Ripening of the tomato fruit is accompanied by an increase in ethylene production and involves color changes, altered sugar metabolism, tissue softening, and the synthesis of aroma volatiles. This study shows that the MADS domain transcription factors FUL1 and FUL2 play a role in the regulation of these ripening processes, but in an ethylene-independent manner.
Abstract
Tomato (Solanum lycopersicum) contains two close homologs of the Arabidopsis thaliana MADS domain transcription factor FRUITFULL (FUL), FUL1 (previously called TDR4) and FUL2 (previously MBP7). Both proteins interact with the ripening regulator RIPENING INHIBITOR (RIN) and are expressed during fruit ripening. To elucidate their function in tomato, we characterized single and double FUL1 and FUL2 knockdown lines. Whereas the single lines only showed very mild alterations in fruit pigmentation, the double silenced lines exhibited an orange-ripe fruit phenotype due to highly reduced lycopene levels, suggesting that FUL1 and FUL2 have a redundant function in fruit ripening. More detailed analyses of the phenotype, transcriptome, and metabolome of the fruits silenced for both FUL1 and FUL2 suggest that the genes are involved in cell wall modification, the production of cuticle components and volatiles, and glutamic acid (Glu) accumulation. Glu is responsible for the characteristic umami taste of the present-day cultivated tomato fruit. In contrast with previously identified ripening regulators, FUL1 and FUL2 do not regulate ethylene biosynthesis but influence ripening in an ethylene-independent manner. Our data combined with those of others suggest that FUL1/2 and TOMATO AGAMOUS-LIKE1 regulate different subsets of the known RIN targets, probably in a protein complex with the latter.
INTRODUCTION
The angiosperms display a broad variety of fruit types, which can be roughly divided into dry fruits and fleshy fruits. Both fruit types evolved independently several times in the different plant lineages, and the occurrence of both dry and fleshy fruits within certain families, like the Solanaceae, suggests that the evolution from one type to the other does not require many diverging steps. Thus, the gene regulatory networks involved in the development of both dry and fleshy fruits may be similar, although the outcomes are morphologically very different. The Arabidopsis thaliana transcription factor APETALA2 (AP2) and its tomato (Solanum lycopersicum) ortholog AP2a, for example, are both involved in fruit development, but At-AP2 regulates dehiscence zone development in the dry silique (Ripoll et al., 2011), whereas Sl-AP2a influences fleshy fruit ripening via regulation of ethylene biosynthesis and signaling (Chung et al., 2010; Karlova et al., 2011).
The gene regulatory networks that are involved in fruit development have been largely unraveled for Arabidopsis, which produces a dry fruit, the silique that dehisces upon maturation to disperse the seeds (Roeder and Yanofsky, 2006). The formation of the valve margin, which separates the valves from the replum, is essential for seed dispersal and was found to be tightly regulated by a network of antagonistically acting transcription factors. The MCM1, AGAMOUS, DEFICIENS, SRF (MADS) box genes SHATTERPROOF1 (SHP1) and SHP2 and the basic helix-loop-helix (bHLH) genes INDEHISCENT (IND) and ALCATRAZ (ALC) are expressed in the valve margin and fulfill both independent and overlapping functions in valve margin development and maturation (Ferrándiz et al., 1999; Liljegren et al., 2000, 2004). In fruitfull (ful) mutant siliques, the valve margin genes become ectopically expressed in the valves and convert the valve cells into valve margin-like cells. As a consequence, ful siliques are very short and bumpy and the dehiscence zone is not specified (Ferrándiz et al., 2000). Thus, Arabidopsis FUL directly or indirectly represses SHP1, SHP2, IND, and ALC to allow seed dispersal.
Fleshy fruits do not form a dehiscence zone but have evolved to attract herbivores for seed dispersal. Fruit maturation typically involves color changes, altered sugar metabolism, tissue softening, and the synthesis of aroma volatiles. Research on fleshy fruit formation has primarily focused on the model system tomato, which produces a true berry derived from the ovary (Barry and Giovannoni, 2007). Early studies on tomato fruit development resulted in the identification of the plant hormone ethylene as an important mediator of ripening. Treatment of other fruits with ethylene inhibitors showed that many fleshy fruits depend on an increased ethylene production to undergo the transition to ripening. These fruits are referred to as climacteric fruits (Yang and Hoffman, 1984; Barry and Giovannoni, 2007).
In tomato, enhanced expression of the ethylene biosynthesis genes ACC synthase 1A (ACS1A), ACS2, ACS4, ACC oxidase1 (ACO1), ACO3, and ACO4 induces the transition from the autoinhibitory ethylene production system 1 to the autocatalytic production system 2, which is required for the onset of ripening (Barry et al., 1996; Nakatsuka et al., 1998). Ethylene-insensitive plants fail to respond to the increased ethylene levels and thus exhibit nonripening phenotypes, as was described for the Never-ripe (Nr) mutant, in which the NR ethylene receptor is impaired (Wilkinson et al., 1995), and for plants that ectopically express the GREEN-RIPE gene, which appears to interact with the ethylene response pathway (Barry and Giovannoni, 2006). Upstream of the ethylene pathway, several transcription factors have been found to regulate the ripening process, including the induction of the autocatalytic system 2, increased respiration, carotenoid biosynthesis, and cell wall softening (Barry and Giovannoni, 2007). The NAM, ATAF1, CUC2 (NAC)-domain family protein NON-RIPENING (NOR), the SQUAMOSA promoter binding protein (SBP) COLORLESS NON-RIPENING (CNR), and the MADS domain protein RIPENING INHIBITOR (RIN) appear to be the most important upstream ripening regulators, and their corresponding mutants, nor, Cnr, and rin, are impaired in many aspects of ripening (Thompson et al., 1999; Vrebalov et al., 2002; Klee and Giovannoni, 2011; Osorio et al., 2011).
The MADS domain transcription factor RIN belongs to the SEPALLATA (SEP) class of MADS domain proteins, of which the members have been found to interact with members from several other classes, mainly involved in flowering, floral organ formation, and reproduction. As such, the SEP proteins are thought to function as cofactors that allow higher order complex formation (Immink et al., 2009). The pleiotropic phenotype of the rin mutant is therefore probably the result of the disturbance of several different MADS complexes that play a role in tomato fruit ripening. Identification of these complexes and dissection of the function of RIN depends on the functional analysis of the interacting MADS box factors that are expressed in the fruit. The MADS domain proteins TOMATO AGAMOUS-LIKE1 (TAGL1), TAGL11, FUL1 (previously TDR4), and FUL2 (previously MBP7) were found to all interact with RIN (Leseberg et al., 2008; Martel et al., 2011) and to be present in ripening fruits (Hileman et al., 2006) and are thus candidates to mediate ripening processes together with RIN. Moreover, the binding of RIN to its target promoters was shown to be dependent on CNR activity, suggesting that CNR or one of its targets is required in a functional complex with RIN (Martel et al., 2011). The SHP1 ortholog TAGL1 has been reported to play an important role in tomato fruit development and ripening (Itkin et al., 2009; Vrebalov et al., 2009). TAGL1 knockdown plants were found to produce yellow-orange fruits with reduced carotenoids and thin pericarps. They have low ethylene levels due to decreased expression of the RIN target ACS2, and TAGL1 thus appears to regulate ripening by inducing the autocatalytic ethylene production system 2 together with RIN. Itkin et al. (2009) studied TAGL1 overexpression in the rin mutant background and found evidence for RIN-dependent and RIN-independent functions of TAGL1.
The fruit-expressed genes FUL1 and FUL2, which encode MADS domain transcription factors that are closely related to Arabidopsis FUL, are the other main candidates to regulate fruit ripening in tomato together with RIN. A FUL homolog of bilberry (Vaccinium myrtillus) was found to regulate color development and anthocyanin-related gene expression during ripening of the berry (Jaakola et al., 2010), indicating that FUL genes play important roles in both dry and fleshy fruits. Understanding the role of the FUL homologs in tomato will provide more information on the divergence and conservation of FUL function in dry and fleshy fruit maturation and contribute to the unraveling of the gene regulatory network involved in tomato fruit ripening.
To study the function of the FUL homologs in tomato, we characterized transgenic lines that were either specifically silenced for FUL1 or FUL2 or silenced for both. Using transcriptomics and metabolomics approaches, we found that the two genes have a redundant function in fruit ripening, downstream of the ripening regulators CNR, NOR, and RIN, and probably independent of the ethylene pathway.
RESULTS
The Tomato FUL Genes Are Predominantly Expressed during Fruit Development
The genome of tomato contains five genes that belong to the SQUAMOSA/AP1 clade of MADS box genes: MACROCALYX (MADS-MC) MBP10, MBP20, MBP7, and TDR4/TM4 (Tomato Genome Consortium, 2012). The orthologs of Arabidopsis AGL79, MBP10, and MBP20 were found to be expressed in vegetative tissues and inflorescences, while the expression pattern of the AP1-like gene MADS-MC mimics that of Arabidopsis AP1, with expression in the inflorescences, sepals, and petals (Hileman et al., 2006). TDR4 and MBP7 are the putative orthologs of Arabidopsis FUL and appear to be predominantly expressed during fruit development (Hileman et al., 2006). Because of the high sequence similarity between Sl-TDR4 and At-FUL (75% on amino acid level) and between Sl-MBP7 and At-FUL (77% on amino acid level), we renamed the genes Sl-FUL1 and Sl-FUL2, respectively.
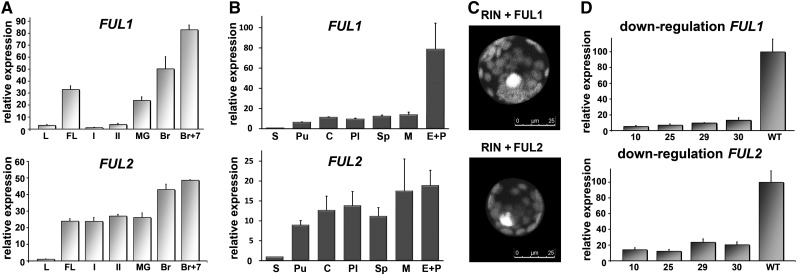
To investigate the activity of FUL1 and FUL2 during fruit development in more detail, we performed quantitative RT-PCR (qRT-PCR) analysis using fruits at different stages from the cultivar Micro Tom (MT) (Figure 1A). FUL1 expression was found to be very low during early stages of fruit development but increased rapidly from the mature green (MG) stage, reaching its maximum in the red ripe stage. The FUL2 transcripts, on the other hand, were abundantly present throughout fruit development and showed only a minor increase during the ripening phase. Both genes also showed considerable expression in the flower, which is in agreement with the data of Hileman et al. (2006), who found that FUL2 is expressed in all floral whorls, while FUL1 is mainly expressed in the stamens. To test where the transcripts of both genes are located in the ripe fruits, we separated the different fruit tissues and performed qRT-PCR analysis. FUL2 appeared to be equally expressed in all tissues, except in developing seeds, while FUL1 expression was clearly most abundant in the exocarp and peel (Figure 1B). Thus, both genes are highly active during fruit development in tomato, but FUL1 expression is especially abundant in the outer layers of the pericarp in ripening fruits.
Figure 1.
FUL1 and FUL2 Expression in Wild-Type Tomato and FUL1 RNAi Lines, and Interaction of the FUL Proteins with RIN.
(A) Relative expression profiles of FUL1 and FUL2 in different MT tissues obtained by qRT-PCR. FL, flower; I, 5-mm fruits; II, 1-cm fruits; L, leaf; FL, flower; I, 5mm fruits; II, 1cm fruits; MG, mature green; Br, breaker; Br+7, 7 days after breaker stage (red ripe).
(B) Relative expression profiles of SlFUL1 and SlFUL2 in separated fruit tissues of red ripe fruits. S = seeds; Pu = pulp; C = columella; Pl = Placenta; Sp = septa; M = mesocarp; E+P = exocarp and peel.
(C) Confocal scanning laser microscopy pictures of Arabidopsis protoplasts transfected with split-YFP constructs. Top panel, RIN-YFP(C) and FUL1-YFP(N); bottom panel, RIN-YFP(C) and FUL2-YFP(N).
(D) Downregulation of FUL1 and FUL2 in the FUL1 RNAi lines R1-10, R1-25, R1-29, and R1-30 compared with the wild type (WT). The relative expression levels were obtained by qRT-PCR.
The error bars in (A) to (C) indicate the se based on two biological replicas.
FUL1 and FUL2 Interact in Vivo with the Ripening Factor RIN
The tomato MADS domain protein RIN, which belongs to the SEP clade, fulfills a key role in fruit ripening by mediating virtually all ripening processes (Vrebalov et al., 2002). RIN can heterodimerize with FUL1 and FUL2 in yeast (Leseberg et al., 2008), suggesting that both FUL homologs may function in the ripening process as interaction partners of RIN. To obtain more evidence for the existence of RIN-FUL1 and RIN-FUL2 heterodimers in planta, we tested the interactions in vivo in Arabidopsis protoplasts using split-yellow fluorescent protein (YFP) (Walter et al., 2004). The combinations RIN-YFP(C)/FUL1-YFP(N) and RIN-YFP(C)/FUL2-YFP(N) yielded a bright yellow fluorescent signal in the nucleus of the protoplast, confirming the interaction of RIN with both FUL1 and FUL2 (Figure 1C). We also tested the combination FUL1-YFP(N)/FUL2-YFP(C) but did not detect YFP signal in the protoplast nuclei (see Supplemental Figure 1 online), indicating that FUL1 does not dimerize with FUL2.
FUL1/2 Silenced Fruits Show an Orange-Ripe Phenotype
To investigate the functions of FUL1 and FUL2 in tomato, we attempted to produce stable transgenic MT lines in which either FUL1 or FUL2 was specifically downregulated. To achieve this, we initially generated a 35S:FUL1 RNA interference (RNAi) construct and transformed it to MT. Because the coding sequences of FUL1 and FUL2 are 78% identical, the RNAi fragment was amplified from the 3′ end of FUL1, including a part of the 3′ untranslated region (UTR), where the identical stretches did not exceed 20 nucleotides (see Supplemental Figure 2 online). The transformation yielded 29 plants containing the 35S:FUL1 RNAi construct, and eight of these lines produced fruits that did not turn red upon ripening.
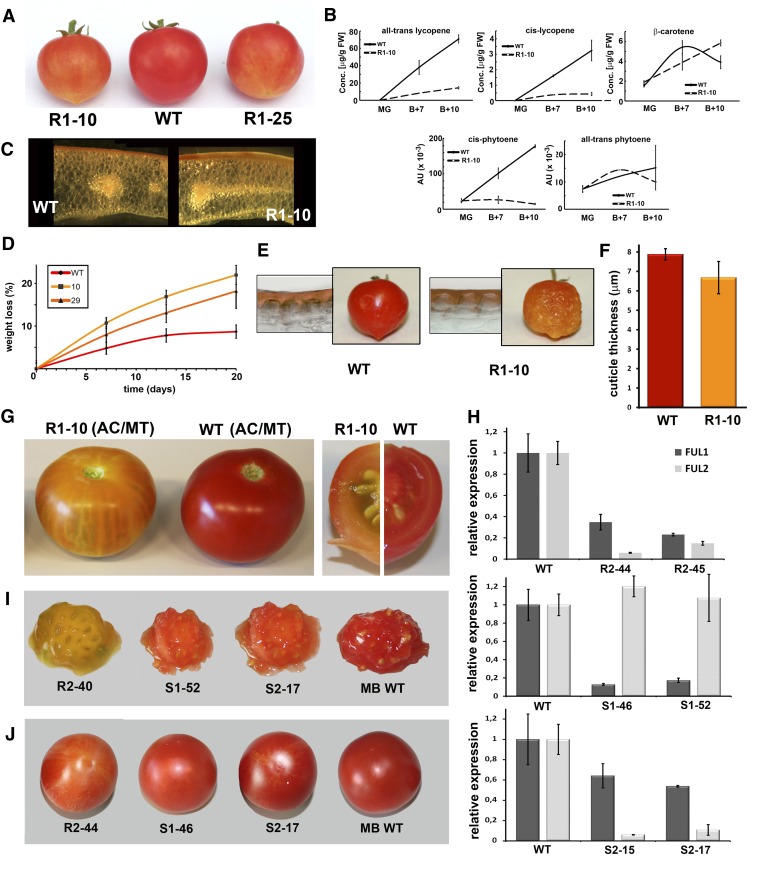
Some lines produced orange-red fruits with orange sections, while others exhibited a more severe phenotype with fruits that remained orange, also often with lighter-colored spots at the blossom end (Figure 2A). We tested the downregulation of Sl-FUL1 in the transgenic lines and found a strong correlation between the fruit phenotype and the endogenous transcript level (see Supplemental Figure 3 online). The four lines with the strongest phenotype, lines R1-10, R1-25, R1-29, and R1-30, were selected for further analysis, and their offspring were raised for segregation analysis. The presence of the construct was tested by PCR in at least 35 T1 plants of each line and was found to cosegregate with the orange-ripe phenotype.
Figure 2.
Phenotypes of the FUL1 and FUL2 Downregulated Fruits.
(A) Fruits of FUL1 RNAi lines R1-10 and R1-25 and wild-type (WT) MT at stage Br + 7.
(B) Comparison of the carotenoid levels in wild-type and R1-10 fruits at stage MG, B+7 (Br + 7 d), and B+10 (Br + 10 d). AU, absorbance units; FW, fresh weight. The complete analysis is presented in Supplemental Table 1 online.
(C) Binocular microscopy pictures of handmade pericarp sections of wild-type and R1-10 fruits at stage Br + 7.
(D) Water loss in wild-type and R1-10 and R1-29 fruits after harvesting at stage Br + 7. The y axis depicts the percentage of weight loss since the day of harvesting.
(E) Cuticle thickness of wild-type and R1-10 Br + 7 fruits is visible in light microscopy pictures of thin pericarp sections stained with Sudan IV. The side panels show the desiccated tomatoes 40 d after harvesting.
(F) Cuticle thickness in wild-type and R1-10 Br + 7 fruits, measured from the top of the epidermal cell to the surface. For both the wild type and R1-10, the cuticles of four different fruits were measured. Error bars indicate the se.
(G) Phenotype of line R1-10 in the Ailsa Craig background. FUL1 RNAi line R1-10 was crossed with Ailsa Craig wild type, and the offspring (harboring the MT mutations recessively) were phenotyped.
(H) qRT-PCR analysis showing the downregulation of FUL1 and FUL2 in transgenic lines carrying the FUL2 RNAi construct (R2-44 and R2-45) or the specific 3′ UTR constructs for FUL1 (S1-46 and S1-52) or FUL2 (S2-15 and S2-17). The error bars depict the se based on two biological replicas.
(I) Jelly of red ripe fruits from lines R2-40, S1-46, S2-17, and Moneyberg wild type.
(J) Red ripe fruits of lines R2-44, S1-46, S2-17, and Moneyberg wild type. Note the variation in pigmentation at the blossom end of the transgenic fruits.
The effect of the FUL1 RNAi construct on the transcript levels of FUL1 and FUL2 in the selected lines was tested by qRT-PCR analysis using three biological replicates of breaker (Br) stage fruits (Figure 1D). In all four lines, endogenous FUL1 levels were <15% of wild-type levels and the highest reduction was found in line R1-10, where only 5% of the transcript was still present. Unexpectedly, the transcript levels of FUL2 were highly reduced as well, ranging from 12% of wild-type levels in line R1-25 up to 24% in line R1-29, suggesting that the FUL1 RNAi fragment targets FUL2 as well. However, it is also possible that FUL1 regulates FUL2 expression in the fruit. To investigate the likelihood of the latter possibility, we tested the downregulation of FUL2 in the fruit stages were FUL1 is hardly expressed, such as in the immature fruit. Also there, FUL2 was considerably silenced (see Supplemental Figure 4 online), suggesting that its expression in the wild type is not induced by FUL1 and that its downregulation is an effect of the RNAi approach. Since both paralogs are silenced in lines R1-10, R1-25, R1-29, and R1-30, we will further refer to these as FUL1/2 RNAi lines. We will first describe the phenotypic characterization of these lines and report thereafter about the successful generation of specifically silenced lines.
Reduced Lycopene Levels Cause the Orange-Ripe Phenotype of the FUL1/2 RNAi Fruits
To investigate the cause of the altered fruit pigmentation in the FUL1/2 RNAi fruits, we performed HPLC analysis and measured the concentrations of the different carotenoids, and their precursor phytoene, in wild-type and line R1-10 fruits (Figure 2B; see Supplemental Table 1 online). Wild-type fruits accumulate different carotenoids in the course of ripening, changing their color from green to red. While the concentration of the yellow pigment lutein decreases during ripening, lycopene (red), mainly in the all-trans configuration, and β-carotene (orange) contents increase to ∼80 and 10% of the total carotenoids in red ripe MT tomatoes, respectively (see Supplemental Table 1 online). Levels of the carotenoid precursor phytoene, predominantly found in the 15-cis configuration, correspondingly increase during ripening in the wild-type fruit. The orange-ripe phenotype of FUL1/2 RNAi fruits was found to result mainly from a major decrease in lycopene and its precursor cis-phytoene, which were both reduced to ∼20% of wild-type levels in the ripe fruit 7 d after Br stage (Br + 7). The lutein content was similar in wild-type and transgenic fruits, but a small increase in the β-carotene content of Br + 10 stage R1-10 fruits was observed, possibly contributing to the orange-ripe phenotype. Finally, we also measured the concentrations of xanthophylls, chlorophylls, and tocopherols but did not find distinct differences in content between wild-type and transgenic fruits (see Supplemental Table 1 online).
FUL1/2 Silenced Fruits Show Normal Pericarp Development but Increased Water Loss
FUL1/2 RNAi tomatoes did not show any visible abnormalities besides the fruit pigmentation. Size and shape of the transgenic fruits resembled those of the wild type, and we did not observe differences in pericarp development in sections of Br + 7 stage fruits (Figure 2C). We also compared the firmness of freshly harvested transgenic tomatoes with the wild type using a fruit hardness meter but found no differences in the softening of the fruits. To investigate if fruit water loss after harvest was altered in the FUL1/2 silenced lines, fruits were harvested 7 d after Br stage, including pedicel (red ripe in the wild type), and the weight of the detached fruits was monitored during 21 d (Figure 2D). The transgenic fruits were found to desiccate much faster than the wild type (Figure 2E), losing up to 25% of their weight, compared with 10% in the average wild-type fruit. Fruit water loss also correlated with FUL1 transcript level and was more severe in line R1-10 than in line R1-29. Because pericarp thickness did not differ between wild-type and transgenic tomatoes, we investigated if altered epidermis characteristics could explain the increased water loss. The epidermal cell layer is the outermost layer of the fruit pericarp and is covered by a waxy cuticle that thickens as the fruit ages, protecting the fruit from desiccation (Mintz-Oron et al., 2008). The thickness of the waxy cuticle can be determined with Sudan staining, which has a high affinity for lipids. To determine cuticle thickness, pericarp sections of Br + 7 stage transgenic fruits and their wild-type siblings were stained with Sudan IV and the cuticle layers were compared (Figures 2E and 2F). We did not find a significant difference in the thickness of the cuticle layers, indicating that the increased water loss has a different cause.
In conclusion, FUL1/2 silenced MT fruits develop normally until Br stage but are impaired in several ripening processes.
The FUL1 RNAi Phenotype Is Similar in a Large-Fruited Background
The tomato variety MT is well suited for research purposes because of its small plants, which have a relatively short generation time. However, MT plants harbor four independent mutations, including one in the brassinosteroid pathway (Martí et al., 2006), which may influence tomato development and ripening. For example, the size of MT fruits is quite variable, and silencing of TAGL1 did not lead to reduced pericarp thickness in MT, but showed this phenotype only in the nondwarf variety Ailsa Craig (Itkin et al., 2009; Vrebalov et al., 2009). To investigate if the phenotype of the MT FUL1/2 silenced fruits was reproducible in a nondwarf variety, we crossed lines R1-10 and R1-29 with Ailsa Craig plants and determined the fruit phenotype of the F1 offspring (Figure 2G), in which the MT mutations are in a recessive state. The wild-type progeny from the cross produced normal red-ripe fruits, but those with the FUL1/2 RNAi construct remained orange with sometimes almost transparent sectors. Both the size of the fruits and the pericarp thickness were similar in the wild-type and transgenic fruits. Thus, the FUL1/2 RNAi phenotype in the large-fruited hybrid offspring is comparable to the MT phenotype, although the pigmentation alterations are more distinct in the former. To investigate the effectiveness of the FUL1/2 RNAi construct in the hybrid background, we determined the expression levels of FUL1 and FUL2 in Br stage fruits by qRT-PCR. The levels in the transgenic fruits were reduced to 20 and 14% of wild-type levels, respectively (see Supplemental Figure 5 online), showing that the construct severely affects the FUL2 expression in the hybrid background as well.
FUL1 and FUL2 Act Redundantly during Fruit Ripening
To be able to differentiate between the functions of FUL1 and FUL2 in tomato fruit ripening, we undertook a second attempt to generate lines that were specifically silenced for either FUL1 or FUL2. Plants of the large-fruited cultivar Moneyberg were either transformed with a FUL2 RNAi construct similar to the previously generated FUL1 RNAi construct (containing 3′ open reading frame and 3′ UTR sequences) or transformed with a synthetically generated construct containing only the 3′ UTR of FUL1 or FUL2. The effect of the constructs was subsequently tested in the ripe fruits of the transgenic lines by qRT-PCR (see Supplemental Figures 6A to 6C online). Similar to what was observed with the FUL1 RNAi construct, plants transformed with the FUL2 RNAi construct were all distinctly silenced for both FUL homologs (five lines were generated and tested with qRT-PCR), although the transcript levels of FUL2 were decreased further than those of FUL1 (see Supplemental Figure 6A online). The qRT-PCR results from the representative lines R2-44 and R2-45 are shown in Figure 2H. The plants all exhibited an orange-ripe fruit phenotype similar to the fruits of the FUL1 RNAi lines (Figures 2I and 2J). Specific silencing was achieved for the plants transformed with the synthetic FUL1 construct, for which we generated 10 transgenic lines. qRT-PCR analysis showed that these lines were in general not downregulated for FUL2, while they showed variable levels of FUL1 silencing (see Supplemental Figure 6C online). The specific silencing of FUL1 was most distinct in lines S1-46 and S1-52, with a reduction in FUL1 transcript level to 13 and 17%, respectively (Figure 2H). The four lines that we analyzed with the specific FUL2 construct showed variable levels of silencing for both FUL2 and FUL1 (see Supplemental Figure 6B online). The lines S2-15 and S2-17 were severely silenced for FUL2, with endogenous FUL2 transcript levels reduced to 6 and 11%, respectively, while the expression of FUL1 was only weakly affected and reduced to ∼60% of wild-type levels (Figure 2H). The gene-specific downregulation of FUL1 or FUL2 in the flowers of the selected lines S2-15, S2-17, S1-46, and S1-52 was found to be comparable to the downregulation in the fruits (see Supplemental Figure 6D online). We analyzed the fruit phenotypes in the lines specifically silenced for FUL1 or FUL2 and observed only very mild alterations in the pigmentation of the ripe fruits. This was most distinct in the jelly of the fruits, which was more orange than that of the wild type (Figure 2I). The alterations in the pericarp pigmentation were most visible at the blossom end of the tomato, where often lighter colored sectors could be observed (Figure 2J). The phenotypes of the specific FUL1 silenced lines did not differ from the FUL2 silenced lines, indicating that they share a role in carotenoid accumulation. However, we cannot exclude that the weak downregulation of FUL1 in lines S2-15 and S2-17 has contributed to the phenotype as well. The fact that the phenotype is much more distinct in lines that have both genes strongly silenced points to redundant roles of FUL1 and FUL2 during tomato fruit ripening.
Microarray Analysis Increases the Insight into the Role of FUL1/2 in Ripening
Tomato fruit ripening involves the regulation of several biochemical pathways associated with pigmentation and the production of aromatic and nutritional compounds, cell wall metabolism, and pathogen susceptibility. To obtain more insight into the roles of FUL1 and FUL2 in mediating these ripening processes, we performed mRNA expression experiments with the double silenced R1-10 and R1-30 lines using microarrays. RNA from Br stage R1-10 and R1-30 fruits and from fruits of their wild-type siblings in the T1 generation was hybridized to Agilent-022270 Tomato Gene Expression microarrays. Analysis of differentially expressed genes in the 44,000 probe set revealed 394 genes that were significantly lower expressed in both transgenic lines (twofold or more) and 501 genes that were twofold or more higher expressed in the transgenic fruits (P < 0.05). A full list of significantly differentially expressed genes can be found in Supplemental Data Set 1 online. Our results confirm downregulation of both FUL1 (log2 = −4.5) and FUL2 (log2 = −2.7) and also yielded a number of genes that were previously reported to be involved in tomato fruit ripening, including RIN and CNR targets (Thompson et al., 1999; Fujisawa et al., 2012; Qin et al., 2012). A short list, containing the up- and downregulated genes discussed in the text, can be found in Table 1. Expression of several known targets of RIN, such as Carbonic Anhydrase3, Gibberellin 20-oxidase-2, Polygalacturonase Ae (PG2A), 1-deoxy-d-xylulose 5-phosphate synthase1 (DXS1), expansin B2, and Phosphoglycerate dehydrogenase, was also affected in the FUL1/2 knockdown plants, suggesting that there is overlap between the targets. Yet, other known targets of RIN, such as the ethylene biosynthesis genes ACO1, ACS2, and ACS4, and the ripening-related genes E4 and E8, lipoxygenase C, or the carotenoid biosynthesis gene Phytoene synthase1 (PSY1) were not significantly affected (Fujisawa et al., 2012). In addition to the earlier identified regulators of tomato fruit ripening, a large number of up- or downregulated genes was not implicated in ripening before but likely play a role in ripening-associated processes as well. These include genes encoding for proteases, pectinases, expansins, and extensins and a high number of genes encoding for enzymes involved in primary or secondary metabolism. In order to identify changed metabolic pathways, we loaded the microarray data into the Plant MetGenMAP database (Joung et al., 2009). Several metabolic pathways that can be linked to ripening were revealed to be regulated by FUL1/2, including Glu degradation, carotenoid biosynthesis, and suberin biosynthesis. The complete list of significantly changed metabolic pathways is available in Supplemental Table 2 online.
Table 1. Short List of Up- and Downregulated Genes in FUL1/2 RNAi Fruits.
| Primary Accession | iTAG Locus | Log2 ratio RNAi/Wild Type | Annotation | Reference |
|---|---|---|---|---|
| BT013224 | Solyc06g069430* | −4.5 | FUL1/TDR4 | Busi et al. (2003) |
| AM949788 | Solyc02g067750* | −3.6 | Carbonic anhydrase3 (CA3) | Fujisawa et al. (2011) |
| TA41663_4081 | Solyc08g082170 | −3.1 | Polygalacturonase (pectinase) homolog (PG) | |
| BP880561 | Solyc10g080210* | −2.9 | PG2A (endopolygalacturonase) | Sheehy et al. (1987) |
| Knapp et al. (1989) | ||||
| Eriksson et al. (2004) | ||||
| Alba et al. (2005) | ||||
| AK327129 | Solyc06g035530* | −2.8 | Gibberellin 20-oxidase-2 (20ox-2) | Fujisawa et al. (2012) |
| BM412293 | Solyc03g114830 | −2.7 | FUL2 / MBP7 | Hileman et al. (2006) |
| BE451418 | Solyc06g061080 | −2.6 | NAM TF homolog | |
| BG126642 | Solyc05g047460 | −2.3 | Auxin response factor ARF19-1 | |
| AF230372 | Solyc07g049690 | −1.8 | Fatty acid hydroperoxide lyase HPL1 | |
| TA56518_4081 | Solyc11g005120 | −1.6 | MADS box TF MBP22 | Hileman et al. (2006) |
| Y14810 | Solyc03g007960 | −1.6 | CHY2/CrtR-b2 (carotene β-hydroxylase) | Galpaz et al. (2006) |
| AF143812 | Solyc01g067890* | −1.6 | DXS1 (1-d-deoxyxylulose 5-phosphate synthase) | Alba et al. (2005) |
| Lois et al. (2000) | ||||
| Fujisawa et al. (2012) | ||||
| AK321432 | Solyc06g084260 | −1.3 | Chalcone isomerase homolog | |
| AK320087 | Solyc06g069760 | −1.3 | AOBP (ascorbate oxidase promoter binding protein) | Alba et al. (2005) |
| AK326929 | Solyc06g060170 | −1.2 | Polygalacturonase (pectinase) homolog | |
| AF195507 | Solyc01g097810 | −1.2 | ζ-Carotene desaturase | Isaacson et al. (2002) |
| AB041811 | Solyc10g047030 | −1.2 | β-d-xylosidase (XYL1) | Itai et al. (2003) |
| AY360170 | Solyc08g076470 | −1.2 | Glycerol-3-P acetyltransferase (GPAT) | |
| AB359914 | Solyc11g011920* | +1.2 | Glu decarboxylase (GAD2) | |
| AY508112 | Solyc02g067180* | +1.4 | Cystathionine γ-synthase (CGS) | |
| DB714856 | Solyc11g044910 | +1.9 | β-d-xylosidase (XYL2) | Itai et al. (2003) |
| BM412023 | Solyc03g098240* | +1.9 | Glu decarboxylase (GAD3) | |
| BP883976 | Solyc10g086480 | +2.0 | Pectin acetyl transferase | Eriksson et al. (2004) |
| AB359913 | Solyc03g098240 | +2.1 | Glu decarboxylase (GAD1) | |
| CK715255 | Solyc06g082550 | +2.3 | Cinnamic acid 4-hydroxylase | |
| AF328785 | Solyc01g009170 | +2.7 | EIN3-like transcription factor EIL2 | |
| BG128054 | Solyc01g079240 | +3.1 | Long-chain acyl-CoA synthetase | |
| BG628578 | Solyc11g065770 | +3.5 | Cytochrome P450-dependent fatty acid hydroxylase (CYP94A25) | |
| AK322239 | Solyc04g063210 | +3.7 | Caffeoyl-CoA 3-O-methyltransferase | |
| AK324919 | Solyc01g099020 | +5.3 | GDSL-like Lipase | |
| AK325197 | Solyc03g123830* | +5.5 | Putative d-3-phosphoglycerate dehydrogenase (PGDH) | Qin et al. (2012) |
| DQ205653 | Solyc03g093390* | +6.6 | Expansin (EXPB2) |
Asterisks indicate genes found to be regulated by RIN, as mentioned in the text.
To test the reliability of the microarray results, we selected five genes and tested their expression in MG, Br, and Br + 7 stage transgenic and wild-type fruits by qRT-PCR. The results are shown in Supplemental Figures 7A and 7B online. The microarray data were confirmed for the upregulated genes GDSL (GDSL-like lipase; log2 = 5.3) and EIL2 (EIN3-like transcription factor; log2 = 2.7) and the downregulated gene PG2A (endopolygalacturonase; log2 = −2.9). The downregulation of PG2A is specific for the Br and Br + 7 stages, in line with its reported RIN-dependent ripening-specific upregulation (Martel et al., 2011). We also confirmed the upregulation of a pectin acetyl transferase gene (PAT; log2 = 2.0) in the transgenic fruits, but it turned out to be less distinct than expected from the microarray data. This difference can presumably be explained by the weak expression of PAT in the fruits (cycle threshold values were around 30), which may result in a bias in either the qRT-PCR or microarray data. Although the microarray results also suggested the downregulation of CrtR-b2 (for carotene β-hydroxylase; log2 = −1.6) this could not be confirmed by qRT-PCR. In conclusion, the qRT-PCR analysis shows that most of the microarray data can be confirmed.
Carotenoid Biosynthesis Genes
A number of genes involved in the carotenoid biosynthesis pathway have been shown to be induced at the MG-to-Br stage transition (Alba et al., 2005; Itkin et al., 2009; Vrebalov et al., 2009). These include PSY1, which regulates the conversion of phytoene and is highly downregulated in TAGL1 silenced plants (Itkin et al., 2009; Vrebalov et al., 2009), and DXS1 (Lois et al., 2000), which is required for the first step in the carotenoid biosynthesis pathway. Unlike in TAGL1 silenced plants, we did not find a reduction of PSY1 expression in the FUL1/2 silenced fruits. Instead, we found a significant downregulation of DXS1 (log2 = −1.6), suggesting that TAGL1 and FUL1/2 act on different steps of the carotenoid pathway to regulate lycopene accumulation in the ripening fruit, while RIN acts on both (Fujisawa et al., 2012).
Cell Wall–Modifying Protein Encoding Genes
Fruit softening as a result of secondary cell wall degradation is an important feature of fruit ripening in general and is regulated by altered expression of genes for polygalacturonases, pectin modification, and degradation enzymes, expansins, and extensins in tomato. In Cnr mutants, expression of a number of these genes is altered during ripening compared with wild-type fruits (Thompson et al., 1999). We found several of the CNR-regulated genes to be also differentially expressed in the FUL1/2 silenced lines. The expression of the major fruit polygalacturonase gene, PG2A, was significantly decreased compared with wild-type fruits (log2 = −2.9, confirmed by qRT-PCR), as well as the expression of the β-xylosidase encoding gene XYL1 (log2 = −1.2), while a pectin acetyl transferase upregulated during ripening in Cnr mutants was also upregulated in the FUL1/2 RNAi fruits (log2 = 2.0) The homolog of XYL1, XYL2, which is normally downregulated during ripening, was increased in expression in FUL1/2 RNAi lines (log2 = 1.9) (Itai et al., 2003). Additionally, the expansin precursor gene EXPB2 was strongly upregulated in FUL1/2 RNAi fruits (log2 = 6.6). Thus, the tomato FUL homologs appear to regulate a number of cell wall–modifying genes, which at least in part overlap with those regulated by CNR and RIN.
Transcriptome Analysis Suggests Altered Lipid and/or Cuticle Metabolism in FUL1/2 Silenced Fruits
The cuticle has been reported to be an important transpiration barrier (Mintz-Oron et al., 2008); therefore, the altered expression of fatty acid biosynthesis genes may contribute to the increased water loss of the FUL1/2 silenced fruits. We found several genes related to cutin and fatty acid synthesis to be downregulated in FUL1/2 silenced fruits, among which were a glycerol-3-P-acyltransferase (log2 = −1.2) and a cytochrome P450 hydroperoxide lyase (HPL) (log2 = −1.8). The latter has also been implemented in lipid-derived volatile production (Schwab et al., 2008). In addition, the suberin pathway was found to be significantly changed in the transgenic fruits by upregulation of the genes encoding for cinnamic acid 4-hydroxylase (log2 = 2.3) and caffeoyl-CoA 3-O-methyltransferase (log2 = 3.7) (Plant MetGenMAP; see Supplemental Table 2 online). Other genes involved in fatty acids metabolism were strongly upregulated in the transgenic fruits, like genes encoding for a cytochrome P450-dependent fatty acid hydroxylase (log2 = 3.5), a long-chain acyl-CoA synthetase family protein (log2 = 3.1), and a GDSL-like lipase (log2 = 5.3).
FUL1/2 Regulate Glu Accumulation during Ripening
The Plant MetGenMAP analysis revealed that the Glu degradation pathway is highly significantly altered in the FUL1/2 RNAi lines (see Supplemental Table 2 online). All three known genes encoding for Glu decarboxylases, which convert l-Glu into γ-amino butyrate (GABA), appeared to be upregulated in the transgenic plants (GAD2, log2 = 1.3; GAD3, log2 = 1.9; GAD1, log2 = 2.1). Interestingly, the Glu content of tomato fruits normally rises strongly upon ripening. While the relative molar content of Glu in MG MT fruits is ∼12.7% of the total free amino acid content, this increases to 55.0% in the red ripe fruit. Concurrently, the relative GABA concentration drops from 40.4% in the green fruit to 8.3% in the red ripe fruit (Sorrequieta et al., 2010). This indicates that the repression of the Glu decarboxylases in wild-type fruits is important for the increase in Glu content in the ripening tomatoes. In rin fruits, the expression of GAD2 and GAD3 is also upregulated, indicating that these genes are also common targets of FUL1/2 and RIN (Fujisawa et al., 2012).
The total free amino acid content of tomato fruits increases approximately fivefold during ripening, which contributes markedly to the taste of the fruit. However, Glu seems particularly important for a tasty tomato, since all cultivated varieties have much higher contents than wild tomatoes (Boggio et al., 2000; Pauliukaite et al., 2006). Glu was also found to be responsible for the characteristic umami taste of tomatoes and other foods like cheese and mushrooms (Bellisle, 1999). Thus, the repression of the Glu decarboxylases by FUL1/2 in the wild type likely results in a higher Glu content and possibly a tastier tomato. To test if the transgenic fruits indeed accumulate less Glu, we determined the concentrations of Glu and GABA, as well as from many other primary metabolites in stage Br + 7 fruits of lines R1-10 and R1-29, and compared the levels to those of wild-type fruits. Table 2 shows all primary metabolites that had significantly lower or higher concentrations in R1-10 fruits than in their wild-type siblings. The complete list of tested primary metabolites of both lines and the wild type is available as Supplemental Table 3 online. The analysis reveals that the Glu content in the transgenic fruits is indeed more than eightfold decreased, while the GABA concentration is increased. The marked decrease in Pro accumulation in FUL1/2 RNAi fruits could also be a result of the reduced Glu levels, since Glu is one of the precursors for Pro biosynthesis in plants. In addition, the contents of n-acetylglutamic acid, putrescine, and 2-oxoglutarate, metabolites that are related to Glu/GABA metabolism, were significantly changed in the transgenics. Also the amino acids Ser, Val, Ile, β-Ala, and Asp showed altered concentrations, indicating that FUL1/2 play a role in the accumulation of certain amino acids during ripening.
Table 2. Primary Metabolites Significantly Changed in FUL1/2 RNAi Fruits.
| Metabolite | Fold Difference to the Wild Type | P Value |
|---|---|---|
| Glu | −8.25 | 0.006901 |
| n-Acetylglutamic acid | −6.81 | 0.002051 |
| Pro | −2.98 | 0.032185 |
| Galacturonate | −2.90 | 0.002637 |
| Glucuronate | −2.64 | 0.002531 |
| Putrescine | −1.84 | 0.019208 |
| Inositol | −1.45 | 0.019968 |
| Xyl | −1.32 | 0.022550 |
| 2-Oxoglutarate | 1.62 | 0.031674 |
| Ser | 1.83 | 0.005587 |
| Val | 2.02 | 0.008840 |
| Ile | 2.14 | 0.018104 |
| GABA | 2.30 | 0.000802 |
| β-Ala | 2.36 | 0.000149 |
| Asp | 2.60 | 0.002157 |
| Hexanoic acid | 3.51 | 0.015746 |
Differences between FUL1/2 RNAi fruits and the fruits of their wild-type siblings are represented as negative or positive fold ratios of the mean gas chromatography–mass spectrometry responses. Significance of the differences is indicated by the P value from the Student’s t test.
In addition to the altered amino acids, a few other primary metabolites were found to differ significantly in concentration between FUL1/2 silenced fruits and those of the wild type (Table 2). The lower concentrations of the primary cell wall component galacturonate and its precursor glucuronate in FUL1/2 RNAi fruits point to a reduced pectin degradation in the transgenics (Mølhøj et al., 2004; Usadel et al., 2004), consistent with the downregulation of two polygalacturonase homologs in FUL1/2 RNAi fruits (log2 = −3.1 and log2 = −1.2). These data provide additional evidence for the role of FUL1/2 in cell wall modification.
FUL1/2 Act Downstream or Independent of the Ethylene Pathway
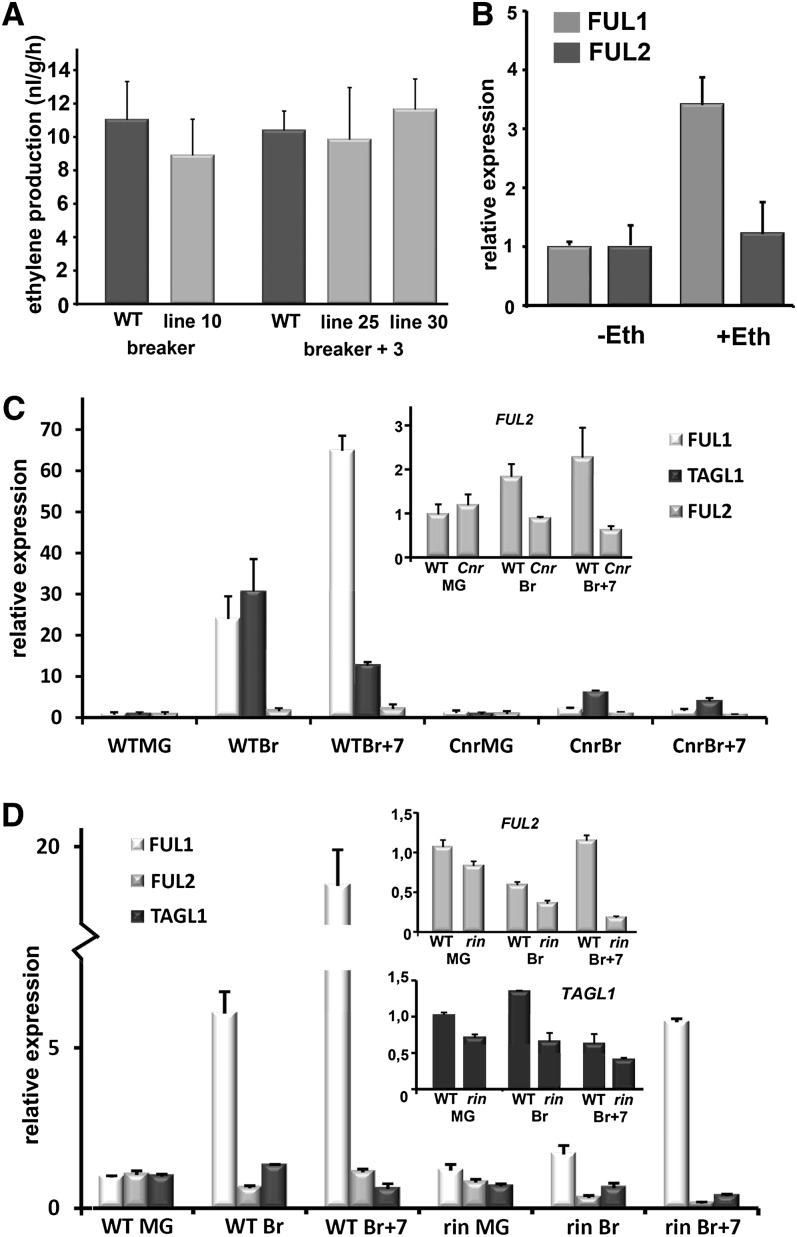
In the climacteric tomato fruit, the onset of ripening is marked by a major increase in ethylene production. In the ripening-deficient fruits of rin and TAGL1 RNAi plants, the autocatalytic ethylene production system is not induced because of decreased expression of the ethylene biosynthesis genes ACS2, ACS4, and ACO1 (Itkin et al., 2009; Vrebalov et al., 2009; Fujisawa et al., 2012). As a consequence, these mutants produce drastically lower ethylene levels and fail to initiate ethylene-dependent ripening processes. Similarly, downregulation of an ACC oxidase gene (ACO) is correlated with reduced ethylene levels in the nonripening fruits of Cnr mutants and hb-1 silenced plants (Thompson et al., 1999; Lin et al., 2008). Unexpectedly, our microarray analysis showed that there was no effect on ACS or ACO gene expression in the FUL1/2 RNAi plants. Only one climacteric ethylene synthesis-related gene was found to be differentially regulated in FUL1/2 silenced plants, encoding cystathionine γ-synthetase (CGS), involved in Met synthesis and as such synthesizing the precursors for ethylene. This gene was found to have increased expression during ripening by Alba et al. (2005) but was slightly upregulated (log2 = 1.4) in FUL1/2 RNAi fruits, suggesting that FUL1/2 downregulates CGS in wild-type fruits. The fact that FUL1/2 RNAi did not significantly affect expression of ethylene biosynthesis genes suggests that ethylene levels are not affected in FUL1/2 RNAi fruits. The upregulation of CGS in the transgenic plants may even point to a repressive or negative feedback function for FUL1/2 in climacteric ethylene synthesis during ripening. To confirm that the rise in ethylene levels of ripening fruits also occurs in the absence of FUL1/2, we harvested Br or Br + 3 stage fruits from lines R1-10, R1-25, and R1-30 and their wild-type siblings and measured the ethylene production. The ethylene levels were similar for wild-type and transgenic fruits (Figure 3A), suggesting that FUL1/2 function downstream or independent of the climacteric ethylene system 2.
Figure 3.
The Link between FUL1/2 and Ethylene, and the Expression of FUL1, FUL2, and TAGL1 in the cnr and rin Mutants.
(A) Ethylene concentrations in wild-type (WT) and FUL1 RNAi Br and Br + 3 stage fruits. Error bars indicate the se based on at least four fruits
(B) Relative expression of FUL1 and FUL2 in MG fruits after 6 h of treatment with or without ethephon.
(C) Relative expression of FUL1, FUL2, and TAGL1 in different stages of Cnr mutant fruits compared with the wild type.
(D) Relative expression of FUL1, FUL2, and TAGL1 in different stages of rin mutant fruits compared with the wild type. In (C) and (D), the expression in the MG wild-type stage is set to 1.0 for each individual gene; the expression levels of the different genes cannot be compared with each other. Because the relative fold differences for FUL2 and TAGL1 ([C] and [D]) are hard to read from the large graphs, these data are also presented in separate inserts in the corresponding panels. Error bars in (B) to (D) indicate the se based on two biological replicates.
In addition to the biosynthesis gene CGS, we also found the ethylene response gene EIL2, encoding a transcription factor homologous to Arabidopsis ETHYLENE-INSENSITIVE 3 (EIN3), to be significantly upregulated (log2 = 2.7) in FUL1/2 RNAi fruits. However, tomato contains four EIL proteins, which appear to act redundantly in ethylene signal transduction, and only downregulation of multiple genes results in a nonripening phenotype (Tieman et al., 2001). The expression of the other three known tomato EIL genes was detected, but not significantly altered, suggesting that the ethylene response is normal in FUL1/2 silenced lines. To confirm this, we applied ethephon to immature R1-10 fruits and immature fruits of their wild-type siblings. Both responded by accelerated ripening in a similar manner, though the transgenic fruits still displayed the orange-ripe phenotype (see Supplemental Figure 8 online). Thus, ethylene perception appears normal in the FUL1/2 silenced fruits, but their mutant phenotype cannot be rescued by ethylene application, providing another indication that FUL1/2 act downstream or independent of the climacteric ethylene system 2.
The ripening regulators CNR, RIN, and TAGL1 were all found to have ethylene-dependent and ethylene-independent functions (Itkin et al., 2009; Vrebalov et al., 2009; Klee and Giovannoni, 2011), and the expression of FUL1 and FUL2 may thus be induced via ethylene or regulated independent of ethylene by the upstream ripening regulators. To discriminate between both possibilities and to obtain more insight into the position of the FUL paralogs in the ripening regulatory network, we studied the expression of FUL1 and FUL2 in wild-type fruits in response to ethylene application and in fruits of the nonripening mutants.
Wild-type fruits were treated with ethephon in the MG stage, harvested 6 h later, and the expression of FUL1 and FUL2 was compared with nontreated fruits (Figure 3B). The expression of FUL2 appeared unchanged after ethephon treatment, but the expression of FUL1 was found to be three to four times upregulated, suggestive of a position downstream of ethylene. However, the expression of FUL1 increases in any case rapidly in wild-type fruits during the ripening induction phase, and we cannot exclude that the observed increase is simply a result of slight variation in the ripening stage.
The expression of FUL1, FUL2, and TAGL1 was investigated in three developmental stages of Cnr mutant fruits by qRT-PCR (Figure 3C). The expression of all three genes was lower in the Cnr mutant than in the wild type in the Br, and Br + 7 stages, indicating that CNR plays a general role in the positive regulation of FUL1, FUL2, and TAGL1. However, a marked effect of the Cnr mutation was found on the rapid induction of FUL1 during the ripening phase, which was completely abolished. This suggests that CNR specifically plays a role in the upregulation of FUL1 during ripening. FUL1 was also found to be a direct target of RIN, and its expression is distinctly reduced in the rin mutant (Martel et al., 2011; Fujisawa et al., 2012; Figure 3D). We found that the expression of FUL2 and TAGL1 is to some degree affected in the rin mutant fruits (Figure 3D). FUL2 was clearly more weakly expressed in the Br + 7 stage, while the expression in the earlier fruit stages was only slightly reduced. The mild reduction in TAGL1 expression was only apparent at the Br stage. In conclusion, the ripening-induced upregulation of FUL1 and, to a lesser extent, of FUL2 depends on CNR and RIN, which may occur in part via ethylene in the case of FUL1. However, FUL1 and FUL2 do not regulate gene expression via their effect on ethylene production or sensitivity.
The Ripening Regulators RIN and TAGL1 Are Upregulated in FUL1/2 RNAi Fruits
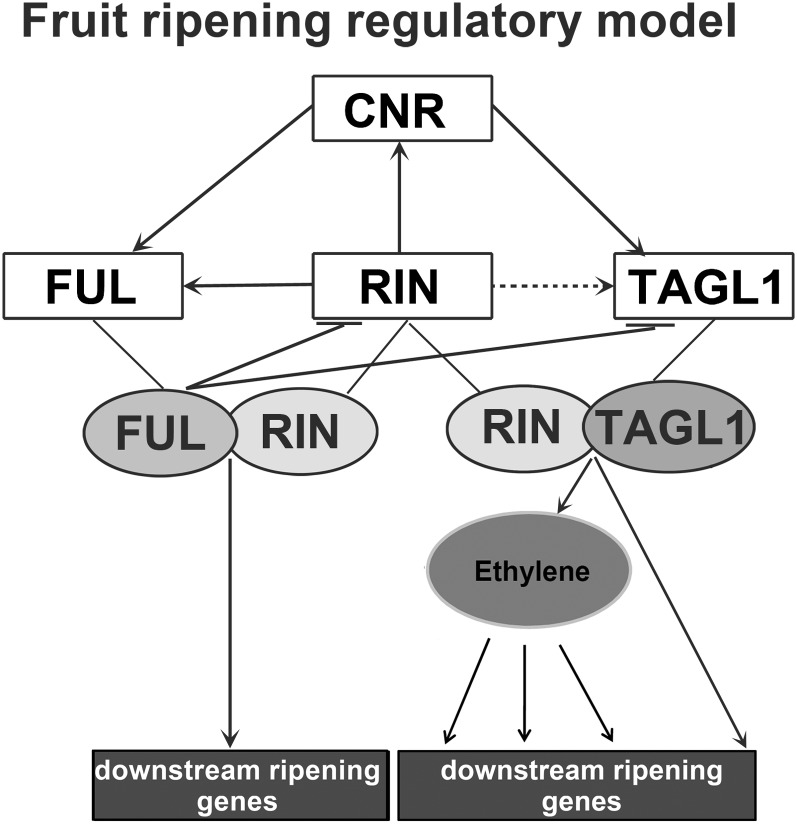
The microarray yielded only very few transcription factor–encoding genes as differentially regulated in FUL1/2 RNAi fruits (1.4%), suggesting that FUL1/2 may function somewhere downstream in the ripening regulatory cascade. The only identified transcription factor previously implicated in ripening is AOBP, encoding an ascorbate oxidase promoter binding protein (log2 = −1.3) (Alba et al., 2005). The other transcription factor genes, encoding among others two MADS domain proteins, two AP2 domain proteins, a NAM family protein (log2 = −2.6), an AUXIN RESPONSE FACTOR (log2 = −2.3), and a YABBY family protein, may execute as yet unidentified functions in ripening (Table 1; see Supplemental Data Set 1 online). The expression of the ripening regulators RIN, CNR, and TAGL1 was not significantly altered in the FUL1/2 RNAi fruits compared with the wild type, but the expression of RIN and TAGL1 did show a nonsignificant twofold increase in the transgenics. Because the microarray was performed with RNA from whole fruits, we decided to extract RNA from pericarp tissues specifically and perform qRT-PCR analysis to examine if the upregulation of the ripening regulators was more distinct in this tissue (see Supplemental Figure 9 online). Indeed, the expression of RIN and TAGL1 was two to three times upregulated in the pericarp of FUL1/2 RNAi Br stage fruits, indicating that they are repressed by FUL1/2 in wild-type fruits, probably via a negative feedback loop. The transcript levels of CNR were unchanged in FUL1/2 silenced fruits compared with the wild type, indicating that the expression of CNR is not regulated by FUL1 or FUL2. We summarized the currently known interactions between the ripening transcription factors RIN, CNR, TAGL1, and FUL1/2 in Figure 4 (Itkin et al., 2009; Vrebalov et al., 2009; Martel et al., 2011; Klee and Giovannoni, 2011).
Figure 4.
Simplified Model of the Fruit Ripening Regulatory Network in Tomato.
The connections between the different regulators are based on expression data from this study and previously published work (Eriksson et al., 2004; Giovannoni, 2007; Vrebalov et al., 2009; Itkin et al., 2009; Klee and Giovannoni, 2011; Martel et al., 2011). RIN functions as a dimer together with either TAGL1 or FUL1/2 to regulate complementary sets of downstream ripening genes. The positive interaction between RIN and TAGL1 is not clear (our expression data show a small downregulation of TAGL1 in the rin mutant only at Br stage). However, this interaction may be indirect via CNR and is therefore depicted with a dashed line. Most likely, RIN is also part of a complex when upregulating CNR, TAGL1, and FUL1/2. However, because the nature of this complex is still unclear, only RIN has been depicted in the white box.
DISCUSSION
In previous publications on tomato fruit ripening, TDR4/FUL1 repeatedly appeared as a gene of which the expression was highly induced at Br stage (Hileman et al., 2006; Vrebalov et al., 2009). Although this pointed toward a role of the gene in fruit ripening, the functions of FUL1 and its paralog MBP7/FUL2 remained so far unknown. We generated plants that are silenced specifically for either FUL1 or FUL2 and plants in which the paralogs are silenced together and demonstrated that FUL1 and FUL2 are both involved in fruit ripening, probably in a redundant manner. However, the differences between their fruit expression patterns suggest that FUL1 and FUL2 do not in all cases contribute equally to the different functions. Downregulation of both FUL1 and FUL2 impairs color development in the fruit and affects the expression of many genes involved in ripening processes, such as cell wall modification, cuticle formation, and aroma production. Interestingly, our data also revealed that Glu accumulates to a lesser extent in the fruits silenced for FUL1 and FUL2 than in the wild type, suggesting a role for FUL1/2 in the production of this major taste component of cultivated tomato fruits (Pauliukaite et al., 2006; Sorrequieta et al., 2010).
FUL1/2 and TAGL1 Regulate Different Aspects of Ripening together with RIN
Different studies have reported that the ripening regulator RIN interacts in yeast with TAGL1, FUL1, and FUL2 (Leseberg et al., 2008; Martel et al., 2011). Because MADS domain proteins often act in higher order complexes (Immink et al., 2009), it is possible that FUL1/2, TAGL1, and RIN interact with each other in a higher order complex to mediate tomato fruit ripening. However, although TAGL1 and FUL1/2 all play important roles in fruit ripening (Itkin et al., 2009; Vrebalov et al., 2009; Pan et al., 2010), they regulate different subsets of RIN target genes (Table 1; see Supplemental Data Set 1 online; Itkin et al., 2009; Vrebalov et al., 2009; Fujisawa et al., 2012), and the phenotypes of the TAGL1 and FUL1/2 silenced lines are distinctly different. Moreover, Leseberg et al. (2008) did not identify interactions between TAGL1 and FUL1/2 in yeast two-hybrid or three-hybrid assays. This all suggests that TAGL1-RIN and FUL1/2-RIN form separate complexes with distinct functions in tomato fruit ripening.
Recently, it was reported that FUL1 is a direct target of RIN and that its upregulation is dependent on the activity of CNR (Martel et al., 2011). Our data confirm that FUL1 depends on CNR and RIN for its upregulation during ripening and show that this applies to FUL2 as well (Figures 3C and 3D). Interestingly, the binding of RIN to its target loci was found to require the presence of CNR, although there is no evidence for a direct interaction between the two proteins. Martel et al. (2011) hypothesize that CNR may induce the expression of a cofactor required for proper DNA binding activity of the RIN-containing complex. FUL1 is mentioned as a likely candidate to encode this cofactor, since the SBP class proteins, of which CNR is a member, are known to preferably bind to promoters of SQUAMOSA class MADS box genes like FUL1. Our expression analyses confirm the CNR-dependent upregulation of FUL1 and show that the induction of FUL2 and TAGL1 is also dependent on CNR. This supports the idea that TAGL1 and FUL1/2 are the CNR-regulated factors that RIN needs to bind to its target promoters, likely regulating different subsets of RIN target genes.
FUL1 and FUL2 Function Largely Independently of the Ethylene Pathway
The fruits of the well-described ripening mutants Cnr and rin have severely reduced ethylene levels but do not ripen in response to ethylene application, although they are responsive to ethylene. This suggests that both CNR and RIN lie upstream of the climacteric ethylene increase but also have ethylene-independent functions (Giovannoni, 2007). We showed that FUL1 and FUL2 do not regulate ethylene biosynthesis and thus function downstream or independent of ethylene. Because the expression of FUL2 is not induced upon ethephon application, this gene appears to act independently of ethylene, while the modest upregulation of FUL1 after ethephon treatment suggests that the induction of FUL1 during ripening may be partly regulated via ethylene. In conclusion, this suggests that the FUL proteins interact with RIN to fulfill ethylene-independent functions in ripening, while the TAGL1-RIN complex regulates a complementary set of target genes via both ethylene-dependent and -independent mechanisms.
The non-ethylene-mediated aspects of ripening are very interesting in the context of evolution, since they may be conserved between climacteric and nonclimacteric species (Giovannoni, 2007). The recent discovery that a FUL homolog regulates anthocyanin accumulation in the nonclimacteric fruits of bilberry (Jaakola et al., 2010) provides strong evidence for the wide conservation of ripening regulators. The role of FUL in fruits may be even more widely conserved, since the gene is also essential for dry fruit development in Arabidopsis (Ferrándiz et al., 2000) and the basal eudicot poppy (Eschscholzia californica; Pabón-Mora et al., 2012). In this context, it will also be very interesting to determine the regulatory network involved in fruit ripening in Solanaceae species that bear dry capsular fruits, like petunia (Petunia hybrida) and tobacco (Nicotiana tabacum). The capsules of petunia and tobacco open by dehiscence at the carpel fusion sites as the Arabidopsis silique does, but in the absence of the specialized replum and dehiscence zones of the latter (Pabón-Mora and Litt, 2011). The close relationship of tobacco and petunia to tomato suggests that the regulatory network involved will show many similarities with the tomato network. Comparing the networks may also elucidate the diverging steps required for the conversion of dry fruits into fleshy fruits.
The Position of FUL1/2 in the Fruit Ripening Regulatory Network
Fruit ripening in tomato starts with the climacteric ethylene increase, induced by the transcription factors CNR, NOR, RIN, and TAGL1. We suggest that RIN and NOR function upstream of CNR or in different pathways, while TAGL1 functions downstream. FUL1/2 appear to have a special place in the tomato ripening regulatory network, since they act at least in part independently of ethylene, but are regulated by CNR and RIN. As such, FUL1/2 may not be important for the onset of ripening but function later in the ripening process by the actual regulation of pigmentation, aroma, and taste. Our microarray analysis supports this idea because many differentially regulated genes are apparently involved in primary or secondary metabolism, while only very few genes that encode transcription factors appear to act downstream of FUL1/2. Moreover, we found the ripening regulators RIN and TAGL1 to be upregulated in the pericarp of FUL1 RNAi fruits, pointing to a negative feedback loop from FUL1/2 on the ripening induction genes. Notably, Arabidopsis FUL also represses the TAGL1 orthologs SHP1/2 (Ferrándiz et al., 2000), suggesting some conservation of the fruit regulatory network between Arabidopsis and tomato. We summarized the current ideas on the relationships between the ripening regulators, FUL1/2, and ethylene in the model in Figure 4.
In conclusion, FUL1/2 function downstream of RIN and CNR in an ethylene-independent manner to regulate several downstream ripening processes and repress with a negative feedback loop the activity of RIN and TAGL1.
METHODS
Growth Conditions
Tomato plants (Solanum lycopersicum) cultivars MT, Ailsa Craig, and Moneyberg were grown in the greenhouse at ambient temperatures (>20°C) under natural light supplemented with artificial sodium lights, according to a 16-h-light/8-h-dark cycle. Seeds of tomato mutant Cnr in the cv Ailsa Craig background where kindly provided by Graham Seymour (University of Nottingham, UK). Seeds of tomato mutant rin in the cv Ailsa Craig background where obtained from the C.M. Rick Tomato Genetics Resource Center (Davis, CA).
qRT-PCR Analysis
For qRT-PCR analysis of FUL1 and FUL2 expression, leaves, flowers, and fruits were harvested from wild-type tomato plant cultivar MT. To test the downregulation in the FUL1 RNAi lines, Br stage fruits were harvested from the transgenics and from wild-type MT plants. For the other transgenic lines, red ripe fruits from cultivar Moneyberg were used. RNA was extracted with Trizol (Sigma-Aldrich), and cDNA was synthesized with the iScript cDNA synthesis kit (Bio-Rad). Real-time RT-PCR was performed with the iQ SYBR Green Supermix from Bio-Rad using primers QSlFUL1for/QSlFUL1rev and QSlFUL2for/QSlFUL2rev, with actin 2/7 as a reference gene (all primer sequences are listed in Supplemental Table 4 online). The following PCR program was used: 1 h at 95°C, 40 cycles (10 min at 95°C; 45 min 57°C). A similar protocol was used to study the expression of FUL1, FUL2, and TAGL1 in Cnr mutant Br stage fruits and the expression of RIN, CNR, and TAGL1 in the pericarp of wild-type and FUL1/2 RNAi Br stage fruits. Primers were as follows: SlFUL1for/SlFUL1rev, SlFUL2for/SlFUL2rev, RINfor/RINrev, CNRfor/CNRrev, and TAGL1for/ TAGL1rev.
Split-YFP
RIN and FUL1 were amplified from Br stage fruit cDNA with primers TOPO SlFUL1for/TOPO SlFUL1rev and TOPO RINfor/TOPO RIN rev. The full-length open reading frames were inserted into the pENTR-D TOPO vector according to the Gateway protocol (Invitrogen). FUL2 was isolated from a cDNA library from mixed fruit stages (CloneMiner cDNA Library construction kit; Invitrogen), which was initially inserted into pDest22 (Invitrogen) and from there recombined back into pDONR207 (Gateway). All plasmids were sequenced (DETT sequence kit; Amersham) and subsequently recombined with the LR reaction into split-YFP destination vectors. For the N-terminal fusions, pARC233 and pARC234 were used, and for the C-terminal fusions, pARC235 and pARC236 were used (Welch et al., 2007).
Arabidopsis thaliana leaf protoplasts were transfected with 15 μg plasmid DNA as described by Aker et al. (2006). The protoplasts were incubated overnight at 25°C before imaging. Confocal laser scanning microscopy images were made with a Leica SPE DM5500 upright microscope using the ×10 0.30 CS ACS APO (air) lens and LAS AF 1.8.2 software (Leica).
Generation of FUL1 and FUL2 Silenced Plants
The FUL1 and FUL2 RNAi constructs were generated using the Gateway system (Invitrogen). For FUL1, a 470-bp fragment was amplified from the 3′end of the cDNA with primers SlFUL1 RNAi for/SlFUL1 RNAi rev. For FUL2, a 437-bp fragment was amplified from the 3′end of the cDNA using primers SlFUL2 RNAi for/SlFUL2 RNAi rev. For the FUL1- and FUL2-specific constructs, the 3′ UTR regions (274-bp fragment for FUL1 and 251-bp fragment for FUL2) were commercially synthesized by Geneart together with the attB1 and attB2 sites for recombination. Fragments were recombined into the pDONR221 or pDONR207 (specific 3′ UTR fragments) vectors using the enzyme BP recombinase. After verification of the inserts, an LR reaction was performed to recombine the fragments into the binary RNAi vector pK7GWIWG2 (Karimi et al., 2002). The resulting vectors were transformed into Agrobacterium tumefaciens strain EHA105 using freeze-thaw transformation (Chen et al., 1994) and transformed to MT wild-type plants by cocultivation with 5-d-old cotyledon explants without a feeding layer, according to a protocol modified from McCormick et al. (1986).
Carotenoid Analysis
Wild-type MT and FUL1 RNAi tomato fruit were extracted as described previously by López-Ráez et al. (2008) with slight modifications. Briefly, freeze-ground material of tomato fruit (500 mg) was extracted with a mixture of methanol/chloroform (2.5/2.0 [v/v]) containing 0.1% butylated hydroxytoluene (BHT) and 50 mM of Tris-chloride, pH 2.5, containing 1 M sodium chloride. Samples were incubated on ice and centrifuged and the chloroform layer transferred to a clean tube. Samples were reextracted twice with chloroform containing 0.1% BHT. Pooled chloroform phases were dried under a nitrogen flow and keep under a nitrogen atmosphere at −20°C until HPLC analysis. Carotenoid extracts were prepared for HPLC analysis by dissolving in ethyl acetate containing 0.1% BHT. Chromatography was performed on a Waters system consisting of a number 600 quaternary pump, number 996 photodiode array detector, and number 2475 fluorescence detector. Carotenoid pigments were separated by HPLC using a YMC-Pack reverse-phase C30 column (250 × 4.6 mm; 5 µm) coupled to a 20- × 4.6-mm C30 guard (YMC), maintained at 40°C, and with ternary gradient elution of water, methanol, and methyl tert-butyl ether (Bino et al., 2005). Flow rate of 1 mL/min was used. Data were collected and analyzed using the Waters Empower software supplied. Carotenoids were detected by setting the photodiode array detector to scan from 220 to 700 nm, and tocopherols were detected by a fluorescence detector at excitation and emission wavelengths of 296 and 405 nm, respectively. Quantitative determination of compounds was conducted by comparison with dose–response curves constructed from authentic standards. Each value was the mean of, at least, two biological replicates.
Sudan IV Staining
A Sudan IV (Sigma-Aldrich) stock solution (0.1% [w/v] in isopropanol) was diluted 3:2 with distilled water, mixed well by rotation for 30 min, and filtered through a syringe filter to remove precipitates (Buda et al., 2009). Thin Br stage pericarp sections were incubated for 10 min in Sudan IV solution, rinsed first with 50% isopropanol and then with water. The sections were mounted in distilled water and observed under a Leitz Orthoplan light microscope.
Microarray Analysis
For microarray analysis of gene expression, RNA was isolated (see above) from whole fruits of Br stage fruits from two transgenic T1 lines and from their wild-type T1 siblings, in duplicate (two fruit samples from line R1-10, two samples from the wild-type siblings of R1-10, two fruit samples from line R1-30 plants, and two samples from the wild-type siblings of R1-30). RNAs were labeled with Cy3 for single-color hybridizations of Agilent-022270 Tomato Gene Expression microarrays at the MicroArray Facility of VIB, Leuven, Belgium. Expression values were obtained from the Agilent Feature Extraction Software version 10.5.1.1. Quantile normalization was performed on the log2-transformed values to normalize intensities between arrays. A total of 5325 spots had no signal above background for any array and were removed, leaving 39,895 probe signals for further analysis. Average expression values were estimated for the wild type versus nontransgenic line using the limma package from Bioconductor. A moderated t-statistic as implemented in limma was used to determine significance of contrast between wild type and transgenic line (Smyth, 2004; Smyth et al., 2005). The resulting P values were corrected for multiple testing with the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995) to control the false discovery rate. Probes with a corrected P value < 0.05 were considered significantly different between the wild type and transgenic line. The qRT-PCR analysis for confirmation of the microarray was performed as described above, using primers PDS4692-CrtR-b2F/R (CrtR-b2, SGN-U568611), PDS4694-GDSL F/R (GDSL, SGN-U573197), PDS4696 PAT F/R (PAT, SGN-U566870), PDS4698-EIL2 F/R (EIL2, AF328785), and PDS4702 SlPG2A F/R (PG2A, SGN-U577423).
Ethylene Content and Response
To measure the ethylene content, fruits from transgenic T1 lines and their wild-type siblings were harvested at stage Br + 3 and placed in a GL32 Schott screw cap vial with rubber ring. Soon after harvesting, a 1-mL gas sample was taken from the vial and analyzed on the Packard gas chromatograph model 437A (t = 0). Approximately 3 h later, another sample was taken and analyzed. The ethylene concentration in the samples was determined by comparing the peak length from the gas chromatograph with a standard, and the ethylene content of the fruit was calculated in milliliters per hour per gram fresh weight.
Ethephon (Acros Organics) treatment of intact MG fruit was a 1-min dip in 50 mM fresh aqueous solution every 2 d, two times.
Primary Metabolite Analysis
Analysis of primary metabolites was performed on the whole fruit as described by Roessner-Tunali et al. (2003). The chromatograms were aligned using the MetAlign software package, and compound mass spectra were extracted according to Tikunov et al. (2005). Metabolites were identified using NIST MSSearch and retention indices using the Golm Metabolite Database as a reference library (http://gmd.mpimp-golm.mpg.de). Significant metabolic differences between nontransgenic and transgenic fruits were detected using the Student’s t test.
Accession Numbers
The primary accession number for Sl-FUL1 (TDR4/TM4) is BT013224. The CompBio (Computational Biology and Functional Genomics Laboratory) and SGN (Solanaceae Genome Network) ID numbers are TC196395 and SGN-U596354, respectively. Sl-FUL1 is located on chromosome 6 (Solyc06g069430). The primary accession number for Sl-FUL2 (MBP7/euFUL2) is BM412293. The CombBio and SGN ID numbers are TC202975 and SGN-U580493, respectively. Sl-FUL2 is located on chromosome 3 (Solyc03g114830). The accession numbers of the genes used in the microarray analysis are provided in Table 1 and Supplemental Data Set 1 online. The microarray data from this study were submitted to the Gene Expression Omnibus repository under accession number GSE41560.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Split-YFP Data from the Combinations RIN-FUL1 and FUL1-FUL2.
Supplemental Figure 2. Alignment of the FUL1 and FUL2 Nucleotide Sequences.
Supplemental Figure 3. Downregulation of FUL1 in Different FUL1 RNAi Lines.
Supplemental Figure 4. Downregulation of FUL1 and FUL2 in Different Fruit Stages of FUL1 RNAi Line R1-10.
Supplemental Figure 5. Downregulation of FUL1 and FUL2 in the Ailsa Craig/Micro Tom F1 Background.
Supplemental Figure 6. Downregulation of FUL1 and FUL2 in the FUL2 RNAi and Specific FUL1 and FUL2 Lines.
Supplemental Figure 7. qRT-PCR Confirmation of a Few Selected Upregulated and Downregulated Genes from the Microarray.
Supplemental Figure 8. Effect of Ethephon Treatment on FUL1/2 RNAi Fruits of Line R1-10.
Supplemental Figure 9. Expression of CNR, RIN, and TAGL1 in FUL1/2 RNAi Fruits.
Supplemental Table 1. Carotenoid Analyses in Wild-Type and FUL1/2 RNAi Fruits.
Supplemental Table 2. Significantly Changed Metabolic Pathways in the FUL1/2 RNAi Tomato Fruits Compared with the Wild Type.
Supplemental Table 3. List of All Analyzed Primary Metabolites in Wild-Type and FUL1/2 RNAi Fruits of Lines R1-10 and R1-29.
Supplemental Table 4. All Primer Sequences Used for the Experiments.
Supplemental Data Set 1. Full List of Genes That Are Significantly Twofold or More Up- or Downregulated in FUL1/2 RNAi Fruits Compared with the Wild Type.
Acknowledgments
The project was financially supported by the Dutch Organization for Scientific research (NWO) in the framework of the ERA-NET on Plant Genomics (ERA-PG) program projects CISCODE (M.B.) and TomQML (R.K.), by the European Union Framework 6 project EU-SOL (Contract FOOD-CT-2006-016214) (G.C.A. and R.A.d.M.), and by a CAPES grant (CAPES/WUR project; P.d.B.R.).
AUTHOR CONTRIBUTIONS
R.A.d.M., G.C.A., A.G.B., R.K., and M.B. designed the research. R.K., M.B., A.R.B., Y.M.T., M.W.-A., and P.d.B.R. performed the research. R.A.d.M. and Y.M.T. analyzed data. M.B. wrote the article.
Glossary
- qRT-PCR
quantitative RT-PCR
- YFP
yellow fluorescent protein
- RNAi
RNA interference
- UTR
untranslated region
- MT
Micro Tom
- MG
mature green
- Br
breaker
- GABA
γ-amino butyrate
- BHT
butylated hydroxytoluene
References
- Aker J., Borst J.W., Karlova R., de Vries S. (2006). The Arabidopsis thaliana AAA protein CDC48A interacts in vivo with the somatic embryogenesis receptor-like kinase 1 receptor at the plasma membrane. J. Struct. Biol. 156: 62–71 [DOI] [PubMed] [Google Scholar]
- Alba R., Payton P., Fei Z., McQuinn R., Debbie P., Martin G.B., Tanksley S.D., Giovannoni J.J. (2005). Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C.S., Blume B., Bouzayen M., Cooper W., Hamilton A.J., Grierson D. (1996). Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 9: 525–535 [DOI] [PubMed] [Google Scholar]
- Barry C.S., Giovannoni J.J. (2006). Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc. Natl. Acad. Sci. USA 103: 7923–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C.S., Giovannoni J.J. (2007). Ethylene and fruit ripening. J. Plant Growth Regul. 26: 143–159 [Google Scholar]
- Bellisle F. (1999). Glutamate and the UMAMI taste: Sensory, metabolic, nutritional and behavioural considerations. A review of the literature published in the last 10 years. Neurosci. Biobehav. Rev. 23: 423–438 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B 57: 289–300 [Google Scholar]
- Bino R.J., Ric de Vos C.H., Lieberman M., Hall R.D., Bovy A., Jonker H.H., Tikunov Y., Lommen A., Moco S., Levin I. (2005). The light-hyperresponsive high pigment-2dg mutation of tomato: alterations in the fruit metabolome. New Phytol. 166: 427–438 [DOI] [PubMed] [Google Scholar]
- Boggio S.B., Palatnik J.F., Heldt H.W., Valle E.M. (2000). Changes in amino acid composition and nitrogen metabolizing enzymes in ripening fruits of Lycopersicon esculentum Mill. Plant Sci. 159: 125–133 [DOI] [PubMed] [Google Scholar]
- Buda G.J., Isaacson T., Matas A.J., Paolillo D.J., Rose J.K. (2009). Three-dimensional imaging of plant cuticle architecture using confocal scanning laser microscopy. Plant J. 60: 378–385 [DOI] [PubMed] [Google Scholar]
- Busi M.V., Bustamante C., D'Angelo C., Hidalgo-Cuevas M., Boggio S.B., Valle E.M., Zabaleta E. (2003). MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 52: 801–815 [DOI] [PubMed] [Google Scholar]
- Chen H., Nelson R.S., Sherwood J.L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16: 664–668, 670 [PubMed] [Google Scholar]
- Chung M.-Y., Vrebalov J., Alba R., Lee J., McQuinn R., Chung J.-D., Klein P., Giovannoni J. (2010). A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Eriksson E.M., Bovy A., Manning K., Harrison L., Andrews J., De Silva J., Tucker G.A., Seymour G.B. (2004). Effect of the colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 136: 4184–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C., Liljegren S.J., Yanofsky M.F. (2000). Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289: 436–438 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C., Pelaz S., Yanofsky M.F. (1999). Control of carpel and fruit development in Arabidopsis. Annu. Rev. Biochem. 68: 321–354 [DOI] [PubMed] [Google Scholar]
- Fujisawa M., Nakano T., Ito Y. (2011). Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biology 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M., Shima Y., Higuchi N., Nakano T., Koyama Y., Kasumi T., Ito Y. (2012). Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 235: 1107–1122 [DOI] [PubMed] [Google Scholar]
- Galpaz N., Ronen G., Khalfa Z., Zamir D., Hirschberg J. (2006). A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 18: 1947–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J.J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Hileman L.C., Sundstrom J.F., Litt A., Chen M., Shumba T., Irish V.F. (2006). Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol. Biol. Evol. 23: 2245–2258 [DOI] [PubMed] [Google Scholar]
- Immink R.G., Tonaco I.A., de Folter S., Shchennikova A., van Dijk A.D., Busscher-Lange J., Borst J.W., Angenent G.C. (2009). SEPALLATA3: The ‘glue’ for MADS box transcription factor complex formation. Genome Biol. 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T., Ronen G., Zamir D., Hirschberg J. (2002). Cloning of tangerine from tomato reveals a Carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 14: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itai A., Ishihara K., Bewley J.D. (2003). Characterization of expression, and cloning, of β-D-xylosidase and α-L-arabinofuranosidase in developing and ripening tomato (Lycopersicon esculentum Mill.) fruit. J. Exp. Bot. 54: 2615–2622 [DOI] [PubMed] [Google Scholar]
- Itkin M., Seybold H., Breitel D., Rogachev I., Meir S., Aharoni A. (2009). TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Jaakola L., Poole M., Jones M.O., Kämäräinen-Karppinen T., Koskimäki J.J., Hohtola A., Häggman H., Fraser P.D., Manning K., King G.J., Thomson H., Seymour G.B. (2010). A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 153: 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J.-G., Corbett A.M., Fellman S.M., Tieman D.M., Klee H.J., Giovannoni J.J., Fei Z. (2009). Plant MetGenMAP: An integrative analysis system for plant systems biology. Plant Physiol. 151: 1758–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karlova R., Rosin F.M., Busscher-Lange J., Parapunova V., Do P.T., Fernie A.R., Fraser P.D., Baxter C., Angenent G.C., de Maagd R.A. (2011). Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee H.J., Giovannoni J.J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Knapp J., Moureau P., Schuch W., Grierson D. (1989). Organization and expression of polygalacturonase and other ripening related genes in Ailsa Craig “Neverripe” and “Ripening inhibitor” tomato mutants. Plant Mol. Biol. 12: 105–116 [DOI] [PubMed] [Google Scholar]
- Leseberg C.H., Eissler C.L., Wang X., Johns M.A., Duvall M.R., Mao L. (2008). Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 59: 2253–2265 [DOI] [PubMed] [Google Scholar]
- Liljegren S.J., Ditta G.S., Eshed Y., Savidge B., Bowman J.L., Yanofsky M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Liljegren S.J., Roeder A.H., Kempin S.A., Gremski K., Østergaard L., Guimil S., Reyes D.K., Yanofsky M.F. (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116: 843–853 [DOI] [PubMed] [Google Scholar]
- Lin Z., Hong Y., Yin M., Li C., Zhang K., Grierson D. (2008). A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois L.M., Rodríguez-Concepción M., Gallego F., Campos N., Boronat A. (2000). Carotenoid biosynthesis during tomato fruit development: regulatory role of 1-deoxy-D-xylulose 5-phosphate synthase. Plant J. 22: 503–513 [DOI] [PubMed] [Google Scholar]
- López-Ráez J.A., Charnikhova T., Gómez-Roldán V., Matusova R., Kohlen W., De Vos R., Verstappen F., Puech-Pages V., Bécard G., Mulder P., Bouwmeester H. (2008). Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 178: 863–874 [DOI] [PubMed] [Google Scholar]
- Martel C., Vrebalov J., Tafelmeyer P., Giovannoni J.J. (2011). The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 157: 1568–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí E., Gisbert C., Bishop G.J., Dixon M.S., García-Martínez J.L. (2006). Genetic and physiological characterization of tomato cv Micro-Tom. J. Exp. Bot. 57: 2037–2047 [DOI] [PubMed] [Google Scholar]
- McCormick S., Niedermeyer J., Fry J., Barnason A., Horsch R., Fraley R. (1986). Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep. 5: 81–84 [DOI] [PubMed] [Google Scholar]
- Mintz-Oron S., Mandel T., Rogachev I., Feldberg L., Lotan O., Yativ M., Wang Z., Jetter R., Venger I., Adato A., Aharoni A. (2008). Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol. 147: 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mølhøj M., Verma R., Reiter W.-D. (2004). The biosynthesis of D-Galacturonate in plants. functional cloning and characterization of a membrane-anchored UDP-D-Glucuronate 4-epimerase from Arabidopsis. Plant Physiol. 135: 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka A., Murachi S., Okunishi H., Shiomi S., Nakano R., Kubo Y., Inaba A. (1998). Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 118: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S., Alba R., Damasceno C.M.B., Lopez-Casado G., Lohse M., Zanor M.I., Tohge T., Usadel B., Rose J.K.C., Fei Z., Giovannoni J.J., Fernie A.R. (2011). Systems biology of tomato fruit development: Combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 157: 405–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón-Mora N., Ambrose B.A., Litt A. (2012). Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol. 158: 1685–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón-Mora N., Litt A. (2011). Comparative anatomical and developmental analysis of dry and fleshy fruits of Solanaceae. Am. J. Bot. 98: 1415–1436 [DOI] [PubMed] [Google Scholar]
- Pan I.L., McQuinn R., Giovannoni J.J., Irish V.F. (2010). Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 61: 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauliukaite R., Zhylyak G., Citterio D., Spichiger-Keller U.E. (2006). L-glutamate biosensor for estimation of the taste of tomato specimens. Anal. Bioanal. Chem. 386: 220–227 [DOI] [PubMed] [Google Scholar]
- Qin G., Wang Y., Cao B., Wang W., Tian S. (2012). Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 70: 243–255 [DOI] [PubMed] [Google Scholar]
- Ripoll J.J., Roeder A.H.K., Ditta G.S., Yanofsky M.F. (2011). A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138: 5167–5176 [DOI] [PubMed] [Google Scholar]
- Roeder A.H.K., Yanofsky M.F. (2006). Fruit development in Arabidopsis. The Arabidopsis Book 4: e0075, doi/10.1199/tab.0075
- Roessner-Tunali U., Hegemann B., Lytovchenko A., Carrari F., Bruedigam C., Granot D., Fernie A.R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab W., Davidovich-Rikanati R., Lewinsohn E. (2008). Biosynthesis of plant-derived flavor compounds. Plant J. 54: 712–732 [DOI] [PubMed] [Google Scholar]
- Sheehy R.E., Pearson J., Brady C.J., Hiatt W.R. (1987). Molecular characterization of tomato fruit polygalacturonase. Mol. Gen. Genet. 208: 30–36 [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article 3
- Smyth G.K., Michaud J., Scott H.S. (2005). Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21: 2067–2075 [DOI] [PubMed] [Google Scholar]
- Sorrequieta A., Ferraro G., Boggio S.B., Valle E.M. (2010). Free amino acid production during tomato fruit ripening: A focus on L-glutamate. Amino Acids 38: 1523–1532 [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Tor M., Barry C.S., Vrebalov J., Orfila C., Jarvis M.C., Giovannoni J.J., Grierson D., Seymour G.B. (1999). Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol. 120: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D.M., Ciardi J.A., Taylor M.G., Klee H.J. (2001). Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J. 26: 47–58 [DOI] [PubMed] [Google Scholar]
- Tikunov Y., Lommen A., de Vos C.H.R., Verhoeven H.A., Bino R.J., Hall R.D., Bovy A.G. (2005). A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 139: 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Schlüter U., Mølhøj M., Gipmans M., Verma R., Kossmann J., Reiter W.-D., Pauly M. (2004). Identification and characterization of a UDP-D-glucuronate 4-epimerase in Arabidopsis. FEBS Lett. 569: 327–331 [DOI] [PubMed] [Google Scholar]
- Vrebalov J., Pan I.L., Arroyo A.J.M., McQuinn R., Chung M., Poole M., Rose J., Seymour G., Grandillo S., Giovannoni J., Irish V.F. (2009). Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., Schuch W., Giovannoni J. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Welch D., Hassan H., Blilou I., Immink R., Heidstra R., Scheres B. (2007). Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 21: 2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J.Q., Lanahan M.B., Yen H.C., Giovannoni J.J., Klee H.J. (1995). An ethylene-inducible component of signal transduction encoded by never-ripe. Science 270: 1807–1809 [DOI] [PubMed] [Google Scholar]
- Yang S.F., Hoffman N.E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35: 155–189 [Google Scholar]