The crystal structure of an indole-3-acetic acid amido synthetase from Vitis vinifera involved in auxin homeostasis is presented. Residues likely to be involved in acyl group, amino acid, and ATP substrate binding have been identified, and this information provides a tool for designing new, effective auxins.
Abstract
Auxins are important for plant growth and development, including the control of fruit ripening. Conjugation to amino acids by indole-3-acetic acid (IAA)-amido synthetases is an important part of auxin homeostasis. The structure of the auxin-conjugating Gretchen Hagen3-1 (GH3-1) enzyme from grapevine (Vitis vinifera), in complex with an inhibitor (adenosine-5′-[2-(1H-indol-3-yl)ethyl]phosphate), is presented. Comparison with a previously published benzoate-conjugating enzyme from Arabidopsis thaliana indicates that grapevine GH3-1 has a highly similar domain structure and also undergoes a large conformational change during catalysis. Mutational analyses and structural comparisons with other proteins have identified residues likely to be involved in acyl group, amino acid, and ATP substrate binding. Vv GH3-1 is a monomer in solution and requires magnesium ions solely for the adenlyation reaction. Modeling of IAA and two synthetic auxins, benzothiazole-2-oxyacetic acid (BTOA) and 1-naphthaleneacetic acid (NAA), into the active site indicates that NAA and BTOA are likely to be poor substrates for this enzyme, confirming previous enzyme kinetic studies. This suggests a reason for the increased effectiveness of NAA and BTOA as auxins in planta and provides a tool for designing new and effective auxins.
INTRODUCTION
Due to their importance in the control of plant development and plant response to external stimuli, plant hormone signaling pathways are under multiple levels of control. Auxins are powerful effectors central to a diverse array of developmental processes (reviewed in Davies, 2010) and so their actions need to be finely regulated. There are a number of ways in which the levels of auxins and their activity can be altered, with increasing evidence that conjugation reactions play an important role in these regulatory processes (reviewed in Wang et al., 2008). In particular, the inactivation of the most abundant auxin in plants, indole-3-acetic acid (IAA), through conjugation to amino acids by IAA-amido synthetases, appears to be significant (reviewed in Ludwig-Müller, 2011). Acyl-amido synthetases of the Gretchen Hagen3 (GH3) family comprise a group of plant-specific proteins with a role in hormone conjugation acting on auxins, jasmonic acid (JA), salicylic acid (SA), and benzoates (Staswick et al., 2002, 2005; Okrent et al., 2009; Westfall et al., 2010). They are members of the ANL superfamily of adenylating enzymes, which are characterized by the employment of an adenylated reaction intermediate to attach organic acids to a variety of substrates (reviewed in Gulick, 2009; Schmelz and Naismith, 2009). The importance of GH3 proteins in plant development is indicated by the presence of families of these proteins throughout the plant kingdom, from mosses to flowering plants (Okrent and Wildermuth, 2011).
GH3 proteins display differing preferences for both amino acid and acyl substrates (Staswick et al., 2005; Ludwig-Müller et al., 2009; Okrent et al., 2009; Böttcher et al., 2010, Böttcher et al., 2011a, Böttcher et al., 2012; Chen et al., 2010). Members of the IAA-amido synthetase family can join IAA to different amino acids, and the particular amino acid used determines the fate of the conjugated auxin: IAA-Asp and IAA-Glu conjugates tend to be converted into oxidized metabolites (Tsurumi and Wada, 1986; Plüss et al., 1989; Östin et al., 1998), whereas, for example, IAA-Ala and IAA-Leu conjugates can be hydrolyzed to release free IAA (LeClere et al., 2002; Savić et al., 2009).
IAA-amido synthetases have been studied in detail in a number of species, including rice (Oryza sativa), Arabidopsis thaliana, and grapevine (Vitis vinifera). Overexpression of IAA-conjugating GH3 proteins produces distinctive low auxin phenotypes, such as reduced growth, increased tiller number, increased leaf angle, and shortened roots (Nakazawa et al., 2001; Takase et al., 2004; Zhang et al., 2007; Ding et al., 2008; Domingo et al., 2009; Zhang et al., 2009). Due to redundancy, decreases in the expression of a single IAA-amido synthetase enzyme do not generate dramatic changes in phenotype (Takase et al., 2004; Staswick et al., 2005; Khan and Stone, 2007). The difficulty in downregulating the activity of a family of enzymes via transgenesis makes a chemical inhibitor approach a valid method for further probing their function in vivo (Böttcher et al., 2012).
IAA-amido synthetases may also play a role in disease development. The overexpression of three different IAA-amido synthases in rice decreased free IAA levels and enhanced resistance to both fungal and bacterial pathogens (Ding et al., 2008; Domingo et al., 2009; Fu et al., 2011b). In contrast with the findings in rice, recent studies in Arabidopsis suggest that the GH3-catalyzed formation of the IAA-Asp conjugate is involved in the promotion of disease development (González-Lamothe et al., 2012).
In grapevine, there are six auxin-conjugating GH3 proteins (Böttcher et al., 2011a). All six are expressed during grape berry development and may be responsible for regulating the initiation of ripening by reducing IAA levels (Böttcher et al., 2010, Böttcher et al., 2011a). This reduction in IAA concentration appears to be required to allow ripening to progress, and the application of auxins delays ripening (Böttcher et al., 2010, Böttcher et al., 2011a, Böttcher et al., 2011b). In field-based experiments, different auxins delayed ripening to different extents when applied to berries preripening. The effectiveness of these auxins in delaying ripening was inversely correlated with their ability to act as substrates for GH3 proteins (Böttcher et al., 2011a). The reaction kinetics of three of the six grapevine family members (GH3-1, GH3-2, and GH3-6) have been analyzed in detail (Böttcher et al., 2011a, Böttcher et al., 2012). This article describes the crystallization of the GH3-1 enzyme that, in terms of acyl substrate, has a preference for IAA over the synthetic auxin 1-naphthaleneacetic acid (NAA) and does not conjugate another synthetic auxin-like compound, benzothiazole-2-oxyacetic acid (BTOA). With regard to amino acid specificity, GH3-1 conjugates Asp and Trp in vitro, but only the IAA-Asp conjugate can be found in developing berries, possibly due to the low levels of Trp present (Böttcher et al., 2010).
A reaction mechanism based on kinetic data has been developed for the auxin-conjugating Os GH3-8 enzyme from rice (Chen et al., 2010), and recently, three-dimensional structures have been determined for two Arabidopsis acyl-amido synthetases: At GH3-12/PBS3, which conjugates benzoate substrates, and the JA-specific At GH3-11/JAR1 (Westfall et al., 2012). Apart from detailing the amino acids in the active site important for substrate binding and catalysis, Westfall et al. (2012) described a conformational change in protein structure. The smaller, C-terminal domain, which acts as a cap over the active site, is seen to move significantly in the different crystal forms, changing between open (ATP-bound) and closed (AMP-bound) forms.
In this article, the three-dimensional structure of the grapevine IAA-amido synthetase GH3-1 is described. Crystallization was achieved through the use of an inhibitor (adenosine-5′-[2-(1H-indol-3-yl)ethyl]phosphate [AIEP]) that mimics the adenylated intermediate of the IAA conjugation reaction (Böttcher et al., 2012). While the grapevine GH3-1 structure is similar in many ways to that reported for the benzoate and JA-conjugating enzymes from Arabidopsis, the active site has unique features related to it using auxin as the acyl substrate. Mutational analysis showed that amino acid specificity can be altered by a single, conservative mutation in the active site. Experimental evidence confirmed that these proteins act as monomers and further details on the reaction mechanism with respect to ATP and magnesium were determined. Modeling of IAA and synthetic auxins into the active site provides an explanation for the relative abilities of these auxins to act as substrates, and this selectivity may explain observations regarding the effectiveness of these molecules as auxins in plants. This structure opens the way for a better understanding of the activity of IAA-amido synthetases and their roles in plant development. It will also assist in the development of more potent inhibitors to characterize and manipulate auxin-related processes and offers the opportunity to develop new auxin-based herbicides and growth regulators.
RESULTS
Crystallization and Structure of Grapevine GH3-1
Crystals of the grapevine GH3-1 protein were obtained in the presence of a tightly binding inhibitor (AIEP), which mimics the transition state of the auxin–amino acid conjugation reaction (Böttcher et al., 2012; see Supplemental Figure 1 online). All attempts to crystallize the uninhibited form of the protein were unsuccessful. The native GH3-1 protein contains 598 residues; the His6-tagged construct used in the structure determination contained an additional 11 expression vector-derived residues at the C terminus. The model had two independent protomers in the asymmetric unit and a 53.5% solvent content. Electron density was observed for residues Ser-16 to Arg-597 in the A protomer and from Ser-16 to Arg-596 in the B protomer. The protein structure (Figures 1A to 1C) consisted of two domains: a large, well-ordered N-terminal domain of almost 440 residues that included most of the inhibitor binding site and a smaller C-terminal domain, less well defined by electron density, which included residues 438 to 598. Separating the two domains and measuring the B-factors independently showed that the N-terminal domain was indeed better ordered with an average B-factor of 30.9 Å2, the C-terminal domain had an average B-factor of 46.0 Å2, and the overall B-factor was 34.5 Å2 (Table 1). In each protomer, there was weak or broken electron density from residues ∼550 to 570 with no density for residues 547 to 570 in chain A and 552 to 560 in chain B. The A and B chains overlaid well (overall 0.57 Å root mean square deviation) except in this area of weak density. Another loop in the C-terminal domain, residues 495 to 499, also displayed weak density.
Figure 1.
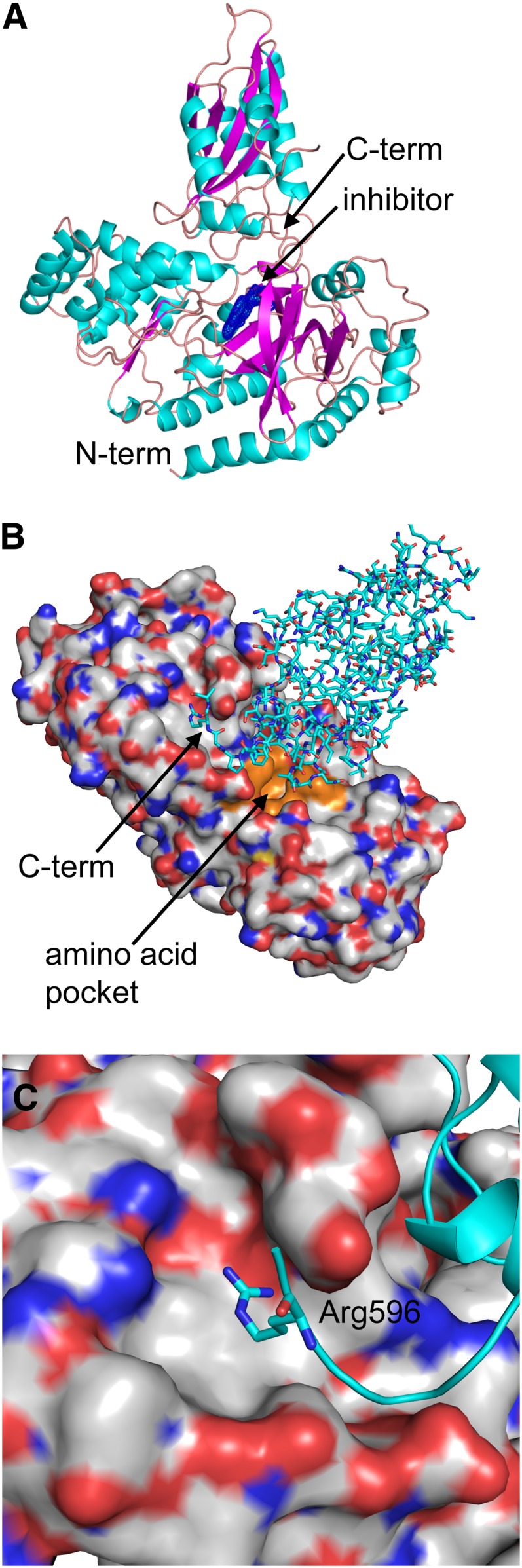
Overall Structure of Grapevine GH3-1.
(A) Cartoon diagram of the GH3-1 monomer with the inhibitor and a 2Fo-Fc map around the inhibitor (blue mesh). α-Helices are in cyan, β-strands are in magenta, and loops are in coral.
(B) The GH3-1 structure with the N-terminal domain shown as a surface representation (carbons are gray, nitrogens are blue, and oxygens are red) with the C-terminal domain shown with the residues in stick representation (carbons are cyan, nitrogens are blue, and oxygens are red). The C-terminal tail, Arg-596, can be seen fitting into a pocket on the surface of the N-terminal domain. The amino acid pocket next to the inhibitor has the surface highlighted in orange; the inhibitor is occluded from view by the C-terminal domain.
(C) Close-up of Arg-596 and the pocket on the N-terminal domain shown in (B). (B) and (C) are approximately a 180° rotation from (A).
Table 1. Crystallographic Statistics.
| GH3-1•AIEP |
GH3-1•AIEP Pt-Derivative |
||
|---|---|---|---|
| Space Group | P41212 | Space Group | P41212 |
| Cell (a = b × c) | 91.17 × 338.07 | Cell (a = b × c) | 91.60 × 339.04 |
| Resolution (Å) | 2.40 | Resolution (Å) | 2.88 |
| Completeness (%) | 99.9 (100) | Completeness (%) | 100 (100) |
| Rmerge (%) | 12.6 (71.6) | Rmerge (%) | 10.8 (64.9) |
| Mean I/sigI | 16.7 (4.0) | Mean I/sigI | 20.0 (5.1) |
| No. of unique reflections | 54,126 | No. of unique reflections | 33,925 |
| Multiplicity | 14.1 | Multiplicity | 13.0 |
| Rwork (%) | 22.4 | Anomalous completeness | 100 |
| Rfree (%) | 26.4 | Anomalous multiplicity | 6.9 |
| No. of protein atoms | 9,096 | RCR anomalous (overall) | 1.56 |
| No. of waters | 150 | Wavelength (Å) | 1.0113 |
| No. of ligand atoms | 66 | ||
| Mean B value overall (Å2) | 34.5 | ||
| Mean B value inhibitor (Å2) | 21.7 | ||
| Mean B value N terminus (Å2) | 30.9 | ||
| Mean B value C terminus (Å2) | 46.0 | ||
| Ramachandran analysis (%) preferred/ allowed/ outliers | 97.2/ 2.7/ 0.2 | ||
| Protein Data Bank code | 4B2G | ||
Values in parentheses are for the high-resolution bin.
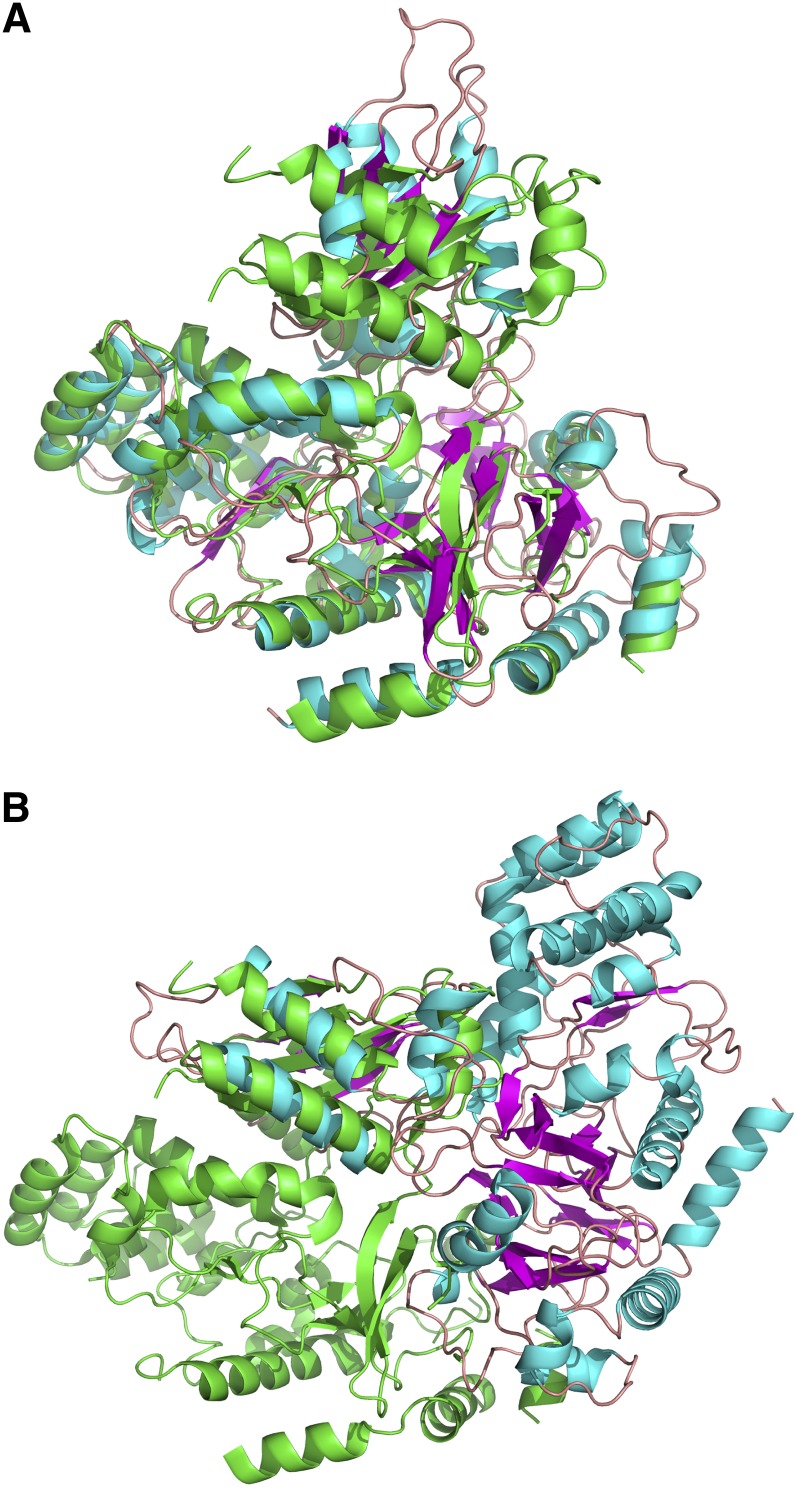
The overall fold of the structure was similar to that of the At GH3-11 and At GH3-12 structures reported recently (Figures 2A, 2B, 3A, and 3B; Westfall et al., 2012) and of the adenylate-forming enzymes in general (reviewed in Gulick, 2009; Schmelz and Naismith, 2009). The N-terminal domain was an α/β fold with two separate β-structures (on either side of the inhibitor/catalytic site) each surrounded by α-helices (Figure 1A). The smaller C-terminal domain had a four-strand β-sheet that was sandwiched between four α-helices (Figure 1A). Additionally, the C-terminal domain had a small β-loop structure between residues 438 and 448, and this structure formed a cap over the inhibitor.
Figure 2.
Comparison of Vv GH3-1 to the At GH3-12 AMP-Bound Structure and the Salicylate-Bound Structure.
(A) Superposition of Vv GH3-1 with 4EPM (At GH3-12 with AMP). The root mean square deviation over 511 aligned residues (out of 554/565) is 1.3 Å with 49.5% sequence identity (as calculated with the SSM algorithm implemented in Coot). Vv GH3-1 is in cyan for α-helices, magenta for β-strands, and coral for the loops; At GH3-12 is in green.
(B) Superposition of Vv GH3-1 with 4EQL (At GH3-12 with SA and AMP). The root mean square deviation over 476 aligned residues (out of 494/565) is 1.3 Å (as calculated with the SSM algorithm implemented in Coot). GH3-1 is shown with cyan-colored carbon atoms (as above) with 4EQL in green. Specific residues are highlighted that are different between the two sequences and that may contribute to the different substrate specificities.
Figure 3.
Comparison of Vv GH3-1 to the At GH3-12 AMPCPP-Bound Structure.
Alignment of the N-terminal domains of GH3-1 and 4EWV (At GH3-12 AMPCPP bound) (A) and alignment of C-terminal domains (4EWV in the same orientation as above) showing the major change needed to align these domains (B) (GH3-1 has reoriented ∼170° from the orientation in [A]). For both alignments, 4EWV is in green, and GH3-1 is in cyan/magenta.
Quaternary Structure of GH3-1 and Metal Binding in the GH3-1 Reaction
The crystal packing of GH3-1 suggested a possible biologically relevant dimer, as there were two protein/protein interfaces having over 1000 Å2 buried surface area as calculated by PISA (Krissinel and Henrick, 2007). To investigate this further, small-angle x-ray scattering (SAXS) was used to determine the solution state of the protein. SAXS data were measured at varying protein concentrations between 3 and 0.1 mg/mL to determine if there was any concentration-dependent oligomerization. The molecular weight and radius of gyration of the species in solution were calculated from the SAXS data at each concentration. These parameters did not vary across the range and were consistent with a monomeric species with a radius of gyration of 28.5 ± 0.5 Å (see Supplemental Table 1 and Supplemental Figures 2A and 2B online).
To validate further the monomeric state of the protein, SAXS data were back-calculated from both the monomeric and putative dimeric crystal structure forms and compared with the experimental data. The measured data were found to fit well with the monomeric species (χ = 1.101) and less well with the dimeric form (χ = 6.442) (see Supplemental Figures 3A to 3C online). This result strongly supports a monomeric solution state of GH3-1.
Although magnesium was present in the protein formulations used in crystallization, it was not seen in the resultant electron density maps. Even after soaking crystals with magnesium mimetics, such as gadolinium or the physiologically active manganese (Chen et al., 2010), no metal ion could be identified in the structure. This suggested that the metal was required only for stabilization of the binding of ATP and was not required for the subsequent steps of the conjugation reaction. To test this hypothesis, activity tests with GH3-1 protein (C- and N-terminal tags) and its known substrates IAA and Asp (Böttcher et al., 2010) were performed where magnesium was present for the entire reaction (control), excluded from the entire reaction (1), or present just for the first part of the reaction (2). Figure 4 shows that without the metal during the first half of the reaction, essentially no product was obtained (∼780-fold decrease when compared with the control). With metal present during the first half of the reaction, but not in the second half, IAA-Asp conjugate was still produced as each intermediate was turned over, but no subsequent reaction occurred (∼40-fold decrease when compared with the control). With metal present during the entire reaction, further turnover events occurred, and both proteins produced ∼2300 nmol IAA-Asp/mg.
Figure 4.
Relevance of Metal Ion Binding for the Two Half Reactions of GH3-1.
The involvement of magnesium ions in the adenylation reaction and the amino acid transfer was tested by adding 20 mM EDTA to a standard reaction mix prior to the addition of GH3-1 protein (1) or after an incubation of protein with a reaction mix lacking Asp (2). After an incubation period of 15 min, either GH3-1 protein (1) or Asp (2) were added to the reaction mix followed by another 20-min incubation period and the quantification of the reaction product (IAA-Asp) by LC-ESI-MS/MS. Control reactions did not contain EDTA. The white bars represent assays with C-terminal His6-tag GH3-1; the black bars show the results for assays with N-terminal His6-tag GH3-1. All data represent mean ± se (n = 3).
Substrate Binding Site of GH3-1
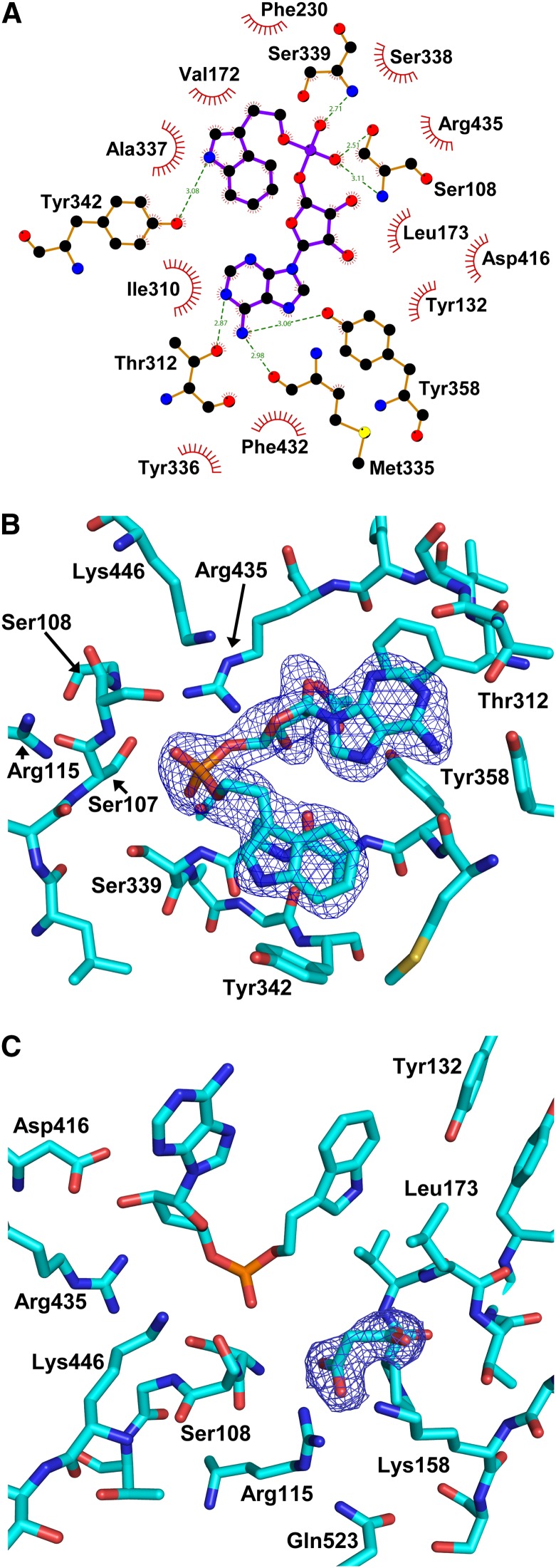
There was clear electron density for the substrate mimetic inhibitor in each of the protomers of the crystallographic asymmetric unit (Figure 5B; see Supplemental Figures 4A and 4B online). Some of the main interactions with the inhibitor (Figures 5A and 5B; see Supplemental Figure 4B online) were with residues Met335-Ser339 (ATP/AMP binding Motif II; Chang et al., 1997). The amino moiety of the inhibitor’s adenine ring (N1) made a hydrogen bond with the backbone carbonyl of Met-335 (∼3 Å). The backbone amino group of Ser-339 was 2.7 to 2.9 Å distant from a phosphate oxygen. Ser-108 (in ATP/AMP binding Motif I; Chang et al., 1997) was located near the phosphate, close enough to hydrogen bond with both side chain (2.5 to 2.7 Å) and backbone atoms (3.1 Å). The importance of Ser-108 for the GH3-1 reaction mechanism was further investigated by exchanging it with Ala (described below). Asp-416 (in ATP/AMP binding Motif III; Chang et al., 1997) made hydrogen bonds with the hydroxyl moieties of the ribose (2.7 to 2.9 Å). Thr-312 made a potential hydrogen bond with the N2 of the adenine ring (2.8 to 3 Å). The adenine ring was further encased in a hydrophobic pocket formed by Tyr-336 and Phe-432 on one side and Ile-310 on the other. The indole ring of the inhibitor also sat within a relatively hydrophobic pocket with Ala-337 on one side and Val-172, Leu-173, Val-229, Met-335, Tyr-342, and Ile-310 (again) forming the rest of the pocket. The hydroxyl of Tyr-342 was within 3.3 Å of the nitrogen in the indole ring.
Figure 5.
Inhibitor Interactions and Electron Density of the Malonate Bound in GH3-1 with Interacting Residues.
(A) Ligplot (produced by LIGPLOT v.4.5.3) of the inhibitor to show the interactions made with surrounding residues of the protein.
(B) Inhibitor with surrounding residues in a stick model with some of the residues mentioned in the text highlighted.
(C) Electron density around the bound malonate molecule in the active site shown as a 2Fo-Fc map at 1.5 sigma (blue mesh) with some of the residues around the malonate highlighted.
The protein was crystallized in the presence of malonate, and this molecule was seen in the structure tightly bound to Arg-115, which was part of ATP/AMP binding Motif I (Figure 5C). The guanidine group of Arg-115 in each protomer made hydrogen bonds of 2.8 and 3.0 Å or 2.7 and 2.8 Å (A and B protomers, respectively) to the bound malonate carboxylate moiety. On the other side of the malonate, the Lys-158 amino group was 2.8 to 2.9 Å from the carboxylate. This configuration suggests that an amino acid, such as Asp, might bind in a similar fashion to the malonate, and the Arg-115 residue was subsequently mutated to Lys to explore this possibility (see below). The amino acid substrate pocket, as determined by the positions of the bound malonate molecule and the indole ring of IAA, was defined by residues Arg-115, Lys-158, Ser-108, Pro-171, Leu-173, and Ser-444 (Figure 1B, orange surface). Arg-115 was held in place by Thr-106, Val-520, and Gln-523, with the Gln-523 head group within 3.5 Å of the epsilon nitrogen of the Arg-115 side chain.
Analyses of Mutated GH3-1 Proteins
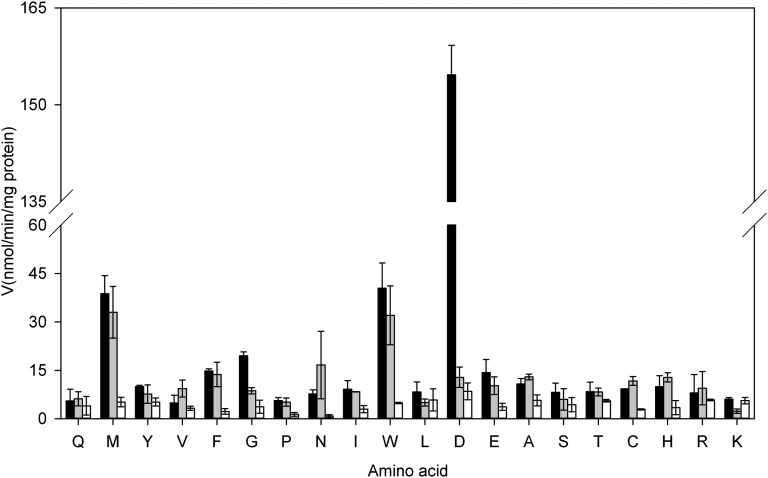
Previous thin layer chromatography (TLC)-based in vitro activity assays with a C-terminal His6-tagged version of grapevine GH3-1 suggested a high selectivity for the amino acid substrate with only Asp and Trp being conjugated (Böttcher et al., 2010). However, a quantitative spectrophotometric assay performed in this study demonstrated that while Asp and Trp were the preferred amino acid substrates of this protein, reduced specific activities could also be observed with the other amino acids tested (see Supplemental Figure 5 online). For all amino acids at least a low level of activity was detected, but Met, Tyr, Phe, Gly, and Glu in particular seemed to be conjugated in addition to Asp and Trp. Changing the His6-tag from the C to the N terminus did not make a difference to the substrate specificity of GH3-1 but increased its activity by about twofold (Figure 6; see Supplemental Figure 5 online). This was also confirmed by TLC analysis where IAA conjugates with Asp, Trp, Met, Tyr, Phe, Gly, and Glu were detected when assaying the N-terminal tagged version of GH3-1 (see Supplemental Figure 6A online). For the C- and N-terminal tagged versions of GH3-1, the identity of the seven main reaction products was further confirmed using liquid chromatography–mass spectrometry (see Supplemental Figures 7 and 8 online). Given the proximity of residues at the very end of the C-terminal domain to a pocket in the N-terminal domain (the Arg-596 side chain was found in a pocket on the surface of the N-terminal domain between residues Glu-114 and Thr-161; Figures 1B and 1C), the addition of 11 additional residues to the C terminus is likely to affect conformational changes during catalysis and thereby cause the observed decrease in activity. Irrespective of the location of the tag, the preferred amino acid substrate for grapevine GH3-1 was found to be Asp (Böttcher et al., 2010, Böttcher et al., 2011a; Figure 6; see Supplemental Figure 5 online), as has been described for many of the auxin-conjugating GH3 enzymes characterized to date (Staswick et al., 2005; Chen et al., 2009; Fu et al., 2011b).
Figure 6.
Amino Acid Specificity of Wild-Type (N-Terminal His6 Tag) and Mutant GH3-1 Proteins.
Initial velocities for wild-type GH3-1 enzyme (black bars), GH3-1-R115K (gray bars), and GH3-1-S108A (white bars) were calculated from the AMP release during the conjugation reaction with 20 amino acids (single letter code). All data represent mean ± se (n = 3).
The N-terminal–tagged version of GH3-1 was further used for the generation of selected mutants via site-directed mutagenesis. The exchange of Ser-108, suggested as essential for the binding of ATP in the first half of the reaction (see above), to Ala led to a mutant protein that showed the same expression and purification behavior as the wild-type GH3-1 protein (see Supplemental Figures 9A and 9C online), but showed a general reduction in specific activities in the spectrophotometric conjugation assay (Figure 6) and accordingly produced no detectable IAA conjugates in a TLC-based activity assay (see Supplemental Figure 6B online).
Based on the binding of malonate in the GH3-1 crystals (see above), Arg-115 was identified as being potentially involved in the selective binding of the preferred amino acid substrate Asp (Figure 1B, orange highlighted surface). An R115K mutant protein was expressed and purified similarly to the wild-type protein (see Supplemental Figure 9B online) and its specific activities determined in the spectrophotometric assay (Figure 6). The specific activity with Asp as the amino acid substrate was reduced by 12-fold, whereas activities with the remaining 19 amino acids were in the range of the wild-type GH3-1 enzyme. In accordance with these data IAA-Met, IAA-Phe, and IAA-Trp were the only three reaction products visible in a TLC screen (see Supplemental Figure 6C online). To confirm further that the binding of Asp was affected in the mutant, a kinetic analysis of IAA-Asp formation by liquid chromatography–electrospray ionization–tandem mass spectrometry (LC-ESI-MS/MS) was performed for R115K and compared with wild-type GH3-1 protein (Table 2). The 14-fold reduction in overall activity of the mutant compared with the wild type was due to a 17-fold decrease in affinity for Asp, as indicated by Km values of 323 μM for wild-type GH3-1 and 5543 μM for R115K, resulting in a 175-fold decrease in catalytic efficiency. The wild-type Km values of 9.89 μM for ATP and 22.04 μM for IAA were in the range of previously published data for GH3-1 (C-terminal His6-tag; Böttcher et al., 2011a,Böttcher et al., 2012). For the R115K mutant, the Asp concentration in standard reaction mixtures was below its Km, which artificially lowered the Km values for ATP and IAA (Table 2).
Table 2. Kinetic Parameters of Wild-Type GH3-1 (N-Terminal His6 Tag) and the R115K Mutant.
| Enzyme | Substrate | Km (μM) | kcat (min−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| GH3-1 | ATP | 9.89 ± 1.92 | 30.2 | 50,863 |
| IAA | 22.04 ± 3.32 | 35.3 | 26,673 | |
| Asp | 323 ± 43 | 40.9 | 2,109 | |
| GH3-1- R115K | ATP | 0.24 ± 0.08 | 2.1 | 144,747 |
| IAA | 14.21 ± 2.15 | 1.3 | 1,507 | |
| Asp | 5,543 ± 897 | 4.0 | 12 |
Km values are expressed as means ± se (n = 3).
Amino Acid Specificities of IAA-Conjugating Grapevine GH3s
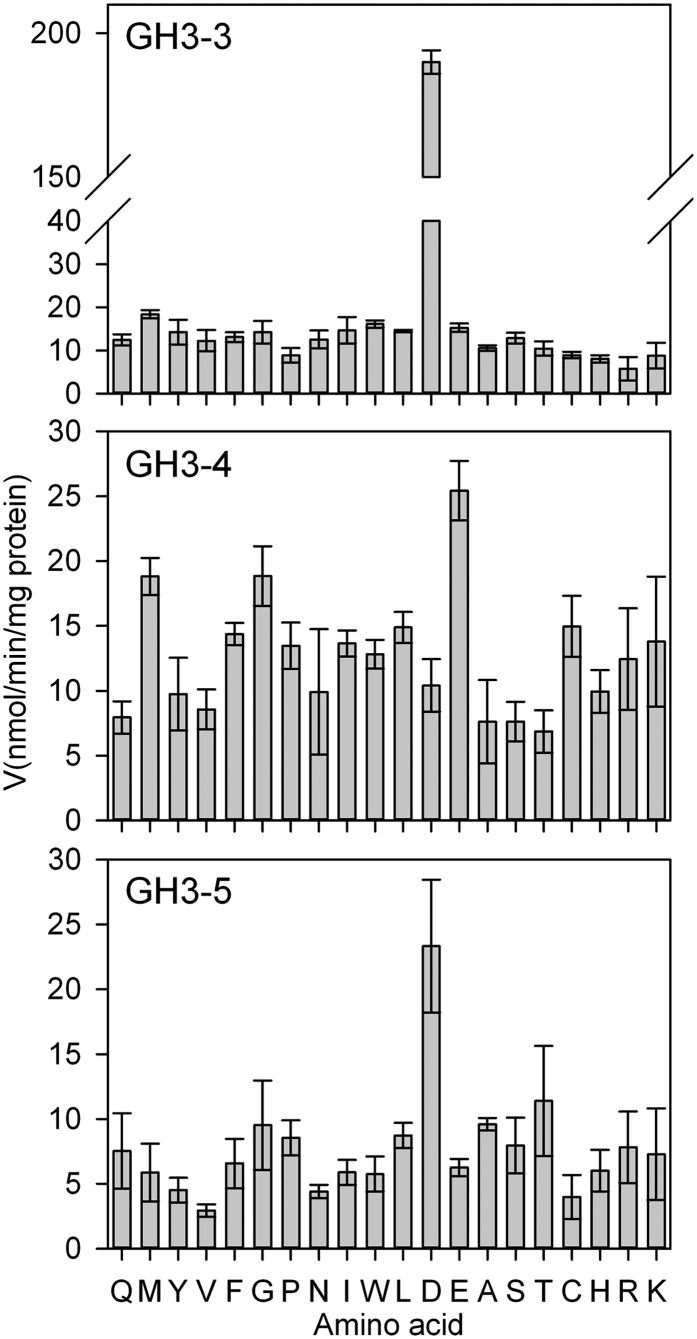
Details of the preference for amino acid substrates for GH3-1, GH3-2, and GH3-6 have been published (Böttcher et al., 2010, Böttcher et al., 2011a, Böttcher et al., 2012). Figure 7 completes this data set by providing the amino acid preference for the GH3-3, GH3-4, and GH3-5 enzymes. The highest specific activity for GH3-3 was with Asp followed by Met and Trp. GH3-5 also preferred Asp, but had an eightfold reduced activity when compared with GH3-3. By contrast, GH3-4 had its highest specific activity with Glu, followed by Met and Gly. As for GH3-5, the highest specific activity observed for GH3-4 was eightfold lower than the highest activity for GH3-3. The results of the spectrophotometric assay were confirmed by TLC screens (see Supplemental Figures 10A to 10C online). The conjugation of Trp was overestimated by the TLC assays since the indole ring of the amino acid enhances the staining of the reaction product.
Figure 7.
Amino Acid Specificity of GH3-3, GH3-4, and GH3-5 Proteins.
Initial velocities for GH3-3, GH3-4, and GH3-5 were calculated from the AMP release during the conjugation reaction with 20 amino acids (single-letter code). All data represent mean ± se (n = 3).
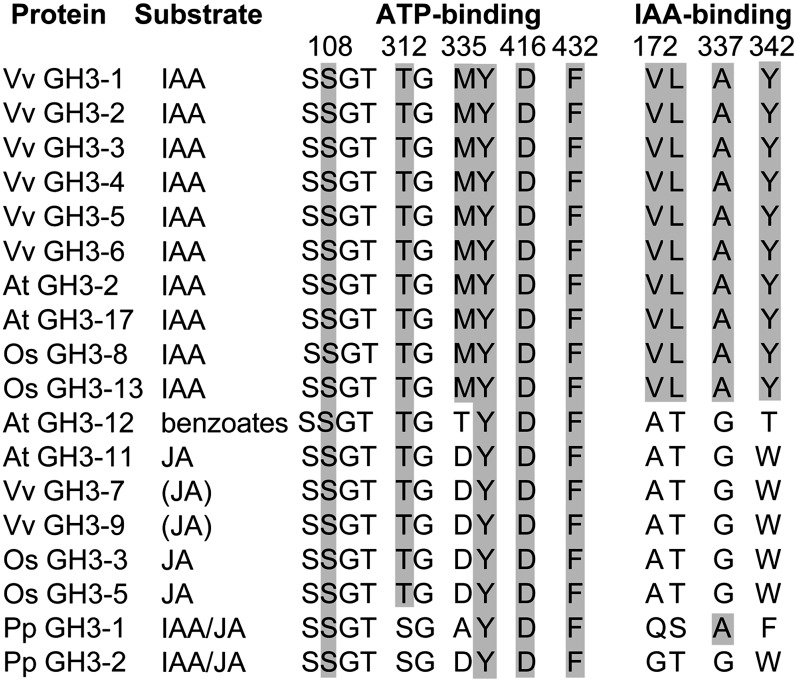
Each of the six IAA-conjugating GH3 enzymes from grapevine displayed their highest in vitro activity with the acidic amino acids Asp (GH3-1, GH3-2, GH3-3, and GH3-5) or Glu (GH3-4 and GH3-6). Interestingly, the preference for either Asp or Glu was reflected in the protein sequences (Figure 8). The residue Arg-115 in GH3-1 was conserved in the four Asp-conjugating grapevine GH3s, whereas Glu-conjugating GH3s had a Pro in that position. The same pattern was also found in IAA-conjugating GH3 enzymes with reported amino acid substrate preferences from Arabidopsis (At GH3-2 to At GH3-6 and At GH3-17) (Staswick et al., 2005) and rice (Os GH3-8 and GH3-13) (Zhang et al., 2009; Chen et al., 2010). In the benzoate- and Glu-conjugating At GH-12 (Okrent et al., 2009; Westfall et al., 2010), Pro was replaced by Gln. Regardless of their substrate preferences, the Ser-108 residue in Vv GH3-1 was conserved in all proteins analyzed (Figure 8), confirming the crucial role of this residue in the binding of ATP (see above).
Figure 8.

Targeted Sequence Comparison of GH3 Proteins with a Known Preference for Asp or Glu as Their Amino Acid Substrate.
Conservation of Ser-108 and Arg-115 was analyzed in GH3 proteins with known preference for Asp and/or Glu. Numbering at the top corresponds to the Vv GH3-1 sequence. Gray boxes highlight the conservation of the two Vv GH3-1 residues. Accession numbers of the sequences used in this analysis are listed in Supplemental Table 2 online. Vv, Vitis vinifera; At, Arabidopsis thaliana; Os, Oryza sativa.
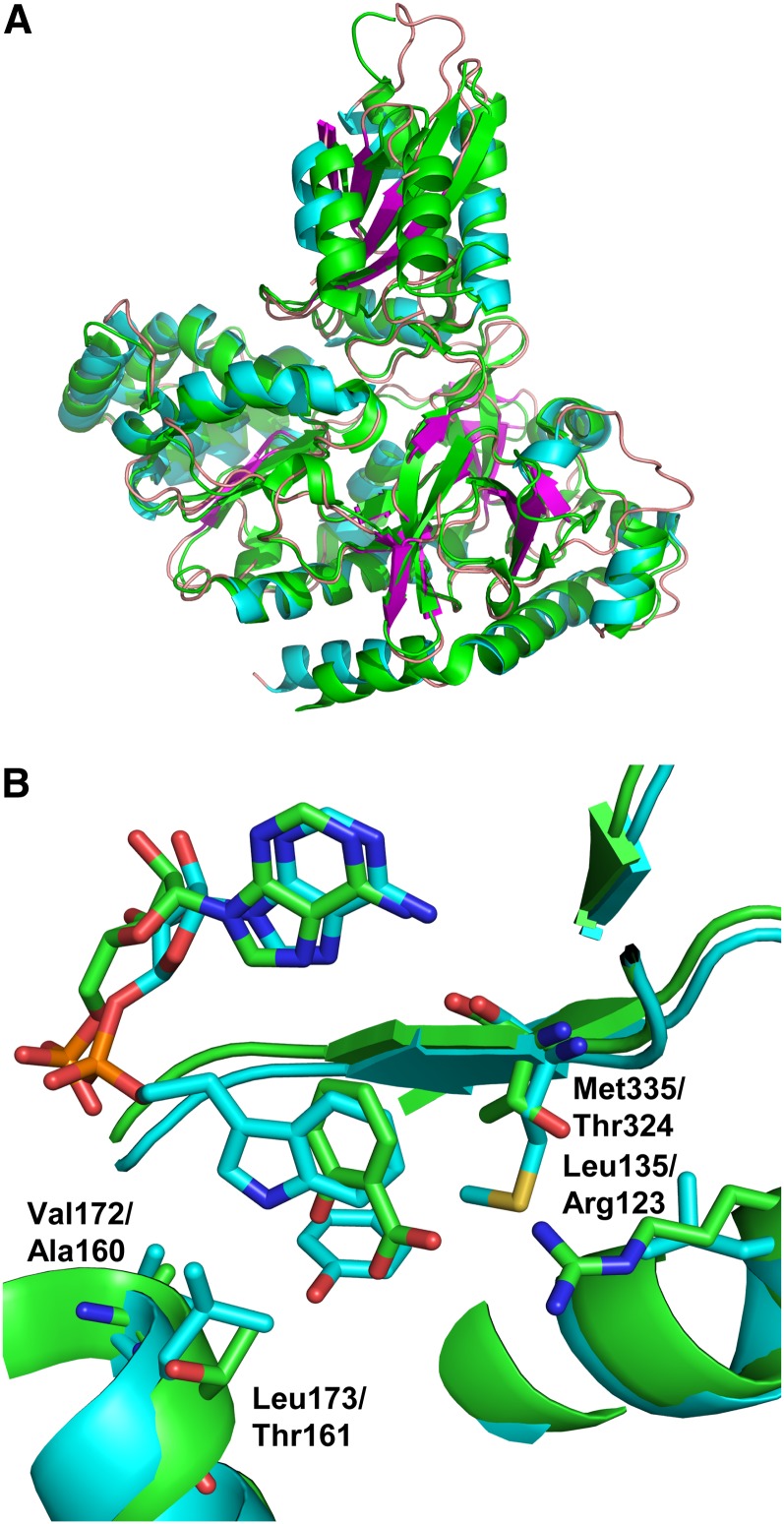
Despite their different acyl- and amino acid substrates, the three-dimensional structures of Vv GH3-1 and At GH3-12 were remarkably similar (Figures 2A, 2B, 3A, and 3B). Differences in their acyl substrate might be explained by some residue exchanges in the substrate binding site. Several residues that are involved in ATP and IAA binding have been identified through their interaction with the inhibitor bound in the GH3-1 crystals (see above) and were analyzed for their conservation in GH3 proteins with known acyl substrates (Figure 9). Vv GH3-7 and Vv GH3-9 were included in this alignment since they cluster with JA-conjugating GH3 proteins (Böttcher et al., 2011a). The phylogenetic position of Vv GH3-8 is unclear (Böttcher et al., 2011a) and its substrates are unknown; it was therefore not included in the analysis. A high degree of conservation was found for residues linked to ATP binding. Irrespective of species and acyl substrate, all selected proteins shared the residues corresponding to Ser-108, Tyr-336, Asp-416, and Phe-432 in Vv GH3-1. Thr-312 was replaced by Ser in the Physcomitrella patens sequences, and Met-335 was found to be restricted to the IAA-conjugating GH3s. A clear distinction between IAA-conjugating and benzoate- or JA-conjugating GH3 enzymes could be made based on four residues that were found to make a likely contribution to IAA binding in Vv GH3-1 (Val-172, Leu-173, Ala-337, and Tyr-342; Figure 9).
Figure 9.
Targeted Sequence Comparison of GH3 Proteins with a Known Preference for Their Acyl Substrate.
Residues likely to be involved in ATP and IAA binding in Vv GH3-1 were analyzed for their conservation in GH3 proteins with the same or different acyl substrates. Numbering at the top corresponds to the Vv GH3-1 sequence. Gray boxes highlight the conservation of the targeted Vv GH3-1 residues. Parentheses indicate the proposed substrate based on sequence similarity. Accession numbers of the sequences used in this analysis are given in Supplemental Table 2 online. Vv, Vitis vinifera; At, Arabidopsis thaliana; Os, Oryza sativa; Pp Physcomitrella patens.
Modeling of Three Auxins into the Active Site of GH3-1
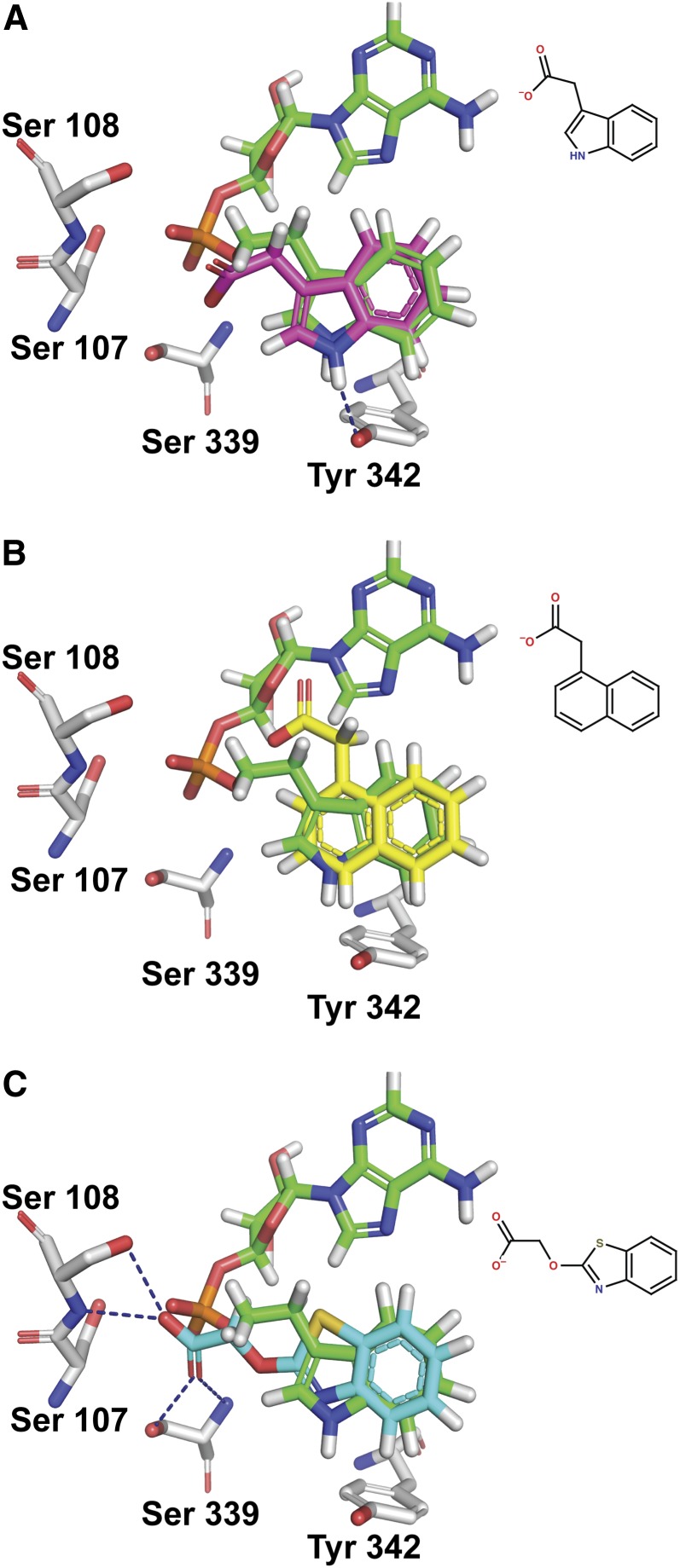
It has been demonstrated previously that two grapevine GH3 proteins display a pronounced preference for IAA as the acyl substrate when compared with NAA and BTOA (Böttcher et al., 2011a). To investigate the possible mechanistic reasons for this, IAA, BTOA, and NAA were docked into the active site of GH3-1 using the molecular docking package FRED (McGann, 2011). The top-scoring conformations of each molecule docked in a way very similar to the indole ring of the inhibitor (Figures 10A to 10C). The indole nitrogen of IAA made a hydrogen bond with Tyr-342, while the carboxylate group was oriented toward Ser-107, Ser-108, and Ser-339 (Figure 10A). The carboxylate groups of NAA and BTOA made similar interactions (Figures 10B and 10C).
Figure 10.
Modeling of IAA, NAA, and BTOA into the Active Site of GH3-1.
IAA with magenta carbons (A), NAA with yellow carbon atoms (B), and BTOA with cyan carbon atoms docked into the GH3-1 active site (C). The inhibitor from the crystal structure is always shown with green carbon atoms, and amino acid residues have gray carbon atoms.
In the case of the top binding conformations for these molecules, it could be seen that despite binding in a similar fashion to IAA, NAA and BTOA could form competitive or nonproductive interactions that may affect adenylation. For NAA, replacing indole with naphthalene eliminated the possibility for a hydrogen bond with Tyr-342 (which in the case of IAA, helps keep the auxin in the bottom of the pocket). This difference manifested as a small but significant change in binding affinity that favored IAA over NAA (see Supplemental Figure 11A online). Additionally, the added steric bulk of the extra carbon in the naphthalene ring caused the carboxylate group to point slightly upwards toward the purine binding portion of the pocket (Figure 10B). As a result, the most stable conformations of NAA may impinge on the binding of ATP.
BTOA bound tightly in the binding pocket predominantly in an orientation with its carboxylate group facing Ser-107 and Ser-108 (Figure 10C). The additional carbon and oxygen leading to the carboxylate group allowed this moiety to form stable hydrogen bonds with these residues. The result of this was that the carboxylate group of the acyl acid was now interacting strongly with the residues responsible for positioning the α-phosphate of ATP for catalysis. NAA and BTOA were most stable when binding in such a way as to interfere with the binding of ATP (see Supplemental Figures 11A to 11D online) and, when AMP was present, had great difficulty binding in the correct orientation (see Supplemental Figures 11C and 11D online). It would seem that NAA can bind in a way that will allow adenylation, but the lack of a hydrogen bond acceptor as well as the added steric bulk leads to potentially nonproductive binding. BTOA, on the other hand, bound quite strongly with residues that are necessary for positioning ATP for catalysis. It can therefore be deduced that NAA and BTOA are less preferred as substrates than IAA for GH3-1 and probably other GH3 proteins.
DISCUSSION
Comparison of the structure presented here for the grapevine GH3-1 enzyme with that published for the Arabidopsis GH3-12 protein (Westfall et al., 2012) shows that their three-dimensional structures are highly conserved despite amino acid sequence divergence (Figures 2A, 2B, 3A, and 3B) and differences in substrate specificities. These proteins have an N-terminal domain (437 amino acids in Vv GH3-1) that contains the bulk of the residues involved in catalysis and a smaller C-terminal domain (161 amino acids in Vv GH3-1). The C-terminal domain of At GH3-12 undergoes a large change in conformation relative to the N-terminal domain during catalysis with a flexible region between the two domains acting as a hinge (Westfall et al., 2012). Such changes in conformation have been observed in other members of the large family of adenylating enzymes (reviewed in Gulick, 2009; Schmelz and Naismith, 2009). Due to the close structural similarities between the grapevine and Arabidopsis proteins, noted above, it can be predicted that the C-terminal domain of the grapevine GH3-1 enzyme also changes in conformation during catalysis.
The large conformational change during catalysis is confirmed by a comparison of the Vv GH3-1 structure to the At GH3-12 structure with an ATP analog bound (Protein Data Bank code 4EWV; Westfall et al., 2012). The AMPCPP-bound At GH3-12 is in the active site open form, while the inhibitor-bound Vv GH3-1 protein is in the active site closed conformation. The two domains in both proteins are similar in secondary structure, but the C-terminal domain is twisted relative to the N-terminal domain in Vv GH3-1 (Figures 3A and 3B). The observed twisting between the two structures shows the considerable movement of the C-terminal domain in relation to the N-terminal domain that occurs during catalysis (Figure 3B). The adenosine moiety of the inhibitor in Vv GH3-1 roughly superposes with the base of ATP and diverges at the α-phosphate, as would be expected. Note that the protein structures do not align in certain regions, particularly in some of the broken regions of the AMPCPP At GH3-12 structure (e.g., Arg-74 to Ser-80 and Gly-378 to Tyr-381).
Similarities and differences in the active sites of the benzoate-conjugating At GH3-12 and the auxin-conjugating Vv GH3-1 were analyzed by comparing the Vv GH3-1 structure to the active site closed At GH3-12 structures with AMP (Figure 2A) or AMP and SA (Figure 2B; Protein Data Bank codes 4EPM and 4EQL; Westfall et al., 2012). The AMP of the At GH3-12 structures superposes well with the AMP moiety of the inhibitor bound in Vv GH3-1. There is a pocket where the superposed indole ring sits and partially superposes with the SA ring that has some residues in common between the two structures: for example, Vv-Phe230 to At-Phe218 and Vv-Val229 to At-Ile217. There are some differences as well: Vv-Val172 to At-Ala160, Vv-Leu173 to At-Thr161, Vv-Tyr342 to At-Thr331, Vv-Leu135 to At-Arg123, and Vv-Met335 to At-Thr324 (Figures 3B and 9). These differences may explain some of the substrate selectivity between the two enzymes. Some of the other residues in common and close to the active site, Arg-435 (At GH3-12 Arg-417) and Lys-446 (At GH3-12 Lys-428), for example, are well defined in the electron density in the Vv GH3-1 structure and do not make direct hydrogen bonds/contacts with the inhibitor.
GH3 proteins, like all adenylate-forming enzymes, require magnesium ions for their activity (Staswick et al., 2005; reviewed in Schmelz and Naismith, 2009). One study on rice GH3-8 reports that manganese ions can substitute for magnesium (Chen et al., 2010). Since all ATP-bound structures published for ANL proteins show at least one magnesium ion in the active site coordinating the phosphate, magnesium is regarded as essential for the adenylation reaction and is hypothesized to leave the active site in conjunction with pyrophosphate (reviewed in Schmelz and Naismith, 2009). In accordance with this hypothesis, no metal ions were identified in the inhibitor-bound, active site closed Vv GH3-1 structure solved in this study. Activity tests with grapevine GH3-1 designed to separately assess the relevance of magnesium ions for the adenylation, and the amino acid transferase reaction demonstrated that magnesium is required only for the adenylation part of the reaction and not for the amino acid transfer (Figure 4).
Chen et al. (2009) suggested that the rice GH3-8 protein functions as a monomeric protein due to its behavior on a size-exclusion column during purification. There are significant (>1000 Å2) surface contacts between protomers in the grapevine GH3-1 crystal structure, but the SAXS data presented here provide proof that oligomers of GH3-1 are unlikely to form in solution.
It has been demonstrated that the naturally occurring auxin IAA and two synthetic auxins, BTOA and NAA, are conjugated with different efficiencies by grapevine GH3-1 and GH3-2 during in vitro assays (Böttcher et al., 2011a). The modeling of these auxin molecules into the GH3-1 protein structure suggests that NAA and BTOA bind in a similar fashion to IAA, but the orientation and affinity are somewhat different, leading mainly to unproductive binding for NAA and potentially competitive binding of ATP with BTOA (Figures 10A to 10C). This demonstrates that quite small changes in the structures of auxin molecules can have large effects on their ability to act as substrates for GH3 proteins, as NAA is conjugated with a greatly reduced efficiency, while BTOA is virtually not conjugated (Böttcher et al., 2011a). Nevertheless, BTOA and NAA can still act as auxins and therefore must be recognized by auxin receptors that presumably have different structural limitations for binding. Indeed, modeling of IAA, NAA, and 2,4-D into the TIR1 receptor binding site has shown that despite their different structures, all three bind well due to the promiscuous nature of the hormone binding site (Tan et al., 2007). The observed order of efficacy as an auxin in grapes (i.e., in increasing order IAA, NAA, and BTOA) (Böttcher et al., 2011a) may therefore be explained by the differences in binding of the auxins in the GH3 protein active site, which in turn influences their longevity in planta.
A study of the evolutionary history of the GH3 family in rosids has indicated that grapevine is a basal rosid whose ancestor had nine GH3 genes (Okrent and Wildermuth, 2011). It was suggested that one of these genes has been lost during evolution to leave grapevine with the eight genes that the authors identified from the genomic sequence. It is interesting to note, however, that another grape GH3-like gene has been identified that clusters with an outlying group of related proteins from rice and poplar (Populus spp; Böttcher et al., 2011a). The high degree of sequence divergence between the nine grapevine GH3 proteins (31 to 86% identity and 51 to 92% similarity; Böttcher et al., 2011a) suggests that the multiple genes arising from duplication have evolved to have different functions, and this is further demonstrated by in vitro activity studies (Böttcher et al., 2010, 2011a, Böttcher et al., 2012; Figures 6 and 7) and sequence comparisons of GH3 proteins with different acyl substrates (Figure 9). In this study, target residues for ATP and IAA binding could be indentified and were examined across a number of GH3 proteins from a moss (P. patens), a monocot (rice), and two dicot species (Arabidopsis and grapevine) (Figure 9). A high degree of conservation was found in residues involved in ATP binding compared with the sequence diversification observed in residues that contribute to the binding of the acyl substrate. It can be concluded that starting from a small number of promiscuous GH3 proteins, as can be found in the moss P. patens (Bierfreund et al., 2004; Ludwig-Müller et al., 2009), proteins with specialized binding sites for IAA, JA, SA, benzoates, and possibly other substrates have evolved. An even higher degree of specialization can be achieved by modifying the affinity for selected amino acid substrates. The grapevine auxin-conjugating GH3 enzymes conjugate a considerable number of amino acids with a wide range of R-group structures (Böttcher et al., 2010, Böttcher et al., 2011a, Böttcher et al., 2012; Figures 6 and 7), but all show a more or less pronounced preference for acidic amino acids. Amino acid residues linked to the preference of apolar and acidic amino acid substrates have previously been reported for a selection of GH3 proteins (Westfall et al., 2012). This study has identified a residue that indicates an even higher degree of discrimination for amino acid substrates. Arg-115 is located in the substrate binding site of Vv GH3-1 and is found exclusively in GH3 proteins preferring Asp over Glu (Figure 8). Furthermore, Arg-115 was shown to be essential for efficient conjugation of Asp by Vv GH3-1 (Figure 6, Table 2). The different preferences for amino acid substrates may have important functional implications for two reasons. The amino acid conjugated can determine the fate of the bound auxin as some conjugates are oxidized (Tsurumi and Wada, 1986; Plüss et al., 1989; Östin et al., 1998), whereas others can release free IAA through hydrolysis (reviewed in Ludwig-Müller, 2011). There is also evidence that some of the conjugates themselves have biological activity in plant development and disease response (Cohen and Baldi, 1983; Magnus et al., 1992; Oetiker and Aeschbacher, 1997; Staswick, 2009; González-Lamothe et al., 2012). It should be kept in mind that the metabolism of IAA conjugates as well as their biological activity may differ from one plant species to another. IAA-Asp is usually regarded as a noncleavable conjugate destined for oxidation, but a number of Medicago truncatula hydrolases have been shown to release IAA from this conjugate in vitro (Campanella et al., 2010). Similarly, IAA-Asp is thought to increase pathogen resistance in rice (Ding et al., 2008) but has been reported to decrease resistance in Arabidopsis (González-Lamothe et al., 2012).
Further evidence for functional divergence within the GH3 family comes from the expression data for the six grape GH3 gene family members that conjugate auxins, which show variation in the transcription of these genes during berry development in response to various inducers and in different tissues (Böttcher et al., 2010, Böttcher et al., 2011a). These data indicate that these genes are under a range of transcriptional controls. Similarly, variations in patterns of transcription have been observed in other species (Gee et al., 1991; Terol et al., 2006; Fu et al., 2011a; Okrent and Wildermuth, 2011; Kumar et al., 2012). If we conclude from the above evidence that there is a degree of specialization among the six grapevine IAA-conjugating enzymes, why are there likely to be only two JA-conjugating enzymes and one enzyme with unknown function? The answer may lie in the importance of auxins throughout plant development. The role of auxins in many developmental processes, including all those that involve growth, means that a range of systems has evolved to provide great flexibility in the regulation of auxin effects. A sophisticated system of auxin conjugation is one way to realize this flexibility. Obviously, biosynthesis, transport, and signaling are additional key points of control providing even further depth to the regulation of auxin-dependent processes.
METHODS
Protein Purification and GH3 Enzyme Assays
Heterologous expression and purification of GH3-1 containing a His6-tag fused to the C terminus were performed as described by Böttcher et al. (2010, Böttcher et al., 2012). The following gene-specific primers were used to clone, express, and purify GH3-3, GH3-4, and GH3-5 in the same way: GH3-3 (5′-TATCATATGCCTGAAGCTCCAAAGAAGTCATTC-3′ and 5′-ATAGCGGCCGCGTTCTTGTTGCACCATTGCT-3′); GH3-4 (5′-TATCATATGGAAGGGAAAAGATTGGAGTACAG-3′ and 5′-ATA-GCGGCCGCAGGGATCCATCTGGGATCTC-3′); GH3-5 (5′-TATATTAATA-TGCCTGAAGCTCCGAAGAACCG-3′ and 5′-ATAGCGGCCGCGGTGCACCATTGCTTATGCC-3′); additional NdeI, AseI, and NotI sites are in bold.
In order to create an N-terminal fusion of the His6-tag with the protein, the coding region of GH3-1 was cloned into the BamHI and NotI restriction sites of a pET-30a(+) vector (Novagen) using gene-specific primers (5′- TATGGATCCATGGCGGTTGATCCGATTCTCTC-3′ and 5′- ATAGCGGCCGCTCACCGCCGGCGTGCCGGAG-3′) with additional BamHI and NotI sites (in bold). Protein expression and purification were performed as described above for the C-terminal fusion protein. Mutated versions of GH3-1 (N-terminal His6-tag) were expressed and purified under the same conditions as the wild-type protein. Spectrophotometric assays of GH3 activity were performed as described by Chen et al. (2010) using 1 mM ATP, 2 mM IAA, 2 mM amino acid, and 20 µg protein as the standard condition. The TLC-based assays for IAA–amino acid conjugate formation were performed as described by Böttcher et al. (2010). Further confirmation of the identity of reaction products via liquid chromatography–mass spectrometry was done as described by Böttcher et al. (2010).
Site-Directed Mutagenesis of GH3-1
The QuikChange PCR method (Stratagene) was used for site-directed mutagenesis of GH3-1. The pET30a(+) construct for the N-terminal His6 fusion protein served as the template in the PCR reactions. Ser at position 108 was exchanged to Ala (primers: 5′-GAATTCCTCACCAGCGCTGGGACATCAGCTG-3′ and 5′-CAGCTGATGTCCCAGCGCTGGTGAGGAATTC-3′) and Arg at position 115 was exchanged to Lys (primers: 5′-CATCAGCTGGTGAGAAGAAACTCATGCCCAC-3′ and 5′-GTGGGCATGAGTTTCTTCTCACCAGCTGATG-3′). Both mutations were confirmed by sequencing, and the proteins were expressed under the conditions described above.
Chemical Synthesis of Labeled IAA-Asp
[Indole-D5]IAA-Asp was synthesized as described previously (Böttcher et al., 2010) using [indole-D5]-labeled IAA as starting substrate.
Determination of Kinetic Parameters
Enzymatic assays were performed as described by Böttcher et al. (2011a). The reactions were initiated by adding 0.5 μg of purified GH3-1 wild-type or mutant protein from glycerol stocks (10% [v/v] glycerol, 1 mM DTT, and 10 to 20 mg/mL protein, stored at −80°C for up to 1 month) and stopped after 10 min by adding 5 μL 50% (v/v) HCl. After the addition of 500 pmol of labeled IAA-Asp as internal standard, the samples were extracted twice with 150 μL ethyl acetate. The extract was dried and the residue resuspended in 30 μL 60% (v/v) methanol and 1% (v/v) acetic acid to be analyzed by LC-ESI-MS/MS as described previously (Böttcher et al., 2011a). Steady state parameters for GH3 enzymes were determined using the standard assay but varying the concentration of one substrate at a time: (0 to 5 mM) Asp, (0 to 1 mM) ATP, and (0 to 1 mM) IAA. All data were fitted to the Michaelis-Menten equation using nonlinear regression (SigmaPlot 12.0).
Metal Binding Assay
Enzymatic assays were performed as described above, but the reaction volume was increased to 250 μL, the MgCl2 concentration was decreased to 1 mM, and reactions were initiated by adding 50 μg of purified GH3-1 wild-type protein (either with C-terminal or N-terminal His6-tag). The importance of magnesium ions for the adenylation reaction as well as the amino acid conjugation was tested by adding 20 mM EDTA to the reaction mix (1) prior to the addition of protein or (2) after a 15-min incubation period of protein with a reaction mix lacking Asp. After an incubation period of 15 min either protein (1) or Asp (2) was added to the reaction mix followed by another 20-min incubation period. Reactions without EDTA served as positive controls. All reactions were stopped, extracted, and analyzed by LC-ESI-MS/MS as described above.
Crystallization and Data Collection
C-terminal His6-tagged GH3-1 was concentrated to ∼6 mg/mL in 50 mM Tris, pH 8.6, in the presence of 10 mM inhibitor and additional magnesium and set up in low-volume crystallization trials. All crystallization experiments were performed in 96-well SD-2 sitting drop plates (IDEX) with drop volumes of 300 to 500 nL at 20°C. Small box-like crystals were found in an in-house salt buffer screen, and these crystals were optimized by varying the salt concentration and precipitant pH. The best crystals were grown from 0.7 to 1.0 M sodium malonate at pH 7.0 and the addition of various buffers between pH 6.0 and 8.0 (or no buffer at all). Crystals grew to a maximum size of 250 × 60 × 60 μm over 30 d. Crystals were cryo-protected by soaking in higher concentrations of sodium malonate and flash-cooled before data collection at the microfocus beamline (MX2) of the Australian Synchrotron. Diffraction patterns were consistent with a tetragonal space group of P41212 or P43212 with a unit cell of ∼91 × 91 × 338 Å (the data were reduced with XDS [Kabsch, 2010] and scaled with SCALA [Collaborative Computational Project, Number 4, 1994]). The c-axis varied considerably from 328 to 341 Å or almost 4% (the a- and b-axes varied from 90.4 to 92.3 Å, ∼2%). To obtain phase information, a variety of heavy atoms, including Pt, were soaked into the crystals. One of the Pt soaks showed some promise, and a further set of Pt compounds was subsequently soaked into a new batch of crystals to obtain sufficient phase information to solve the structure. The diamino platinum dinitrate soak proved to be the best diffracting crystal with anomalous scattering present, with the diffraction extending to ∼2.9 Å resolution at wavelength 1.01126 Å.
Structure Solution and Refinement
AutoRickshaw (Panjikar et al., 2009) was used to obtain 14 Pt sites, to refine 12 of the sites, to identify the correct enantiomer of the space group (P41212), and to build a partial structure using the MR-SAD technique. The correlation coefficient for the best SHELXD solution was CC(All) = 29.08 and CC(weak) = 14.98. Density modification gave a figure of merit = 0.596 and a real space FreeR of 0.196, providing evidence that the true space group was P41212. The partial structure from AutoRickshaw was extended by hand using Coot (Emsley et al., 2010) and refined using Refmac (Murshudov et al., 1997). A higher resolution native data set was then used to extend the resolution limit to 2.40 Å (the partially refined Pt structure was used as a model in CCP4 Phaser for molecular replacement; McCoy et al., 2007). Soaks and cocrystallization with Mn were done to see whether a metal binding site could be located in the structure. These data sets did not give a clear indication of metal binding, and no Mg site was obvious in the density of any of the native structures. The inhibitor showed clear density in both the A and B chains and made similar contacts in each; the average B-factor for the inhibitor in both protomers was 21.7 Å2.
SAXS Data Measurements
SAXS measurements were made at the SAXS-WAXS beamline of the Australian Synchrotron, Melbourne, Australia. For each SAXS measurement, 10 1-s exposures were measured and averaged. During collection, the sample was flowing through a 1-mm quartz capillary to control for radiation damage. Measurements were performed on a dilution series of GH3-1 protein (N-terminal His6-tagged protein was cleaved with Enterokinase overnight and was subsequently diluted and reconcentrated in an Amicon 30-kD filter in 125 mM NaCl and 50 mM HEPES, pH 7.5, to remove the cleaved His6 tag) from 3 to 0.1 mg/mL. The molecular weights of the scattering species were estimated from the total forward scatter of the SAXS measurements that were normalized by comparison to water scatter and with reference to the measured protein concentrations. The monomeric state of GH3 protein was inferred by comparison of the theoretical molecular weight of the protein sequence with the calculated molecular weight from the SAXS experiment. A 1.6-m camera was used and a beam energy of 12 keV. Measurements were made at room temperature (∼25°C).
SAXS Data Analysis
Data analysis was performed using version 2.3.1 of the ATSAS suite of software. The program CRYSOL (Svergun et al., 1995) was used to calculate scatter from both monomeric and putative dimeric forms of GH3-1 from the x-ray crystal structures and to compare the calculated scatter with the measured data (see Supplemental Figures 2A, 2B, and 3A to 3C online). As there was no discernable change in the SAXS data as a function of concentration (see Supplemental Table 1 and Supplemental Figures 2A and 2B online), the highest protein concentration data was used for modeling (3 mg/mL) giving the highest signal-to-noise ratio.
Modeling of Auxins into the Active Site
IAA, BTOA, and NAA were docked into the active site of GH3-1 using the molecular docking package FRED (OpenEye Scientific Software). The structure with the inhibitor bound was used for the docking, and auxins were docked into both apo and holo (AMP bound) pockets. Protonation states and partial atomic charges were set for BTOA, IAA, and NAA using a pH 7.0 protonation-state model (these should not be significantly different at pH 8.5 as no His residues are in the vicinity in the protein and the calculated pKas of the substrates are not in this range either) and the AM1BCC Hamiltonian (Jakalian et al., 2002) as implemented in Quacpac (OpenEye Scientific Software). Conformations were generated with Omega2 (OpenEye Scientific Software). Hydrogens were added to GH3-1 using Reduce (Word et al., 1999). All docking calculations were performed with FRED 2.2.5. The top 100 poses were ranked using a consensus method based on the following scoring functions: Chemgauss3, Chemscore, OEChemscore, PLP, Shapegauss, and CGO (McGann, 2011). The top 10 poses were rescored using the PBSA method (Zapbind as implemented in FRED) to estimate binding affinity (Nicholls et al., 2009).
Figure Preparation
All crystallographic and modeling figures were produced with PyMol.
Accession Numbers
Coordinates and structure factors can be found in the Protein Data Bank under accession code 4B2G. Accession numbers for the proteins used in the experiments are as follows: Vv GH3-1, XP_002271252; Vv GH3-3, XP_002283229; Vv GH3-4, XP_002263353; and Vv GH3-5, XP_002276241. Accession numbers for sequences used in targeted sequence comparisons are listed in Supplemental Table 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Crystals of GH3-1.
Supplemental Figure 2. SAXS Raw Data and Guinier Plots.
Supplemental Figure 3. GH3-1 Dilution Series and Calculated SAXS Curves.
Supplemental Figure 4. Difference Fourier Maps of the GH3 Inhibitor Bound to the GH3-1 Protein.
Supplemental Figure 5. Amino Acid Specificity of GH3-1 (C-Terminal His6 Tag).
Supplemental Figure 6. TLC screen for in Vitro Function of Wild-type and Mutant GH3-1 Proteins.
Supplemental Figure 7. LC-MS Analysis of Amino Acid Conjugates Synthesized by Wild-Type GH3-1 Protein (N-Terminal His6 Tag).
Supplemental Figure 8. LC-MS Analysis of Amino Acid Conjugates Synthesized by Wild-Type GH3-1 Protein (C-Terminal His6 Tag).
Supplemental Figure 9. Expression of Recombinant GH3-1 Proteins.
Supplemental Figure 10. TLC Screen for in Vitro Function of GH3-3, GH3-4, and GH3-5 Proteins.
Supplemental Figure 11. Box-Whisker Plots of Binding Energies and Conformational Preferences for Auxins to GH3-1.
Supplemental Table 1. SAXS Data.
Supplemental Table 2. Accession Numbers for Sequences Used in Targeted Sequence Comparisons.
Acknowledgments
We thank the beamline scientists at the Australian Synchrotron for their help with data collection and in particular Santosh Panjikar for his help with AutoRickshaw. The crystallization trials were all performed at the CSIRO Collaborative Crystallization Centre (C3). We also thank Crista Burbidge for assistance with the article and OpenEye Scientific Software for an academic license to use their software.
AUTHOR CONTRIBUTIONS
T.S.P., C.B., J.N., and C.D. designed the research. T.S.P., C.B., J.N., D.L., and N.C. performed the research. T.S.P., C.B., J.N., D.L., and N.C. analyzed data. T.S.P, C.B, J.N., and C.D. wrote the article with contributions from D.L. and N.C.
Glossary
- IAA
indole-3-acetic acid
- JA
jasmonic acid
- SA
salicylic acid
- NAA
1-naphthaleneacetic acid
- BTOA
benzothiazole-2-oxyacetic acid
- AIEP
adenosine-5′-[2-(1H-indol-3-yl)ethyl]phosphate
- SAXS
small-angle x-ray scattering
- TLC
thin layer chromatography
- LC-ESI-MS/MS
liquid chromatography–electrospray ionization–tandem mass spectrometry
References
- Bierfreund N.M., Tintelnot S., Reski R., Decker E.L. (2004). Loss of GH3 function does not affect phytochrome-mediated development in a moss, Physcomitrella patens. J. Plant Physiol. 161: 823–835 [DOI] [PubMed] [Google Scholar]
- Böttcher C., Boss P.K., Davies C. (2011a). Acyl substrate preferences of an IAA-amido synthetase account for variations in grape (Vitis vinifera L.) berry ripening caused by different auxinic compounds indicating the importance of auxin conjugation in plant development. J. Exp. Bot. 62: 4267–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C., Dennis E.G., Booker G.W., Polyak S.W., Boss P.K., Davies C. (2012). A novel tool for studying auxin-metabolism: the inhibition of grapevine indole-3-acetic acid-amido synthetases by a reaction intermediate analogue. PLoS ONE 7: e37632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C., Harvey K., Forde C.G., Boss P.K., Davies C. (2011b). Auxin treatment of pre-veraison grape (Vitis vinifera L.) berries both delays ripening and increases the synchronicity of sugar accumulation. Aust. J. Grape Wine Res. 17: 1–8 [Google Scholar]
- Böttcher C., Keyzers R.A., Boss P.K., Davies C. (2010). Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J. Exp. Bot. 61: 3615–3625 [DOI] [PubMed] [Google Scholar]
- Campanella J.J., Sigethy S., Ludwig-Müller J. (2010). Truncation of Medicago truncatula auxin conjugate hydrolases alters substrate specificity. Plant Mol. Biol. Rep. 29: 745–752 [Google Scholar]
- Chang K.-H., Xiang H., Dunaway-Mariano D. (1997). Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site-directed mutagenesis study with the Pseudomonas sp. strain CBS3 4-chlorobenzoate:coenzyme A ligase. Biochemistry 36: 15650–15659 [DOI] [PubMed] [Google Scholar]
- Chen Q., Westfall C.S., Hicks L.M., Wang S., Jez J.M. (2010). Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J. Biol. Chem. 285: 29780–29786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zhang B., Hicks L.M., Wang S., Jez J.M. (2009). A liquid chromatography-tandem mass spectrometry-based assay for indole-3-acetic acid-amido synthetase. Anal. Biochem. 390: 149–154 [DOI] [PubMed] [Google Scholar]
- Cohen J.D., Baldi B.G. (1983). Studies of endogenous indole-3-acetyl-L-aspartate during germination of soybeans. In Proceedings of Plant Growth Regulator Society of America, A.R. Cooke, ed (Lake Alfred, Florida: Plant Growth Regulator Society of America), pp. 117–122 [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Davies P.J. (2010). Plant Hormones: Biosynthesis, Signal Transduction, Action! (Dordrecht, The Netherlands: Springer) [Google Scholar]
- Ding X., Cao Y., Huang L., Zhao J., Xu C., Li X., Wang S. (2008). Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo C., Andrés F., Tharreau D., Iglesias D.J., Talón M. (2009). Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant Microbe Interact. 22: 201–210 [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Liu H., Li Y., Yu H., Li X., Xiao J., Wang S. (2011b). Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 155: 589–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Yu H., Li X., Xiao J., Wang S. (2011a). Rice GH3 gene family: Regulators of growth and development. Plant Signal. Behav. 6: 570–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee M.A., Hagen G., Guilfoyle T.J. (1991). Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs. Plant Cell 3: 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe R., El Oirdi M., Brisson N., Bouarab K. (2012). The conjugated auxin indole-3-acetic acid-aspartic acid promotes plant disease development. Plant Cell 24: 762–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick A.M. (2009). Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4: 811–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakalian A., Jack D.B., Bayly C.I. (2002). Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 23: 1623–1641 [DOI] [PubMed] [Google Scholar]
- Kabsch W. (2010). XDS. Acta Crystallogr. D Biol. Crystallogr. 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Stone J.M. (2007). Arabidopsis thaliana GH3.9 influences primary root growth. Planta 226: 21–34 [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Kumar R., Agarwal P., Tyagi A.K., Sharma A.K. (2012). Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum). Mol. Genet. Genomics 287: 221–235 [DOI] [PubMed] [Google Scholar]
- LeClere S., Tellez R., Rampey R.A., Matsuda S.P.T., Bartel B. (2002). Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277: 20446–20452 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J. (2011). Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 62: 1757–1773 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J., Jülke S., Bierfreund N.M., Decker E.L., Reski R. (2009). Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol. 181: 323–338 [DOI] [PubMed] [Google Scholar]
- Magnus V., Hangarter R., Good N. (1992). Interaction of free indole-3-acetic acid and its amino acid conjugates in tomato hypocotyl cultures. J. Plant Growth Regul. 11: 67–75 [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. (2007). Phaser crystallographic software. J. Appl. Cryst. 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGann M. (2011). FRED pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 51: 578–596 [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nakazawa M., Yabe N., Ichikawa T., Yamamoto Y.Y., Yoshizumi T., Hasunuma K., Matsui M. (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25: 213–221 [DOI] [PubMed] [Google Scholar]
- Nicholls A., Wlodek S., Grant J.A. (2009). The SAMP1 solvation challenge: Further lessons regarding the pitfalls of parametrization. J. Phys. Chem. B 113: 4521–4532 [DOI] [PubMed] [Google Scholar]
- Oetiker J.H., Aeschbacher G. (1997). Temperature-sensitive plant cells with shunted indole-3-acetic acid conjugation. Plant Physiol. 114: 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okrent R.A., Brooks M.D., Wildermuth M.C. (2009). Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 284: 9742–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okrent R.A., Wildermuth M.C. (2011). Evolutionary history of the GH3 family of acyl adenylases in rosids. Plant Mol. Biol. 76: 489–505 [DOI] [PubMed] [Google Scholar]
- Östin A., Kowalyczk M., Bhalerao R.P., Sandberg G. (1998). Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 118: 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panjikar S., Parthasarathy V., Lamzin V.S., Weiss M.S., Tucker P.A. (2009). On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination. Acta Crystallogr. D Biol. Crystallogr. 65: 1089–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plüss R., Jenny T., Meier H. (1989). IAA-induced adventitious root formation in greenwood cuttings of Populus tremula and formation of 2-indolone-3-acetylaspartic acid, a new metabolite of exogeneously applied indole-3-acetic acid. Physiol. Plant. 75: 89–96 [Google Scholar]
- Savić B., Tomić S., Magnus V., Gruden K., Barle K., Grenković R., Ludwig-Müller J., Salopek-Sondi B. (2009). Auxin amidohydrolases from Brassica rapa cleave the alanine conjugate of indolepropionic acid as a preferable substrate: A biochemical and modeling approach. Plant Cell Physiol. 50: 1587–1599 [DOI] [PubMed] [Google Scholar]
- Schmelz S., Naismith J.H. (2009). Adenylate-forming enzymes. Curr. Opin. Struct. Biol. 19: 666–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E. (2009). The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 150: 1310–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Serban B., Rowe M., Tiryaki I., Maldonado M.T., Maldonado M.C., Suza W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I., Rowe M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D., Barberato C., Koch M.H.J. (1995). CRYSOL - A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 28: 768–773 [Google Scholar]
- Takase T., Nakazawa M., Ishikawa A., Kawashima M., Ichikawa T., Takahashi N., Shimada H., Manabe K., Matsui M. (2004). ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 37: 471–483 [DOI] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I.A., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Terol J., Domingo C., Talón M. (2006). The GH3 family in plants: Genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371: 279–290 [DOI] [PubMed] [Google Scholar]
- Tsurumi S., Wada S. (1986). Dioxindole-3-acetic acid conjugates formation from indole-3-acetylaspartic acid in Vicia seedlings. Plant Cell Physiol. 27: 1513–1522 [Google Scholar]
- Wang H., Tian C.-E., Duan J., Wu K. (2008). Research progresses on GH3s, one family of primary auxin-responsive genes. Plant Growth Regul. 56: 225–232 [Google Scholar]
- Westfall C.S., Herrmann J., Chen Q., Wang S., Jez J.M. (2010). Modulating plant hormones by enzyme action: The GH3 family of acyl acid amido synthetases. Plant Signal. Behav. 5: 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall C.S., Zubieta C., Herrmann J., Kapp U., Nanao M.H., Jez J.M. (2012). Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 336: 1708–1711 [DOI] [PubMed] [Google Scholar]
- Word J.M., Lovell S.C., Richardson J.S., Richardson D.C. (1999). Asparagine and glutamine: Using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 285: 1735–1747 [DOI] [PubMed] [Google Scholar]
- Zhang S.-W., Li C.-H., Cao J., Zhang Y.-C., Zhang S.-Q., Xia Y.-F., Sun D.-Y., Sun Y. (2009). Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 151: 1889–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li Q., Li Z., Staswick P.E., Wang M., Zhu Y., He Z. (2007). Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 145: 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]