR-RT, DNA TE, and other repetitive sequences, whereas 74 and 26% of the DNA TE insertion sites was found within LTR-RT and DNA TE sequences (Table 6).
The portions of the genic sequences flanking the nonreference LTR-RT insertion sites and DNA TE insertion sites were also analyzed. Of the 1594 nonreference TE insertions sites identified as genic sequences, 152 (9.6%), 573 (35.9%), and 869 (54.5%) were found within untranslated regions (UTRs), exons, and introns, respectively (Table 7). These percentages of insertion sites in the three components of genic sequences were consistent with the proportions of UTRs (11.2%), exons (34.1%), and introns (54.6%) of the 46,430 genes predicted in the soybean genome (r = 0.99, P = 0.04), indicating similar frequencies or densities of nonreference TE insertions retained in the three genic components. However, nonreference LTR-RTs and DNA TEs showed distinct distribution patterns regarding their frequencies in the three genic components. As shown in Table 7, the frequencies of nonreference DNA TE insertions in UTRs relative to DNA TE insertions in other genic portions were found to be considerably higher than those of nonreference LTR-RT insertions in UTR regions relative to nonreference LTR-RT insertions in other genic portions.
A total of 32,370 LTR-RTs, including 14,106 intact elements and 18,264 solo LTRs with clear boundaries, were previously identified in the Williams82 reference genome (Du et al., 2010a). Of these reference LTR-RTs, 21,266 (65.33%) were harbored in TE sequences. Of the 14,106 intact elements, 256 contain two identical LTRs, 142 (55%) of which were found in TE sequences. LTR-RTs with two identical LTRs were generally more difficult to assemble by the WGS approach, particularly when they were harbored in repetitive DNA; thus, their relative enrichment in TE sequences may be underestimated. At the first glimpse, the overall organizational pattern of LTR-RTs in the reference genome seemed to be contrasting with the insertional pattern of the nonreference LTR-RTs described in this study; however, our bioinformatics strategy filtered out nonreference TE insertions flanked by individual sequences with multiple matches that were not able to be mapped to unique sites of the reference genome; thus, the insertional pattern of mappable nonreference TE insertions alone would not reflect the insertional pattern of all nonreference TEs, including both mappable and unmappable ones. To elucidate the general patterns of nonreference TE insertions, we manually examined all NGS reads of the accession C27 that were retained from the step 2 of the bioinformatics pipeline. After combining redundant reads into unique ones, 2024 TE-flanking sequence junction reads, each of which represents a unique putative nonreference TE insertion in C27, were identified. Of these insertions, 1920 were LTR-RTs and 104 were DNA TEs. As shown in Supplemental Figure 6 online, 76.40, 16.88, and 6.72% of the nonreference LTR-RT insertions were found in TEs, unclassified sequences, and genic sequences, respectively. By contrast, 16.35, 75.96, and 7.69% of the nonreference DNA TEs were found in TEs, unclassified sequences, and genic sequences, respectively. These observations obtained from a relatively unbiased data set further demonstrated the clear distinction of insertional preference between LTR-RTs and DNA TEs.
DISCUSSION
Genome Resequencing: A High-Throughput Approach to Study TE-Driven Genetic Diversity
High-throughput genome resequencing has become an important approach to characterize genome-wide structural variations, such as single nucleotide polymorphisms, small insertions/deletions, and copy number variations, in higher eukaryotic genomes (DePristo et al., 2011). Recently, this approach has been employed to profile new TE insertion sites or insertion polymorphisms of a few active TE families, including a DNA transposon family mPing among individual rice plants regenerated from tissue culture (Naito et al., 2009) and a RT family L1 among humans (Ewing and Kazazian, 2011). In this study, we identified 34,154 unique nonredundant nonreference TE insertion sites in the resequenced soybean population that were mapped to the reference genome, providing a valuable addition to structural variations among these soybean genomes previously revealed by analyses of single nucleotide polymorphisms and small insertions/deletions (Lam et al., 2010). Although a proportion (∼50%) of the existing mappable nonreference TE insertion sites in a particular resequenced soybean accession were not detected due to the low depth of genome sequencing, as indicated by PCR analysis (see Supplemental Figure 2 and Supplemental Table 2 online), the majority of the nonredundant nonreference TE insertion sites in the population with the current genome coverage of short reads have been found by our bioinformatics pipeline, providing a relatively complete picture of genome-wide nonreference TE insertions in the resequenced soybean population. This pipeline can be modified and used to investigate haplotype variations of TEs in other sequenced plant species and facilitate the functional study of TE-mediated genetic and epigenetic variations. Recently, population resequencing of Arabidopsis, rice, and maize (Zea mays) with the NGS platforms has been performed (Cao et al., 2011; Jiao et al., 2012; Xu et al., 2012), and sequencing and resquencing of many other plant species are underway. It will be interesting to compare the distribution patterns of nonreference TE integration sites, as well as reference TE insertion sites in multiple species to draw a more comprehensive picture regarding the evolutionary propensities of TEs in flowering plants.
The Distribution Patterns of Nonreference TEs Reflect the Preferences of TE Insertions
We infer that the majority of nonreference TEs are relatively young elements on the basis of following observations. First, most nonreference TEs were predicted only in one or two accessions or one of the two subpopulations, suggesting that insertions occurred during the diversification of these accessions. Second, the TE portions of TE junction sequences perfectly match structurally intact elements in the reference genome, suggesting the detected nonreference TEs would not be old ones that were generally highly degenerated. Third, the distribution pattern of nonreference DNA transposons was significantly different from that of accumulated DNA transposons, suggesting that these two sets of DNA TEs have been evolving within two distinct time frames. Fourth, we observed similar frequencies of nonreference TE insertions (i.e., the number of insertions per kilobase nucleotides) in exonic and intronic sequences. Given that TE insertions in exonic sequences are generally more deleterious than in intronic sequences and tend to be eliminated more easily from the former, these observations would be interpreted as evidence that the nonreference TEs have been under limited selection pressure due to the relatively short evolutionary time they have experienced. Nevertheless, only 5% of the nonreference TE insertions were detected in genic sequences, which make up 18% of the soybean genomes. This lower than expected frequency of nonreference TE insertions in genic sequences may be primarily caused by nonrandom insertions.
We would like to point out that, although strict parameters, such as perfect matches between the non-TE portions of the TE junction sequences and the reference genome, were used to map the nonreference TE insertion sites to the reference genome sequence, some of the nonreference TEs, particularly the ones flanked by repetitive sequences, may not be precisely mapped solely based on sequence matches. In addition, DNA rearrangements, such as translocation, deletion/insertion, and duplication, if any, between the G. soja and G. max genomes would further complicate attempts at precisely mapping the nonreference insertion sites. In particular, a large fraction of nonreference TEs were unmappable to single sites in the reference genome sequence and excluded in the analyses described in this study. Thus, the relative abundance of nonreference TE insertions in repetitive sequences, primarily a large proportion of TEs in pericentromeric regions, was underestimated. Nevertheless, the relationships between the distribution of the mapped nonreference TEs and genomic features along chromosomes of the soybean genome remain unchanged and the distribution patterns of the mapped nonreference LTR-RTs and DNA TEs remain distinct, regardless of whether the pericentromeric regions were excluded in the comparisons, suggesting that the distribution patterns of nonreference insertions of LTR-RTs and DNA TEs revealed in this study would be reflective of their insertional preferences.
Our data revealed a clear distinction of insertional preferences between LTR-RTs and DNA TEs. Overall, LTR-RTs exhibited a lower level of insertional bias for unclassified sequences and genic sequences and a higher level of insertional bias for TE sequences than nonreference DNA transposons did (Figure 5). This distinction was much clearer when unmappable insertions were included in the comparison between the two classes of nonreference TEs (see Supplemental Figure 6 online). Because the proportions of TEs, genes, and unclassified sequences vary significantly between pericentromeric regions and chromosomal arms (Table 4; Du et al., 2012), the relative abundance of the nonreference LTR-RT and DNA TE insertions in these three categories of DNA components also differ substantially (Figure 5). It is notable that, in chromosomal arms, the relative abundance of nonreference LTR-RTs in genic sequences (17.7%) was even higher than that of nonreference DNA TEs in genic regions (9.2%) (Figure 5), suggesting that the insertional biases of either LTR-RTs or DNA TEs vary between these two distinct chromatin environments.
The Distribution Patterns of Accumulated TEs Reflect the Effects of Natural Selection
Regardless of the categories of DNA sequences hosting nonreference TE insertions, the nonreference LTR-RT insertions were overwhelmingly more frequent in pericentromeric regions than in chromosomal arms. Although a large fraction of nonreference LTR-RT insertions were unmappable due to the repetitive nature of their flanking sequences (see Supplemental Figure 6 online), it is reasonable to deduce that the majority of those unmappable LTR-RT insertions were located in pericentromeric regions where TEs are preferentially accumulated. This distribution pattern of nonreference LTR-RTs was consistent with the distribution pattern of the accumulated LTR-RTs in the soybean reference genome (Table 4), suggesting that the physical distribution pattern of LTR-RTs between pericentromeric regions and chromosomal arms was not reshaped much or was only slightly reshaped. In other words, the biased accumulation of LTR-RTs in pericentromeric regions was largely caused by preferential insertions. Different from the distribution pattern and insertional bias of LTR-RTs, there was no significant difference in the frequency of nonreference DNA TE insertions between pericentromeric regions and chromosomal arms. Of the 104 nonreference DNA TE insertions identified in C27, 63 were mapped to chromosomal arms, 18 were mapped to pericentromeric regions, and 23 were unmappable. Thus, even all these unmappable insertions assumingly occurred in pericentromeric regions, and there were still more nonreference DNA TE insertions in chromosomal arms than in pericentromeric regions. By contrast, the accumulated DNA TEs are significantly more enriched in pericentromeric regions than in chromosomal arms. These observations suggest that the distribution pattern of DNA TEs has been substantially reshaped by selection against insertions in chromosomal arms. These findings also suggest that LTR-RTs and DNA TEs in the soybean genomes were under different levels of selection pressure primarily due to their distinct site insertion preferences.
Pericentromeric Effects on TE Integration and Accumulation
Pericentromeric regions have several unique biological properties, including heterochromatic states and severe suppression of local GR (Gaut et al., 2007; Tian et al., 2009). It seems that these properties are associated with TE integration and accumulation. As described above, overall, the nonreference LTR-RTs detected in this study showed preferential insertions in pericentromeric regions (Figure 3, Table 4), although their distribution patterns vary among different families (see Supplemental Figure 5 online). These observations are echoed by a previous study of a few LTR-RT families with chromodomains at their integrase C termini (referred to as chromoviruses) in yeast, which demonstrated that the representative chromoviruses from each family recognize histone H3 K9 methylation, an epigenetic mark characteristic of heterochromatin at the time of integration, and then perpetuate the heterochromatic mark by triggering epigenetic modification (Gao et al., 2008). The targeted integration of LTR-RTs into heterochromatin may partially explain the preferential insertions of nonreference LTR-RTs in pericentromeric regions. However, the mechanisms that underlie preferential integration of DNA TEs into chromosomal arms remain largely unknown.
Comparative analysis of distribution patterns of nonreference TEs and accumulated TEs in chromosomal arms and pericentromeric regions revealed a lower level of fixation of DNA TEs in the former regions than in the latter regions. This could be explained by the evolutionary models (Charlesworth and Charlesworth, 1983; Charlesworth et al., 1994, 1997; Biémont et al., 1997) that suggest that (1) TE insertions in gene-rich euchromatic chromosomal arms are more deleterious than in gene-poor heterochromatic pericentromeric regions; and (2) purifying selection against TE insertions in pericentromeric regions is less efficient than in chromosomal arms due to severely suppressed meiotic recombination in the former regions. Indeed, the distribution of accumulated TEs along the 20 chromosomes of the soybean genome were found to correlate negatively with local GR rates and gene densities, no matter if pericentromeric regions were excluded or not from the analyses, suggesting that, in addition to the biased TE insertions, the suppression of meiotic recombination in pericentromeric regions may be the major cause of preferential accumulation of both LTR-RTs and DNA TEs in the regions.
METHODS
Prediction of Nonreference TE Insertions Using Genome-Resequencing Short Reads
A semiautomated bioinformatics pipeline, as illustrated in Supplemental Figure 1 online, was developed to identify nonreference TE insertions in the 31 soybean (Glycine max) genomes using the NGS reads (Lam et al., 2010). To identify nonreference TE insertion sites in the resequenced genomes, we first established a database of TE ends composed of 100-bp sequences extracted from the two ends (i.e., S2 and S3) of each of the 38,581 TEs with clear boundaries identified in the soybean reference genome (Du et al., 2010a) and then searched against this database by BLAST using the 75-bp NGS reads from the 31 resequenced genomes as query sequences. NGS reads with perfect matches (i.e., 100% sequence identity) in the TE ends database were discarded. Reads perfectly (100%) matching 20 to 55 bp of one or multiple TE ends in the TE ends database with clearly defined TE end boundaries, as shown in Supplemental Figure 1 online, were thought to contain TE insertion junction sites and were kept for further analyses. These retained short reads were then used as queries to BLAST search against the Williams82 reference genome sequence as well as the complete set of WGS sequences used to assemble the soybean reference genome sequence. In this BLAST search above, the reads showing ≥65-bp matches to the Williams82 reference genome sequence or the WGS sequences with ≥95% sequence identity were thought to contain TE insertion sites shared by Williams82 and the resequenced accession(s) and excluded from further analyses. This step of sequence comparison with relatively lower stringent criterion ensured that the TEs shared between the reference genome and each of the resequenced genome were maximally or completely removed, when small insertions/deletions or point mutations exist at the same TE-flanking junction sites between compared genomes. Reads with 20 to 55 bp perfect matches starting from detected TE junction sites in the Williams82 genome were thought to contain insertion sites of the predicted nonreference TEs. When a nonreference TE insertion was detected by both TE end junctions, target site duplication at the insertion site was examined. The portions of NGS sequences flanking the identified nonreference TE insertion sites were searched against the Williams82 reference genome sequence to map these TE insertions. To minimize potential inaccuracy of mapping result, a stringent criterion (i.e., full-length perfect matches of the non-TE portions of short reads with unique sites in the reference genome) was employed. Putative nonreference insertions flanked by individual sequences with multiple matches in the reference genome sequence were unmappable and filtered out from the pipeline. All the nonreference insertions retained from the pipeline were manually inspected, and the retained TE insertion junction reads that match the reference genome sequence at the insertion junction sites at relatively lower stringency were further removed. Only the 2,024 unmappable nonreference TE insertions from the accession C27 were manually examined and analyzed in detail.
Validation of Nonreference TE Insertions in the Resequenced Soybean Genomes by PCR
A sample of predicted nonreference insertions were validated by PCR amplification of TE junctions. As shown in Supplemental Figure 2A online, three primers were used to check one predicted nonreference TE insertion in the resequenced population. P1 and P3 were designed based on two sequences flanking a predicted TE insertion, and P2 was designed based on one terminal sequence of the predicted TE. As illustrated in Supplemental Figures 2B and 2C online, the sizes of the amplicons in the population were used to determine whether the predicted nonreference TE insertions exist in the resequenced accessions.
Estimation of Local GR Rates
The local GR rates were estimated using MareyMap (Rezvoy et al., 2007). A total of 3873 markers from the genetic map of soybean (http://soybase.org) were used in this analysis. The GR-suppressed pericentromeric regions were defined based on the comparison of soybean genetic and physical maps as previously described (Schmutz et al., 2010).
The Distribution of TE Insertions and Genes and Subsequent Statistical Analyses
Each chromosome was split into contiguous 1-Mb regions (called windows) from the end of the long arm to the end of the short arm of the chromosome. GR rates were obtained for each window and plotted on the basis of their midpoints. The distributions and densities of genes were obtained from the latest annotation of soybean genome (Schmutz et al., 2010). The TE and gene contents/densities were calculated base on their proportions within each window. For the data set of arm windows, the first window, the last window, and the windows covered pericentromeric regions were not included in the analysis. The correlations among investigated parameters were assessed using Pearson’s correlation by 10,000 bootstrap resamplings as described previously (Tian et al., 2009).
Accession Number
The genome sequences used in this study were deposited in the National Center for Biotechnology Information Short Read Archive under accession number SRA020131 (Lam et al., 2010).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Diagram of the Strategy for Identification of Nonreference TEs Using Genome Resequencing Short Reads.
Supplemental Figure 2. PCR-Based Validation of Presence or Absence of Nonreference TEs in the Resequenced Genomes.
Supplemental Figure 3. Nonreference TE Insertion Sites and Genomic Features along the 20 Soybean Chromosomes.
Supplemental Figure 4. Distribution of Nonreference TEs between Pericentromeric Regions and Chromosomal Arms.
Supplemental Figure 5. Chromosomal Distribution of Nonreference LTR-RTs That Belong to the 10 Most Abundant Families Accumulated in the reference genome.
Supplemental Figure 6. Proportions of Nonreference TE Insertions in Different Categories of Sequence Components in C27 According to the Reference Genome.
Supplemental Table 1. Prediction and Validation of Nonreference TE Insertions in the 31 Resequenced Genomes.
Supplemental Table 2. Primers Used for Validation of Nonreference TE Insertions in the Resequenced Population.
Supplemental Data Set 1. Nonreference TE Insertions Identified in the Resequenced Genomes.
Acknowledgments
We thank Phillip SanMiguel and Jeff Bennetzen for their thoughtful comments on this article. This work was mainly supported by the United Soybean Board (Project 1249) to J.M. and partly supported by the National Science Foundation Plant Genome Research Program (Grant IOS-0822258) and Purdue Agricultural Research Program to J.M., the National Natural Science Foundation of China (Grant 91131005) and Chinese Academy of Sciences Startup Funds to Z.T., the Hong Kong Research Grants Council General Research Fund (Grant 468610) to H.-M.L. and X.X., and the Lo Kwee Seong Biomedical Research Fund and Lee Hysan Foundation to H.-M.L.
AUTHOR CONTRIBUTIONS
Z.T. and J.M. designed research. Z.T., M.Z., and M.S. performed research. Z.T., M.Z., M.S., J.D., X.L., X.X., S.B.C., X.Q., M.-W.L., H.-M.L., and J.M. generated and analyzed data. Z.T. and J.M. wrote the article.
Glossary
- TE
transposable element
- RT
retrotransposon
- LTR
long terminal repeat
- GR
genetic recombination
- NGS
next-generation sequencing
- WGS
whole-genome sequence
- UTR
untranslated region
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bennetzen J.L., Ma J., Devos K.M. (2005). Mechanisms of recent genome size variation in flowering plants. Ann. Bot. (Lond.) 95: 127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biémont C., Vieira C., Hoogland C., Cizeron G., Loevenbruck C., Arnault C., Carante J.P. (1997). Maintenance of transposable element copy number in natural populations of Drosophila melanogaster and D. simulans. Genetica 100: 161–166 [PubMed] [Google Scholar]
- Cao J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Carter T., Nelson R.L., Sneller C., Cui Z. (2004). Soybeans: Improvement Production and Uses. (Madison, WI: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America) [Google Scholar]
- Charlesworth B., Charlesworth D. (1983). The population dynamics of transposable elements. Genet. Res. 42: 1–27 [Google Scholar]
- Charlesworth B., Langley C.H., Sniegowski P.D. (1997). Transposable element distributions in Drosophila. Genetics 147: 1993–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Sniegowski P., Stephan W. (1994). The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220 [DOI] [PubMed] [Google Scholar]
- DePristo M.A., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K.M., Brown J.K., Bennetzen J.L. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K.M., Ma J., Pontaroli A.C., Pratt L.H., Bennetzen J.L. (2005). Analysis and mapping of randomly chosen bacterial artificial chromosome clones from hexaploid bread wheat. Proc. Natl. Acad. Sci. USA 102: 19243–19248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Grant D., Tian Z., Nelson R.T., Zhu L., Shoemaker R.C., Ma J. (2010a). SoyTEdb: A comprehensive database of transposable elements in the soybean genome. BMC Genomics 11: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Tian Z., Hans C.S., Laten H.M., Cannon S.B., Jackson S.A., Shoemaker R.C., Ma J. (2010b). Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: Insights from genome-wide analysis and multi-specific comparison. Plant J. 63: 584–598 [DOI] [PubMed] [Google Scholar]
- Du J., Tian Z., Sui Y., Zhao M., Song Q., Cannon S.B., Cregan P., Ma J. (2012). Pericentromeric effects shape the patterns of divergence, retention, and expression of duplicated genes in the paleopolyploid soybean. Plant Cell 24: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Marais G., Biémont C. (2000). Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics 156: 1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing A.D., Kazazian H.H., Jr (2011). Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 21: 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Hou Y., Ebina H., Levin H.L., Voytas D.F. (2008). Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 18: 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B.S., Wright S.I., Rizzon C., Dvorak J., Anderson L.K. (2007). Recombination: An underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 8: 77–84 [DOI] [PubMed] [Google Scholar]
- Hirochika H., Sugimoto K., Otsuki Y., Tsugawa H., Kanda M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93: 7783–7788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.T., Dooner H.K. (2008). Macrotransposition and other complex chromosomal restructuring in maize by closely linked transposons in direct orientation. Plant Cell 20: 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2012). Genome-wide genetic changes during modern breeding of maize. Nat. Genet. 44: 812–815 [DOI] [PubMed] [Google Scholar]
- Kumar A., Bennetzen J.L. (1999). Plant retrotransposons. Annu. Rev. Genet. 33: 479–532 [DOI] [PubMed] [Google Scholar]
- Lam H.M., et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42: 1053–1059 [DOI] [PubMed] [Google Scholar]
- Luce A.C., Sharma A., Mollere O.S., Wolfgruber T.K., Nagaki K., Jiang J., Presting G.G., Dawe R.K. (2006). Precise centromere mapping using a combination of repeat junction markers and chromatin immunoprecipitation-polymerase chain reaction. Genetics 174: 1057–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bennetzen J.L. (2004). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Devos K.M., Bennetzen J.L. (2004). Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14: 860–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A., Tanaka K., Murata K., Sawaki H., Takeda S., Abe K., Shinozuka Y., Onosato K., Hirochika H. (2003). Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K., Zhang F., Tsukiyama T., Saito H., Hancock C.N., Richardson A.O., Okumoto Y., Tanisaka T., Wessler S.R. (2009). Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Piegu B., Guyot R., Picault N., Roulin A., Sanyal A., Kim H., Collura K., Brar D.S., Jackson S., Wing R.A., Panaud O. (2006). Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 16: 1262–1269 Erratum. Genome Res. 21: 1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvoy C., Charif D., Guéguen L., Marais G.A. (2007). MareyMap: An R-based tool with graphical interface for estimating recombination rates. Bioinformatics 23: 2188–2189 [DOI] [PubMed] [Google Scholar]
- Rizzon C., Marais G., Gouy M., Biémont C. (2002). Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 12: 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C., Ponger L., Gaut B.S. (2006). Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput. Biol. 2: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Tian Z., Rizzon C., Du J., Zhu L., Bennetzen J.L., Jackson S.A., Gaut B.S., Ma J. (2009). Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res. 19: 2221–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z., Yu Y., Lin F., Yu Y.-S., SanMiguel P., Wing R.A., McCouch S., Ma J., Jackson S.A. (2011). Exceptional lability of a genomic complex of rice and its close relatives. BMC Genomics 12: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Dooner H.K. (2006). Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc. Natl. Acad. Sci. USA 103: 17644–17649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8: 973–982 [DOI] [PubMed] [Google Scholar]
- Wicker T., Yahiaoui N., Guyot R., Schlagenhauf E., Liu Z.D., Dubcovsky J., Keller B. (2003). Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell 15: 1186–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S.I., Agrawal N., Bureau T.E. (2003). Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 13: 1897–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., et al. (2012). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30: 105–111 [DOI] [PubMed] [Google Scholar]
- You F.M., Wanjugi H., Huo N., Lazo G.R., Luo M.C., Anderson O.D., Dvorak J., Gu Y.Q. (2010). RJPrimers: Unique transposable element insertion junction discovery and PCR primer design for marker development. Nucleic Acids Res. 38(Web Server issue): W313–W320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Gaut B.S. (2003). Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res. 13: 2533–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]