Analysis of the supramolecular organization of enzymes in the lignin pathway shows that cytochrome P450s oligomerize and move along with the very mobile plant endoplasmic reticulum. Their expression favors relocalization of their soluble partner proteins nearer the membrane and association of sequential enzymes in the pathway.
Abstract
Supramolecular organization of enzymes is proposed to orchestrate metabolic complexity and help channel intermediates in different pathways. Phenylpropanoid metabolism has to direct up to 30% of the carbon fixed by plants to the biosynthesis of lignin precursors. Effective coupling of the enzymes in the pathway thus seems to be required. Subcellular localization, mobility, protein–protein, and protein–membrane interactions of four consecutive enzymes around the main branch point leading to lignin precursors was investigated in leaf tissues of Nicotiana benthamiana and cells of Arabidopsis thaliana. CYP73A5 and CYP98A3, the two Arabidopsis cytochrome P450s (P450s) catalyzing para- and meta-hydroxylations of the phenolic ring of monolignols were found to colocalize in the endoplasmic reticulum (ER) and to form homo- and heteromers. They moved along with the fast remodeling plant ER, but their lateral diffusion on the ER surface was restricted, likely due to association with other ER proteins. The connecting soluble enzyme hydroxycinnamoyltransferase (HCT), was found partially associated with the ER. Both HCT and the 4-coumaroyl-CoA ligase relocalized closer to the membrane upon P450 expression. Fluorescence lifetime imaging microscopy supports P450 colocalization and interaction with the soluble proteins, enhanced by the expression of the partner proteins. Protein relocalization was further enhanced in tissues undergoing wound repair. CYP98A3 was the most effective in driving protein association.
INTRODUCTION
Metabolons have been defined as supramolecular complexes of sequential metabolic enzymes and cellular structural elements (Srere, 1985). This organization of metabolic pathways at the molecular level is expected to have several advantages, such as increasing local concentrations of the enzymes and their substrates, sequestering reactive toxic intermediates, and improving channeling of intermediates between consecutive enzymes (Jørgensen et al., 2005; Ralston and Yu, 2006).
Protein association was proposed to support the formation of the huge diversity of plant natural products by a limited number of enzymes, some of which have broad substrate specificity. Formation of a metabolon is expected to be controlled by the availability of its constituents to match developmental and environmental requirements. It might be further favored by a local membrane composition or local solvent environment that may favor protein and substrate colocalization via extraction from a cytosolic aqueous phase (Choi et al., 2011). According to the latter hypothesis, colocalization may result from the concentration of proteins in microdomains rather than from specific interactions.
In the case of plant secondary metabolism, membrane-bound proteins, in particular cytochrome P450 enzymes, were proposed to serve for nucleation of metabolons (Jørgensen et al., 2005; Ralston and Yu, 2006). P450 enzymes are usually anchored via their N terminus on the cytoplasmic surface of the endoplasmic reticulum (ER) with the main protein fold protruding on the surface of the membrane (Bayburt and Sligar, 2002). Mobility of P450s is thus restricted to two dimensions. They can be associated with membrane domains of specific lipid composition or membrane-structuring proteins. Recently, some evidence for the formation of drug-metabolizing complexes involving P450 enzymes in the mouse liver has been reported (Li et al., 2011). P450 anchoring of plant metabolons has also found support in flavonoid and cyanogenic glucoside metabolism (Winkel, 2004; Jørgensen et al., 2005), but the best documented example is at the entry of the phenylpropanoid pathway (reviewed in Ralston and Yu, 2006).
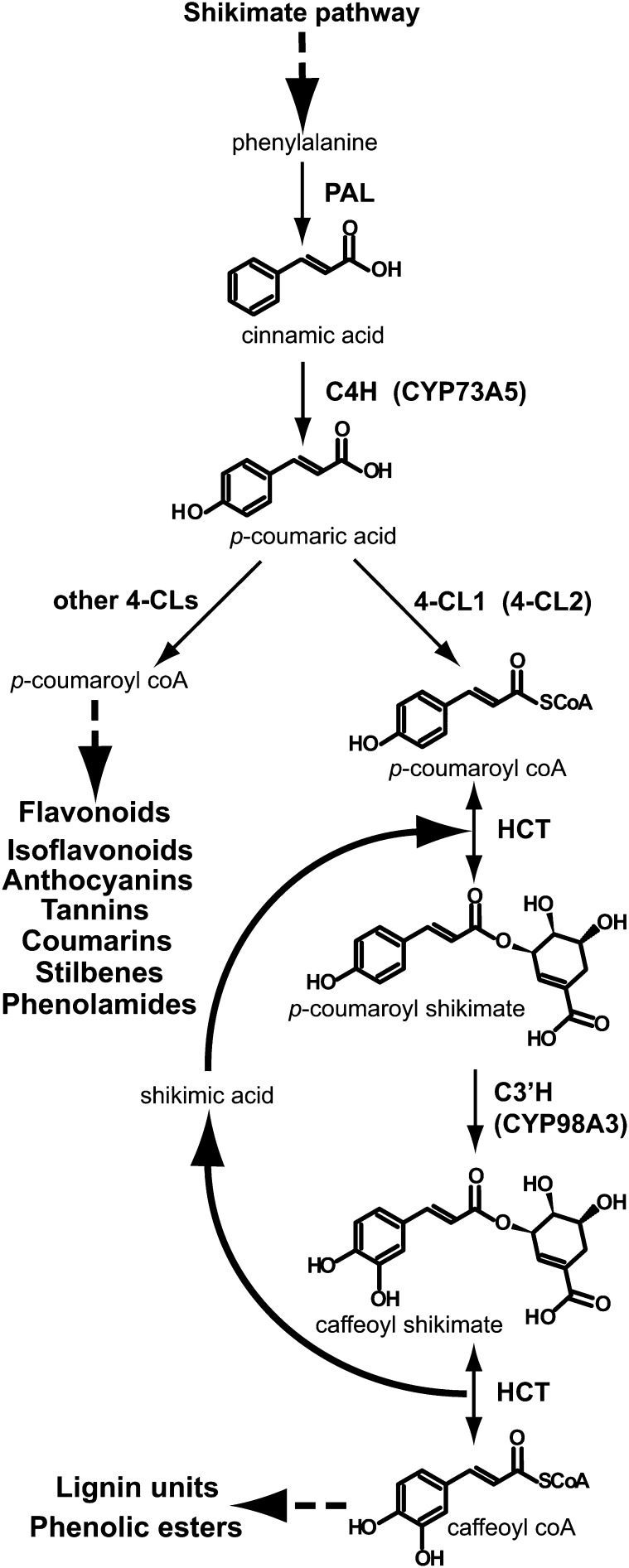
Phenylpropanoid metabolism (Figure 1) mobilizes up to 30% of the carbon fixed by photosynthesis for the synthesis of biopolymers in woody plants. It also provides precursors of soluble metabolites for UV screening, defense, and communication with other organisms. Such soluble compounds accumulate to high levels in leaf tissues (Vogt, 2010). Gene expression is tightly coordinated in each branch of the pathway. The upstream core segment involves one P450 and two soluble proteins, the Phe ammonia lyase (PAL) that converts Phe into cinnamic acid, the P450 cinnamate 4-hydroxylase (C4H), CYP73A5 in Arabidopsis thaliana that forms p-coumaric acid, and the 4-coumaric acid CoA ligase (4-CL) that generates 4-coumaroyl CoA. This activated CoA ester is then distributed into a number of branch pathways, leading to the formation of flavonoids, coumarins, phenolic esters, amides, and monolignols, to which the main flux is directed in lignified cells. In Arabidopsis, 4-CL is encoded by four paralogs (Hamberger and Hahlbrock, 2004; Costa et al., 2005). Given its strong expression in stem and root and high catalytic efficiency for conversion of p-coumaric acid into its CoA ester (Ehlting et al., 1999; Costa et al., 2005), 4-CL1 was proposed to be the best candidate to drive the flux of metabolites into monolignols. Nonetheless, contribution of other paralogs, in particular 4-CL2, cannot be excluded. CYP98A3 is the p-coumaroylshikimate 3′-hydroxylase (C3′H), with a more restricted commitment to lignin synthesis. Its direct upstream and downstream partner is the hydroxycinnamoylCoA:shikimate hydroxycinnamoyl transferase (HCT) that reversibly converts hydroxycinnamoyl CoA into hydroxycinnamoyl shikimate. Together, C3′H and HCT form a loop sequence at the branch point to lignin synthesis (Schoch et al., 2001; Hoffmann et al., 2004). The two enzymes seem to be tightly coupled since shikimate derivatives never accumulate to UV-detectable levels in wild-type nor in C3′H-null mutant plants (Schoch et al., 2006).
Figure 1.
Lignin Branch Point of Phenolic Metabolism.
Dashed arrows indicate multistep reactions.
The first evidence of metabolic channeling in the phenylpropanoid pathway was reported for the PAL-C4H couple. Czichi and Kindl (1975, 1977) detected a partial association of PAL with a purified microsomal fraction from potato tuber. This fraction was found to convert Phe more efficiently than exogenously supplied cinnamate into p-coumaric acid, which supported a channeling hypothesis. Further work differentiated the behavior of PAL1 and PAL2 in tobacco (Nicotiana tabacum), where PAL1, but not PAL2, was found to localize to the ER (Rasmussen and Dixon, 1999). In tobacco cell cultures and microsomal membranes, exogenous radiolabeled cinnamate did not equilibrate with the pool of cinnamate directly produced from PAL. Both green fluorescent protein (GFP)–tagged PALs were partially relocalized to the ER upon C4H overexpression (Achnine et al., 2004), and competition experiments supported stronger affinity of PAL1 for ER binding sites. Colocalization with C4H was confirmed by double immunolabeling and fluorescence resonance energy transfer (FRET), although data were indicative of loose interaction. PAL1 localization near the ER membrane was more recently confirmed in Nicotiana benthamiana (Bassard et al., 2012). Despite the fact that these data converge, channeling between poplar (Populus trichocarpa) PAL and C4H could not be confirmed in yeast (Ro and Douglas, 2004). 4-CL is expected to be the downstream partner of C4H. So far, the only experimental indication for the participation of 4-CL in a protein complex is the detection of activity in the membrane-associated enzyme aggregates isolated by gel filtration reported by Hrazdina and Wagner (1985). Recently, Chen et al. (2011) also reported interaction between poplar C4H and C3′H, which positively affects the activity of both proteins.
The aim of this work was to investigate protein–membrane and protein–protein association at the entry point of the lignin pathway with a main focus on Arabidopsis HCT and C3′H in a plant cellular environment. Our data point to fast P450 streaming with the ER membranes and hampered lateral diffusion on the surface of the ER. We show P450 aggregation into homomers and heteromers in vivo and binding to other membrane constituents, such as reticulons. Partial ER association and relocalization of the soluble proteins in the presence of P450 proteins is detected. Finally, our data indicate potential interaction of P450 and soluble proteins, enhanced by the other partner proteins. The CYP98A3/HCT couple appears most closely associated and seems to enhance further protein association.
RESULTS
CYP98A3 and CYP73A5 Are Mobile with and within the ER
Based on data from animal systems, P450s are pictured as proteins with free lateral mobility in a slowly remodeling ER (Szczesna-Skorupa et al., 1998). To test if this applies to plant enzymes, CYP73A5 and CYP98A3 were expressed as enhanced GFP (eGFP) fusion proteins in plant membranes by transfection of N. benthamiana leaves. Since these P450s are anchored to the ER through an N-terminal signal peptide, they were eGFP tagged at the C terminus. Both CYP73A5 and CYP98A3 behaved as typical ER proteins when compared with free and ER-retained eGFP (Figures 2A to 2H). Determination of C4H and C3′H activities in the microsomal membranes of transformed leaves indicated that the NADPH:cytochrome P450 reductase (CPR) from N. benthamiana successfully reduced the P450 enzymes from Arabidopsis. Based on relative activities detected in microsomes from leaves expressing the native and tagged proteins, eGFP tagging caused 57 and 65% reduction in enzyme activity for CYP73A5 and CYP98A3, respectively (see Supplemental Table 1 online). The presence of the eGFP tag at the C terminus is expected to produce a steric hindrance for P450 interaction with its electron donor CPR, since the C terminus and P450 regions involved in electron transfer (Sevrioukova et al., 1999; Jensen et al., 2011) are located on the same face of the P450 protein. However, the near 40% residual activity of the tagged enzymes indicates that the presence of the eGFP does not completely prevent the P450–CPR interaction.
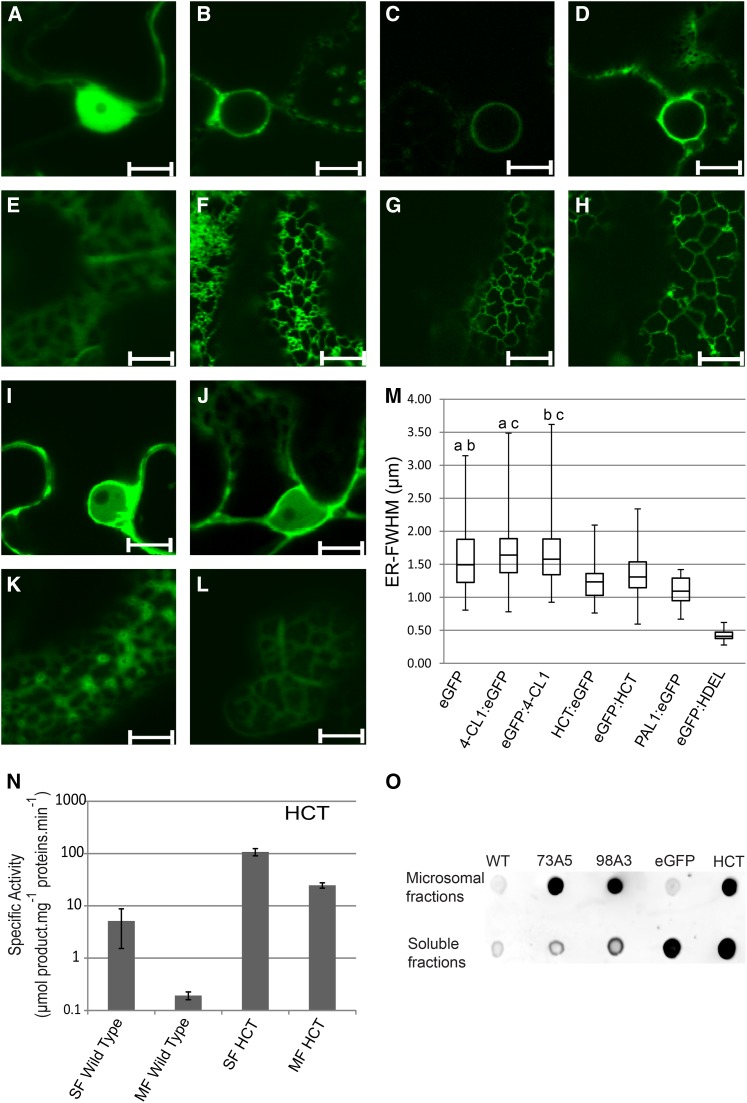
Figure 2.
Subcellular Localization of 4-CL1, HCT, CYP73A5, and CYP98A3.
Confocal images, microsomes, and soluble fractions were collected 5 d after agroinfiltration of the N. benthamiana leaves.
(A) to (L) Confocal Laser Scanning Microscopy (CLSM) images of the nucleus ([A] to [D], [I], and [J]) and parietal ER ([E] to [H], [K], and [L]) of leaves expressing eGFP ([A] and [E]), eGFP:HDEL ([B] and [F]), CYP73A5:eGFP ([C] and [G]), CYP98A3:eGFP ([D] and [H]), 4-CL1:eGFP ([I] and [K]), or HCT:eGFP ([J] and [L]). Soluble enzymes diffuse into the nucleus and fill in gaps between organelles and plasma membranes. ER proteins are confined to nuclear membranes and to a well-defined membrane network. 4-CL1:eGFP and HCT:eGFP localization usually appears less diffuse than eGFP on confocal images. Latrunculin B (20 µM) was used to stop the movement of the ER. Similar images were obtained with mRFP1 fusion constructs or N-terminal fluorescent fusion constructs for HCT and 4-CL1. Bars = 10 µm.
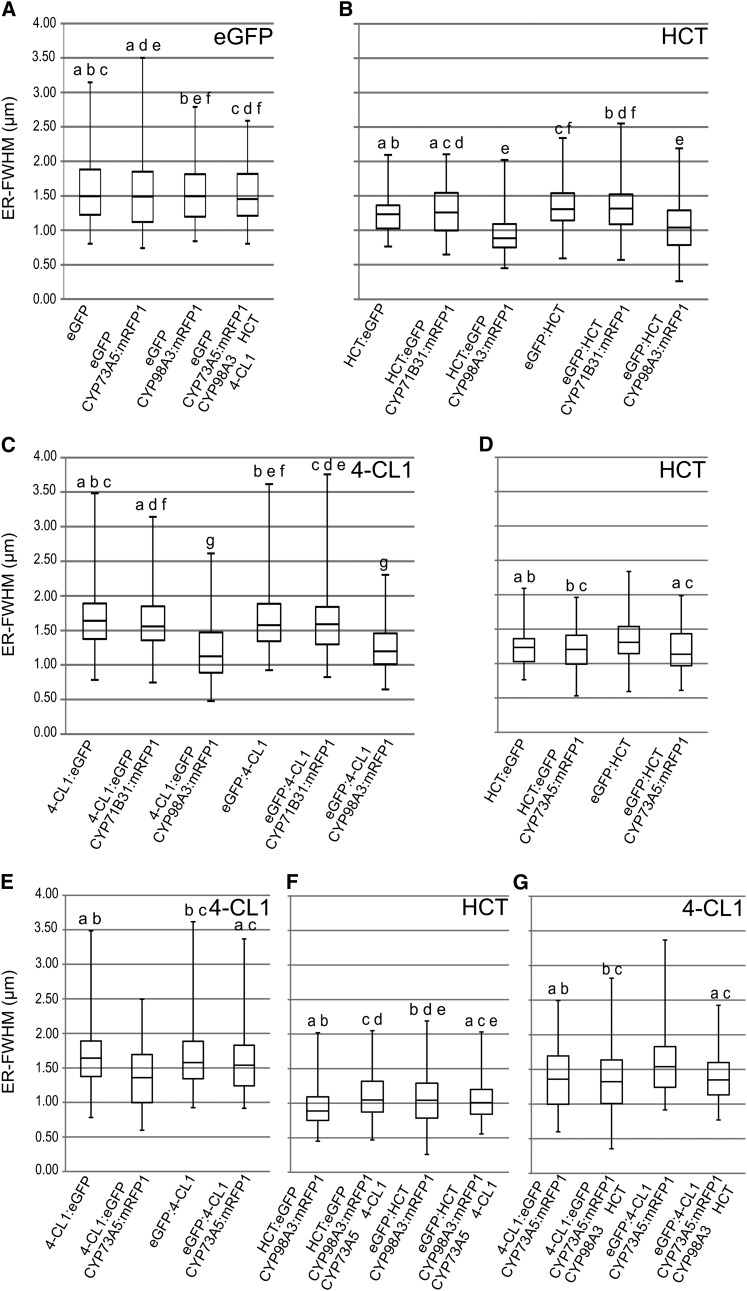
(M) Box plot representing the distribution of HCT, 4-CL1, eGFP, NtPAL1:eGFP, and eGFP:HDEL detected by CLSM around ER tubules evaluated by confocal microscopy based on confocal images such as those shown in (E), (F), (K), and (L). The box plot representation provides information on the distribution of a population of proteins near the membrane (median, maximum, and minimum distances, as well as first and third quartile). For each experimental condition, 100 measurements were randomly recorded from independent images. a, b, and c indicate pairs with similar distribution according to ANOVA analysis (see Supplemental Data Set 2 online). ER-FWHM, ER-full width at half maximum expressed in micrometers. The box plot shows the median distance and the protein located farther (above) or closer (below) to the membrane.
(N) HCT activities detected in the soluble fraction and associated with washed microsomal membranes. Specific activities are expressed in µmol of product per milligram of total proteins per minute. MF, microsomal fraction; SF, soluble fraction. Mean and sd (indicated by error bars) are determined from three independent experiments and three technical replicates. See Supplemental Table 1 online for details on eGFP-fusion enzyme activities.
(O) Dot blot of soluble fractions or microsomes from N. benthamiana plants expressing eGFP-tagged proteins detected by eGFP antibodies. WT, the wild type N. benthamiana (control); eGFP, eGFP alone (control); 73A5, CYP73A5:eGFP; 98A3, CYP98A3:eGFP; HCT, HCT:eGFP.
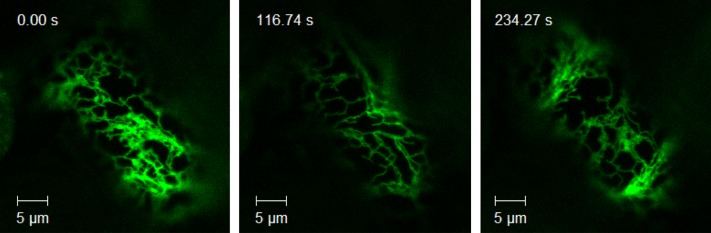
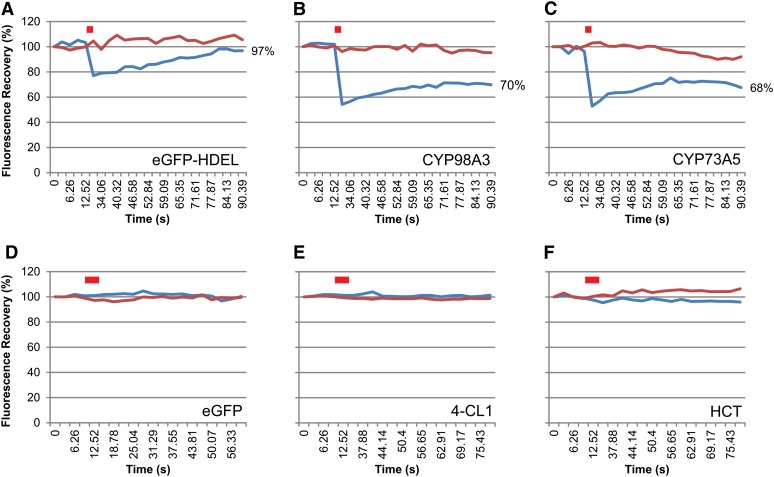
The plant ER is described as being very dynamic and constantly remodeling (Griffing, 2010; Sparkes et al., 2011). Time-lapse imaging of CYP73A5 and CYP98A3 as eGFP-fused proteins (see CYP98A3 in Figure 3 and Supplemental Movie 1 online) illustrates their fast movement in the plant cell together with the ER network. The samples were then treated with the actin-depolymerizing agent Latrunculin B that limits movement to diffusion but preserves the ER structure (Coué et al., 1987; Runions et al., 2006; Griffing, 2010). Lamellar ER regions were then selected for fluorescence recovery after photobleaching (FRAP) experiments to evaluate lateral protein diffusion on the surface of the ER. Figure 4 compares bleaching and recovery recorded with plant leaves expressing free and ER-retained eGFP, P450:eGFP, 4-CL1:eGFP, and HCT:eGFP constructs. Bleaching was close to 20%, and recovery was nearly complete within 90 s for eGFP-HDEL (Figure 4A). Bleaching was above 40% and recovery very low and limited for both P450:eGFP constructs (Figures 4B and 4C; see Supplemental Figure 1 online). No bleaching was observed for soluble free eGFP, nor for 4-CL1:eGFP and HCT:eGFP (Figures 4D to 4F). The lateral diffusion of both CYP98A3 and CYP73A5 enzymes on the surface of the ER appears thus restricted, either due to supramolecular organization or the anchoring on a membrane or cytoskeleton component.
Figure 3.
Mobility of CYP98A3 with the Plant ER.
Five days after agroinfiltration of N. benthamiana leaves with the CYP98A3:eGFP construct, movies of the GFP fluorescence were taken. Representative images from three times as indicated are shown. The full movie is available as Supplemental Movie 1 online. Movies showing the behavior of eGFP (soluble enzyme control) and eGFP:HDEL (ER protein control) are available for comparison as Supplemental Movies 2 and 3 online. Bar = 5 µm.
Figure 4.
FRAP.
FRAP experiments were performed 5 d after agroinfiltration of N. benthamiana leaves with eGFP-HDEL (or ER-anchored eGFP) (A), CYP98A3:eGFP (B), CYP73A5:eGFP (C), eGFP (D), 4-CL1:eGFP (E), and HCT:eGFP (F) constructs. Latrunculin B (20 µM) was used to stop the movement of the ER tubules. Red bar indicates bleaching. Blue and red lines represent fluorescence recorded in bleached and control areas, respectively, for one representative experiment. See Supplemental Figure 1 for details about FRAP statistics.
CYP98A3 and CYP73A5 Interact with Other ER-Resident Proteins
To investigate interaction of CYP98A3 and CYP73A5 with other membrane proteins, both of them were used as baits in tandem affinity purification (TAP) experiments performed with P450s that were C-terminally tagged with the protein G - Streptavidin (GS) tag (Bürckstümmer et al., 2006) and expressed in Arabidopsis cell suspension cultures (Van Leene et al., 2007). TAP tagging was performed in cells grown under standard conditions and under conditions inducing differentiation to tracheary elements and secondary wall deposition, which enhances expression of the lignin pathway (Oda et al., 2005). Using CYP98A3 as bait, CYP73A5 was copurified two times in three independent TAP experiments, and this occurred only under inducing conditions (Table 1; see Supplemental Data Set 1 online). Two other proteins, the P450 PHYTOALEXIN DEFICIENT3 (PAD3 or CYP71B15), reported to catalyze the final steps in camalexin synthesis (Schuhegger et al., 2006; Böttcher et al., 2009) and an esterase of unknown function (At5g22460) copurified with both CYP73A5 and CYP98A3 only under inducing conditions.
Table 1. Interactors of CYP73A5 and CYP98A3 Determined by TAP Screens.
| Baits | |||||
|---|---|---|---|---|---|
| Identified Proteins |
CYP73A5 |
CYP98A3 |
|||
| Accession No. | Prey Annotation | Induced | Noninduced | Induced | Noninduced |
| AT2G30490 | CYP73A5 | 0 | 0 | 2 | 0 |
| AT3G26830 | PAD3 | 1 | 0 | 3 | 0 |
| AT5G22460 | Esterase/lipase/thioesterase family protein | 1 | 0 | 3 | 0 |
| AT4G22710/AT4G22690* | CYP706A2/CYP706A1 | 1 | 2 | 2 | 1 |
| AT5G53560 | Cytochrome b5 isoform 1 | 3 | 2 | 1 | 1 |
| AT4G23630/AT4G11220* | Reticulon family protein (RTNLB1/RTLNB2) | 2 | 2 | 3 | 1 |
| AT1G64090 | Reticulon family protein (RTNLB3) | 0 | 1 | 2 | 1 |
| AT5G41600 | Reticulon family protein (RTNLB4) | 0 | 2 | 2 | 0 |
| AT2G46170 | Reticulon family protein (RTNLB5) | 0 | 1 | 2 | 0 |
| AT2G03510 | Band 7 family protein | 1 | 1 | 3 | 0 |
| AT1G51570 | C2 domain-containing protein | 2 | 0 | 1 | 2 |
| AT2G20990 | SYNAPTOTAGMIN A (SYTA) | 0 | 1 | 0 | 2 |
| AT1G20330 | SMT2 | 3 | 2 | 3 | 2 |
| AT5G52240 | MSBP1 | 2 | 0 | 0 | 0 |
| AT3G48890 | MSBP2 | 2 | 1 | 0 | 0 |
| AT2G45140 | Vesicle-associated membrane protein (PVA12) | 1 | 1 | 0 | 0 |
| Repeats | 3 | 2 | 3 | 2 | |
The left column shows all the proteins (accession no. and annotation) that copurified more than once with CYP73A5:GS and CYP98A3:GS expressed in Arabidopsis cell suspension cultures (PSB-D), either under induced or noninduced conditions for tracheary element and secondary wall formation. Numbers within the table indicate the number of positive results for each combination of prey/bait in three (for the induced condition) or two (for the noninduced condition) independent TAP experiments, respectively. Asterisk indicates that mass spectrometry cannot distinguish between these two isoforms because no specific peptides were generated. See Supplemental Data Set 1 online for MS identification of CYP73A5 and CYP98A3 interactors.
Furthermore, the NADH-cytochrome b5 reductase (At5g53560), another P450, CYP706A1 (and/or its very close paralog CYP706A2), with as yet unknown activity, as well as a number of structural proteins copurified with both CYP73A5 and CYP98A3, but independent from the induction conditions. Among them were several members of the reticulon family that were shown to contribute to ER tubule shaping and to interact with ER resident proteins (Sparkes et al., 2010, 2011). Other CYP73A5/98A3 ligands included a band7 family protein that is the Arabidopsis homolog of the ER lipid raft-associated proteins (erlins) found associated with high molecular weight protein complexes in mammalian cells (Browman et al., 2006; Hoegg et al., 2009) and two proteins containing a C2-domain (At1g51570 and SYTA) that is thought to be involved in calcium-dependent phospholipid binding and in membrane targeting processes (Davletov and Südhof, 1993). Finally, MEMBRANE STEROID BINDING PROTEIN1 (MSBP1) and MSBP2, a plant VAMP-associated protein (PVA12), and STEROL C24-METHYL TRANSFERASE2 (SMT2) interacted with both CYPs in all repeats and conditions. MSBP1 and MSBP2 can bind to progesterone, brassinolide, and stigmasterol with different affinities and presumably have a role in steroid signaling (Yang et al., 2005). PVA12 was demonstrated to be important for the ER localization of sterol binding proteins (Saravanan et al., 2009). SMT2 expression impacts sterol composition of the membrane (Schaeffer et al., 2001) and is associated with vascular patterning (Carland et al., 2002). Erg6p, the yeast homolog of SMT2, has been shown to belong to an ER complex of sterol biosynthetic enzymes in Saccharomyces cerevisiae (Mo et al., 2004; Mo and Bard, 2005).
A similar TAP experiment using as bait an unrelated P450 enzyme of unknown function (At1g13710; CYP78A5) did not identify any of the interactors in Table 1. Such a robust set of proteins interacting with both CYP73A5 and CYP98A3 thus supports the hypothesis of a specific membrane-anchoring complex limiting P450 mobility on the membrane surface.
HCT Is Partially Associated with the ER Membranes
4-CL1 and HCT are described as soluble proteins (Ehlting et al., 2001; Hoffmann et al., 2004). Their subcellular localization was compared with that of their membrane-bound partner P450s CYP73A5 and CYP98A3 in transfected N. benthamiana. 4-CL1 and HCT were expressed as both N-terminal and C-terminal eGFP fusions to allow all possible interactions with the membranes and membrane-bound proteins. eGFP tagging of the 4-CL1 or HCT did not hamper their catalytic activities indicating that the fusion proteins were functional (see Supplemental Table 1 online). Solubility of 4-CL1 and HCT (Figures 2I and 2J) was attested by their transport into the nucleus and further supported by high mobility observed in FRAP analysis (Figures 4E and 4F). Free diffusion of both enzymes is comparable to that of free eGFP and too fast for detectable photobleaching. However, both 4-CL1 and HCT generate a fuzzy reticulate pattern in the parietal regions of the cell (Figures 2K and 2L). This reticulate pattern is due to the squeezing of the cytoplasm between the turgescent vacuole and the cell wall but might also be indicative of a partial association of the soluble enzymes with the ER as previously reported for tobacco PAL1 (Bassard et al., 2012). We thus analyzed the distribution of the eGFP-tagged 4-CL1 and HCT proteins near the ER tubules and compared it to the distribution of the free eGFP, eGFP fused to the HDEL ER retention signal, and to NtPAL1:eGFP by image analysis of fluorescence as recently described by Bassard et al. (2012). The statistical analysis of the fluorescent pixels around the ER tubules indicated a similar distribution for eGFP and 4-CL1, whereas HCT was found more closely associated with the ER (Figure 2M).
To confirm membrane association of HCT, soluble fractions and washed microsomal membranes were prepared from leaves of N. benthamiana expressing the soluble tag-free protein before testing enzyme activity. Significant activity was found associated with the microsome fraction (Figure 2N). HCT activity was also detected in washed microsomes isolated from control noninfiltrated plants. Membrane association was further confirmed by immunodetection of GFP in the membrane fraction prepared after transient expression of the eGFP fusion protein (Figure 2O).
Taken together, our data indicate partial association of HCT with the ER membranes, which is reminiscent of what was previously reported for PAL1 (Rasmussen and Dixon, 1999; Achnine et al., 2004; Bassard et al., 2012). This association can be detected with isolated membranes and in vivo.
Expression of CYP98A3 Redistributes HCT and 4-CL Closer to the ER
To test if P450 proteins may influence membrane association of their soluble partners, we evaluated the impact of P450 expression on the localization of HCT and 4-CL1. In a first step, each soluble protein was coexpressed with each P450. Soluble proteins were tagged with eGFP and P450s with monomeric Red Fluorescent Protein 1 (mRFP1) for confirmation of their expression in the cells selected for measurement. After optimization in standard experiments, visual assessment indicated that nearly 100% of the epidermal cells expressed the fluorescent proteins.
Coexpression with CYP98A3 triggered a significant relocalization of both HCT and 4-CL1 nearer to the ER, independent of the position of the tag (Figures 5A to 5C). CYP98A3 did not influence the distribution of free eGFP, and expression of CYP71B31, a P450 enzyme involved in isoprenoid metabolism (Ginglinger, 2010), did not modify the distribution of both HCT and 4-CL1. Localization of the soluble proteins was little affected by the coexpression of CYP73A5, with redistribution of 4-CL1 closer to the ER detected only when eGFP was fused at its C terminus (Figures 5D and 5E). We next tested the effect of addition of the two additional partner enzymes, the latter expressed without tags to keep the steric interference of the fluorescent tags minimal (Figures 5F and 5G). Significant redistribution in the presence of CYP98A3 and HCT was observed only in the case of eGFP:4-CL1, which was not pulled closer to the ER by CYP73A5:mRFP1 expressed alone (Figure 5G). No further change in protein distribution was detected when 4-CL1 or HCT were already drawn closer to the membrane by the expression of a single P450 protein.
Figure 5.
Distribution of HCT and 4-CL1 around ER Tubules Detected by CLSM.
Confocal images were recorded 5 d after agroinfiltration of the N. benthamiana leaves. Box plots are shown. For each experimental condition, 100 measurements were randomly recorded from independent images. a, b, c, d, e, and f indicate pairs with similar distribution according to ANOVA analysis (see Supplemental Data Set 2 online). ER-FWHM, ER-full width at half maximum expressed in micrometers. See Supplemental Figure 4 online for dot blot confirmation of protein expression and Supplemental Figure 5 online for comparison of the expression of the CYP71B31, CYP98A3, and CYP73A5 constructs.
(A) Distribution of eGFP (control).
(B) Distribution of HCT coexpressed with CYP98A3 or CYP71B31 (negative control).
(C) Distribution of 4-CL1 coexpressed with CYP98A3 or CYP71B31 (negative control).
(D) Distribution of HCT coexpressed with CYP73A5.
(E) Distribution of 4-CL1 coexpressed with CYP73A5.
(F) Distribution of HCT when coexpressed with the three other enzymes.
(G) Distribution of 4-CL1 when coexpressed with the three other enzymes.
CYP98A3 expression thus seems to drive redistribution of HCT and 4-CL1 near the ER membrane, while CYP73A5 only has a minor effect.
CYP98A3 and CYP73A5 Associate to Form Homo- and Heteromers
Supramolecular organization of the lignin pathway would imply protein–protein interaction or at least local concentration of the partners involved. Indeed, TAP-tagging experiments performed with CYP98A3 led to repeated copurification of CYP73A5 (Table 1). To further corroborate protein interaction or association, the different target proteins were coexpressed as fusion constructs with different fluorescent proteins suitable for fluorescence lifetime imaging microscopy (FLIM).
First we investigated the P450-P450 self- and cross-interactions. FRET detected upon expression of different P450 combinations was compared with FRET obtained upon P450 expression with free mRFP and mRFP anchored into the cytoplasmic surface of ER via the N-terminal signal peptide of the plant sterol 14α-demethylase CYP51G1 (51-mRFP) as a negative control (Bassard et al., 2012).
Background FRET values (between 3.6 and 5.5%) were detected between soluble free mRFP and P450:eGFP proteins (Table 2). FRET between anchored 51-mRFP and P450s yielded comparable values (between 0 and 2.7%). By contrast, a high FRET value of 22.2% between eGFP- and mRFP-tagged CYP98A3s indicated a high propensity to dimerize or oligomerize. The FRET value was also significant although lower (9.3%) upon coexpression of CYP73A5:eGFP and CYP73A5:mRFP. Tests for cross-interactions between CYP73 and CYP98 provided FRET values ranging from 9.1 to 17.2% depending on the donor and acceptor protein, which most likely reflects a variable environment caused by the fusion with the fluorescent probes. As an additional negative control, we determined the FRET values resulting from the coexpression of CYP73A5 and CYP98A3 with CYP71B31, a P450 enzyme involved in isoprenoid metabolism (Ginglinger, 2010), which never exceeded 6%.
Table 2. Measures of Fluorescence Lifetime of P450:eGFP and FRET Evaluation of P450:eGFP/P450:mRFP1 Associations.
| Expressed Proteins |
Fluorescence Lifetime (ns) |
FRET Efficiency |
||||
|---|---|---|---|---|---|---|
| Donor | Acceptor | n | Average | sd | % | sd |
| CYP73A5:eGFP | / | 121 | 2.05 | 0.09 | – | – |
| mRFP1 | 30 | 1.97 | 0.08 | 3.6 | 0.2 | |
| Anchored-mRFP1 | 20 | 1.99 | 0.31 | 2.7 | 0.4 | |
| CYP71B31:mRFP1 | 17 | 1.96 | 0.07 | 4.5 | 0.3 | |
| CYP98A3:mRFP1 | 22 | 1.70 | 0.17 | 17.2 | 1.8 | |
| CYP73A5:mRFP1 | 26 | 1.86 | 0.07 | 9.3 | 0.5 | |
| CYP98A3:eGFP | / | 112 | 2.12 | 0.15 | – | – |
| mRFP1 | 17 | 2.00 | 0.14 | 5.5 | 0.5 | |
| Anchored-mRFP1 | 18 | 2.13 | 0.07 | 0.0 | 0.0 | |
| CYP71B31:mRFP1 | 8 | 2.13 | 0.23 | 0.0 | 0.1 | |
| CYP98A3:mRFP1 | 20 | 1.65 | 0.09 | 22.2 | 2.0 | |
| CYP73A5:mRFP1 | 26 | 1.92 | 0.12 | 9.1 | 0.9 | |
| CYP71B31:eGFP | / | 11 | 2.33 | 0.16 | – | – |
| CYP98A3:mRFP1 | 10 | 2.22 | 0.13 | 4.4 | 0.4 | |
| CYP73A5:mRFP1 | 8 | 2.18 | 0.12 | 6.4 | 0.6 | |
Values of P450:eGFP fluorescence lifetimes were obtained by FLIM. FRET efficiency is calculated by comparison with eGFP fluorescence lifetime measured in a control experiment in absence of the mRFP1 acceptor protein. n, number of independent cells analyzed. Slash indicates no acceptor; dash indicates no value; bold value indicates value higher than controls.
Taken together, our data support the formation of homo- and heteromers (dimers or higher order oligomers) of CYP98A3 and CYP73A5. Tighter protein interaction is observed with CYP98A3, alone or in combination with CYP73A5.
P450 Association with Soluble Partners Is Enhanced by Coexpression of More Partner Enzymes
The association of CYP98A3 and CYP73A5 with HCT and 4-CL1 was tested in the presence or absence of the other two partner enzymes (Table 3). FLIM analysis was first performed to investigate interaction of CYP73A5 (eGFP at the C terminus) with 4-CL1 and HCT (mRFP at the N or C terminus). Low FRET values (0 to 5.3%) were indicative of no or poor interaction of CYP73A5 with 4-CL1 or HCT (Table 3). However, association of CYP73A5 with HCT was significantly increased upon coexpression of CYP98A3 and 4-CL1, with FRET values reaching 8.6 to 10.3%.
Table 3. Measures of Fluorescence Lifetime of P450:eGFP and FRET Evaluation of Associations between P450:eGFP and Soluble Enzymes:mRFP1.
| Expressed Proteins |
Fluorescence Lifetime (ns) |
FRET Efficiency |
|||||
|---|---|---|---|---|---|---|---|
| Donor | Acceptor | n | Untagged Proteins | Average | sd | % | sd |
| CYP73A5:eGFP |
/ | / | 121 | 2.05 | 0.09 | – | – |
| mRFP1 | / | 30 | 1.97 | 0.08 | 3.6 | 0.2 | |
| mRFP1 | CYP98A3/HCT /4-CL1 | 10 | 2.20 | 0.08 | 0.0 | 0.4 | |
| mRFP1:4-CL1 | / | 17 | 2.10 | 0.05 | 0.0 | 0.1 | |
| CYP98A3/HCT | 20 | 1.93 | 0.07 | 5.7 | 0.3 | ||
| 4-CL1:mRFP1 | / | 41 | 1.94 | 0.07 | 5.3 | 0.3 | |
| CYP98A3/HCT | 16 | 2.02 | 0.05 | 1.4 | 0.1 | ||
| HCT:mRFP1 | / | 17 | 1.99 | 0.03 | 2.9 | 0.1 | |
| CYP98A3/4-CL1 | 17 | 1.88 | 0.06 | 8.6 | 0.5 | ||
| mRFP1:HCT | / | 17 | 2.06 | 0.07 | 0.0 | 0.0 | |
| CYP98A3/4-CL1 | 17 | 1.84 | 0.07 | 10.3 | 0.6 | ||
| CYP98A3:eGFP | / | / | 112 | 2.12 | 0.15 | – | – |
| mRFP1 | / | 17 | 2.00 | 0.14 | 5.5 | 0.5 | |
| mRFP1 | CYP73A5/HCT/4-CL1 | 11 | 2.33 | 0.08 | 0.0 | 0.8 | |
| HCT:mRFP1 | / | 25 | 2.18 | 0.12 | 0.0 | 0.3 | |
| CYP73A5/4-CL1 | 15 | 1.73 | 0.09 | 18.2 | 1.6 | ||
| mRFP1:HCT | / | 26 | 1.90 | 0.12 | 10.3 | 1.0 | |
| CYP73A5/4-CL1 | 22 | 1.89 | 0.12 | 10.5 | 1.0 | ||
| 4-CL1:mRFP1 | / | 37 | 2.06 | 0.14 | 2.7 | 0.3 | |
| CYP73A5/HCT | 26 | 1.92 | 0.16 | 9.4 | 1.0 | ||
| mRFP1:4-CL1 | / | 27 | 2.06 | 0.19 | 2.4 | 0.3 | |
| CYP73A5/HCT | 22 | 1.86 | 0.21 | 12.2 | 1.6 | ||
Values of P450:eGFP fluorescence lifetimes were obtained by FLIM. FRET efficiency is calculated by comparison with eGFP fluorescence lifetime measured in a control experiment in absence of the mRFP1 acceptor protein. To determine the contribution of the different partners to the interaction between each P450 and soluble protein, the other proteins were expressed without fluorescent tags. n, number of independent cells analyzed. Slash indicates no acceptor; dash indicates no value; bold value indicates value higher than controls. See Supplemental Table 2 online for FLIM measured between soluble proteins and Supplemental Figure 4 online for dot blot confirmation of protein expression.
A similar FLIM analysis was performed to test CYP98A3 association with HCT or 4-CL1. FRET values of 10.3% indicated proximity of HCT to CYP98A3 in the absence of the other partner enzymes. This association seemed to involve specific regions of the proteins since it occurred only with the N-terminal tagged mRFP:HCT. In the latter configuration, interaction was not further enhanced by the coexpression of CYP73A5 and 4-CL1. Conversely, interaction of CYP98A3 with the C-terminal tagged HCT:mRFP required the coexpression of partner protein(s) that favored close proximity of the eGFP and mRFP labels in the complex as indicated by a high FRET value of 18.2%. No interaction of CYP98A3 with 4-CL1 was detected in the absence of CYP73A5 and HCT. However, coexpression of the latter proteins drove CYP98A3/4-CL1 association, independent of the position of the mRFP-tag on 4-CL1.
Finally, we tested if it was possible to detect 4-CL1 and HCT association (see Supplemental Table 2 online). Background FRET values between 0 and 2.3% were measured between the soluble proteins with all possible combinations of fusions with fluorescent probes. Upon coexpression of the P450 enzymes, an average FRET of 6.2% was detected with the 4-CL1:eGFP and HCT:mRFP combination. While this suggests possible spatial reorganization and closer proximity of the soluble proteins in the presence of partner P450s, it remains too low to conclude. However, values reached close to 20% in a small number of cells where relocalization was clearly visible in confocal images. The factor (cell type and stress) responsible for this conditional interaction could not be identified from the inspection of the leaf tissues.
Local Wounding Enhances Protein Association
The conditional interactions observed in the differential TAP tagging of the P450 membrane proteins upon induced lignification in plant cells (Table 1) raised the question whether protein interaction or association can be influenced by the physiological status of the plant and/or the metabolic flux in the phenylpropanoid pathway.
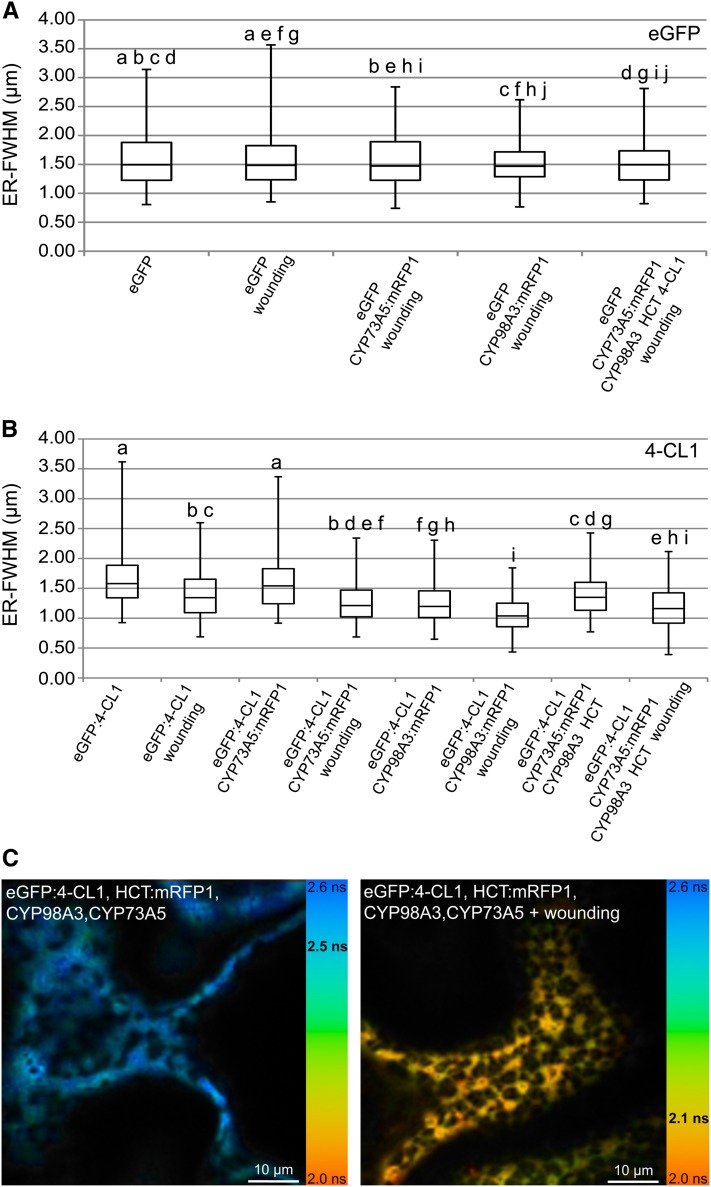
Mechanical injury is known to activate the expression of the genes of the phenolic pathway to support lignin and suberin deposition in the wound periderm near the wound site (Hawkins and Boudet, 1996; Moura et al., 2010), and activated expression of CYP73A5 and CYP98A3 around wounded leaf tissues was reported in Arabidopsis (Nair et al., 2002). Injury might thus provide a natural context favoring protein association. Hence, we investigated enzyme localization in the cell layers located in a wound healing zone. Transfected leaves of N. benthamiana were injured with a nail board 3 h before evaluation of protein relocalization in the healing cell layer (see Supplemental Figure 2A online). No wounding effect was detected with eGFP used as negative control (Figure 6A). Relocalization was observed with eGFP:4-CL1 expressed alone or coexpressed with CYP73A5, CYP98A3, or with a combination of CYP98A3, CYP73A5, and HCT (Figure 6B). Relocalization of HCT was detected only when HCT:eGFP was coexpressed with CYP73A5 (see Supplemental Figure 2B online).
Figure 6.
Effect of Wounding on Fluorescence Lifetime Values and Relocalization of Soluble Proteins.
(A) Box plot comparing distribution of eGFP (control) in the wound-healing zone upon coexpression of proteins of interest.
(B) Box plot comparing distribution of 4-CL1 in the wound-healing zone upon coexpression of partner proteins.
For each experimental condition, 100 measurements were randomly recorded from independent images. a, b, c, d, e, f, g, h, i, and j indicate pairs significantly similar according to ANOVA analysis. See Supplemental Data Set 2 online for details about ANOVA results. ER-FWHM, ER-full width at half maximum expressed in micrometers. The transfected leaves were wounded with a nail board 3 h before acquisition of the images in the healing zone.
(C) Effect of wounding on protein–protein interactions. Images were recorded 5 d after agroinfiltration of the N. benthamiana leaves. Fluorescence lifetime images acquired by FLIM are pseudo-colorized according to the color code ranging from 2.0 ns (orange) to 2.6 ns (light blue). The mean lifetime value measured in the image is indicated in bold black on the color scale. Bars = 10 µm.
See Supplemental Figure 2 online for details about relocalization analysis procedure, HCT relocalization, and FLIM after wounding.
FLIM analysis was then performed to determine if this resulted in enhanced protein association (see Supplemental Figure 2C online). Wounding did not result in significant changes in FRET values upon expression of eGFP-tagged P450s in any combination with other potential partners. By contrast, a clear wound effect was observed on the 4-CL1/HCT combinations with an average FRET value of 12.0% measured in the healing zone compared with 2.1% in healthy tissues. As already mentioned above, relocalization of the soluble proteins was visible in only a fraction of the cells when all four enzymes were present (Figure 6C). This fraction increased to 50% (instead of 20%) of the cells in the wound periphery. FRET values reached 19.5% between fluorescent 4-CL and HCT in these cells (see Supplemental Figure 2C online). The wound-healing context thus seems to favor association of 4-CL1 and HCT in the presence of CYP73A5 and CYP98A3.
DISCUSSION
This work provides an extensive analysis of the localization and dynamic association of four sequential enzymes around the first branch point in phenylpropanoid metabolism. Our investigation was performed in a natural plant context to provide the best possible membrane lipid and protein environment and to ensure the presence of metabolites at physiological concentrations, since these factors are likely to influence protein-membrane organization. An optimal context would be provided by vascular tissues where all relevant metabolites and partner proteins would be found. Vascular cells are currently not accessible for confocal live imaging. This analysis was thus performed on epidermal tissues. Whereas their protein and metabolite contents do not perfectly match the content of actively lignifying tissues, the overexpression of rate-limiting P450 enzymes of the monolignol pathway likely modify cellular homeostasis in a favorable way for our analysis. Wounding and subsequent cell repair provide a further contextual improvement. To limit the bias potentially resulting from protein overexpression, the analyses were performed at the early stages of protein expression, before the appearance of vesicular material and detectable modifications of the ER membranes.
P450s Move with the Fast Remodeling Plant ER but Their Lateral Mobility Is Restricted
In contrast with the animal cell system in which CYP2C2 was shown to be freely mobile throughout the ER (Szczesna-Skorupa et al., 1998), the plant CYP73A5 and CYP98A3 appeared to be fast streaming together with the highly dynamic plant ER, while their lateral diffusion on the surface of the membrane was restricted. The mobility of the plant P450 enzymes thus seems to essentially rely on the actin/myosin cytoskeleton and on the high ER plasticity, which was recently described to be a remarkable characteristic of plant cells (Sparkes et al., 2009). Fast streaming of the plant ER provides an opportunity for accelerated exchanges between membrane-anchored enzymes and the cytosolic proteins and metabolites. It may thus be an important factor to enhance plant metabolism.
An explanation for the restricted lateral mobility on the membrane surface could be direct or indirect anchoring to a cytoskeleton-associated element or their association with larger membrane structures. Both CYP73A5 and CYP98A3 interacted in TAP-tagging experiments with several ER-resident proteins, such as reticulons that contribute to ER shaping and to the formation of high molecular weight protein complexes (Table 1). In a similar way, the mammalian CYP2C2 was shown to interact with the ubiquitous BAP31 integral membrane protein (Szczesna-Skorupa and Kemper, 2006) reported to associate with the cytoskeletal components actin and myosin (Ducret et al., 2003). BAP31 was shown to be important to maintain expression levels of CYP2C2 and to support its ER retention (Szczesna-Skorupa and Kemper, 2006). In animal cells, however, this CYP2C2–BAP31 association did not seem to restrict the P450 mobility. More recently, tagged P450 protein complexes were isolated from mouse liver that, in addition to soluble drug-metabolizing enzymes, contained chaperones and reticulons (Li et al., 2011). The latter are integral ER proteins reported to shape cortical ER tubules and proposed to contribute to protein retention and anchoring into the ER and cytoskeleton (Sparkes et al., 2009; Nziengui and Schoefs, 2009; Tolley et al., 2010).
Another factor potentially restricting CYP73A5 and CYP98A3 mobility could be their involvement in large homo/heteropolymers and/or in more complex supramolecular structures. Such multimeric complexes could have reduced mobility simply because of their size but could, in addition, be associated with membrane microdomains, or membrane protein(s). Strikingly, a large number of the CYP73A5/CYP98A3 interactors identified by TAP (Table 1) are related in some way to lipids or sterols. This suggests that lipid microenvironment could be an important factor of protein association and activity. A small transmembrane protein Erg28p was reported to interact with several enzymes involved in sterol biosynthesis, including CYP51 (or Erg11p), catalyzing sterol 14α-demethylation, and the homolog of plant SMT2 sterol C-24 methyltransferase, in yeast. The Erg28p protein with no reported catalytic activity was proposed to tether a large complex of yeast sterol biosynthetic enzymes (Mo and Bard, 2005). No plant homolog to Erg28p has been reported so far, but several of the proteins identified by TAP might play a role similar to Erg28p and attempts to reveal protein complexes using blue native gel blots of microsomes isolated from plant leaves transfected with CYP98A3 revealed the presence of the CYP98A3 protein in several protein aggregates with molecular masses ranging up to 700 kD (see Supplemental Figure 3 online), which seems to confirm tight association with other membrane proteins.
Why Do P450s Form Oligomers?
In support of the existence of multimeric protein organization, our FLIM analyses clearly indicate that CYP98A3 self-associates to form homodimers or oligomers. Lower FRET values observed for CYP73A5 seem to indicate looser self-association. Both TAP tagging and FRET concur to show that CYP73A5 and CYP98A3 also interact to form heteromers. Conversely, selectivity of TAP tagging and poor interaction with a control fluorescent probe anchored on the cytoplasmic surface of the ER via the N-terminal transmembrane helix of the sterol 14α-demethylase CYP51G1 or with the ER-localized monoterpene-metabolizing CYP71B31 indicate that this interaction is specific and does not just result from a crowding effect.
P450 aggregation has been mainly documented for mammalian enzymes, and the formation of homo- and heterooligomers was detected in solution, in reconstituted systems in vitro (Myasoedova and Berndt, 1990; Cawley et al., 2001; Davydov et al., 2005; Hazai and Kupfer, 2005; Subramanian et al., 2010; Reed and Backes, 2012), but also in natural membranes or living cells (Backes and Kelley, 2003; Szczesna-Skorupa et al., 2003; Ozalp et al., 2005; Praporski et al., 2009; Hu et al., 2010). Furthermore, P450 proteins, such as human CYP2B4 and CYP2C8, crystallized as homodimers (Scott et al., 2003; Schoch et al., 2004). The size of the reported oligomers ranges from dimers up to 40-mers (Backes and Kelley, 2003). Oligomers have been reported to be more active than monomers. Homo- and heterotypic interaction seems P450 specific, selectively modulates P450 activities, and was shown in many instances to be mediated by the membrane-anchored hydrophobic N terminus (Szczesna-Skorupa et al., 2003; Ozalp et al., 2005; Subramanian et al., 2010). It is interesting to note that our TAP-tagging experiments not only detected CYP98A3–CYP73A5 interaction, but also that the orphan CYP706A1/2 bound both CYP73A5 and CYP98A3 under induced and noninduced conditions, while CYP71B15, in the indole pathway, bound to them only under induced conditions. This observation might give a hint to the function(s) of CYP706A1/2 (phylogenetically most closely related to the flavonoid-metabolizing CYP75) and might point to some form of crosstalk between lignin and indole-derived metabolism.
The impact of multimerization on CYP73/CYP98 enzyme activity could not be properly evaluated in the context of plant tissues with background activity but, during the preparation of this article, Chen et al. (2011) reported heteromerization of the poplar homologs of CYP73A5 and CYP98A3 that led to a drastic enhancement of their catalytic functions in yeast microsomes. However, in that report, the activities of the poplar enzymes measured in the absence of each partner P450 were very low, and those observed upon coexpression of both enzymes were closer to those described for Arabidopsis CYP73A5 and CYP98A3 individually expressed in yeast (Schalk et al., 1997; Schoch et al., 2001). Heteromerization thus seems to be less critical for activation of Arabidopsis CYP73A5 and CYP98A3.
In addition to the potential allosteric effect, P450 association in oligomers would increase the local concentration of P450 enzymes and of their products. This might be required to match metabolic demand and to feed soluble enzymes since P450s usually catalyze the slow steps in the pathway. The work of Davydov et al. (2000), based on high-pressure spectroscopy, suggested that P450 aggregation might contribute to their stability. Interestingly, in the case of human CYP2C8, it has been shown that the formation of homodimers relies on both signal anchor and F-G loop interfaces (Hu et al., 2010). In such a model, the monomers must be tilted so that the F-G loop is no longer in the membrane and the surface of contact with the ER is decreased. In the case of CYP73s and especially of CYP98s with very hydrophilic substrates, dimerization may thus favor substrate access to the active site. P450 activity in addition requires electrons provided by CPR also anchored via its N terminus in the ER. P450s are usually present in 10- to 20-fold excess over CPR (Backes and Kelley, 2003). It has therefore been proposed that P450s could be organized as clusters around CPRs (Peterson et al., 1976). With such organization, clustering would favor P450 reduction when CPR is in subsaturating amounts. In favor of such a model, P450 was reported to be more active in an aggregated state, and increasing CPR concentration was reported to disrupt P450 aggregation and to increase its mobility (Backes and Kelley, 2003). While a free diffusion model was recently considered as more likely, P450–CPR association would be required in the case of restricted lateral mobility.
Cytochrome b5 is an alternative electron donor for P450 enzymes. It was shown to favor P450-catalyzed reactions by allowing faster transfer of the second electron in the catalytic cycle compared with NADPH-P450 reductase and to induce conformational changes in P450 catalytic sites (Schenkman and Jansson, 2003). Quite interestingly, TAP tagging, mainly in the case of CYP73A5, indicates interaction with a NADH-cytochrome b5 reductase. While cytochrome b5 is usually considered as nonessential for P450 activity in vitro, a genetic approach pointed to its importance for P450 function in phenolic metabolism, to achieve effective flavonoid 3′-5′-hydroxylation and to a lesser extent cinnamic acid hydroxylation in petunia (Petunia hybrida; de Vetten et al., 1999). Another recent report also points the importance of NADH-cytochrome b5 reductase for effective fatty acid hydroxylation in Arabidopsis (Kumar et al., 2006). A potential structuring role of NADH-cytochrome b5 reductase may thus deserve closer investigation.
The Expression of CYP98A3 Favors Partner Localization near the ER and Protein Association
Formation of P450 oligomers could also favor flux transfer of metabolites and provide a platform to bridge soluble enzymes. At this stage, it seems unlikely that P450s form stable complexes with their soluble partner enzymes since neither TAP tagging nor blue native gel experiments detected P450 interaction with HCT or 4-CL. However, we observed consistent increases in their relocalization upon P450 expression and increased FRET among partners upon coexpression of selected proteins, reminiscent of the tobacco PAL2 relocalization observed upon C4H expression by Achnine et al. (2004). The highest FRET values between P450s and soluble enzymes were obtained upon coexpression of the four proteins both in intact and wounded tissues. It was not due to a crowding effect since it was not observed with free eGFP and for all tested combinations. The most consistently high FRET values were observed for the couple CYP98A3-HCT. This is in agreement with the following facts: (1) HCT is found associated with the membrane and readily relocalizes to the ER upon coexpression of CYP98A3, (2) HCT is both the donor of substrate and acceptor of product of CYP98A3 (Hoffmann et al., 2003), and (3) the shikimate conjugates of p-coumaric and caffeic acid that are the metabolic intermediates between the two enzymes are usually not detected in tissues of wild-type Arabidopsis (Schoch et al., 2006). The latter observation implies a tight coupling between CYP98A3 and HCT that might require close enzyme association. Less expected was the influence of CYP98A3 on 4-CL1 redistribution closer to the ER (Figure 5C) and the association of CYP98A3 with 4-CL1 detected upon coexpression of other partner enzymes (Table 3). The reciprocal effect of CYP98A3/4-CL1 expression on the association of CYP73A5 and HCT would suggest that the formation of P450 heteromers could be driving such associations. It is interesting to note that the CYP98A3–HCT association is influenced by the fluorescent tag localization at the N or C terminus of HCT (Table 3) and that CYP73A5/4-CL1 association with the tagged enzymes could not be detected, which supports some supramolecular organization rather than just protein concentration at the surface of the ER.
The presence of partner enzymes investigated in this study does not seem to be the only factor affecting protein localization and association. We observed a strong enhancement of protein association located in the healing zone after mechanical injury. General activation of the lignin pathway and resulting expression of other partner proteins and/or modification of the metabolome in the healing cells could be driving forces of such protein association. This suggests that beyond direct protein interactions, the cellular and membrane contexts should also be investigated.
Conclusion
CYP73A5 and CYP98A3 associate to form homomers and heteromers but also strongly and specifically interact with other membrane proteins potentially associated with membrane structure, mobility, and lipid composition. Looser association of both P450s with HCT and 4-CL1 is also observed, with CYP98A3 playing a prominent nucleation role in this association. In particular, the CYP98A3/HCT couple seems to readily associate in vivo, but this association is not stable enough to be detected by TAP tagging or blue native gel blot experiments that require use of mild detergents. Interactions among membrane proteins are thus stronger than those observed between P450s and their soluble partner enzymes. Our data suggest that clusters of membrane proteins might provide a platform for further more loose association of soluble partners, which may well be considered a dynamic metabolon. A more extensive characterization of associated membrane proteins will provide a helpful background to reveal supramolecular structures or local environments that harbor metabolic enzymes and likely favor protein associations.
METHODS
Construction of Candidate Protein Fusions with Fluorescent Proteins
Construction of the pCAMBIA2300 (for eGFP fusion) and pCAMBIA3300 (for mRFP fusion) plasmids with tagged CYP73A5 and CYP98A3 and anchored-eGFP and mRFP1 constructs was described by Bassard et al. (2012). Construction of the pCAMBIA2300 and pCAMBIA3300 plasmid with the CYP71B31:eGFP/mRFP1 fusion was described by Ginglinger (2010). The full-length coding sequences of HCT and 4-CL1 were amplified with appropriate primers (see Supplemental Table 3 online) and fused in 5′ or 3′ with the amplified mRFP1 or eGFP coding sequences in the pCAMBIA230035Su or pCAMBIA330035Su vectors with the PCR-based USER fusion cloning technique (Geu-Flores et al., 2007). The plasmids for expression of untagged CYP73A5, CYP98A3, HCT, and 4-CL1 were constructed by amplification of the coding sequences of the respective genes and insertion of the amplicons in the vector pCAMBIA230035Su or pCAMBIA330035Su with the single insert USER cloning technique (Nour-Eldin et al., 2006).
A comprehensive list of the primers used for the constructions is provided in Supplemental Table 3 online. All PCR-based constructs were sequence verified for proper in-frame insertion and absence of mutations.
Transient Expression in Nicotiana benthamiana Leaf Epidermal Cells
The cauliflower mosaic virus 35S promoter-driven genes were introduced into N. benthamiana leaves by agroinfiltration for transient expression as described by Bassard et al. (2012). At 4 to 5 d after infiltration, leaf disks were excised for observation by confocal microscopy, or leaves were collected to prepare soluble extracts and microsomal membranes.
Agrobacterium tumefaciens strains harboring vectors for the expression of eGFP:HDEL and mRFP1:HDEL were kindly provided by C. Ritzenthaler (Centre National de la Recherche Scientifique, Institut de Biologie Moléculaire des Plantes, Strasbourg, France). Expression of untagged proteins was confirmed by dot blot (see Supplemental Figure 4 online).
Preparation of Microsomal Membranes
Fresh tissues were ground in extraction buffer (40 mM ascorbic acid, 15 mM β-mercapthoethanol, 1 mM phenylmethanesulfonyl fluoride, and 10% glycerol in 0.1 M sodium phosphate, pH 7.4) in a ratio of 10 mL of buffer to 1 g of fresh tissue. Grinding was performed with an Ultra-turrax T25 (IKA) by two successive runs of 1 min at 7000 rpm. The extract was filtered on Miracloth (Calbiochem) and centrifuged for 10 min at 10,000g. The resulting supernatant was filtered on Miracloth and ultracentrifuged for 1 h at 100,000g at 4°C. For 4-CL1 and HCT activity tests and control dot blot with eGFP, the supernatant was kept and the resulting pellet of microsomes was resuspended in extraction buffer in a ratio of 10 mL of buffer to 1 g of fresh tissue. The mixture was ultracentrifuged for 1 h at 100,000g at 4°C. The pellet was washed with 3 × 2 mL of recovery buffer (30% glycerol in 0.1 M sodium phosphate, pH 7.4) and carefully resuspended in recovery buffer (10 μL per gram of initial fresh tissues). Microsomes and supernatant were stored at −30°C until analysis. Total protein concentration was determined using the Bio-Rad protein assay.
Test of 4-CL1 and HCT Activities
The 4-CL1 assay was performed in a microplate reader (Synergy HT; Biotek), monitoring the absorbance of the product at 333 nm. The assay mixture contained 4 μL of fraction to analyze, 1 mM DTT, 2.5 mM ATP, 2.5 mM MgCl2, 0.2 mM p-coumaric acid, and 50 mM potassium phosphate, pH 7.4. Reaction was started by the addition of 0.4 mM reduced CoA (Sigma-Aldrich). Kinetics were recorded at 25°C. Specific activities (SA) were calculated from initial velocity of product formation using the extinction coefficient for p-coumaroyl CoA: ε333 nm = 21 mM−1⋅cm−1 (Stöckigt and Zenk, 1975).
The HCT assay was performed in UV-star microplates (Greiner Bio-One), monitoring the absorbance of the product at 310 nm with a microplate reader (Synergy HT). The assay mixture contained the fraction to analyze (20 to 80 µg of total proteins), 1 mM DTT, 0.68 mM shikimic acid (Sigma-Aldrich), and 50 mM potassium phosphate, pH 7.4. Reactions were started by addition of 0.14 mM coumaroyl CoA (see Supplemental Methods 1 online for details about the preparation). Kinetics were recorded at 30°C. Specific activities were calculated from initial velocity of product formation using the extinction coefficient for p-coumaroyl shikimate: ε310 nm = 21.7 mM−1⋅cm−1.
Confocal Laser Scanning Microscopy
Cell imaging was performed using a Zeiss LSM510 laser scanning confocal microscope equipped with an inverted Zeiss Axiovert 200 microscope (Carl Zeiss). Images were recorded using a C-apochromat (×63/ 1.2 W Korr; Carl Zeiss) water immersion objective lens. For wide-field observations, a ×20 Zeiss objective was used. Excitation/emission wavelengths were 488/505 to 550 nm for eGFP and 561/575 to 615 nm for mRFP1. The images were acquired using AIM version 2.8 software (Carl Zeiss) and directly analyzed with ImageJ version 1.43n3 (Wayne Rasband, National Institutes of Health). The movies were acquired using a Zeiss LSM700 laser scanning confocal microscope with a Plan-Apochromat (×63/1.4 oil differential interference contrast M27) oil immersion objective lens. Excitation/emission wavelengths were 488/SP555 nm for eGFP. The movies were acquired and prepared using the Zen version 2009 software (Carl Zeiss). Figures were assembled with Illustrator CS4 (Adobe Systems).
Image Acquisition and Processing
Image acquisition and processing was performed as described by Bassard et al. (2012). An analysis of variance (ANOVA) with posthoc analysis was performed with the Bonferroni-Holm posthoc test to determine consistency of significant difference of the values recorded in different conditions. Calculations were performed with the XL toolbox add-in for excel (XL toolbox v3.10; http://xltoolbox.sourceforge.net/). Figures were prepared with Excel 2010 (Microsoft) and assembled with Illustrator CS4.
FRAP
The tissues were mounted and cell imaging was performed by confocal laser scanning microscopy (CLSM) as above. FRAP was performed on ER regions of 90 pixels (x) over 10 pixels (y) using a high-intensity bleach mode (with argon laser at 488 nm), unidirectional scans, intensity at 40 mA, line average 2, and a scan time of 3.15 s. Five prebleaching images were recorded, and area to bleach was photobleached during 200 scans (400 during tests with soluble enzymes). After the bleach, a series of postbleaching scans were performed. The postbleach fluorescence was sampled every 3.15 s. Figures were prepared with Excel 2010 and assembled with Illustrator CS4.
FLIM
The tissues were mounted as described above. FLIM was performed by time-correlated single-photon counting as described by Brandner et al. (2008). Measurements were done on a homebuilt two-photon system based on an Olympus IX70 microscope with an Olympus ×60 1.2–numerical aperture water immersion objective (Azoulay et al., 2003). Two-photon excitation was provided by a titanium:sapphire laser (Tsunami; Newport) that was setup to an emission wavelength of 900 nm (for eGFP excitation). Imaging was performed with a laser scanning system using two fast galvo mirrors (Model 6210; Cambridge Technology). Photons were collected using a two-photon short-pass filter with a cutoff wavelength of 680 nm (F75-680; AHF), and a band-pass filter 520 ± 17 nm (F37-520; AHF). Fluorescence was analyzed by a fiber-coupled avalanche photodiode detector (SPCM-AQR-14-FC; Perkin-Elmer), which was connected to a time-correlated single-photon-counting module (SPC830; Becker and Hickl) operated in reversed start-stop mode.
The samples were scanned continuously for 1 to 5 min to obtain appropriate photon numbers for reliable statistics for the fluorescence decays. Data were analyzed using SPCimage V2.9 (Becker and Hickl), which uses an iterative reconvolution method to recover the lifetimes from the fluorescence decays. Measurements were done on the whole image with a threshold setup to 50 and with an average of two pixels. For each condition, average values of all images recorded were used to calculate FRET efficiency. FRET was calculated using the equation: FRET = 1 – (τASSAY/τALONE) = (R06/(R06 + R)) with R0 the Förster radius, R the distance between donor and acceptor, τASSAY, the lifetime of the donor in the presence of the acceptor, and τALONE the lifetime of the donor in the absence of acceptor. Standard deviation was calculated as follows: sdFRET = %FRET √((sdASSAY/averageASSAY)2 + (sdALONE/averageALONE)2).
TAP
Cloning of transgenes encoding tag fusions under control of the cauliflower mosaic virus 35S promoter and transformation of Arabidopsis thaliana cell suspension cultures were performed as previously described (Van Leene et al., 2007). To scale up production, cells were subcultured in 50 mL of fresh medium without antibiotics at a 1:10 dilution and grown under standard conditions. After 1 week, cells were either harvested (non-=induced condition) or washed three times with 200 mL of adapted MSMO medium (4.43 g/L MSMO [Sigma-Aldrich], 30 g/L Suc, and 520 mg/L KH2PO4, pH 5.7) without hormones (induced condition). Washed cells were subsequently transferred to a one-liter shaker flask, diluted in 400 mL of the same medium used for washing, and incubated for 40 h at standard conditions. TAP of protein complexes was performed using the GS tag (Bürckstümmer et al., 2006) with the following protocol modifications. For all protein extractions prior to the affinity purification steps, the detergent Nonidet P-40 was replaced by digitonin (high purity; Calbiochem, Merck). Crude protein extracts were prepared in extraction buffer without detergent. After the mixing step, digitonin was added to a final concentration of 1% (w/v) and extracts were incubated for 1 h at 4°C under gentle rotation. A soluble protein fraction was obtained by centrifugation at 36,900g for two times 20 min at 4°C. In all further steps, the detergent 0.1% (v/v) Nonidet P-40 was replaced by 0.2% (w/v) digitonin. Protein precipitation and separation were done according to Van Leene et al. (2008). For the protocols of proteolysis and peptide isolation, acquisition of mass spectra by a 4800 Proteomics Analyzer (Applied Biosystems), and mass spectrometry–based protein homology identification based on The Arabidopsis Information Resource genomic database, we refer to Van Leene et al. (2010). Experimental background proteins were subtracted based on ∼40 TAP experiments on wild-type cultures and cultures expressing TAP-tagged mock proteins β-glucuronidase, RFP, and GFP (Van Leene et al., 2010).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: CYP98A3 (At2g40890), CYP73A5 (At2g30490), HCT (At5g48930), 4-CL1 (At1g51680), CYP71B31 (At3g53300), eGFP (GenBank DQ768212), mRFP1 (GenBank AF506027), and CYP51G1 (At1g11680).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Fluorescence Recovery after Photobleaching.
Supplemental Figure 2. Effect of Wounding on Protein Relocalization and Protein Interaction.
Supplemental Figure 3. 2D Blue Native/SDS-PAGE.
Supplemental Figure 4. Dot Blot of Crude Extracts or Microsomes from N. benthamiana Plants Expressing Untagged Proteins.
Supplemental Figure 5. Control of the Coexpression of CYP71B31 with CYP73A5 and CYP98A3.
Supplemental Table 1. eGFP Fusion Constructs Encode Functional Enzymes.
Supplemental Table 2. Interactions among Soluble Proteins.
Supplemental Table 3. List of Primers.
Supplemental Methods 1. Details of Methods Used in This Work.
Supplemental Movie 1. CYP98A3:eGFP Fluorescence in N. benthamiana Leaves 5 d after Agroinfiltration.
Supplemental Movie 2. eGFP Fluorescence in N. benthamiana Leaves 5 d after Agroinfiltration.
Supplemental Movie 3. eGFP:HDEL Fluorescence in N. benthamiana Leaves 5 d after Agroinfiltration.
Supplemental Data Set 1. MS Identification of CYP73A5 and CYP98A3 Interactors Determined by TAP-Tag.
Supplemental Data Set 2. ANOVA Tests of Confocal Analyses.
Acknowledgments
J.-E.B. and J.G. thank the Ministère de l’Enseignement Supérieur de la Recherche et de la Technologie and the Agency for Innovation by Science and Technology in Flanders, respectively, for doctoral fellowships. This project was initiated thanks to the Human Frontier of Science Program RGP0065/2005-C. M.S. and D.W.-R. are grateful for the funding of the Agence Nationale de la Recherche for the PHENOWALL project. We also acknowledge the European Community’s Framework VII Program FP7/2007-2013 for funding from the SMARTCELL and RENEWALL projects (Grants 222716 and KBBE-2007-3-1-01, respectively). We thank Leon Otten for critical reading of the article and Christophe Ritzenthaler for helpful discussions.
AUTHOR CONTRIBUTIONS
J.-E.B. prepared constructs, designed and performed confocal microscopy, enzyme analysis, and purification, and wrote the first draft. H.R. and F.D. contributed to fusion constructs and to the control experiments. P.U. synthesized coumaroyl-CoA and contributed to supervision. M.S. synthesized coumaroyl quinate. L.R. and J.M. supervised confocal microscopy. E.M. ran the blue native two-dimensional analyses. Y.M. implemented FLIM setup. G.D.J. and A.G. designed TAP analysis. J.G. performed TAP. J.G., W.B., G.D.J., and A.G. analyzed TAP data. A.G. and Y.M. contributed to article redaction. D.W.-R. supervised work and article redaction.
Glossary
- PAL
Phe ammonia lyase
- C4H
cinnamate 4-hydroxylase
- 4-CL
4-coumaric acid CoA ligase
- C3′H
p-coumaroylshikimate 3′-hydroxylase
- HCT
hydroxycinnamoylCoA:shikimate hydroxycinnamoyl transferase
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- FRET
fluorescence resonance energy transfer
- eGFP
enhanced GFP
- CPR
to be defined
- FRAP
fluorescence recovery after photobleaching
- TAP
tandem affinity purification
- GS
to be defined
- FLIM
fluorescence lifetime imaging microscopy
- CLSM
confocal laser scanning microscopy
- ANOVA
analysis of variance
References
- Achnine L., Blancaflor E.B., Rasmussen S., Dixon R.A. (2004). Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16: 3098–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoulay J., Clamme J.P., Darlix J.L., Roques B.P., Mély Y. (2003). Destabilization of the HIV-1 complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J. Mol. Biol. 326: 691–700 [DOI] [PubMed] [Google Scholar]
- Backes W.L., Kelley R.W. (2003). Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharmacol. Ther. 98: 221–233 [DOI] [PubMed] [Google Scholar]
- Bassard J.-E., Mutterer J., Duval F., Werck-Reichhart D. (2012). A novel method for monitoring the localization of cytochromes P450 and other endoplasmic reticulum membrane associated proteins: A tool for investigating the formation of metabolons. FEBS J. 279: 1576–1583 [DOI] [PubMed] [Google Scholar]
- Bayburt T.H., Sligar S.G. (2002). Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc. Natl. Acad. Sci. USA 99: 6725–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C., Westphal L., Schmotz C., Prade E., Scheel D., Glawischnig E. (2009). The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21: 1830–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner K., Sambade A., Boutant E., Didier P., Mély Y., Ritzenthaler C., Heinlein M. (2008). Tobacco mosaic virus movement protein interacts with green fluorescent protein-tagged microtubule end-binding protein 1. Plant Physiol. 147: 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browman D.T., Resek M.E., Zajchowski L.D., Robbins S.M. (2006). Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 119: 3149–3160 [DOI] [PubMed] [Google Scholar]
- Bürckstümmer T., Bennett K.L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. (2006). An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Carland F.M., Fujioka S., Takatsuto S., Yoshida S., Nelson T. (2002). The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14: 2045–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley G.F., Zhang S., Kelley R.W., Backes W.L. (2001). Evidence supporting the interaction of CYP2B4 and CYP1A2 in microsomal preparations. Drug Metab. Dispos. 29: 1529–1534 [PubMed] [Google Scholar]
- Chen H.C., Li Q., Shuford C.M., Liu J., Muddiman D.C., Sederoff R.R., Chiang V.L. (2011). Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. USA 108: 21253–21258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.H., van Spronsen J., Dai Y., Verberne M., Hollmann F., Arends I.W., Witkamp G.J., Verpoorte R. (2011). Are natural deep eutectic solvents the missing link in understanding cellular metabolism and physiology? Plant Physiol. 156: 1701–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M.A., et al. (2005). Characterization in vitro and in vivo of the putative multigene 4-coumarate:CoA ligase network in Arabidopsis: Syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry 66: 2072–2091 [DOI] [PubMed] [Google Scholar]
- Coué M., Brenner S.L., Spector I., Korn E.D. (1987). Inhibition of actin polymerization by latrunculin A. FEBS Lett. 213: 316–318 [DOI] [PubMed] [Google Scholar]
- Czichi U., Kindl H. (1975). Formation of p-coumaric acid and o-coumaric acid from L-phenylalanine by microsomal membrane fractions from potato: Evidence of membrane-bound enzyme complexes. Planta 125: 115–125 [DOI] [PubMed] [Google Scholar]
- Czichi U., Kindl H. (1977). Phenylalanine ammonia-lyase and cinnamic acid hydroxylase as assembled consecutive enzymes on microsomal membranes of cucumber cotyledons: Cooperation and subcellular distribution. Planta 134: 133–143 [DOI] [PubMed] [Google Scholar]
- Davletov B.A., Südhof T.C. (1993). A single C2 domain from synaptotagmin I is sufficient for high affinity Ca2+/phospholipid binding. J. Biol. Chem. 268: 26386–26390 [PubMed] [Google Scholar]
- Davydov D.R., Fernando H., Baas B.J., Sligar S.G., Halpert J.R. (2005). Kinetics of dithionite-dependent reduction of cytochrome P450 3A4: Heterogeneity of the enzyme caused by its oligomerization. Biochemistry 44: 13902–13913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov D.R., Petushkova N.A., Archakov A.I., Hoa G.H. (2000). Stabilization of P450 2B4 by its association with P450 1A2 revealed by high-pressure spectroscopy. Biochem. Biophys. Res. Commun. 276: 1005–1012 [DOI] [PubMed] [Google Scholar]
- de Vetten N., ter Horst J., van Schaik H.P., de Boer A., Mol J., Koes R. (1999). A cytochrome b5 is required for full activity of flavonoid 3′,5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl. Acad. Sci. USA 96: 778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret A., Nguyen M., Breckenridge D.G., Shore G.C. (2003). The resident endoplasmic reticulum protein, BAP31, associates with gamma-actin and myosin B heavy chain. Eur. J. Biochem. 270: 342–349 [DOI] [PubMed] [Google Scholar]
- Ehlting J., Büttner D., Wang Q., Douglas C.J., Somssich I.E., Kombrink E. (1999). Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 19: 9–20 [DOI] [PubMed] [Google Scholar]
- Ehlting J., Shin J.J., Douglas C.J. (2001). Identification of 4-coumarate:coenzyme A ligase (4CL) substrate recognition domains. Plant J. 27: 455–465 [DOI] [PubMed] [Google Scholar]
- Geu-Flores F., Nour-Eldin H.H., Nielsen M.T., Halkier B.A. (2007). USER fusion: A rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 35: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginglinger J.F. (2010). Functional Analysis of Cytochromes P450 Involved in the Biosynthesis of Monoterpenoids in Arabidopsis thaliana. PhD dissertation (Strasbourg, France: University of Strasbourg)
- Griffing L.R. (2010). Networking in the endoplasmic reticulum. Biochem. Soc. Trans. 38: 747–753 [DOI] [PubMed] [Google Scholar]
- Hamberger B., Hahlbrock K. (2004). The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc. Natl. Acad. Sci. USA 101: 2209–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazai E., Kupfer D. (2005). Interactions between CYP2C9 and CYP2C19 in reconstituted binary systems influence their catalytic activity: Possible rationale for the inability of CYP2C19 to catalyze methoxychlor demethylation in human liver microsomes. Drug Metab. Dispos. 33: 157–164 [DOI] [PubMed] [Google Scholar]
- Hawkins S., Boudet A. (1996). Wound-induced lignin and suberin deposition in a woody angiosperm (Eucalyptus gunnii Hook.): Histochemistry of early changes in young plants. Protoplasma 191: 96–104 [Google Scholar]
- Hoegg M.B., Browman D.T., Resek M.E., Robbins S.M. (2009). Distinct regions within the erlins are required for oligomerization and association with high molecular weight complexes. J. Biol. Chem. 284: 7766–7776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L., Besseau S., Geoffroy P., Ritzenthaler C., Meyer D., Lapierre C., Pollet B., Legrand M. (2004). Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16: 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L., Maury S., Martz F., Geoffroy P., Legrand M. (2003). Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J. Biol. Chem. 278: 95–103 [DOI] [PubMed] [Google Scholar]
- Hrazdina G., Wagner G.J. (1985). Metabolic pathways as enzyme complexes: Evidence for the synthesis of phenylpropanoids and flavonoids on membrane associated enzyme complexes. Arch. Biochem. Biophys. 237: 88–100 [DOI] [PubMed] [Google Scholar]
- Hu G., Johnson E.F., Kemper B. (2010). CYP2C8 exists as a dimer in natural membranes. Drug Metab. Dispos. 38: 1976–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K., Osmani S.A., Hamann T., Naur P., Møller B.L. (2011). Homology modeling of the three membrane proteins of the dhurrin metabolon: Catalytic sites, membrane surface association and protein-protein interactions. Phytochemistry 72: 2113–2123 [DOI] [PubMed] [Google Scholar]
- Jørgensen K., Rasmussen A.V., Morant M., Nielsen A.H., Bjarnholt N., Zagrobelny M., Bak S., Møller B.L. (2005). Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8: 280–291 [DOI] [PubMed] [Google Scholar]
- Kumar R., Wallis J.G., Skidmore C., Browse J. (2006). A mutation in Arabidopsis cytochrome b5 reductase identified by high-throughput screening differentially affects hydroxylation and desaturation. Plant J. 48: 920–932 [DOI] [PubMed] [Google Scholar]
- Li B., Yau P., Kemper B. (2011). Identification of cytochrome P450 2C2 protein complexes in mouse liver. Proteomics 11: 3359–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo C., Bard M. (2005). Erg28p is a key protein in the yeast sterol biosynthetic enzyme complex. J. Lipid Res. 46: 1991–1998 [DOI] [PubMed] [Google Scholar]
- Mo C., Valachovic M., Bard M. (2004). The ERG28-encoded protein, Erg28p, interacts with both the sterol C-4 demethylation enzyme complex as well as the late biosynthetic protein, the C-24 sterol methyltransferase (Erg6p). Biochim. Biophys. Acta 1686: 30–36 [DOI] [PubMed] [Google Scholar]
- Moura J.C., Bonine C.A., de Oliveira Fernandes Viana J., Dornelas M.C., Mazzafera P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52: 360–376 [DOI] [PubMed] [Google Scholar]
- Myasoedova K.N., Berndt P. (1990). Immobilized cytochrome P-450LM2. Dissociation and reassociation of oligomers. FEBS Lett. 270: 177–180 [DOI] [PubMed] [Google Scholar]
- Nair R.B., Xia Q., Kartha C.J., Kurylo E., Hirji R.N., Datla R., Selvaraj G. (2002). Arabidopsis CYP98A3 mediating aromatic 3-hydroxylation. Developmental regulation of the gene, and expression in yeast. Plant Physiol. 130: 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Hansen B.G., Nørholm M.H., Jensen J.K., Halkier B.A. (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nziengui H., Schoefs B. (2009). Functions of reticulons in plants: What we can learn from animals and yeasts. Cell. Mol. Life Sci. 66: 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Mimura T., Hasezawa S. (2005). Regulation of secondary cell wall development by cortical microtubules during tracheary element differentiation in Arabidopsis cell suspensions. Plant Physiol. 137: 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozalp C., Szczesna-Skorupa E., Kemper B. (2005). Bimolecular fluorescence complementation analysis of cytochrome p450 2c2, 2e1, and NADPH-cytochrome p450 reductase molecular interactions in living cells. Drug Metab. Dispos. 33: 1382–1390 [DOI] [PubMed] [Google Scholar]
- Peterson J.A., Ebel R.E., O’Keeffe D.H., Matsubara T., Estabrook R.W. (1976). Temperature dependence of cytochrome P-450 reduction. A model for NADPH-cytochrome P-450 reductase:cytochrome P-450 interaction. J. Biol. Chem. 251: 4010–4016 [PubMed] [Google Scholar]
- Praporski S., Ng S.M., Nguyen A.D., Corbin C.J., Mechler A., Zheng J., Conley A.J., Martin L.L. (2009). Organization of cytochrome P450 enzymes involved in sex steroid synthesis: PROTEIN-PROTEIN INTERACTIONS IN LIPID MEMBRANES. J. Biol. Chem. 284: 33224–33232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston L., Yu O. (2006). Metabolons involving plant cytochrome P450. Phytochem. Rev. 5: 459–472 [Google Scholar]
- Rasmussen S., Dixon R.A. (1999). Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11: 1537–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.R., Backes W.L. (2012). Formation of P450 · P450 complexes and their effect on P450 function. Pharmacol. Ther. 133: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro D.K., Douglas C.J. (2004). Reconstitution of the entry point of plant phenylpropanoid metabolism in yeast (Saccharomyces cerevisiae): Implications for control of metabolic flux into the phenylpropanoid pathway. J. Biol. Chem. 279: 2600–2607 [DOI] [PubMed] [Google Scholar]
- Runions J., Brach T., Kühner S., Hawes C. (2006). Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J. Exp. Bot. 57: 43–50 [DOI] [PubMed] [Google Scholar]
- Saravanan R.S., Slabaugh E., Singh V.R., Lapidus L.J., Haas T., Brandizzi F. (2009). The targeting of the oxysterol-binding protein ORP3a to the endoplasmic reticulum relies on the plant VAP33 homolog PVA12. Plant J. 58: 817–830 [DOI] [PubMed] [Google Scholar]
- Schaeffer A., Bronner R., Benveniste P., Schaller H. (2001). The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J. 25: 605–615 [DOI] [PubMed] [Google Scholar]
- Schalk M., Batard Y., Seyer A., Nedelkina S., Durst F., Werck-Reichhart D. (1997). Design of fluorescent substrates and potent inhibitors of CYP73As, P450s that catalyze 4-hydroxylation of cinnamic acid in higher plants. Biochemistry 36: 15253–15261 [DOI] [PubMed] [Google Scholar]
- Schenkman J.B., Jansson I. (2003). The many roles of cytochrome b5. Pharmacol. Ther. 97: 139–152 [DOI] [PubMed] [Google Scholar]
- Schoch G., Goepfert S., Morant M., Hehn A., Meyer D., Ullmann P., Werck-Reichhart D. (2001). CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 276: 36566–36574 [DOI] [PubMed] [Google Scholar]
- Schoch G.A., Morant M., Abdulrazzak N., Asnaghi C., Goepfert S., Petersen M., Ullmann P., Werck-Reichhart D. (2006). The meta-hydroxylation step in the phenylpropanoid pathway: A new level of complexity in the pathway and its regulation. Environ. Chem. Lett. 4: 127–136 [Google Scholar]
- Schoch G.A., Yano J.K., Wester M.R., Griffin K.J., Stout C.D., Johnson E.F. (2004). Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site. J. Biol. Chem. 279: 9497–9503 [DOI] [PubMed] [Google Scholar]
- Schuhegger R., Nafisi M., Mansourova M., Petersen B.L., Olsen C.E., Svatos A., Halkier B.A., Glawischnig E. (2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141: 1248–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E.E., He Y.A., Wester M.R., White M.A., Chin C.C., Halpert J.R., Johnson E.F., Stout C.D. (2003). An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Proc. Natl. Acad. Sci. USA 100: 13196–13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova I.F., Li H., Zhang H., Peterson J.A., Poulos T.L. (1999). Structure of a cytochrome P450-redox partner electron-transfer complex. Proc. Natl. Acad. Sci. USA 96: 1863–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I., Hawes C., Frigerio L. (2011). FrontiERs: Movers and shapers of the higher plant cortical endoplasmic reticulum. Curr. Opin. Plant Biol. 14: 658–665 [DOI] [PubMed] [Google Scholar]
- Sparkes I., Tolley N., Aller I., Svozil J., Osterrieder A., Botchway S., Mueller C., Frigerio L., Hawes C. (2010). Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 22: 1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Frigerio L., Tolley N., Hawes C. (2009). The plant endoplasmic reticulum: A cell-wide web. Biochem. J. 423: 145–155 [DOI] [PubMed] [Google Scholar]
- Srere P.A. (1985). The metabolon. Trends Biochem. Sci. 10: 109–110 [Google Scholar]
- Stöckigt J., Zenk M.H. (1975). Chemical synthesis and properties of hydroxycinnamoyl coenzyme A derivatives. Z. Naturforsch. C 30c: 352–358 [DOI] [PubMed] [Google Scholar]
- Subramanian M., Tam H., Zheng H., Tracy T.S. (2010). CYP2C9-CYP3A4 protein-protein interactions: Role of the hydrophobic N terminus. Drug Metab. Dispos. 38: 1003–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesna-Skorupa E., Chen C.D., Rogers S., Kemper B. (1998). Mobility of cytochrome P450 in the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA 95: 14793–14798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesna-Skorupa E., Kemper B. (2006). BAP31 is involved in the retention of cytochrome P450 2C2 in the endoplasmic reticulum. J. Biol. Chem. 281: 4142–4148 [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa E., Mallah B., Kemper B. (2003). Fluorescence resonance energy transfer analysis of cytochromes P450 2C2 and 2E1 molecular interactions in living cells. J. Biol. Chem. 278: 31269–31276 [DOI] [PubMed] [Google Scholar]
- Tolley N., Sparkes I., Craddock C.P., Eastmond P.J., Runions J., Hawes C., Frigerio L. (2010). Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant J. 64: 411–418 [DOI] [PubMed] [Google Scholar]
- Van Leene J., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Van Leene J., Witters E., Inzé D., De Jaeger G. (2008). Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 13: 517–520 [DOI] [PubMed] [Google Scholar]
- Vogt T. (2010). Phenylpropanoid biosynthesis. Mol. Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Winkel B.S. (2004). Metabolic channeling in plants. Annu. Rev. Plant Biol. 55: 85–107 [DOI] [PubMed] [Google Scholar]
- Yang X.H., Xu Z.H., Xue H.W. (2005). Arabidopsis membrane steroid binding protein 1 is involved in inhibition of cell elongation. Plant Cell 17: 116–131 [DOI] [PMC free article] [PubMed] [Google Scholar]