Pollen grains are protected by beautiful and elaborate cell walls, exines. This work examines the formation of one distinct patterning element of pollen surfaces, apertures, or areas where exine is not deposited. It demonstrates that the formation of apertures depends on the novel plant protein INP1, which is directed to aperture areas and regulates aperture length in a dosage-dependent manner.
Abstract
Pollen grains protect the sperm cells inside them with the help of the unique cell wall, the exine, which exhibits enormous morphological variation across plant taxa, assembling into intricate and diverse species-specific patterns. How this complex extracellular structure is faithfully deposited at precise sites and acquires precise shape within a species is not understood. Here, we describe the isolation and characterization of the novel Arabidopsis thaliana gene INAPERTURATE POLLEN1 (INP1), which is specifically involved in formation of the pollen surface apertures, which arise by restriction of exine deposition at specific sites. Loss of INP1 leads to the loss of all three apertures in Arabidopsis pollen, and INP1 protein exhibits a unique tripartite localization in developing pollen, indicative of its direct involvement in specification of aperture positions. We also show that aperture length appears to be sensitive to INP1 dosage and INP1 misexpression can affect global exine patterning. Phenotypes of some inp1 mutants indicate that Arabidopsis apertures are initiated at three nonrandom positions around the pollen equator. The identification of INP1 opens up new avenues for studies of how formation of distinct cellular domains results in the production of different extracellular morphologies.
INTRODUCTION
Cells rely on a regulated deposition of extracellular materials to control their shapes, growth, and motility, to promote tissue formation, and to protect themselves (Szymanski and Cosgrove, 2009; Rosario and DeSimone, 2011; Underwood, 2012). In the tissues and organisms in which cells are surrounded by the extracellular matrix or by cell walls, the ultimate cell morphology often depends on the production of such extracellular structures at precisely the right places at the right time. Despite the importance of extracellular structures in development and disease, the question of how cells decide when, where, and in which manner they should be produced and deposited is far from being understood in any system (Rosario and DeSimone, 2011).
Exine, the outer cell wall of pollen grains, represents a unique model for synthesis and assembly of complex extracellular structures. In addition to its remarkable physicochemical properties that allow it to successfully protect sperm cells inside pollen (Scott, 1994; Wiermann et al., 2001), exine is also one of the most diverse structures in nature and can assume thousands of elaborate, species-specific patterns (Kesseler and Harley, 2004). Such species-specific differences in patterns may facilitate pollen–pollinator and pollen–pistil interactions. How exine’s complex extracellular structures are faithfully placed at exact sites and acquire certain shape within a species is an intriguing cell biological question. The molecular players and cellular mechanisms guiding the development of exine patterns in pollen and the alterations in developmental programs leading to interspecific differences in exine morphology are virtually unknown.
Some of the most common and well-defined elements of exine patterning are apertures, the areas on pollen surface where exine is not deposited or is reduced (Furness and Rudall, 2004). They are expected to facilitate the exit of pollen tubes during germination and to accommodate volume changes during shifts in the pollen hydration state (Wodehouse, 1935; Heslop-Harrison, 1979a; Muller, 1979; Blackmore and Barnes, 1986; Katifori et al., 2010). Although aperture number, positions, and morphology vary greatly across different species, within a given species apertures usually have precise shape, size, and decorations and are formed at exact locations. Presence of apertures suggests the existence of cellular mechanisms that reliably define the areas where apertures will be formed and that prevent exine deposition at these positions.
Here, we describe the isolation and characterization of the gene INAPERTURATE POLLEN1 (INP1) from Arabidopsis thaliana that is specifically involved in aperture formation and encodes a novel protein. We demonstrate that the loss of this gene leads to the loss of all three apertures in Arabidopsis pollen grains and that its protein product exhibits a unique tripartite localization in developing pollen, indicative of its direct involvement in specification of aperture positions. We also show that the aperture length appears to be sensitive to the INP1 dosage and that INP1 misexpression can have an effect on the global exine patterning. Phenotypes of some inp1 mutants indicate that Arabidopsis apertures are initiated at three nonrandom positions around the pollen equator. The identification of INP1 opens up new avenues for the studies of how formation of the distinct cellular domains results in the production of extracellular differences.
RESULTS
Isolation of the INP1 Gene
Exine of Arabidopsis pollen has a characteristic reticulate pattern, interrupted at three equidistant positions by long and narrow apertures (Figure 1A). Previously, in a forward genetic screen, we isolated a large number of Arabidopsis mutants defective in the production of pollen exine (Dobritsa et al., 2011). Among them were 14 inaperturate (inp) mutants, whose pollen appeared round, rather than oval, in a dry state and had normal reticulate exine patterning but completely lacked apertures (Figures 1B and 1C) (Dobritsa et al., 2011). Although these mutants were independently isolated from different batches of the mutagenized population, they belonged to a single complementation group, with the mutation linked to the molecular marker ciw7 on chromosome 4 (Dobritsa et al., 2011). We now performed positional cloning with additional molecular markers on two of the lines (25-1-1 and 31-1-2) and mapped the mutation to a 53-kb interval between the markers F7K2-9 and T12H17-6. We sequenced the 17 genes located in this interval and found that one of them, At4g22600, had a single nucleotide (A262) missing in both mapping lines. We called the gene INP1 and named this mutant allele inp1-1 (Figures 1B and 1G). Sequencing of the other inp mutant lines revealed that all but one carried an identical mutation in At4g22600; the remaining line (118-2-2) was missing the nucleotide C74 (allele inp1-2; Figures 1C and 1G). In both cases, the mutations cause a frame shift in the beginning of the open reading frame (ORF) and are expected to completely inactivate the gene.
Figure 1.
INP1 Is Necessary for Aperture Formation in Pollen Exine.
(A) Wild-type (WT) Arabidopsis pollen grains have three apertures (arrows).
(B) to (D) All three inp1 mutants have normal reticulate exine pattern but no apertures.
(E) and (F) INP1pro:INP1 and INP1pro:INP1-YFP transgenes restore apertures (arrows) in the inp1-1 mutant. Bars = 10 μm.
(G) Map of the structure of the INP1 gene. Positions of mutations in the three inp1 alleles are indicated.
A T-DNA line with an insertion in the At4g22600 ORF (SALK_093834c) was available through the ABRC. The pollen of this mutant had an identical inp phenotype (allele inp1-3; Figures 1D and 1G). All three mutants produced otherwise normal and fertile plants. Transformation of the inp1-1 mutant with the INP1 gene under its own promoter conferred the wild-type pollen phenotype (oval shape of dry pollen and presence of apertures; Figure 1E) in all 20 T1 transformants, further confirming that At4g22600 is INP1 and is required for the formation of pollen apertures.
INP1 Is a Novel Plant-Specific Gene
INP1 encodes a 273–amino acid protein. BLAST searches with the INP1 protein sequence as a query and application of programs designed to identify protein motifs and specific sequences revealed that it is a novel protein with no predicted sorting signal sequences. No other similar proteins exist in Arabidopsis. Whereas most domain search algorithms did not identify any recognizable domains in INP1, Pfam 26.0 (http://pfam.sanger.ac.uk/) predicted a plant-specific DELAY OF GERMINATION1 (DOG1) domain (PF14144) in the INP1 N terminus (amino acids 31 to 108; see Supplemental Figure 1 online). The founding member of this domain family is associated with the control of seed dormancy (Bentsink et al., 2006).
Homologs of INP1 exist in different plant species, including a variety of eudicots, monocots (Poaceae), and, possibly, gymnosperms (Pinus pinaster; see below) (see Supplemental Figure 1 online), but are absent from species outside the plant kingdom. Also, notably, similar proteins are absent from the genomes of early land plant lineages, such as the lycophyte Selaginella moellendorfii (spikemoss) and the bryophyte Physcomitrella patens (moss), which lack pollen but generate spores that are also covered by sporopollenin, the major component of exine. Absence of INP1 in these lineages was in contrast with the other previously described, mostly biosynthetic, exine genes—e.g., cytochromes P450 CYP704B1 and CYP703A2, MALE STERILITY2 (MS2), LESS ADHESIVE POLLEN3 (LAP3), LAP5, LAP6, ACYL-CoA SYNTHASE5 (ACOS5), NO EXINE FORMATION1 (NEF1), DEFECTIVE EXINE1 (DEX1), and THIN EXINE2 (TEX2)—all of which are present throughout the land plant lineages and some are even found in algae (Aarts et al., 1997; Paxson-Sowders et al., 2001; Ariizumi et al., 2004; Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2009a, 2009b, 2010, 2011; Kim et al., 2010; Chen et al., 2011; Colpitts et al., 2011). This suggests that the INP1 gene was acquired relatively recently in plant evolutionary history, after plants developed the ability to synthesize sporopollenin. Similarly, unlike the other exine genes that exhibit a high degree of sequence conservation in diverse species (on average sharing ∼60% sequence identity between proteins from Arabidopsis and moss), INP1 is generally much less conserved. The amino acid sequence identity that Arabidopsis INP1 shares with its homologs ranges from ∼85 to 95% for very closely related taxa (see Supplemental Figure 2B online) to ∼44 to 58% (species from various eudicot families) and to ∼37 to 40% (monocotyledonous family of grasses). Interestingly, in general, the INP1 proteins of eudicots are quite divergent (e.g., INP1s from three species in Fabaceae share only ∼70% sequence identity with each other), whereas INP1 homologs from the monocot family Poaceae (grasses) are very conserved (90 to 97% sequence identity), despite the fairly long evolutionary history of this family (Kellogg, 2001). Sequence alignment of INP1 proteins highlights the existence of generally conserved regions and of several highly divergent regions, as well as the presence of the distinct areas of grass-specific amino acids (see Supplemental Figure 1 online).
We used the Phyre2 algorithm (Kelley and Sternberg, 2009) to predict secondary structures of the INP1 proteins from Arabidopsis, cassava (Manihot esculenta), columbine (Aquilegia coerulea), monkey flower (Mimulus guttatus), and rice (Oryza sativa). All of these proteins were predicted to have a similar fold with several (three or more) α-helical regions and four to five short loop-forming structurally disordered regions. Interestingly, all of the predicted disordered regions (framed in green in Supplemental Figure 1 online) in each of the five INP1s fell within the areas of high sequence divergence.
Although INP1 from Arabidopsis is expected to be cytoplasmic, the EMBL-EBI Phobius and TMpred algorithms predicted the existence of a C-terminal transmembrane domain in many of the INP1 homologs from other plant families (see Supplemental Table 1 and Supplemental Figure 1 online). Therefore, these proteins may be anchored to membranes.
All INP1 homologs have a predicted DOG1 domain in their N termini. The only exception is a protein from the gymnosperm P. pinaster, whose sequence (derived from the single available EST BX250600) may be incomplete and possibly misses its N terminus. Another characteristic of the INP1 proteins is a rather high content of Leu residues, which is 13.8% on average, and as high as 16.5% in some species (see Supplemental Table 1 online). This is comparable to the Leu content of Leu-rich repeat Arabidopsis proteins of similar size (14.6% ± 2.5% in 20 random Leu-rich repeat proteins versus 9.1% ± 2.4% in 50 randomly chosen Arabidopsis proteins of similar size), although the Leu residues in INP1 proteins do not form recognizable Leu-rich domains. However, they are often conserved and present in the same relative positions in a variety of species: 13 of the 43 most highly conserved amino acids in INP1 are Leu (see Supplemental Figure 1 online).
INP1 Has Multiplied and Diversified in Some Species of Brassicaceae
In the genomes of members of the Brassica genus, in addition to a gene product very similar to INP1 (87% identity), we identified three INP1-like (IPL) genes encoding more divergent proteins (see Supplemental Figure 2 online). Transcripts of two of them were found in flower buds and anthers (EST accession numbers EV146860, EX089388, and EE392336). These three predicted proteins share only 40 to 43% identity with INP1 and differ from it about as much or more as its homologs from other plant families (see Supplemental Figure 2 online). However, when their sequences were used as queries in BLAST against the Arabidopsis genome, INP1 was pulled out as their only related protein. Two of these proteins (IPL1 and IPL2) are more similar to each other and are quite different from IPL3 (see Supplemental Figure 2 online). Also, the genomes of two other Brassicaceae species, Eutrema parvulum and Capsella rubella, in addition to the predicted INP1 orthologs, each encode an IPL protein. These proteins are very divergent from the bona fide INP1 orthologs, from each other and from the IPL proteins from Brassica rapa (see Supplemental Figure 2 online). Therefore, in some lineages of the Brassicaceae family, INP1 has multiplied and diversified very rapidly.
INP1 Is Expressed in Anthers and Acts in a Sporophytic Manner
Many of the genes involved in exine production, particularly in sporopollenin synthesis, exhibit strong coexpression (Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2009a, 2010, 2011; Grienenberger et al., 2010; Kim et al., 2010; Quilichini et al., 2010). Therefore, we used coexpression algorithms (BAR Expression Angler [Toufighi et al., 2005], ATTED-II [Obayashi et al., 2007], and PRiME [http://prime.psc.riken.jp/?action=coexpression_index]) to check if INP1 is also expressed together with these genes. However, INP1 does not appear to be coexpressed with the genes known to be involved in exine formation. In fact, no gene exhibits strong coexpression correlation with INP1 (Pierson correlation coefficient r values were below 0.5). Based on the microarray transcription data (AtGenExpress ATGE_31_A2-C2), it is expected that INP1 is a weakly expressed gene that has a somewhat increased expression in developing flower buds and in pollen. According to the microarray analysis, INP1 is expressed at very low levels: Its highest expression values in stage 9 buds were only ∼18, whereas the values for several sporopollenin synthesis genes also expressed at that stage ranged from 500 to 1700.
We investigated INP1 expression by placing a β-glucuronidase (GUS) gene under the INP1 promoter. The INP1pro:GUS construct was expressed only in anthers, with the earliest detectable signal in stage 9 buds (Figure 2). The early signal was very weak and appeared to be confined to microspores. This signal continued through anther development, becoming very prominent in the pollen of mature anthers (Figure 2F). Tapetum, the secretory tissue in which components of exine and pollen coat are synthesized, also exhibited a weak GUS staining (Figure 2D), although at slightly later stages than microspores. No signal was detected in other flower tissues, stems, leaves, and siliques.
Figure 2.
INP1 Is Expressed in Anthers and Acts Sporophytically.
(A) to (F) The earliest INP1pro:GUS expression was visible in the anthers of stage 9 buds (B) and continued throughout flower development. Anthers from the following flower development stages are shown: early stage 9 bud (no GUS staining) (A), stage 9 bud (weak GUS staining) (B), stage 10 bud (C), stage 12 bud (E), and mature flower (F). In (D), a magnified anther from a stage 9 bud exhibits GUS staining in tetrads of microspores (several areas of microspore signal are marked with arrowheads) as well as weak staining in the tapetum cell layer (T, delineated by dashed lines).
(G) All four microspores in the tetrad from the INP1/inp1; qrt1/qrt1 plant have normal apertures.
Bars = 100 μm in (A) to (C), (E) and (F), 20 μm in (D), and 10 μm in (G).
Pollen characteristics are influenced by both sporophytic and gametophytic genomes that function in diploid microspore mother cells (MMCs)/tapetum and in haploid microspores, respectively (McCormick, 1993). To check whether INP1 determines aperture formation in a sporophytic or gametophytic manner, we looked at the pollen of heterozygous INP1/inp1 plants. INP1/inp1 plants produced pollen with normal apertures, suggesting that INP1 functions sporophytically. This conclusion was confirmed by the analysis of the pollen tetrads generated by the INP1/inp1; qrt1/qrt1 mutant. In these tetrads, all four products of male meiosis, held together due to the qrt1 mutation (Preuss et al., 1994), had normal apertures (Figure 2G), indicating that INP1 acts in a sporophytic manner. Given that the early transcript signal is located in microspores and that apertures are formed at the time when tetrads of microspores are enclosed by thick callose walls, the sporophytic INP1 function necessary for aperture formation is likely internal, coming from the diploid MMC, and not provided externally by the diploid tapetum.
To determine if expression of INP1 during male sporogenesis was sufficient to rescue inp1 phenotypes, the INP1 gene was expressed in inp1-1 plants under a male sporogenic line–specific promoter, MSP1, active in MMCs and microspores (Honys et al., 2006). The pollen of the transgenic plants had apertures restored (see Supplemental Figure 3 online), indicating that INP1 expression during sporogenesis is sufficient to promote aperture development.
INP1 Protein Exhibits Distinct Tripartite Subcellular Localization in Microspores and Marks Positions of Future Apertures
To monitor the INP1 subcellular localization, we created a translational reporter INP1pro:INP1-YFP (for yellow fluorescent protein) and introduced it into the inp1-1 mutant. Importantly, this reporter rescued the inaperturate phenotype of the mutant (Figure 1F). We could not detect any INP1-YFP fluorescence in undivided MMCs, MMCs undergoing meiosis and cytokinesis, or in tapetum (Figure 3). Yet, INP1-YFP exhibited distinct punctate fluorescence in the tetrad stage and early free microspores (Figures 3 and 4). Strikingly, the fluorescence corresponded to three equidistant areas, reminiscent of the positions of three apertures (Figures 4A to 4D). At the tetrad stage, fluorescence was restricted to the directly opposing areas of the neighboring microspores arranged in a tetrahedral formation (Figures 4C and 4D); those correspond to the positions where apertures develop (Fischer, 1890; Wodehouse, 1935; Ressayre et al., 2003). Although single optical sections of microspores in tetrads or at the early free stage often revealed only three distinct YFP spots per microspore, the reconstruction of serial z-stacks demonstrated that YFP signal was restricted not just to three spots, but to three long and narrow regions. Those long areas of fluorescence could occasionally be observed in single sections as well, depending on a microspore’s positioning during a preparation (Figures 4E and 4F). The three-dimensional reconstruction and surface rendering from confocal z-stacks of tetrads confirmed that the INP1-YFP puncta were underlying the positions of future long and narrow apertures, which were already sometimes recognizable at the tetrad stage (Figure 4H; see Supplemental Movie 1 online). These findings indicate that INP1 is directly involved in specifying aperture positions. The aperture areas delineated by the INP1 proteins had their centers at the contact points between microspores arranged in a tetrahedral tetrad conformation.
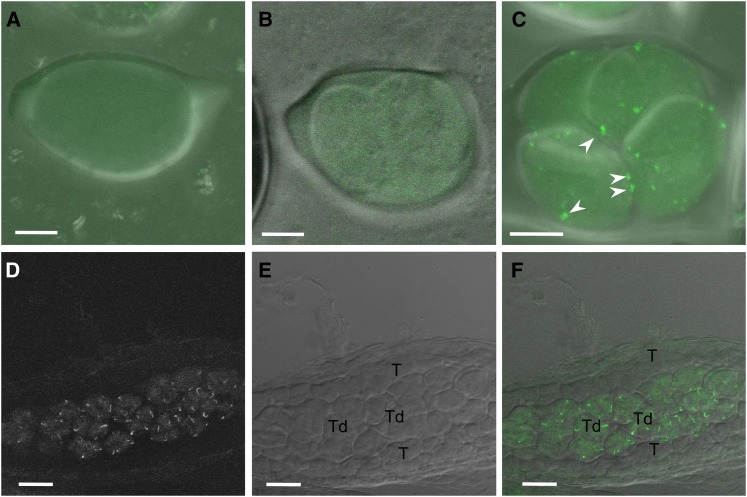
Figure 3.
INP1-YFP Protein Is Expressed in Tetrads but Not in MMC, Dividing MMC, or in Tapetum.
(A) and (B) Maximum intensity projections of z-stack images of an undivided MMC (A) and an MMC that undergoes cytokinesis (B). No YFP signal is present.
(C) Maximum intensity projection of z-stack images of a tetrad. Punctate YFP signal is readily observable (several puncta are labeled with arrowheads).
(D) to (F) In anthers, INP1-YFP fluorescence is present in tetrads, but not in the tapetum layer (T).
(D) YFP fluorescence in the anther is restricted to the tetrad-containing central part.
(E) Bright-field image of the same anther. T, tapetum; Td, tetrads of microspores.
(F) Overlay of (D) and (E) showing absence of the fluorescent signal in the tapetum (T).
Bars = 5 μm in (A) to (C) and 20 μm in (D) to (F).
[See online article for color version of this figure.]
Figure 4.
INP1-YFP Is Localized to Three Areas per Microspore That Underlie Three Aperture Positions.
(A) to (F) YFP expression in single optical sections of a free microspore ([A] and [B]) or tetrads of microspores ([C] to [F]). Overlay of the YFP fluorescence and bright-field views ([B], [D], and [F]).
(G) An illustration of four microspores held together in a tetrad. Each microspore has three contacts with its sisters. Positions of apertures are shown in yellow.
(H) A three-dimensional surface rendering from a confocal z-stack of a tetrad of microspores. Surface was transiently labeled by DAPI (blue), rendered with IMARIS, and shown partially transparent to visualize the internal INP1-YFP fluorescence. INP1-YFP shows distinct punctate signals (yellow-gray) surrounding the apertures. See Supplemental Movie 1 online.
Bars = 10 μm.
In contrast with the transcriptional reporter (INP1pro:GUS) results, we did not observe the expression of the INP1 translational reporter beyond the early free microspore stage (after exine formation). This discrepancy could be due to the high sensitivity of the GUS reporter and the high stability of the GUS protein (Jefferson et al., 1987; De Block and Debrouwer, 1992; Kavita and Burma, 2008).
A Gradient of Aperture Lengths in INP1pro:INP1-myc Lines Suggests That Apertures Originate at Three Equidistant Spots Close to the Pollen Equator
In parallel with INP1pro:INP1-YFP, we also introduced an INP1pro:INP1-myc construct in the inp1-1 plants with the idea to use it for future biochemical characterization of the protein. Unexpectedly, this construct provided an important insight into the mechanism of aperture formation. When unhydrated, the wild-type and the inp1 inaperturate pollen can be easily distinguished at a low magnification under a dissection microscope, with wild-type pollen having an oval shape and inaperturate pollen having a round shape (Dobritsa et al., 2011). Surprisingly, when we examined pollen generated by the INP1pro:INP1-myc transgenic plants, we found some plants that produced pollen that appeared spheroidal, intermediate between round and oval. Out of the 40 T1 plants, six plants were classified as having oval pollen, 15 as round, and 19 as having intermediate pollen. This prompted us to carefully examine the exine morphology in these lines with confocal microscopy. We found that the majority (15/16) of the examined INP1pro:INP1-myc; inp1-1 plants generated pollen in which apertures, although restored, were abnormal to some degree. Most of these plants produced pollen with apertures that were shorter than in the wild type (Figure 5) and in extreme cases were reduced to holes (Figures 5D, 5F, and 5F’). Therefore, a gradient of aperture lengths could be envisioned across the transgenic lines.
Figure 5.
A Gradient of Aperture Lengths in the INP1pro:INP1-myc; inp1-1 Transgenic Lines.
The apertures formed in the pollen of these lines ranged from the wild-type-like ones (A) to the hole-like ones ([D], [F], and [F’]).
(A) Pollen from line 14 with apertures of the wild-type length.
(B) Pollen from line 5 with shortened apertures.
(C) Pollen from line 8 with further shortened apertures.
(D) Pollen from line 40 with very short apertures.
(E), (E’), (F), and (F’) Examples of individual grains from the lines 8 (medium apertures; [E] and [E’]) and 34 (very short apertures; [F] and [F’]) that demonstrate the presence of three apertures per microspore even in the pollen with shortened apertures. (E) and (E’) are top and bottom views of the same pollen grain (line 8). (F) and (F’) are top and bottom views of the same pollen grain (line 34). Arrowheads point to the extremely short apertures.
Bars = 10 μm.
In these transgenic lines, there were still three apertures present per pollen grain. The exceptional grains with one or two apertures were found on rare occasions only among the pollen of the lines with the hole-like apertures. Notably, the three shortened apertures were located not randomly, but were placed equidistantly, with their centers close to the equators of the grains (Figures 5E to 5F’). This finding implies that three equidistant positions at the equator of microspores serve as the initiation points for formation and spreading of apertures. Consistent with this prediction, we found that in some tetrads that had thin intersporal callose walls and, therefore, likely had just recently undergone cytokinesis, the INP1-YFP fluorescence was limited to approximately three spots per microspore (see Supplemental Figure 4 online).
INP1 Quantitatively Controls Aperture Length
Since all the INP1pro:INP1-myc; inp1-1 transgenic lines contained the same construct, the most likely explanation for the differences in the aperture lengths is the variations in the transgene expression due to its insertions in different locations. This suggests an interpretation that aperture length is sensitive to the INP1 dosage. The specific differences among the lines were present in both T1 and T2 generations and were therefore heritable.
To test if aperture length varies in plants from the same line that differ in the number of INP1-myc copies, we used line 28 that had short apertures in T1 and a 3:1 segregation of Bastar:Bastas plants in T2 (70:25, χ2 = 0.054, P > 0.9), consistent with the presence of a single insertion in the T1 generation. We reasoned that potential low levels of functional INP1 in that line might provide a sensitized background, sufficient to see the difference in aperture length between one and two transgene insertions in T2. Indeed, among the 33 Bastar T2 plants that were examined under a microscope, we found some that produced pollen with short (S) apertures (22 plants) and some that produced pollen with medium (M) apertures (11 plants) (Figures 6A and 6B). Although the number of tested plants was small, the segregation ratio of S-to-M plants was 2:1 (χ2 = 0, P = 0.975), which would be expected if S plants were hemizygous and M plants were homozygous for transgene insertions. We measured the size of the apertures for several of these plants and found that plants classified as S had, on average, aperture length of ∼3 to 4 μm, whereas plants classified as M had, on average, aperture length of ∼6 to 7 μm (see Supplemental Table 2 online). Similar differences between the aperture lengths of T2 plants was also observed in an independent short-aperturate line 34 (see Supplemental Table 2 online) that also had a 3:1 Bastar:Bastas segregation (71:24, χ2 = 0, P = 0.975).
Figure 6.
INP1 Controls Aperture Length in a Quantitative Manner.
(A) and (B) Examples of pollen grains produced by T2 plants from the INP1pro:INP1-myc line 28.
(A) Pollen with short apertures (S) generated by plant #8.
(B) Pollen with medium apertures (M) generated by plant #35. Bars = 10 μm.
(C) INP1-myc transcript levels were measured in three INP1pro:INP1-myc; inp1-1 lines by qRT-PCR and normalized to the levels of the inp1-1 transcript.
S, short apertures (line 28); M, medium apertures (line 10); L, long apertures (line 14). Error bars indicate se (n = 4 biological replicates). Student's t test P values = 0.061 (S versus M), 0.057 (S versus L), and 0.073 (M versus L).
To confirm that the T2 S plants from line 28 had one transgene insertion and the M plants had two insertions, we tested Basta sensitivity of their progeny (see Supplemental Table 3 online). All T3 progeny of the M plants were resistant to Basta, whereas in the progeny of all S plants, there was a segregation of Bastar and Bastas plants (see Supplemental Table 3 online), indicating that differences in aperture length in M and S plants can, in fact, be attributed to the differences in the number of transgenes.
We also compared the levels of the INP1-myc transcripts in three lines that had different aperture lengths (S, line 28; M, line 10; long, line 14) by performing quantitative RT-PCR (qRT-PCR) on RNA extracted from stage 9 buds. The INP1-myc lines generated from the inp1-1 background have both INP1-myc and inp1-1 alleles transcribed (see Supplemental Figure 5A online). Therefore, to ensure that the INP1-myc transcript is specifically detected, we used primer pairs that could distinguish between the endogenous mutant and transgenic wild-type copies of INP1 (see Supplemental Figure 5B online). Different samples of stage 9 buds were expected to have unequal combinations of different developmental stages of male germ line cells (MMCs, tetrad, and free microspores) and, therefore, to likely vary in their INP1 transcript levels. To compensate for stage differences between the samples, we decided to use the endogenous inp1-1 transcript, rather than a housekeeping reference gene, for normalization. With qRT-PCR, we observed a trend for correlation between the aperture length and the amount of the transcript, with higher INP1 levels present in the samples with longer apertures (Figure 6C).
Taken together, the results of the genetic analysis and qRT-PCR strongly suggest that at low levels of INP1 expression, aperture length depends on the INP1 dosage. Interestingly, although we also observed differences in aperture length between individual lines in the case of other transgenes, such as MSP1pro:INP1 and INP1pro:INP1-YFP, the variation among those lines was smaller than with INP1-myc, with most lines having wild-type aperture length and some having medium-sized apertures (out of the 24 tested INP1pro:INP1-YFP lines, 16 had long apertures, six had apertures that were slightly shorter than the wild-type, and two had medium-sized apertures). The very short apertures were only observed in the case of the INP1-myc lines, suggesting that the addition of the myc tag at the C terminus makes the effects of dosage dependence especially pronounced, possibly by destabilizing, interfering with transport, or partially inactivating the INP1 protein.
Abnormalities in INP1 Expression May Affect Global Exine Patterning or Cause Formation of Ectopic Apertures
Although the vast majority of pollen grains in any given transgenic line had normal reticulate exine patterning and apertures of a relatively uniform length, in multiple transgenic lines, we also observed rare grains with global exine patterning defects and/or defective aperture margins. Such abnormal pollen grains represented not more than a few percent of pollen populations (∼1 to 3%, n = 200 to 300 pollen grains/line), but they were never observed in the morphologically uniform wild-type or inp1-1 pollen populations (n > 300). For example, grains with large abnormal holes and disrupted exine patterns, like the ones shown on Figures 7A to 7E, were seen among the pollen with shortened apertures. Such abnormal pollen grains were more often found among the lines with extremely short apertures. One possible explanation for the origin of these abnormalities is that microspores may be very sensitive to INP1 concentrations above and/or below certain thresholds and that perhaps unequal distribution of the limited amounts of the INP1 protein or mRNA into microspores during meiosis contributes to differences in exine and aperture phenotypes.
Figure 7.
Pollen Grains with Abnormal Exine Patterning and Ectopic Apertures Are Occasionally Produced by the INP1-Expressing Transgenic Lines.
(A) to (E’) Pollen grains with abnormal exines produced by the INP1pro:INP1-myc; inp1-1 lines 32 ([A] and [A’]), 38 ([B], [B’], [C], and [C’]), and 39 ([D] and [E]). Top and bottom views of the same grains are indicated, when appropriate.
(F) to (G’) Ectopic apertures were observed in pollen that had 35Spro:INP1 in the wild-type background ([F] and [F’]; line 15) and in the inp1-1 background ([G] and [G’]; line 22).
Bars = 10 μm.
To test the effects of excessive INP1 on aperture and exine formation, we created 35Spro:INP1 transgenic lines in the wild-type or inp1-1 backgrounds. 35Spro:INP1 was sufficient to restore apertures in the inp1-1 plants, although like the INP1pro:INP1-myc and MSP1pro:INP1 transgenic lines, the 35Spro:INP1; inp1-1 lines exhibited some variations in terms of aperture lengths, which varied from the wild-type-like to medium sized. The overwhelming majority of pollen grains in the 35Spro:INP1 lines looked normal. Although cauliflower mosaic virus 35S is a strong constitutive promoter commonly used to overexpress genes in plants, its expression in anther tissues is somewhat controversial (McCormick et al., 1991; de Mesa et al., 2004; Hraška et al., 2008; Zhang et al., 2008). The levels of INP1 transcript in stage 9 buds were significantly higher in the 35Spro:INP1 lines than in the wild type (see Supplemental Figure 6 online); however, to prove that the construct is similarly overexpressed in tetrads is more challenging. Therefore, we cannot rule out a possibility that levels of INP1 within tetrads of microspores were not as high as typically expected with the 35S construct.
However, it is of note that among the pollen of the 35Spro:INP1 lines, we also observed some that had ectopic (more than three) apertures (Figures 7F to 7G’), suggesting that excessive INP1 might be sufficient, at least to a partial extent, for aperture formation. Pollen with such defects was not seen in every line and, when observed, did not constitute more than 2 to 5% of pollen (n = 200 to 300 pollen grains/line). Defects in aperture margins or exine patterns, similar to the ones shown for INP1pro:INP1-myc in Figures 7A to 7E, were also observed. Abnormalities in aperture number, morphology, and/or exine structures were seen in different plants of the same line and found both in lines with the inp1-1 and the wild-type backgrounds. Although it is not clear at the moment why some grains, but not others, generated by the same plant had these phenotypes, we speculate that variations in the INP1 dosage among microspores, in particular, too much INP1 loaded in a microspore, can result in pollen grains with ectopic apertures, irregular aperture margins, or abnormal exine patterning. More sensitive detection methods that can measure RNA or protein concentrations in individual microspores will be required to test this hypothesis.
Grass INP1 Homolog Has No Effect on Arabidopsis Apertures
Apertures in pollen of different species often differ dramatically in their number and morphology. While many eudicots have a tricolpate configuration of apertures (three long furrows, like in Arabidopsis), monocots often have a single aperture, which in grasses has a shape of a round pore (Furness and Rudall, 2004) (see Supplemental Figure 7A online). The significant differences in the INP1 sequences between eudicots and grasses (see Supplemental Figure 1 online) led us to hypothesize that such sequence variations may be directly responsible for differences in aperture morphologies. To test this hypothesis, we cloned the INP1 homolog from the grass Brachypodium distachyon (Bradi3g50820) and placed it under the Arabidopsis INP1 promoter that had already allowed the successful expression of GUS, INP1, INP1-myc, and INP1-YFP. The resulting INP1pro:Bd-INP1 construct was transformed into the inp1-1 plants and into the wild-type Arabidopsis, and we monitored the aperture phenotypes in a large number of independent transgenic lines (n = 18 for T1 plants with inp1-1 background; n = 14 for T1 plants with the wild-type background). We confirmed the Bd-INP1 expression in stage 9 buds in these transgenic plants (see Supplemental Figure 7D online). However, the Bd-INP1 transgenic plants had the same aperture phenotypes as their respective background lines (see Supplemental Figures 7B to 7C online), indicating that Bd-INP1 has no effect on apertures in Arabidopsis pollen. We cannot yet formally exclude the possibility that INP1 homologs from grasses are simply not involved in aperture formation. However, given (1) the fact that Bd-INP1 is the only INP1-related gene in the Brachypodium genome, (2) the high degree of conservation of the INP1 homologs within the grass family, and (3) the distinct shape of apertures in the pollen of grasses, the more likely interpretation of this result is that additional factors have coevolved with the INP1 homologs and are also required to control aperture formation and/or specify aperture morphology.
DISCUSSION
The beauty and the dramatic variety of pollen exine patterns have been long admired (Grew, 1682; Fritzsche, 1837; Wodehouse, 1935; Kesseler and Harley, 2004), and it has been suggested that these patterns may have an adaptive significance in plant reproduction (Lee, 1978; Heslop-Harrison, 1979b). Yet, how these patterns develop at the molecular and cellular level remains unknown. Here, we describe a gene that specifically controls formation of a distinct and common element of exine patterning, the apertures. INP1 encodes a novel protein with a sporophytic function that is present in microspores after meiosis and marks positions of the future apertures. We demonstrate that apertures appear to be sensitive to the levels of INP1, suggesting a possibility that variations in aperture lengths observed in nature, sometimes among species belonging to the same genus (Dickison et al., 1982), may be achieved through regulating INP1 at the expression level.
The results described in this article are consistent with the following working model for aperture specification and the role of INP1 in this process (Figure 8). INP1 transcript or protein is produced by a MMC (Figure 8A) and is inherited by microspores after meiosis (Figure 8B). The sensitivity of microspores to INP1 dosage, which is indicated both by the production of lines with short apertures and by pollen with abnormal apertures and defective exine patterning, implies that a mechanism must exist that ensures equal INP1 distribution among the four products of male meiosis. The tendency of extremely short apertures to be distributed equidistantly and close to the equators of grains suggests that three equidistant positions at the equator serve as points from which apertures originate (Figure 8C). Arabidopsis microspores are arranged in a tetrahedral conformation in a tetrad, and each has three places of contact with its sisters. The centers of these places of contact correspond to the regions where centers of apertures are formed (Figures 4G to 4H). Almost 80 years ago, R.P. Wodehouse hypothesized that in eudicots with tricolpate apertures and a tetrahedral arrangement of microspores in tetrads, like in Arabidopsis, apertures develop at the last points of contact persisting between the cytoplasm of future microspores at the end of the simultaneous cytokinesis (Wodehouse, 1935). Our results demonstrate that INP1 is located in the vicinity of the areas of last contact between microspores. Consistent with this idea is the identification of young tetrads, in which only about three INP1-YFP fluorescent spots per microspore were present close to such equatorial positions. The INP1 protein or transcript may be brought to these positions by the direction of cytokinesis and centripetal growth of cell walls between the sister microspores (Otegui and Staehelin, 2004). INP1 localization may depend on the distribution of microtubules that form radial systems around four postmeiotic nuclei and define four cytoplasmic domains of future microspores (Brown and Lemmon, 1988, 2001; Yang et al., 2003; Otegui and Staehelin, 2004). Alternatively, INP1 may be specifically stabilized in these three areas. From those equatorial points, INP1 spreads longitudinally in two directions to underlie and specify positions of future apertures (Figure 8D).
Figure 8.
Working Model for the INP1-Controlled Specification of Aperture Positions in Arabidopsis Pollen.
(A) and (B) INP1 transcript or protein (black dots) is produced in a MMC (A) and is equally distributed among four products of meiosis (B).
(C) The simultaneous cytokinesis of MMC generates four microspores arranged in a tetrahedral tetrad and held together by a callose wall (gray). For simplicity, only three microspores lying in the same plane are shown here. INP1 protein localizes to three equidistant equatorial points shortly after cytokinesis (shown as black dots for the two in-plane positions per microspore and as gray dots for the one out-of-plane position per microspore).
(D) At a later tetrad stage, INP1 spreads out longitudinally to mark positions of apertures. Here, positions of two in-plane half apertures (black dots) and one out-of-plane aperture (gray dots) are visible per microspore.
In pollen of most plant species, apertures represent a well-defined, tightly controlled, and species-specific element of pollen surface patterning. Therefore, exine deposition machinery must reliably recognize some areas on pollen surface as different from others and not deposit exine onto these areas. The discovery of INP1 signifies the first step in understanding at the molecular level of how such functionally distinct cellular domains are produced in microspores and how these cellular asymmetries are then translated into extracellular differences between surface areas that will be covered with exine and those that will not be. It also immediately generates a number of exciting cell biological questions that await further investigation: (1) How is the postulated equal distribution of INP1 among four products of meiosis actually achieved? (2) What brings INP1 to the three equidistant equatorial positions and which sequence elements of INP1 are required for this? (3) What controls the length, shape, and direction of INP1 spreading, which likely determines the length, shape, and relative positions of apertures? (4) How does the generation of three distinct cellular domains prevent exine deposition machinery from placing extracellular exine at the regions above those domains? (5) Do the variations in the INP1 sequences in different species contribute to the differences in aperture morphologies?
METHODS
Cloning of the INP1 Gene
Positional cloning was used to map INP1 in two independently isolated inp Arabidopsis thaliana lines (25-1-1 and 31-1-2), using F2 populations generated by crossing the mutants (in Columbia background) with Landsberg erecta wild-type plants. Bulked segregant analysis (Michelmore et al., 1991) revealed linkage to the marker ciw7 on chromosome 4 (Dobritsa et al., 2011). Fine mapping of inp1 in 399 F2 plants with additional markers (see Supplemental Table 4 online) placed it in a 53-kb region between the markers F7K2-9 and T12H17-6. ORFs of the 17 genes from this interval (At4g22580 to At4g22730) were sequenced, and a polymorphism relative to the wild-type Columbia was found in At4g22600 (a single nucleotide, A262, was absent). This polymorphism was present in all, but one, lines forming the inp1 complementation group. It resulted in the creation of a SacI site and allowed us to generate a cleaved amplified polymorphic sequence marker, which was used to verify the presence of the inp1-1 mutation in the subsequent experiments with transgenic constructs (primers 22600-DF and 22600-DR, SacI cuts the inp1-1 allele). For the list of primers used in this study, see Supplemental Table 5 online. The remaining mutant line (118-2-2) had a different nucleotide (C74) absent in At4g22600.
Transgenic Constructs
The INP1 complementation construct was produced by amplifying the INP1 genomic region, including the ORF, 1060 nucleotides upstream of ORF, and 239 nucleotides downstream of the stop codon (primers INP1-CF and INP1-ER). PCR product was digested with BamHI-AgeI and incorporated in a modified pGreenII02229 binary vector (pGR111) (Hellens et al., 2000; von Besser et al., 2006; Dobritsa et al., 2010). To create the INP1pro:INP1-YFP construct, the INP1 gene was amplified with primers INP1-CF and INP1-FR, digested with BamHI-AgeI, and placed into pGR111 upstream of the YFP gene.
The INP1pro:GUS construct was produced by amplifying the 1060-nucleotide region upstream of INP1 ORF (primers INP1-CF and INP1-CR), digesting with BamHI/AgeI, and incorporating it into the GUS-pGR111 vector (Dobritsa et al., 2010). The MSP1pro:INP1 construct was produced by amplifying the INP1 gene with primers INP1-EF and INP1-GR, digesting the product with AgeI-NcoI, and incorporating it into AgeI-NcoI–digested pGR111; this was followed by amplification of the MSP1 promoter with primers MSP1-F and MSP1-R, digestion of the product with SacI-AgeI, and incorporation into the SacI-AgeI–digested INP1-pGR111. To create the 35Spro:INP1 construct, the AgeI-NcoI–digested INP1 fragment was placed into a modified pGreenII02229 binary vector containing the cauliflower mosaic virus 35S promoter (pGR117). To create the myc-tagged INP1, the INP1 gene was amplified with primers Pst-INP1-CF1 and Not-INP1-HR, digested with PstI-NotI, and subcloned into PstI-NotI–digested pCMV/myc/mito vector (Invitrogen). The resulting INP1pro:INP1-myc was cut out with SacI-AgeI and incorporated into pGR111.
To create the INP1pro:Bd-INP1 construct, the INP1 homolog from Brachypodium distachyon was amplified using primers BdINP1-AF1 and BdINP1-AR1 in the presence of 3% DMSO (PCR program: 98°C, 1 min; 30 × [98°C, 15 s; 63°C, 30 s; 72°C, 30 s]; 72°C, 10 min), digested with NcoI-SpeI, and replaced the GUS gene in the INP1pro:GUS-pGR111 construct.
All constructs were verified by sequencing and transformed into the Agrobacterium strain GV3101. Wild-type Columbia or inp1-1 plants were then transformed by floral dip method (Clough and Bent, 1998), transgenic plants were selected with BASTA, and the presence of transgenes was confirmed with specific primers.
Phylogenetics, Bioinformatics, and Protein Sequence Analysis
To identify INP1 homologs in other species, we used BLAST searches against the National Center for Biotechnology Information databases (nr/nt, EST_others, whole-genome short gun contigs, and high-throughput genomic sequences), multiple plant species genomes available in the Phytozome v.7.0 database (http://www.phytozome.net/) (Goodstein et al., 2012), and genomes of Theobroma cacao (http://cocoagendb.cirad.fr/) and Fragaria vesca (http://www.strawberrygenome.org/). Protein sequences were aligned using ClustalW (MegAlign Company, DNAStar program, Lasergene subprogram). SignalP 3.0, Phobius, TMpred, TMHMM2.0, PROSITE, InterProScan, Pfam, and LRRFinder were used to search for domains or signal sequences. Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/) was used to predict protein secondary structures. In addition to the true INP1 gene present on chromosome 1 of Brassica rapa (identified in clone AENI01000155), three other INP1-like genes clustered on chromosome 3 were found. For B. rapa and Brassica napus, BLAST identified three ESTs of two types (accession numbers EE392336, EX089388, and EV146860) expressed in floral buds and anthers that encoded putative proteins with similarity to Arabidopsis INP1. By searching the sequenced genomic clones of B. rapa (AC241197, AENI01002719, and AENI01002720), we were able to identify a third protein sequence with similarity to INP1. In the recently released sequenced genome of B. rapa, these three genes are annotated as Bra019371, Bra019372, and Bra019373, respectively. The annotation predicts second exons in each of these INP1-like genes, which encode the C-terminal sequences unrelated to INP1. The presence of these 3′ exons is also suggested by the existence of the EST EV146719 from B. napus. To check for INP1 coexpression with other genes, BAR Expression Angler (Toufighi et al., 2005), ATTED-II (Obayashi et al., 2007), and PRiME (http://prime.psc.riken.jp/?action=coexpression_index) coexpression algorithms were used.
Microscopy
To visualize aperture and exine morphology, pollen was stained with auramine O and observed with an Olympus Fluoview FV1000 confocal laser scanning microscope as previously described (Dobritsa et al., 2011). For aperture length measurements in the INP1pro:INP1-myc T2 plants, photographs of 20 to 30 pollen grains per plant were taken and lengths of short to medium apertures were measured using the straight-line tool in NIH ImageJ. To visualize YFP fluorescence, MMCs, tetrads, and free microspores were dissected from anthers and placed in a drop of Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (H-1200; Vector Laboratories) under a cover slip and observed with a Leica SP2 confocal microscope (×63 oil immersion objective). DAPI transiently stained the outsides of the microspores (see Supplemental Movie 1 online), and this staining was used for surface rendering. A total of 45 to 65 0.3-μm z-sections were collected. Z-stack images were processed with NIH ImageJ or, for three-dimensional surface rendering, with IMARIS software (Bitplane). GUS staining was performed as previously described (Dobritsa et al., 2009a).
RT-PCR and qRT-PCR
Total RNA was extracted from 130 stage 9 buds (∼0.3- to 0.5-mm long) with TRIzol reagent (Invitrogen) following the manufacturer’s instructions. RNA concentration was determined via spectrophotometry using Nanodrop (Thermo Scientific). To remove any contaminating genomic DNA, 5 μg of RNA was treated with 1.5 units of RQ1 DNase (Promega) in 20-μL reactions at 37°C for 20 min, followed by addition of 0.8 μL of DNase stop solution and DNase inactivation at 75°C for 10 min. The DNase-treated RNA (an equivalent of initial 2.5 μg) was reverse transcribed with the help of the Masterscript kit (5PRIME) in a 20-μL reaction in the presence of 0.5 μM deoxynucleotide triphosphate, 3′ rapid amplification of cDNA ends (RACE) adapter (60 ng/μL), Prime RNase inhibitor (0.05 units/μL), and Masterscript reverse transcriptase (0.75 units/μL). The reactions were performed at 42°C for 1 h. For each cDNA sample, the absence of genomic DNA was verified by PCR with the intron-flanking ACT2 primers. To detect the INP1 transcript by traditional RT-PCR, first-strand cDNA (an equivalent of 375 ng of initial RNA) was amplified in 50-μL reactions in Qiagen PCR buffer in the presence of 0.2 mM deoxynucleotide triphosphate, 0.4 μM each of primers INP1-EF and 22600-DR, and 1.5 unit of Taq polymerase (Qiagen). To distinguish between the INP1 transcripts from the endogenous mutant copies and from the INP1-myc transgene, PCR reactions were diluted twofold with water and digested with SacI that cuts the inp1-1 version of INP1 but not the wild-type one. Although we were able to amplify INP1 transcripts several times, the process was challenging and required a lot of optimization, likely due to INP1 being expressed at very low level in a limited number of cells and in a dynamic fashion. To measure INP1 expression in 35S lines, primer pairs INP1-EF and 22600-DR, as well as INP1-GF and 3′RACE adapter, were used. To confirm Bd-INP1 expression, primers BdINP1-BF and BdINP1-AR1 were used.
For qRT-PCR, cDNA samples were prepared as above and equivalents of 375 ng of initial RNA were amplified in 10-μL reactions using FastStart Universal SYBR Green Master Mix (with Rox as a reference dye) (Roche) on AB7900 real-time PCR system (Applied Biosystems). To amplify the wild-type transcript of the INP1-myc transgene, the primers INP1-KF1 and INP1-LR were used. To amplify the transcript of the endogenous mutant inp1-1 gene, the primers INP1-KF1mut and INP1-LR were used. The specificity of the primers was confirmed with PCR (see Supplemental Figure 5B online) and qPCR reactions done on the wild-type and inp1-1 genomic DNA with both sets of primers. qRT-PCR cycling conditions included an initial incubation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. For each INP1-myc line, four biological replicates and two technical replicates were used. After qRT-PCR, the amounts of both transcripts were calculated relative to standard curves obtained for serial dilutions of the wild-type and inp1-1 genomic DNAs, respectively. For each sample, the amounts of the INP1-myc transgenic transcripts were then normalized to the amounts of the endogenous inp1-1 transcripts.
Accession Numbers
Accession numbers for sequences from this article can be found in Supplemental Table 6 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Alignment of INP1 Protein Sequences from Multiple Species.
Supplemental Figure 2. INP1 Gene Has Multiplied and Diversified in Some Members of Brassicaceae.
Supplemental Figure 3. Expression of INP1 during Male Sporogenesis Is Sufficient for Aperture Formation.
Supplemental Figure 4. Some Tetrads Have INP1-YFP Signal Confined to Approximately Three Spots per Microspore and Not to Three Long Areas.
Supplemental Figure 5. Both the INP1-myc Transgene and the Endogenous inp1-1 Allele Are Transcribed in the INP1pro:INP1-myc; inp1-1 Lines.
Supplemental Figure 6. INP1 Is Overexpressed in stage-9 Buds of 35Spro:INP1 Lines.
Supplemental Figure 7. The INP1 ortholog from B. distachyon Has No Effect on Arabidopsis Pollen Apertures.
Supplemental Table 1. Leucine Content and Positions of a C-Terminal Transmembrane Domain Predicted by TMpred in INP1 Proteins of Different Species.
Supplemental Table 2. Aperture Lengths in Selected T2 Plants in the INP1pro:INP1-myc Lines 28 and 34.
Supplemental Table 3. Segregation of Sensitivity to Basta in the Progeny of 33 T2 Plants from Line 28.
Supplemental Table 4. Genomic Markers Used for Positional Cloning of the INP1 Gene.
Supplemental Table 5. Primers Used in This Study.
Supplemental Table 6. Accession numbers for INP1 and INP1-like (IPL) Sequences from Different Species.
Supplemental Movie 1. Z-Stack of Confocal Sections through a Tetrad of Microspores Expressing INP1-YFP (Green).
Acknowledgments
We thank Christine Labno for microscopy expertise, Artem Abanov for help with generating a three-dimensional tetrad model, and Jean Greenberg for her continuous support, for providing safe haven to A.A.D. in her lab, and for critical reading of the article. The work was partially supported by the National Science Foundation Arabidopsis 2010 grant (MSB-0520283).
AUTHOR CONTRIBUTIONS
A.A.D. designed and performed the experiments, analyzed the data, and wrote the article. D.C. participated in the positional cloning of INP1 and created several transgenic lines.
Glossary
- ORF
open reading frame
- GUS
β-glucuronidase
- MMC
microspore mother cell
- YFP
yellow fluorescent protein
- qRT-PCR
quantitative RT-PCR
- DAPI
4′,6-diamidino-2-phenylindole
References
- Aarts M.G., Hodge R., Kalantidis K., Florack D., Wilson Z.A., Mulligan B.J., Stiekema W.J., Scott R., Pereira A. (1997). The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Hatakeyama K., Hinata K., Inatsugi R., Nishida I., Sato S., Kato T., Tabata S., Toriyama K. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39: 170–181 [DOI] [PubMed] [Google Scholar]
- Bentsink L., Jowett J., Hanhart C.J., Koornneef M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore S., Barnes S.H. (1986). Harmomegathic mechanisms in pollen grains. In Pollen and Spores: Form and Function, S. Blackmore and I.K. Ferguson, eds (London: Academic Press), pp. 137–149 [Google Scholar]
- Brown R.C., Lemmon B.E. (1988). Microtubules associated with simultaneous cytokinesis of coenocytic microsporocytes. Am. J. Bot. 75: 1848–1856 [Google Scholar]
- Brown R.C., Lemmon B.E. (2001). The cytoskeleton and spatial control of cytokinesis in the plant life cycle. Protoplasma 215: 35–49 [DOI] [PubMed] [Google Scholar]
- Chen W., Yu X.H., Zhang K., Shi J., De Oliveira S., Schreiber L., Shanklin J., Zhang D. (2011). Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 157: 842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colpitts C.C., Kim S.S., Posehn S.E., Jepson C., Kim S.Y., Wiedemann G., Reski R., Wee A.G., Douglas C.J., Suh D.Y. (2011). PpASCL, a moss ortholog of anther-specific chalcone synthase-like enzymes, is a hydroxyalkylpyrone synthase involved in an evolutionarily conserved sporopollenin biosynthesis pathway. New Phytol. 192: 855–868 [DOI] [PubMed] [Google Scholar]
- de Azevedo Souza C., Kim S.S., Koch S., Kienow L., Schneider K., McKim S.M., Haughn G.W., Kombrink E., Douglas C.J. (2009). A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mesa M.C., Santiago-Doménech N., Pliego-Alfaro F., Quesada M.A., Mercado J.A. (2004). The CaMV 35S promoter is highly active on floral organs and pollen of transgenic strawberry plants. Plant Cell Rep. 23: 32–38 [DOI] [PubMed] [Google Scholar]
- De Block M., Debrouwer D. (1992). In-situ enzyme histochemistry on plastic-embedded plant material. The development of an artefact-free β-glucuronidase assay. Plant J. 2: 261–266 [Google Scholar]
- Dickison W.C., Nowicke J.W., Skvarla J.J. (1982). Pollen morphology of the Dilleniaceae and Actinidiaceae. Am. J. Bot. 69: 1055–1073 [Google Scholar]
- Dobritsa A.A., Geanconteri A., Shrestha J., Carlson A., Kooyers N., Coerper D., Urbanczyk-Wochniak E., Bench B.J., Sumner L.W., Swanson R., Preuss D. (2011). A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiol. 157: 947–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Lei Z., Nishikawa S., Urbanczyk-Wochniak E., Huhman D.V., Preuss D., Sumner L.W. (2010). LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol. 153: 937–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Nishikawa S., Preuss D., Urbanczyk-Wochniak E., Sumner L.W., Hammond A., Carlson A.L., Swanson R.J. (2009b). LAP3, a novel plant protein required for pollen development, is essential for proper exine formation. Sex. Plant Reprod. 22: 167–177 [DOI] [PubMed] [Google Scholar]
- Dobritsa A.A., Shrestha J., Morant M., Pinot F., Matsuno M., Swanson R., Møller B.L., Preuss D. (2009a). CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H. (1890). Beiträge zur vergleichenden Morphologie der Pollenkörner. (Breslau, Germany: J. U. Kern's Verlag) [Google Scholar]
- Fritzsche J. (1837). Über den Pollen. (St. Petersburg, Germany: Academie der Wissenschaften) [Google Scholar]
- Furness C.A., Rudall P.J. (2004). Pollen aperture evolution—A crucial factor for eudicot success? Trends Plant Sci. 9: 154–158 [DOI] [PubMed] [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40(Database issue): D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grew N. (1682). The Anatomy of Flowers, Prosecuted with the Bare Eye, and the Microscope. (London: W. Rawlins) [Google Scholar]
- Grienenberger E., Kim S.S., Lallemand B., Geoffroy P., Heintz D., Souza Cde.A., Heitz T., Douglas C.J., Legrand M. (2010). Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22: 4067–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1979a). An interpretation of the hydrodynamics of pollen. Am. J. Bot. 66: 737–743 [Google Scholar]
- Heslop-Harrison J. (1979b). Pollen walls as adaptive systems. Ann. Miss. Bot. Gard. 66: 813–829 [Google Scholar]
- Honys D., Oh S.-A., Renák D., Donders M., Solcová B., Johnson J.A., Boudová R., Twell D. (2006). Identification of microspore-active promoters that allow targeted manipulation of gene expression at early stages of microgametogenesis in Arabidopsis. BMC Plant Biol. 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraška M., Rakoský S., Čurn V. (2008). Tracking of the CaMV-35S promoter performance in GFP transgenic tobacco, with a special emphasis on flowers and reproductive organs, confirmed its predominant activity in vascular tissues. Plant Cell Tiss. Org. Cult. 94: 239–251 [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katifori E., Alben S., Cerda E., Nelson D.R., Dumais J. (2010). Foldable structures and the natural design of pollen grains. Proc. Natl. Acad. Sci. USA 107: 7635–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavita P., Burma P.K. (2008). A comparative analysis of green fluorescent protein and β-glucuronidase protein-encoding genes as a reporter system for studying the temporal expression profiles of promoters. J. Biosci. 33: 337–343 [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J.E. (2009). Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kellogg E.A. (2001). Evolutionary history of the grasses. Plant Physiol. 125: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesseler R., Harley M. (2004). Pollen: The Hidden Sexuality of Flowers. (London: Papadakis) [Google Scholar]
- Kim S.S., et al. (2010). LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. (1978). A factor analysis study of the functional significance of angiosperm pollen. Syst. Bot. 3: 1–19 [Google Scholar]
- McCormick S. (1993). Male gametophyte development. Plant Cell 5: 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S., Yamaguchi J., Twell D. (1991). Deletion analysis of pollen-expressed promoters. In Vitro Cell. Dev. Biol. 27P: 15–20 [Google Scholar]
- Michelmore R.W., Paran I., Kesseli R.V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M., Jørgensen K., Schaller H., Pinot F., Møller B.L., Werck-Reichhart D., Bak S. (2007). CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J. (1979). Form and function in angiosperm pollen. Ann. Miss. Bot. Gard. 66: 593–632 [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. (2007). ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35(Database issue): D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M.S., Staehelin L.A. (2004). Electron tomographic analysis of post-meiotic cytokinesis during pollen development in Arabidopsis thaliana. Planta 218: 501–515 [DOI] [PubMed] [Google Scholar]
- Paxson-Sowders D.M., Dodrill C.H., Owen H.A., Makaroff C.A. (2001). DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol. 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Rhee S.Y., Davis R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Quilichini T.D., Friedmann M.C., Samuels A.L., Douglas C.J. (2010). ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol. 154: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressayre A., Mignot A., Siljak-Yakovlev S., Raquin C. (2003). Postmeiotic cytokinesis and pollen aperture number determination in eudicots: Effect of the cleavage wall number. Protoplasma 221: 257–268 [DOI] [PubMed] [Google Scholar]
- Rosario T., DeSimone D.W. (2011). The extracellular matrix in development and morphogenesis: A dynamic view. Dev. Biol. 341: 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.J. (1994). Pollen exine: The sporopollenin enigma and the physics of pattern. In Molecular and Cellular Aspects of Plant Reproduction, R.J. Scott and A.D. Stead, eds (Cambridge, UK: University Press), pp. 49–81 [Google Scholar]
- Szymanski D.B., Cosgrove D.J. (2009). Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr. Biol. 19: R800–R811 [DOI] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The Botany Array Resource: e-Northerns, expression angling, and promoter analyses. Plant J. 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Underwood W. (2012). The plant cell wall: A dynamic barrier against pathogen invasion. Front Plant Sci. 3: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Besser K., Frank A.C., Johnson M.A., Preuss D. (2006). Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133: 4761–4769 [DOI] [PubMed] [Google Scholar]
- Wiermann R., Ahlers F., Schmitz-Thom I. (2001). Sporopollenin. In Biopolymers, A. Stenbüchel and M. Hofrichter, eds (Weinheim, Germany: Wiley-VCH Verlag), pp. 209–227 [Google Scholar]
- Wodehouse R.P. (1935). Pollen Grains: Their Structure, Identification and Significance in Science and Medicine. (New York: McGraw-Hill) [Google Scholar]
- Yang C.Y., Spielman M., Coles J.P., Li Y., Ghelani S., Bourdon V., Brown R.C., Lemmon B.E., Scott R.J., Dickinson H.G. (2003). TETRASPORE encodes a kinesin required for male meiotic cytokinesis in Arabidopsis. Plant J. 34: 229–240 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Huang L., Liu T., Yu X., Cao J. (2008). Functional analysis of a pollen-expressed polygalacturonase gene BcMF6 in Chinese cabbage (Brassica campestris L. ssp. chinensis Makino). Plant Cell Rep. 27: 1207–1215 [DOI] [PubMed] [Google Scholar]