PAN2 functions with PAN1, a Leu-rich repeat–receptor-like kinase (LRR-RLK) to polarize the divisions that form stomatal subsidiary cells in maize. Quantitative proteomics was used to identify PAN2 as a second LRR-RLK. PAN2 functions upstream of PAN1, potentially perceiving extracellular cues that initiate or amplify premitotic subsidiary mother cell polarity.
Abstract
Mechanisms governing the polarization of plant cell division are poorly understood. Previously, we identified pangloss1 (PAN1) as a leucine-rich repeat–receptor-like kinase (LRR-RLK) that promotes the polarization of subsidiary mother cell (SMC) divisions toward the adjacent guard mother cell (GMC) during stomatal development in maize (Zea mays). Here, we identify pangloss2 (PAN2) as a second LRR-RLK promoting SMC polarization. Quantitative proteomic analysis identified a PAN2 candidate by its depletion from membranes of pan2 single and pan1;pan2 double mutants. Genetic mapping and sequencing of mutant alleles confirmed the identity of this protein as PAN2. Like PAN1, PAN2 has a catalytically inactive kinase domain and accumulates in SMCs at sites of GMC contact before nuclear polarization. The timing of polarized PAN1 and PAN2 localization is very similar, but PAN2 acts upstream because it is required for polarized accumulation of PAN1 but is independent of PAN1 for its own localization. We find no evidence that PAN2 recruits PAN1 to the GMC contact site via a direct or indirect physical interaction, but PAN2 interacts with itself. Together, these results place PAN2 at the top of a cascade of events promoting the polarization of SMC divisions, potentially functioning to perceive or amplify GMC-derived polarizing cues.
INTRODUCTION
Asymmetric cell divisions, which give rise to daughters with distinct developmental fates, are an important mechanism for the generation of cell diversity during plant development (Abrash and Bergmann, 2009; Menke and Scheres, 2009). Such divisions are often physically asymmetric as well, producing daughters with distinct sizes and/or shapes. Many observations suggest mechanistic links between physical and developmental asymmetry (Gallagher and Smith, 2000; Song et al., 2008; Dong et al., 2009). Moreover, orientation of division polarity is crucial for proper placement of the daughter cells within developing tissues to create functional cellular arrangements. Thus, polarization of cell division is a process of fundamental importance for plant development.
In preparation for a physically asymmetric plant cell division, the mother cell polarizes, which involves actin-dependent migration of the premitotic nucleus into the future division plane where the preprophase band later forms (Rasmussen et al., 2011). Premitotic division polarity may be determined by intrinsic cues (preexisting spatial landmarks within the mother cell) or extrinsic cues (spatial cues originating from outside the mother cell; Facette and Smith, 2012). After entry into mitosis, the dividing nucleus is retained within the future division plane, and the cell plate is ultimately attached there at the conclusion of cytokinesis through interactions between the cortical division site and the expanding phragmoplast/cell plate (Rasmussen et al., 2011).
In plants, where pathways and most proteins known to govern division polarity in animal cells (reviewed in Gönczy, 2008) appear to be lacking, relatively little is known in mechanistic terms about how division asymmetry is achieved. Stomatal development has provided a useful focus for studies of division asymmetry. In Arabidopsis thaliana, asymmetric divisions create stomatal precursor cells while generating a pattern that ensures a minimum of one nonstomatal cell separating neighboring stomata. Ligand–receptor interactions act through a mitogen-activated protein kinase signaling cascade to regulate the occurrence and orientation of stomate-forming asymmetric divisions (Pillitteri and Torii, 2012). BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) and POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION (POLAR) act downstream to promote division polarity (Dong et al., 2009; Pillitteri et al., 2011).
In maize (Zea mays), an invariant sequence of asymmetric and symmetric divisions generates stomatal complexes consisting of a pair of guard cells flanked by a pair of subsidiary cells that regulate stomatal aperture. The first asymmetric division generates a guard mother cell (GMC), which is believed to signal its lateral neighbors, the subsidiary mother cells (SMCs), to divide asymmetrically in an orientation that positions the smaller daughter (the subsidiary cell) adjacent to the GMC (Farquharson, 2012; Stebbins and Shah, 1960). Asymmetric SMC divisions are preceded by localized accumulation of cortical F-actin at the SMC-GMC interface and migration of the premitotic SMC nucleus to that site (Galatis and Apostolakos, 2004). Subsequently, the GMC divides symmetrically to form a pair of guard cells flanked by the subsidiary cells.
Prior work has identified a leucine-rich repeat–receptor-like kinase (LRR-RLK), pangloss1 (PAN1), which promotes the polarization of SMC divisions and thus might function as a receptor for GMC-derived polarizing cues (Cartwright et al., 2009). Shortly after GMC formation, PAN1 localizes asymmetrically in SMCs, accumulating at the site of GMC contact prior to nuclear polarization to that site. More recently, Type I rho of plants (ROP) GTPases were shown to act downstream of PAN1 to promote SMC polarization (Humphries et al., 2011). Partial loss of Type I ROP function results in mild SMC polarization defects, and rop mutations dramatically enhance the pan1 phenotype. Like PAN1, Type I ROPs localize at the SMC surface as a patch at the site of GMC contact but ROP patches form later than PAN1 patches. PAN1 appears to recruit ROPs through a physical interaction as indicated by coimmunoprecipitation of PAN1 and ROPs. Phenotypes resulting from partial loss of ROP function or depolarization of ROP indicate that polarized accumulation of ROPs leads to localized accumulation of F-actin and nuclear polarization, but the links between ROPs and these downstream events are unclear.
Here, we use a quantitative proteomic approach to identify a second LRR-RLK promoting SMC polarization, pangloss2 (PAN2). Analysis of PAN2 reveals that its localization and function are similar to PAN1, but PAN2 acts upstream of PAN1. Thus, PAN2 is the earliest acting component of the SMC-polarizing mechanism identified to date.
RESULTS
PAN1 and PAN2 Interact Genetically
As described previously (Cartwright et al., 2009), pan2 mutations have similar effects on stomatal subsidiary cell formation compared with pan1. In this study, 22 to 32% of stomatal subsidiaries formed aberrantly in mutants homozygous for either of two different mutant alleles of each gene (Figures 1A, arrowheads, and 1B). To explore the functional relationship between pan1 and pan2, we generated double mutants homozygous for three different combinations of pan1 and pan2 alleles. Like the single mutants, the overall morphology of double mutant plants was not markedly different from the wild type. However, as shown in Figures 1A and 1B, all double mutants had a synergistic phenotype with a high frequency of aberrant subsidiary cells that was far more than the sum of the frequencies seen in the corresponding single mutants.
Figure 1.
Analysis of pan Single and Double Mutant Phenotypes.
(A) Fixed epidermal preparations from leaf 3 (counting leaf 1 as the first leaf made by the plant) of the indicated genotypes (B73 wild type, pan1-ems, pan2-O, and pan1-Mu;pan2-O) were stained with concentrated propidium iodide to visualize cell walls and nuclei. Arrowheads point to examples of aberrant subsidiary cells in mutants. Bar = 100 µm.
(B) Cyanoacrylate glue impressions of the abaxial surface of leaf 3 from four to six plants of each of the indicated genotypes were analyzed to determine the percentage of abnormal stomatal subsidiary cells (n > 75 subsidiary cells analyzed per individual). Error bars indicate sd.
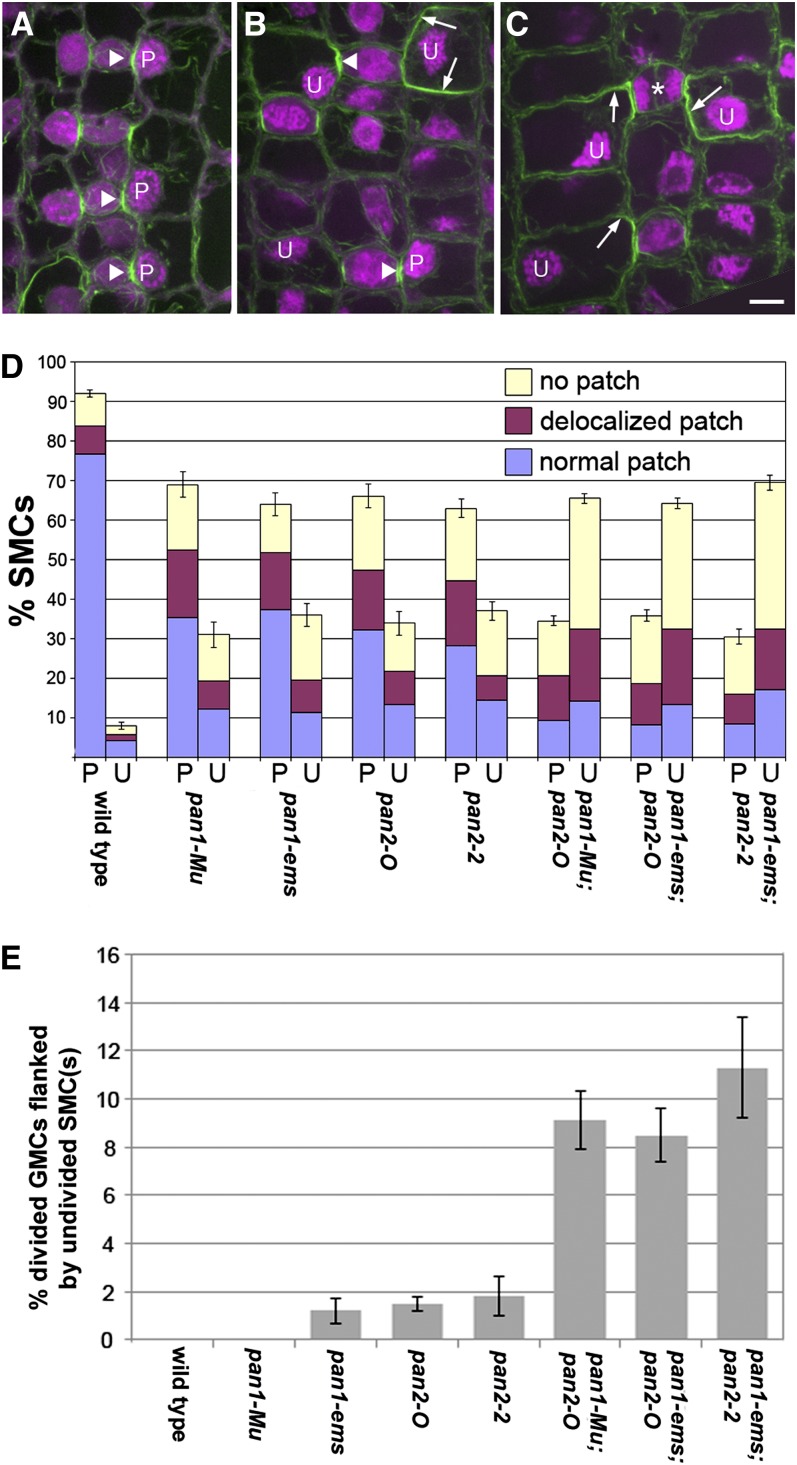
We extended this analysis to consider the developmental origins of aberrant stomatal subsidiaries in pan1;pan2 double mutants. Similar to what we reported previously (Cartwright et al., 2009; Humphries et al., 2011), 76% of wild-type SMCs at the developmental stage chosen for analysis are conspicuously polarized with nuclei localized at the GMC interface and a tightly focused patch of cortical F-actin at that site (Figures 2A and 2D). Although the function of this actin patch is unknown, it serves as a useful marker of polarity in these cells. Also as described previously (Cartwright et al., 2009), pan1 and pan2 single mutants exhibit defects in SMC polarization, including an elevated frequency of SMCs with unpolarized nuclei and/or a delocalized F-actin patch or no actin patch (Figures 2B and 2D). These SMC polarity defects were greatly exaggerated in pan1;pan2 double mutants. The frequency of SMCs with polarized nuclei and a properly localized F-actin patch was reduced from 76% in the wild type and 28 to 37% in single mutants to <10% in all three double mutant combinations tested with corresponding increases in the frequency of nuclear polarity and F-actin patch defects (Figures 2C and 2D). Thus, synergism between pan1 and pan2 mutations is also observed at the level of SMC polarization. Unexpectedly, we found that a substantial proportion of double mutant SMCs appeared to be delayed in polarization and division as indicated by the finding that 8 to 11% of recently divided GMCs were flanked by SMCs that had not yet divided (Figures 2C, asterisk, and 2E). This was not observed in wild-type leaves, and rarely if ever (<2% of GMCs) in any of the pan single mutants (Figure 2E). This finding further indicates synergism between pan1 and pan2 in double mutants and suggests a previously unrecognized role for both genes in regulating the timing of SMC division. The synergistic phenotypes observed in pan1;pan2 double mutants indicate that pan1 and pan2 act cooperatively to promote subsidiary cell formation or are partially redundant.
Figure 2.
Analysis of SMC Polarization and Division Defects in pan Single and Double Mutants.
(A) to (C) F-actin (green) and propidium iodide–stained nuclei (magenta) in developing stomata of B73 wild type (A), pan2-2 single mutants (B), and pan1-ems:pan2-2 double mutants (C). Arrowheads in (A) and (B) lie on top of GMCs and point to normal actin patches in adjacent SMCs; arrows in (B) and (C) point to areas of ectopic cortical actin accumulation in mutant SMCs. Asterisk marks a divided GMC flanked by undivided SMCs. Polarized and unpolarized SMC nuclei are marked P and U, respectively. Bar = 10 µm.
(D) Quantification of the proportion of SMCs with polarized (P) or unpolarized (U) nuclei and actin patch status (>300 cells and four plants analyzed for each genotype). Error bars represent sd.
(E) Quantification of the occurrence of divided GMCs flanked by undivided SMCs (as illustrated by the example in [C]) in plants of each genotype indicated. This analysis used the same collection of images as that presented in (D); error bars represent sd.
Quantitative Proteomic Analysis Reveals Changes in Membrane Protein Accumulation in pan Single and Double Mutants
To identify proteins potentially functioning in common pathways with PAN1 and PAN2, we performed a comparative proteomic analysis using the isobaric tags for relative and absolute quantitation (iTRAQ) method (see Supplemental Figure 1 online). Reasoning that proteins of greatest interest were likely to be membrane localized, we isolated membrane proteins from the basal portions of unexpanded maize leaves (enriched in dividing cells, including those dividing to form stomata) from wild- type, pan1, pan2, and pan1;pan2 mutant plants. Tryptic peptides from each of the four membrane protein preparations were labeled with distinct iTRAQ tags and then mixed. The tags have identical masses, but after fragmentation they can be distinguished from one another by mass spectrometry, permitting assessment of the proportion of each peptide in the mixture derived from each genotype.
Peptides mapping to a total of 13,101 possible proteins were identified by mass spectrometry (see Supplemental Data Set 1 online). These proteins were assigned to 5438 groups representing a minimal set of proteins to which the identified peptides could belong (see Methods for further explanation of groups). These protein groups (henceforth referred to simply as “proteins”) are listed in Supplemental Data Set 2 online. A mutant:wild type ratio of spectral counts was calculated for each protein (mean ratio for six biological replicates) to identify those whose abundance was reproducibly altered in mutant extracts relative to the wild type. Notably, PAN1 ratios were the most reduced of any protein in both pan1 single and pan1;pan2 double mutants (mutant:wild type ratios 0.11 and 0.05, respectively; Table 2; also see Supplemental Data Set 3 online). This is consistent with our earlier finding that PAN1 is undetectable in pan1 mutants by immunoblotting (Cartwright et al., 2009) and demonstrates that our methods are capable of revealing differences in protein abundance in pan mutants relative to the wild type. Defining changed proteins as those showing at least a 1.5-fold increase or decrease with an associated P value (determined from a Student’s t test on the ln-transformed values) ≤ 0.1 in at least one of the mutants, we found that the abundance of 253 of the 5438 identified proteins was changed (see Supplemental Data Set 3 online).
Table 2. Selected Examples of Proteins Involved in Signaling That Were Reduced (as Defined in the Text) in One or More pan Mutants as Indicated.
| Accession | pan1 Ratio | pan2 Ratio | pan1; pan2 Ratio | Map Man Bin No. | Map Man Bin/ Sub-Bin Description | Description |
|---|---|---|---|---|---|---|
| Selected signaling proteins decreased only in pan1;pan2 | ||||||
| GRMZM2G375002_P01 | 0.81 | 0.96 | 0.52* | 30.5 | Signaling/G-proteins | Zm-ROP4 |
| GRMZM2G120657_P03 | 0.75 | 0.96 | 0.64* | 30.2.6 | Signaling/receptor kinases/leucine-rich repeat VI | Highly similar to AT3G03770, LRR-RLK |
| GRMZM2G072573_P03 | 0.74 | 0.85 | 0.55* | 29.4 | Protein/ posttranslational modification | Moderately similar to AT2G46920, POL protein phosphatase |
| GRMZM2G104125_P01 | 1.40 | 1.02 | 0.61* | 30.3 | Signaling/calcium | Highly similar to AT5G12480, calmodulin-domain protein kinase |
| Selected signaling proteins decreased in pan1;pan2 and either pan1 or pan2 | ||||||
| GRMZM5G836190_P02 | 0.11* | 0.95 | 0.05* | 30.2.3 | Signaling/receptor kinases/leucine-rich repeat III | PAN1 |
| GRMZM2G034572_P01 | 0.91 | 0.26* | 0.31* | 30.2.3 | Signaling/receptor kinases/leucine-rich repeat III | PAN2 |
| GRMZM2G102088_P02 | 0.55* | 0.80 | 0.55* | 29.4 | Protein/ posttranslational modification | Highly similar to AT2G24360, raf-type MAPKKK |
Mean ratios significantly differing from 1 (P value <0.1) are marked with an asterisk. Detailed information for each protein, including means, P values, and ion intensities for each replicate are found Supplemental Data Set 3 online. MAPKKK, mitogen-activated protein kinase kinase kinase.
Since pan1 and pan2 are loss-of-function mutants, we reasoned that proteins whose abundance is reduced in pan mutants are more likely to be closely linked functionally to PANs than those whose abundance is increased and therefore focused subsequent analyses on the reduced proteins. The 120 proteins from Supplemental Data Set 3 online that are reduced at least 1.5-fold are classified in the Venn diagram in Figure 3 according to the genotypes in which they were reduced. Not surprisingly, in view of the synergistic phenotype of pan1;pan2 double mutants, the majority of the proteins that are decreased in double mutants are not decreased in either of the single mutants. Unexpectedly, in spite of their very similar stomatal development defects, there is only modest overlap between the set of proteins reduced in pan1 versus pan2 single mutants, even when one considers only those that are also depleted in the double mutant (Figure 3). This finding suggests that PAN1 and PAN2 have many distinct functions but does not reveal whether they have distinct functions in the SMC, since that is only one of many cell types included in the tissue sample analyzed.
Figure 3.

Venn Diagram Illustrating the Overlaps in Sets of Proteins Depleted in pan Mutant Membrane Preparations.
Proteins included in this analysis are those listed in Supplemental Data Set 3 online that were found to be reduced at least 1.5-fold relative to the wild type (regardless of P value) in one or more pan mutants. However, depletion of at least 1.5-fold with an associated P value ≤0.1 in at least one mutant genotype was required for inclusion of the protein in Supplemental Data Set 3 online.
[See online article for color version of this figure.]
A hypergeometric test was performed to determine whether proteins depleted in the mutants belong predominantly to particular functional classes, using Map Man bins (Thimm et al., 2004) to assign each protein to a functional class (see Supplemental Table 1 online for a comprehensive report for all Map Man bins and genotypes). Proteins in the Map Man bin “Miscellaneous” were significantly overrepresented among those depleted in pan1 single mutants relative to all proteins identified (Table 1). Notably, five of the seven “miscellaneous” proteins depleted in pan1 are glycosyltransferases (possibly involved in pectin or hemicellulose biosynthesis) or callose-degrading glucanases, suggesting a possible unique role for PAN1 in cell wall assembly. Proteins in the Map Man bins “Transport” and the “Cell” sub-bin “vesicle transport” were significantly overrepresented among those depleted in pan1;pan2 double mutants compared with all proteins identified (Table 1), suggesting defects in membrane transport and vesicle trafficking in double mutants. Interestingly, targeted vesicle trafficking is important for achieving polarized distribution of a variety of membrane transport proteins (Dettmer and Friml, 2011). Thus, PAN1 and PAN2 may be important for polarized trafficking of membrane transport proteins, and this may be related to the cell polarity defects seen in pan mutants. Proteins in the Map Man bin “Signaling” were also overrepresented among proteins depleted in pan single and double mutants compared with all proteins identified, although the enrichment was only statistically significant for pan1;pan2 double mutants (Table 1). This observation suggests roles for PAN proteins in signaling, which is of great interest in view of the identity of PAN1 as a receptor-like protein.
Table 1. Map Man Bins (Functional Categories) in Which Proteins Reduced in pan Mutants Were Found to Be Significantly Overrepresented (Indicated by Upward Arrow) Relative to All Proteins Identified.
| Percentage of Proteins Decreased at Least 1.5-Fold |
|||||
|---|---|---|---|---|---|
| Map Man Bin No. | Map Man Bin Description | Percentage of Proteins Identified | pan1 | pan2 | pan1;pan2 |
| 26 | Miscellaneous | 4.2% | ↑ 14.3% | 7.1% | 4.5% |
| 30 | Signaling | 7.0% | 8.2% | 9.5% | ↑ 10.6% |
| 31.4 | Cell vesicle transport | 1.7% | 2.0% | 0.0% | ↑ 6.1% |
| 34 | Transport | 3.7% | 4.1% | 4.8% | ↑ 9.1% |
Table 2 presents selected examples of individual proteins with signaling functions decreased in various combinations of pan mutants. Interestingly, ROP4 (the only type I ROP identified in our analysis) was significantly depleted in pan1;pan2 double mutants. We previously showed that type I ROPs function with PAN1 to establish SMC polarity, and by immunoblotting we demonstrated that ROPs are depleted from a Triton-insoluble membrane fraction of pan1 leaf extracts compared with the wild type (Humphries et al., 2011). Thus, our findings for ROP4 in the proteomic analysis (where a different nonionic detergent was used to wash the membrane fraction prior to analysis) parallel the immunoblotting results, albeit with significant depletion detected only in double mutants; the degree of depletion in pan1 single mutants may simply be too small to detect by iTRAQ comparisons. As shown in Table 2, other signaling proteins depleted in pan single and double mutants include a variety of kinases, a phosphatase related to POLTERGEIST, which is required for polarization of asymmetric cell division in Arabidopsis (Song et al., 2008), a protein kinase C substrate, and a transducin family protein. Investigating the functional relationships of these proteins to PANs will be interesting topics for future work.
Identification of PAN2 as a Catalytically Inactive LRR-RLK Family Protein
In pan2 mutants, the most significantly depleted protein identified by the quantitative proteomic analysis was a LRR-RLK encoded by GRMZM2G034572_T01 (Table 2); this LRR-RLK was also among the most depleted proteins in pan1;pan2 double mutants. This gene encoding this protein is located near the tip of chromosome 2 in bin 2.10, closely matching the location where pan2 was mapped using a mass spectrometry–based single nucleotide polymorphism (SNP) genotyping method (see Supplemental Data Set 4 online). To test the possibility that GRMZM2G034572_T01 is pan2, we sequenced the exons of this gene in pan2 mutants. The pan2-O allele described previously (Cartwright et al., 2009) was found to contain a missense mutation changing a conserved Ser to Phe (S211 > F) near the N terminus of the protein (Figure 4A). Three independent ethyl methanesulfonate–induced alleles of pan2 that we isolated in a screen for noncomplementation of pan2-O were all found to contain premature stop codon mutations at different sites within the gene (Figure 4A). These findings establish that GRMZM2G034572_T01 is pan2.
Figure 4.
PAN2 Domain Structure and Expression.
(A) Schematic illustration (drawn to scale) of predicted domains of PAN2. The GRMZM2G034572_T01 gene model underlying this diagram was confirmed via sequencing of PCR products amplified from B73 immature leaf cDNA with primers Pan2ex1ex2Fa + Ra and Fb + Rb, and Pan2ex2ex3Fa + Ra and FB + Rb (see Supplemental Table 1 online). Phosphorylation sites shown were identified via mass spectrometry analysis of phosphopeptides enriched from various maize tissues and reported at http://maizeproteome.ucsd.edu/.
(B) Summary of transcript abundance data for pan1 and pan2 obtained via short read sequencing and reported by Li et al. (2010), accompanied by diagram illustrating segments of the developing maize leaf 3 (counting the first leaf made by the plant as leaf 1) that were analyzed. Transcript abundance data for highlighted segments are reported in the table. Gene models employed for study of Li et al. (2010) were from the 4a maize genome release at www.maizesequence.org; the AC234203.1_FG002 pan1 model is incorrect but wholly contained within our experimentally validated pan1 gene model corresponding to release 5b.60 model GRMZM5G836190_T02. The highest expression levels for pan1 and pan2 are found in segment 1, which contains a high frequency of dividing cells, including those dividing to form stomata.
[See online article for color version of this figure.]
A previously published transcription profiling study employing short read sequencing demonstrated that like pan1, maximum pan2 mRNA levels are observed at the base of the developing leaf where cell divisions occur (including SMC divisions) (Figure 4B; Li et al., 2010). A microarray study surveying many maize tissues and developmental stages (Sekhon et al., 2011) confirmed that in developing leaves, pan2 is most highly expressed in the basal cell division zone (see Supplemental Figure 2 online). Sekhon et al. (2011) further demonstrated that like pan1 (Cartwright et al., 2009), pan2 is expressed in many other maize tissues in addition to developing leaves, including expanding stems, immature tassels and ears, and developing seeds (see Supplemental Figure 2 online). In general, pan2 is expressed in tissues where cells are dividing and expanding, suggesting that pan2 has other functions in addition to promoting SMC polarization.
Like PAN1, PAN2 belongs to the LRR-RLK subfamily III (Shiu and Bleecker, 2001), but to a different major clade within this subfamily. PAN2 is larger (predicted molecular weight of 115 kD) than PAN1 (68 kD) due mainly to a much larger extracellular domain with 20 predicted LRR motifs compared with five for PAN1 (Figure 4A). There are no published analyses of the functions or properties of the closest relative of PAN2 in Arabidopsis or other plants. PAN1 lacks several amino acids that are conserved in catalytically active kinases and is catalytically inactive in vitro (Cartwright et al., 2009). As illustrated in Supplemental Figure 3 online, PAN2 also lacks key features expected of a catalytically active kinase (Manning et al., 2002), in particular a GXGXXG consensus sequence in the G-loop region of subdomain I (GRSSHG in PAN2), an HRD motif in subdomain VI (HGN in PAN2), and a DFG motif in subdomain 7 (DYC in PAN2). Interestingly, the kinase domain of the closest relative of PAN2 in Arabidopsis (At4g20940) shares these same deviations from the expectations for a catalytically active kinase (see Supplemental Figure 3 online).
To test the enzymatic activity of the PAN2 kinase domain, in vitro kinase assays were performed with the intracellular domain of PAN2 fused to glutathione S-transferase (GST; expressed and purified from Escherichia coli). A GST-PAN1 intracellular domain fusion protein was included as a negative control (Cartwright et al., 2009) and GST-BRI1 as a positive control (Friedrichsen et al., 2000). Under conditions in which the BRI1 kinase domain phosphorylated itself and the artificial substrate myelin basic protein, the PAN2 intracellular domain exhibited no kinase activity (Figure 5). Inspection of an atlas of proteotypes (http://maizeproteome.ucsd.edu/) revealed that PAN2 has several sites of phosphorylation between its transmembrane and kinase domains (Figure 4A), suggesting that this juxtamembrane region has a regulatory function, possibly negatively regulating the enzymatic activity of the kinase domain. Therefore, we tested the kinase domain of PAN2 alone (lacking the juxtamembrane region) along with the corresponding fragment of PAN1 but found that these, too, exhibited no kinase activity in vitro (Figure 5). Thus, consistent with analysis of the amino acid sequence, the kinase domain of PAN2 lacks detectable kinase activity.
Figure 5.
Analysis of in the Vitro Kinase Activity of Intracellular Portions of PAN2.
The left panel shows Coomassie blue staining of the proteins used in each reaction: myelin basic protein (MBP) plus the kinase domain (KD) of BRI1 (lane 1), PAN2 (lane 4), or PAN1 (lane 5) or the entire intracellular domain (IC) of PAN2 (lane 2) or PAN1 (lane 3) fused to GST. In the right panel, autoradiography of the kinase assay reaction products reveals that only the BRI1 kinase domain phosphorylates itself and myelin basic protein.
PAN2 Localizes Asymmetrically in SMCs in a PAN1-Independent Manner
To investigate where in the cell PAN2 functions to promote SMC polarization, we raised a polyclonal antibody against a unique 96–amino acid region of PAN2 between the transmembrane and kinase domains. In extracts from maize leaf division zones, the affinity-purified anti-PAN2 antibody predominantly recognizes a protein whose mobility is consistent with the expected mass for PAN2 (115 kD), which is found exclusively in the membrane fraction as expected for a LRR-RLK (Figure 6A; see Supplemental Figure 4 online). This membrane protein is undetectable In membrane fractions of leaf division zone extracts from plants homozygous for the premature stop codon alleles pan2-1, pan2-2, and pan2-3, and its abundance is greatly reduced in plants homozygous for the pan2-O missense allele (Figure 6A), confirming its identity as PAN2. However, the abundance of this protein is unchanged in both pan1 mutants tested (Figure 6A).
Figure 6.
Immunodetection and Localization of PAN2.
(A) Gel blot of proteins extracted from leaf division zones of plants of the indicated genotypes probed with affinity-purified anti-PAN2 or anti-α-tubulin as a loading control. Comparison of results for wild-type high-speed supernatant and pellet shows that PAN2 is detectable only in the pellet fractions and thus only pellet fractions were analyzed for mutants. PAN2 abundance in pan1 mutants is equivalent to that in the wild type but reduced in pan2-O mutants (containing a missense mutation in pan2) and undetectable in pan2-1, pan2-2, and pan2-3 (containing premature stop codons in pan2).
(B) to (E) PAN2 (green) visualized via immunolocalization with affinity-purified anti-PAN2 and nuclei (magenta) visualized via propidium iodide staining. (B) and (C) illustrate earlier and later stages of stomatal development in wild-type leaves, respectively; (D) and (E) illustrate developing stomata in pan1 and pan2 mutant leaves, respectively. Arrowheads lie on top of GMCs and point to sites of PAN2 accumulation in adjacent SMCs where they contact the GMC. Arrows in (B) point out incipient PAN2 patches forming in SMCs with unpolarized nuclei adjacent to recently formed GMCs. In (C), “d” marks a divided subsidiary cell nucleus, illustrating persistence of the PAN2 patch at the GMC contact site after division. Bar = 10 µm.
(F) and (G) Analysis of the timing of PAN1 versus PAN2 patch formation. GMCs elongate as SMC polarization proceeds, so GMC length can be used as a measure of the developmental stage of the adjacent SMC (Cartwright et al., 2009). Analysis of wild-type leaf areas where some SMCs lack PAN patches and/or polarized nuclei showed that the average lengths of GMCs flanking SMCs with PAN1 versus PAN2 patches are not significantly different (n > 200 SMCs analyzed for each antibody; error bars represent sd). The proportion of SMCs with PAN1 versus PAN2 patches that have polarized nuclei is also similar.
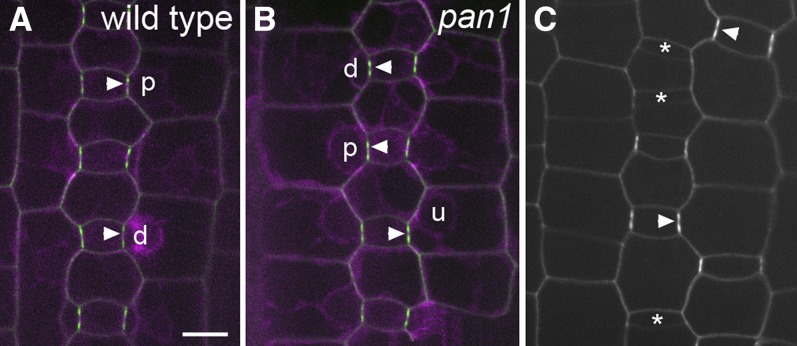
Immunolocalization with anti-PAN2 reveals labeling in the wild type (Figures 6B and 6C), but not in pan2-2 mutant (Figure 6E) leaves, confirming the specificity of the labeling for PAN2. Polarized accumulation of PAN2 is detectable at the surfaces of wild-type SMCs, where they contact GMCs. This polarized distribution of PAN2 is observed in SMCs flanking recently formed (very short) GMCs prior to polarization of SMC nuclei toward the GMC (Figure 6B, arrows and arrowheads), persisting as SMC nuclei polarize and after completion of the SMC division (Figure 6C, arrowheads). Thus, PAN2 localization is very similar to that of PAN1, which also localizes as a patch in SMCs at sites of contact with GMCs prior to nuclear polarization (Cartwright et al., 2009). Double labeling experiments were precluded by the fact that PAN1 and PAN2 antibodies were both raised in rabbits. Instead, we performed indirect comparisons of the timing of PAN1 versus PAN2 patch formation by taking advantage of the gradual increase in GMC length that occurs over time as SMCs polarize and divide (Cartwright et al., 2009). This approach provided evidence that polarized patches of Type I ROP GTPase form in SMCs after PAN1 patches: SMCs with ROP patches have a higher proportion of polarized nuclei and their adjacent GMCs are longer on average compared to SMCs with PAN1 patches (Humphries et al., 2011). However, as shown in Figures 6F and 6G, such comparisons revealed no difference in the timing of PAN1 versus PAN2 patch formation: The lengths of adjacent GMCs and the proportion of polarized nuclei were equivalent for SMCs with PAN1 and PAN2 patches. This finding suggests that PAN1 and PAN2 patches form at approximately the same time.
To further investigate PAN2 localization, we expressed a native promoter-driven PAN2-YFP (for yellow fluorescent protein) fusion protein in transgenic maize. Introduction of PAN2-YFP into pan2 mutants via crossing demonstrated that PAN2-YFP is functional because it rescues the pan2 mutant phenotype (see Supplemental Figure 5 online). In several independent transgenic lines examined, PAN2-YFP exhibits polarized localization in SMCs (Figure 7A, arrowheads), similar to what was observed via immunolocalization. While immunostaining reveals no PAN2 labeling in non-SMCs or in early SMCs prior to the appearance of a polarized PAN2 patch (Figure 6B), PAN2-YFP signal is detected at the periphery of all cells in the area where stomatal divisions are occurring (Figure 7C). PAN2-YFP is distributed around the entire SMC periphery when GMCs first form (see cells flanking GMCs marked with asterisks, Figure 7C) and later becomes concentrated at the GMC contact site (Figure 7C, arrowheads).
Figure 7.
Localization of a Functional PAN2-YFP Fusion Protein Expressed from Its Native Promoter in Transgenic Maize.
(A) and (B) PAN2-YFP (green) coexpressed with CFP-tubulin (magenta) in the wild type (A) and pan1 mutant (B), demonstrating equivalent PAN2-YFP localization in both genotypes. Nuclei are marked “u” if unpolarized, “p” if polarized, and “d” if divided.
(C) PAN2-YFP only in the wild type. Arrowheads lie on top of GMCs and point to sites of PAN2-YFP accumulation in adjacent SMCs where they contact the GMC. Asterisks mark newly formed GMCs whose flanking SMCs do not yet have PAN2-YFP patches but have PAN2-YFP distributed around the entire SMC periphery. Bar = 10 µm.
Previously, we showed by immunolocalization that the polarized accumulation of PAN1 in SMCs requires PAN2 because it was not observed in pan2 mutants (Cartwright et al., 2009). In this study, we confirmed that finding using a native promoter-driven PAN1-YFP fusion protein, which localizes similarly to endogenous PAN1 detected by immunolocalization (Humphries et al., 2011). As shown in Supplemental Figure 6 online, PAN1-YFP forms patches at the GMC contact sites of the wild type but not pan2 SMCs. Conversely, to investigate a possible role for PAN1 in PAN2 localization, anti-PAN2 labeling was examined in pan1 mutant leaves and found to be equivalent to the wild type (Figure 6D). Consistent with that observation, PAN2-YFP localization in pan1 mutant SMCs is indistinguishable from the wild type (cf. Figures 7A and 7B).
In summary, PAN2 exhibits polarized localization in SMCs at sites of contact with GMCs, similar to PAN1. Polarized distribution of PAN2 is detected in SMCs shortly after GMC formation but prior to nuclear polarization and does not depend on PAN1, whereas polarized accumulation of PAN1 requires PAN2. Thus, although we find no difference in the timing of PAN2 versus PAN1 patch formation by the indirect comparison method employed, PAN2 acts genetically upstream of PAN1.
PAN2 Interacts with Itself but Not with PAN1
Our findings that PAN1 and PAN2 colocalize, that PAN2 promotes the polarized accumulation of PAN1, and that PAN1 and PAN2 interact genetically suggest the possibility that these two proteins might physically interact. We tested this possibility in several ways. First, the intracellular domains of PAN1 and PAN2 were tested for their ability to interact in the GAL4-based yeast two-hybrid system with interactions taking place inside the nucleus. As shown in Figure 8A, PAN1 and PAN2 intracellular domains failed to interact in both combinations tested (pAS-PAN1 + pACT-PAN2 and vice versa), although immunoblotting confirmed that the fusion proteins were expressed in yeast transformed with the fusion protein constructs (see Supplemental Figure 7 online). Full-length PAN1 and PAN2 proteins also showed no interaction in the split-ubiquitin yeast two-hybrid system testing the ability of proteins to interact at the plasma membrane (see Supplemental Figure 8 online). To investigate whether PAN1 and PAN2 physically interact in plant cells (directly or indirectly), we conducted reciprocal coimmunoprecipitation experiments. Membrane proteins from wild-type maize leaf division zones were solubilized and immunoprecipitated with either anti-PAN1 (Cartwright et al., 2009) or anti-PAN2 antibodies. As illustrated in Figure 8B (left panel), anti-PAN1 immunoprecipitates PAN1 itself and Type I ROP GTPases as previously described (Humphries et al., 2011). Although PAN2 was readily detected in the input sample, no PAN2 coimmunoprecipitated with PAN1. Conversely, anti-PAN2 precipitated PAN2 itself, and although PAN1 was detectable in the input sample it did not coimmunoprecipitate with PAN2 (Figure 8B, right panel).
Figure 8.
Analysis of PAN1–PAN2 Interaction via Multiple Methods.
(A) GAL4-based yeast two-hybrid analysis; left panel illustrates growth of colonies in the presence of His (demonstrating viability of cotransformed colonies) and growth of the same colonies in the absence of His (indicating interaction between bait and prey constructs). Black arrowheads point to sites on the plate lacking His where colony growth would indicate an interaction between PAN1 and PAN2 but no growth was observed; gray arrowhead points to colonies whose growth in the absence of His indicates an interaction of PAN2 with itself.
(B) Coimmunoprecipitation experiments using anti-PAN1 or anti-PAN2 antibodies for precipitation from solubilized wild-type membrane proteins (input) and probing duplicate gel blots with anti-PAN1, anti-PAN2, or anti-ROP as indicated (left). The band detected by anti-PAN1 antibody in the anti-PAN2 precipitate is a nonspecifically precipitated protein larger than PAN1 itself (arrowhead indicates the position of PAN1, which is seen in the input sample only). No coimmunoprecipitation of PAN1 and PAN2 is observed.
(C) Coimmunoprecipitation experiments using anti-GFP for precipitation from solubilized membrane proteins of PAN2-YFP transgenic or nontransgenic leaves (input) and probing duplicate gel blots with anti-GFP or anti-PAN2 as indicated. Anti-PAN2 detects both PAN2-YFP (larger) and endogenous PAN2 (smaller), revealing coimmunoprecipitation of endogenous PAN2 with PAN2-YFP.
Although none of the methods employed revealed evidence of a physical interaction between PAN1 and PAN2, PAN2 does display an interaction with itself. As shown in Figure 8A, the cytoplasmic domain of PAN2 interacts with itself in the GAL4-based yeast two-hybrid system, as indicated by growth of yeast cotransformed with pAS-PAN2 and pACT-PAN2 in the absence of His. To investigate whether PAN2 interacts with itself in plant cells, we used PAN2-YFP transgenic plants for coimmunoprecipitation experiments with anti-GFP antibodies. PAN2-YFP was immunoprecipitated with anti-GFP from PAN2-YFP transgenic plants (Figure 8C, top panel). Probing duplicate gel blots with anti-PAN2 revealed that endogenous PAN2 was present in anti-GFP precipitates from PAN2-YFP transgenics but not from nontransgenic control plants. Taken together, these findings suggest that PAN2 forms homodimers by direct interaction with itself.
DISCUSSION
In this study, we identified PAN2 as a second receptor-like protein that promotes the polarization of maize SMCs (nuclear polarization and polarized actin accumulation at the GMC contact site) in preparation for their asymmetric division to form subsidiary cells of stomatal complexes. Indeed, the roles of PAN1 and PAN2 in SMC polarization appear to be similar and interrelated. The identities of PAN1 and PAN2 as receptor-like proteins leads naturally to the hypothesis that they function as receptors for the GMC-derived polarizing cues that are thought to orient SMC divisions (Stebbins and Shah, 1960), possibly working as a coreceptor pair. Some of our findings are consistent with this hypothesis, but others call for modifications of this simple idea.
Typically, RLKs function in signaling by phosphorylating themselves and other substrates in a ligand-dependent manner. However, we found that the kinase domain of PAN2, like that of PAN1, is catalytically inactive in vitro, consistent with the absence of several residues in the kinase domain that are typically required for catalytic activity. While we cannot rule out the possibility that there are some conditions in vitro or in vivo in which PAN2 is catalytically active as shown for other proteins with atypical kinase domains (Abe et al., 2001; Min et al., 2004), PAN2 is inactive in vitro under a wide variety of conditions tested. Approximately 10% of all kinases in both plant and animal genomes and almost 20% of Arabidopsis RLKs are predicted to lack catalytic activity based on their sequences (Manning et al., 2002; Castells and Casacuberta, 2007). In addition to PAN1 and PAN2, other plant RLKs functioning in a wide variety of processes have been demonstrated to lack catalytic activity in vitro (e.g., maize atypical receptor kinase [MARK], Llompart et al., 2003; STRUBBELIG/SCRAMBLED, Chevalier et al., 2005; CORYNE, Nimchuk et al., 2011). Some pseudokinases function as scaffolding proteins, interacting with other proteins via their kinase domains to mediate the assembly or stabilization of multiprotein complexes (Boudeau et al., 2006). Others function in signal transduction by interacting with and regulating the activities of active kinases, at least in some cases in a ligand-dependent manner (Llompart et al., 2003; Boudeau et al., 2006; Rajakulendran and Sicheri, 2010). In this regard, it is interesting that PAN2 interacts with itself, since ligand-induced dimerization is a common feature of receptor kinases functioning in signaling, including some that are catalytically inactive such as human epidermal growth factor 3 (Boudeau et al., 2006). The presence of multiple phosphorylation sites in the PAN2 intracellular domain and the depletion of proteins with signaling functions from membranes of pan2 single and pan1;pan2 double mutants further support the possibility of a function for PAN2 in signaling. However, to function in signaling PAN2 would need to partner with one or more active kinases yet to be identified. Identification of PAN2 binding partners along with comprehensive phosphopeptide comparisons between the wild type and pan2 mutants using mass spectrometry will facilitate the determination of whether and how PAN2 may participate in signaling.
Polarized localization of PAN2 within SMCs at sites of GMC contact supports the view that it functions in a GMC–SMC interaction important for SMC polarization but raises questions about the timing and nature of its function. By immunolocalization, PAN2 is undetectable at the SMC surface prior to the appearance of polarized PAN2 patches at GMC contact sites, suggesting its direct recruitment to these sites by an earlier acting sensor of the GMC position. By contrast, PAN2-YFP is initially detected around the entire SMC periphery, later becoming concentrated at the GMC contact site. This localization is consistent with the hypothesis that PAN2 acts as a primary sensor of GMC position, interacting with GMC-derived ligands and becoming concentrated at the site of ligand binding like many other cell surface receptors (Wülfing et al., 2002). We cannot presently determine whether immunlocalization or YFP fusion protein localization gives a more accurate picture of endogenous PAN2 distribution in early SMCs prior to patch formation: Our immunolocalization method may lack the sensitivity to reveal low levels of uniformly distributed PAN2 at the early SMC surface, or PAN2-YFP overexpression may lead to artifactual uniform localization in early SMCs (although PAN2-YFP is expressed from its native promoter and exhibited the reported distribution in all transgenic lines, including those with the lowest overall expression levels). Thus, PAN2 may perceive GMC-derived ligands that initiate SMC polarization or may function farther downstream to amplify SMC polarity. Alternatively, if PAN2 is recruited directly to GMC contact sites by earlier acting sensors of GMC position, then its ligands need not be spatially localized or derived specifically from the GMC to promote localized scaffolding or signaling events at the SMC-GMC contact site. Clearly, identification and analysis of PAN2 ligands would be of great interest in relation to these models.
Although the timing of polarized PAN2 and PAN1 accumulation in SMCs is very similar, PAN2 appears to act earlier because it is required for polarized accumulation of PAN1, whereas PAN2 accumulation at the GMC contact site is PAN1 independent. How PAN2 promotes the polarized accumulation of PAN1 is unknown. We found no evidence of a physical interaction between PAN2 and PAN1, though we cannot rule out the possibility that these proteins interact in a spatially or temporally restricted manner in vivo such that a small fraction of each protein is bound to the other. However, the most obvious interpretation of our findings is that the connection between PAN2 and PAN1 is indirect, albeit rapid. If the only function of PAN2 were to promote the polarized accumulation and function of PAN1, then we would expect the pan1;pan2 double mutant phenotype to be equivalent to the pan single mutant phenotypes, but instead it is synergistic. This finding implies that PAN2 must have other functions important for SMC polarization. The notion that PAN2 and PAN1 have both distinct and common functions is further supported by the results of our proteomic profiling of membrane fractions of pan mutants, which identified some proteins depleted only in one pan single mutant or the other along with others depleted in both single mutants.
In summary, we identified PAN2 as a LRR-RLK promoting the polarization of SMC divisions, whose function is closely related to that of PAN1. PAN2 lacks characteristics that would be expected for a PAN1 coreceptor (e.g., simultaneous rather than the observed sequential action, presence of an active kinase domain, and physical interaction with PAN1). Instead, our findings place PAN2 at the top of a cascade of events leading to SMC polarization in part by promoting polarized accumulation of PAN1, which in turn physically associates with and polarizes ROP GTPases (Humphries et al., 2011), leading to polarized actin accumulation and nuclear polarization in SMCs in preparation for their asymmetric division. Future work will concentrate on further elucidation of the pathway(s) in which PAN2 and PAN1 function to promote the premitotic polarization of SMCs.
METHODS
Primers
The sequences of all primers used in this study are provided in Supplemental Table 2 online.
Plants and Genetics
All mutants analyzed in this study segregate as single gene recessives, and all alleles were backcrossed into the B73 inbred wild-type background at least three times for use in the experiments presented. pan1-Mu and pan1-ems were previously described; both are null alleles with no PAN1 detectable via immunoblot analysis of homozygous mutants (Cartwright et al., 2009). pan2-O arose from an ethyl methanesulfonate mutagenesis with previous phenotypic description by Gallagher and Smith (2000) and Cartwright et al. (2009). pan2 was mapped to the tip of chromosome 2 (bin 2.10) via bulked segregant analysis of F2 families segregating pan2-O after a single backcross to B73, Mo17, and W22 inbred backgrounds. Genomic DNA extracted from pools of mutant and wild-type sibling tissue were used for PCR with a panel of 2076 primer pairs, and the products analyzed on an automated Sequenom mass spectrometry platform to identify markers linked to pan2 as described by Liu et al. (2010). Results for all markers and genetic backgrounds are presented in Supplemental Data Set 4 online. An F1 noncomplementation screen was performed to isolate additional pan2 mutant alleles. Briefly, ethyl methanesulfonate–mutagenized pollen was crossed onto pan2-O homozygous female ears as previously described (Neuffer, 1994), and the F1 progeny were screened for the presence of the pan2 mutant phenotype. Rare plants displaying the mutant phenotype were outcrossed to B73 wild-type plants. F1 progeny not inheriting pan2-O (as determined by analysis of genetic markers closely linked to pan2) were selfed to produce plants homozygous for the new pan2 alleles (pan2-1, -2, and -3), which all exhibited the pan2 phenotype. To identify mutations in pan2, exons of GRMZM2G034572_T01 were amplified from genomic DNA of plants homozygous for pan2-O, pan2-1, pan2-2, and pan2-3 via PCR with M2g34572 series primers listed in Supplemental Table 2 online, and PCR products were directly sequenced.
Phenotypic Analysis
For routine scoring of mutant phenotypes and quantitative analyses of subsidiary cell defects, imprints of the abaxial surfaces of mature leaf 3 or 4 (calling the first leaf made by the plant leaf 1) were made in cyanoacrylate glue and examined on a stereomicroscope or on a compound microscope at ×10 magnification with bright-field or differential interference contrast optics. To visualize nuclei and cell walls (Figure 1A), mature leaf 3 tissue was fixed and stained with 100 µg/mL propidium iodide as described previously (Hunter et al., 2012), mounted in water, and imaged via confocal microscopy. Actin and nuclei were visualized via confocal microscopy of fixed tissues excised from the basal 1 cm of unexpanded leaf 3 or 4 stained with Alexafluor 488-phalloidin (Molecular Probes/Invitrogen) and 10 µg/mL propidium iodide as previously described (Cartwright et al., 2009). Images were analyzed in a double blind manner for actin patches and nuclear position in SMCs.
Confocal Microscopy and Image Processing
Confocal microscopy was performed using a custom-assembled spinning disk microscope system described previously (Walker et al., 2007). Image processing was performed using Metamorph version 7.0r1, NIH Image J, or Adobe Photoshop, applying only linear adjustments to pixel values.
Preparation and Analysis of Membrane Proteins via Mass Spectrometry
A detailed description of peptide preparation, mass spectrometry, and analysis of mass spectra is provided in Supplemental Methods 1 (in Supplemental Data online). Briefly, membrane proteins were isolated from the cell division zone at the bases of unexpanded leaves, detergent extracted, reduced, alkylated, and trypsin digested. Following detergent removal via chromatography, peptides were labeled with iTRAQ reagents (AB SCIEX) and separated via online two-dimensional HPLC prior to acquisition of tandem mass spectra on a LTQ linear ion trap tandem mass spectrometer (Thermo Electron). Spectra were searched using Spectrum Mill vB04.00 (Agilent) against the maize (Zea mays) 5a.59 working gene set database (136,770 protein sequences). A 1:1 concatenated forward-reverse database of 273,540 proteins was constructed to calculate the false discovery rate (FDR). Cutoff scores were set to obtain a FDR of <0.1% at the unique peptide level.
Peptides mapping to a total of 13,101 proteins were identified (protein level FDR = 0.3%, reported in Supplemental Data Set 1 online) from the maize 5a working gene set, but many of these peptides could not be uniquely assigned to a single protein. This total of 13,101 includes all proteins to which such shared peptides could belong and is almost certainly an overestimate of the true number of different proteins detected in a given sample. To obtain a more realistic estimate, groups of proteins to which individual shared peptides mapped were identified. Within such a group (collection of proteins sharing the peptide sequence), the protein having the highest number of different peptides mapped to it (or, in the case of ties, the longest protein) was assigned as a group leader. Alternatively, a group consists of one protein if it shares no peptides with any other protein. Groups, identified by their leaders, are listed in Supplemental Data Set 2 online and correspond to the 5438 proteins discussed in Results.
Generation and Purification of PAN2-Specific Antibody
A 96–amino acid fragment of PAN2 corresponding to amino acids 675 to 770 (the region between the transmembrane and kinase domains), which displayed low sequence identity with other LRR-LRKs or other proteins in maize, was used to generate a PAN2-specific antibody. The corresponding coding sequence was amplified from B73 leaf division zone cDNA with PCR primers PAN2pip96F and R (see Supplemental Table 1 online) and cloned into the BamHI and XhoI sites of pET28a (Novagen) in frame with the 6HIS tag. The fusion protein was induced in Escherichia coli strain BL21 with 0.5 mM isopropyl-β-d-thio-galactoside at 28°C for 6 h and purified on nickel-nitrilotriacetic acid (Ni-NTA) resin (Novagen) according to the manufacturer’s protocols. The His tag was removed using a Thrombin CleanCleave kit (Sigma-Aldrich) according to the manufacturer’s instructions. Three milligrams of the purified PAN2 fragment was used to produce polyclonal antibodies in rabbits by Pacific Immunology. For affinity purification, the same protein fragment was coupled to beads using an AminoLink Plus immobilization kit (Pierce), and purification was performed according to the manufacturer’s protocols.
Immunolocalization
Immunolocalization of PAN2 was performed in leaf tissue excised from the basal 1 to 3 cm of unexpanded leaves of 2-to 4-week-old plants as described previously (Cartwright et al., 2009) using affinity-purified anti-PAN2 at 0.5 to 2 µg/mL and tyramide-based signal amplification with Invitrogen TSA kit #12 (Alexa Fluor 488) according to the manufacturer’s protocol. Following antibody staining, tissues were stained with 10 µg/mL propidium iodide (Sigma-Aldrich) in PBS to label nuclei prior to mounting in Vectashield (Vector Laboratories) for confocal microscopy.
Production of PAN2-YFP Transgenic Plants
A 10.5-kb genomic DNA fragment including the entire pan2 coding region (minus the stop codon) and 3.5 kb of 5′ sequence was amplified from B73 genomic DNA with primers PAN2-3GWp1 and PAN2-3GWp4. A 1.5-kb fragment immediately 3′ of the pan2 coding region was amplified from B73 genomic DNA with PAN2-3GWp3 and PAN2-3GWp2. Citrine YFP was amplified as described previously (Mohanty et al., 2009). These three fragments were assembled in pDONR221 (Invitrogen) to insert YFP in frame with PAN2 at its C terminus with the 3′ pan2 flanking sequence downstream using a MultiSite Gateway three-fragment vector construction kit (Invitrogen) following the manufacturer’s instructions. The sequence of the PAN2-YFP coding region was verified using primers of the PAN2seq series listed in Supplemental Table 2 online. An error-free PAN2-YFP construct was recombined into the binary vector pAM1006 and introduced into maize via Agrobacterium tumefaciens–mediated transformation at the Iowa State University Plant Transformation Facility (http://www.agron.iastate.edu/ptf/) as described at http://maize.jcvi.org/cellgenomics/protocol.shtml. Primary transformants were crossed to B73 to produce T1 progeny used for immunoprecipitation experiments, and T1s were crossed with plants expressing CFP-TUB (described at http://maize.jcvi.org/cellgenomics/protocol.shtml) to produce T2 progeny used for the imaging experiments presented in Figure 7. Primary transformants were also crossed to pan1-ems and pan2-2 mutants, and the T1 progeny backcrossed again to mutants, to produce homozygous pan1 and pan2 mutants expressing PAN2-YFP. Images presented are representative of multiple independent events that were examined by microscopy.
Yeast Two-Hybrid Analysis
For use in the split ubiquitin yeast two-hybrid interaction system (Grefen et al., 2007), full-length pan1 cDNA was amplified from a maize leaf division zone cDNA preparation with primers PAN1-GWF and -GWR and cloned into pDONR207 using BP Clonase (Invitrogen) to generate pDONR-PAN1. Full-length pan2 was amplified with PAN2-TOPO-F and -R from full-length pan2 cDNA clone pSK-PAN2 constructed as illustrated in Supplemental Figure 9 online. The PCR product was cloned into vector pENTR/TOPO-D (Invitrogen) according to manufacturer’s directions to generate pENTR-PAN2. Using the Gateway recombination system, pMetYC_GW (Lalonde et al., 2010) was used to generate PAN1-CUB and PAN2-CUB, and pXN25_GW, a close relative of pXN22_GW (Lalonde et al., 2010), was used to generate PAN1-NUB and PAN2-NUB. Yeast transformation was performed as described previously (Gietz and Woods, 2002), and the split ubiquitin assay was performed according to Lalonde et al. (2010).
For use in the GAL4-based yeast two-hybrid interaction system, cDNA fragments encoding intracellular portions of PAN1 and PAN2 were cloned into yeast two-hybrid vectors via Gateway cloning. Primers P1SOL_pENTR_F and P1_STOP_pENTR_R were used to amplify the cDNA segment encoding amino acids 308 to 662 of PAN1 from pDONR-PAN1 (described above). Primers P2SOL_pENTR_F and P2STOP_pENTR_R were used to amplify the cDNA segment encoding amino acids 675 to 1075 of PAN2 from pSK-PAN2 (described above). PCR products were cloned to the entry vector pENTR/D-TOPO (Invitrogen). Gateway recombination was used to transfer the inserts from these entry clones to destination vectors pASGW-attR and pACTGW-attR (Nakayama et al., 2002). Transformation of these plasmids into yeast strain AH109 was performed as described by Gietz and Woods (2002). At least four independent cotransformed colonies from two separate experiments were tested for each construct to determine their ability to grow in the absence of His.
Protein Gel Blot Analysis and Immunoprecipitation
Immunoblotting experiments used maize leaf tissues enriched in dividing cells (the basal 2 cm of leaves remaining on 3- to 4-week-old maize plants after removal of all leaves with fully or partially expanded sheaths). Membrane fractions of extracts from these tissues were prepared, separated via SDS-PAGE, and analyzed via immunoblotting as previously described (Cartwright et al., 2009) with the following modifications: microsomal fractions were resuspended in PBS containing 0.1% SDS and sonicated for 10 s to aid resuspension prior to freezing aliquots at −80°C; aliquots were boiled in SDS loading buffer with 100 mM DTT for 10 min prior to loading on 4 to 20% gradient polyacrylamide gels (Mini-Protean TGX, Bio-Rad); and 0.5 g SDS was added per liter of transfer buffer to facilitate transfer of PAN2 to the membrane. PAN2 was detected with affinity-purified anti-PAN2 at 1 µg/mL; tubulin was detected with mouse monoclonal anti-α-tubulin clone B-5-1-2 (Sigma-Aldrich) at 0.2 µg/mL.
Yeast protein extracts were prepared for immunoblotting (see Supplemental Figure 7 online) from cotransformed colonies as described in the Clontech Yeast Protocols Handbook (Protocol number PT3024-1, version PR973283, July 2009; http://www.clontech.com/xxclt_ibcGetAttachment.jsp?cItemId=17602andminisite=10020andsecItmId=14852). Separation, transfer, and detection of proteins was performed as described by Cartwright et al. (2009) using rabbit anti-GAL4 binding domain (SC-577; Santa Cruz Biotechnology) diluted 1:600, rabbit anti-HA epitope tag polyclonal antibody (Pierce PA1-985) diluted 1:500, and affinity-purified anti-PAN2 at 1 µg/mL.
Immunoprecipitation experiments were performed using membrane fractions of maize leaf tissue enriched in dividing cells prepared as above, after solubilizing the membrane proteins as described previously (Chinchilla et al., 2007). Immunoprecipitation with anti-PAN1 (Cartwright et al., 2009) and anti-PAN2 was performed as previously described (Humphries et al., 2011). Immunoprecipitation of PAN2-YFP was performed using magnetic beads covalently coupled to anti-GFP antibody (Miltenyi µMACS system) according to the manufacturer’s instructions. Precipitated proteins were removed from the beads by boiling in SDS loading buffer, separated via SDS-PAGE, and analyzed via immunoblotting as described above, detecting with anti-PAN1 at 2.5 µg/mL, anti-PAN2 at 1 µg/mL, anti-ROP2/4/9 (described in Humphries et al., 2011) at 1 µg/mL, and rabbit anti-GFP serum (Invitrogen A-6455) diluted 1:1000.
Analysis of in Vitro Kinase Activity
Portions of pan1 encoding the intracellular region (Arg-301 to Gly-662) and kinase domain (Arg-421 to Gly-662) were amplified by PCR using the primers PAN1-KD-F and -R, and PAN1-IC-F and -R, respectively. Similarly, portions of the pan2 gene encoding the intracellular region (Lys-676 to Ile-1080) and kinase domain (Arg-759 to Ile-1080) were amplified using the primers PAN2-KD-F and -R and PAN2-IC-F and -R, respectively (see Supplemental Table 2 online). The amplified fragments were cloned into BamHI and XhoI sites of pGEX-4T3 (GE Life Sciences) to generate N-terminal GST fusions. The positive control construct (BRI1-JKC GST fusion) was provided by Joanne Chory (Salk Institute). GST fusion proteins were induced in E. coli strain BL21 with 0.5 mM isopropyl-β-d-thio-galactoside at 28°C and purified from cell lysates using GST bind columns (Novagen) according to the manufacturer’s protocol. To measure kinase activity, purified GST fusion proteins were mixed with 0.5 mg/mL myelin basic protein (Sigma-Aldrich), incubated for 1 h at room temperature in kinase buffer (50 mM Tris-Cl, 0 to 20 mM MgCl2, 0 to 20 mM MnCl2, 0 to 40 mM CaCl2, 2 mM DTT, 1% glycerol, and 10 µCi [γ-32P]ATP). Reaction products were separated on NuPAGE 4 to 12% Bis-Tris gels (Invitrogen) and visualized via Coomassie Brilliant Blue staining and autoradiography.
Accession Numbers
pan gene and PAN protein sequences can be found at http://www.maizesequence.org as GRMZM2G034572_T01 and _P01 (PAN2) and GRMZM5G836190_T02 and _P02 (PAN1) (release 5b.60).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Illustration of Proteomics Workflow.
Supplemental Figure 2. Summary of pan2 Transcript Abundance in Various Maize Tissues Determined by Microarray Analysis.
Supplemental Figure 3. Alignment of the Kinase Domains of PAN2 and Other LRR-RLKs.
Supplemental Figure 4. Full Extent of Protein Gel Blot Probed with Anti-PAN2 Antibody Displayed in Cropped View in Figure 6A.
Supplemental Figure 5. Rescue of pan2 Mutant Phenotype by PAN2-YFP.
Supplemental Figure 6. Localization of PAN1-YFP Expressed from the Native pan1 Promoter in Wild-Type and pan2 Mutant Leaves.
Supplemental Figure 7. Protein Gel Blot Analysis of Yeast Transformed with PAN1 and PAN2 Intracellular Domain Constructs Used for Two-Hybrid Analysis.
Supplemental Figure 8. Analysis of PAN1 and PAN2 Interaction at the Yeast Plasma Membrane Using the Split-Ubiquitin Two-Hybrid System.
Supplemental Figure 9. Schematic Illustration of Construction of a Full-Length pan2 cDNA Clone.
Supplemental Table 1. Distribution of Changed Proteins among Map Man Bins.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. All 13,143 Possible Proteins Identified in Six Biological Replicates of iTRAQ Experiments.
Supplemental Data Set 2. Subset of Supplemental Data Set 1 Composed of 5438 Group Leaders.
Supplemental Data Set 3. Proteins Changed in pan1, pan2, and/or pan1 pan2 Mutants.
Supplemental Data Set 4. pan2 Mapping Data Generated via Bulked Segregant Analysis of Alleles at 2076 Marker Loci Analyzed on a Sequenom Mass Spectrometry Platform.
Acknowledgments
We thank Travis Alexander (University of California San Diego) for help with quantitative analysis of double mutants, Lauren Clark for isolation of pan2-1, pan2-2, and pan2-3, Adam Zona (University of California San Diego) for analysis of complementation of the pan2 phenotype by PAN2-YFP, Heather Cartwright (University of California San Diego) for initial analysis of pan1;pan2 mutants, Erik Vollbrecht and Pat Schnable (Iowa State University) for Sequenom mapping, and Anding Luo (University of Wyoming) for help with construction of PAN2-YFP. This work was funded by National Science Foundation Grants IOS-0843704 and IOS-1147265 to L.G.S., National Science Foundation Grant DBI-0924023 to S.P.B., and National Science Foundation Grant DBI-0501862 to A.W.S.
AUTHOR CONTRIBUTIONS
X.Z., M.F., J.A.H., Z.S., Y.P., and D.S. performed the research and analyzed the data. X.Z., M.F., J.A.H., L.G.S., Z.S., S.P.B., and A.W.S. designed the research. L.G.S. wrote the article with input from the coauthors.
Glossary
- GMC
guard mother cell
- SMC
subsidiary mother cell
- LRR-RLK
Leu-rich repeat–receptor-like kinase
- iTRAQ
isobaric tags for relative and absolute quantitation
- GST
glutathione S-transferase
- YFP
yellow fluorescent protein
- FDR
false discovery rate
References
- Abe Y., Matsumoto S., Wei S., Nezu K., Miyoshi A., Kito K., Ueda N., Shigemoto K., Hitsumoto Y., Nikawa J., Enomoto Y. (2001). Cloning and characterization of a p53-related protein kinase expressed in interleukin-2-activated cytotoxic T-cells, epithelial tumor cell lines, and the testes. J. Biol. Chem. 276: 44003–44011 [DOI] [PubMed] [Google Scholar]
- Abrash E.B., Bergmann D.C. (2009). Asymmetric cell divisions: A view from plant development. Dev. Cell 16: 783–796 [DOI] [PubMed] [Google Scholar]
- Boudeau J., Miranda-Saavedra D., Barton G.J., Alessi D.R. (2006). Emerging roles of pseudokinases. Trends Cell Biol. 16: 443–452 [DOI] [PubMed] [Google Scholar]
- Cartwright H.N., Humphries J.A., Smith L.G. (2009). PAN1: A receptor-like protein that promotes polarization of an asymmetric cell division in maize. Science 323: 649–651 [DOI] [PubMed] [Google Scholar]
- Castells E., Casacuberta J.M. (2007). Signalling through kinase-defective domains: The prevalence of atypical receptor-like kinases in plants. J. Exp. Bot. 58: 3503–3511 [DOI] [PubMed] [Google Scholar]
- Chevalier D., Batoux M., Fulton L., Pfister K., Yadav R.K., Schellenberg M., Schneitz K. (2005). STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 9074–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Dettmer J., Friml J. (2011). Cell polarity in plants: When two do the same, it is not the same. Curr. Opin. Cell Biol. 23: 686–696 [DOI] [PubMed] [Google Scholar]
- Dong J., MacAlister C.A., Bergmann D.C. (2009). BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facette M.R., Smith L.G. (2012). Division polarity in developing stomata. Curr. Opin. Plant Biol., vol. 15 (http://dx.doi.org/10.1016/j.pbi.2012.09.013) [DOI] [PubMed] [Google Scholar]
- Farquharson K. (2012). Polarization of Subsidiary Cell Division in Maize Stomatal Complexes. Plant Cell 24: 4313. [DOI] [PMC free article] [Google Scholar]
- Friedrichsen D.M., Joazeiro C.A., Li J., Hunter T., Chory J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatis B., Apostolakos P. (2004). The role of the cytoskeleton in the morphogenesis and function of stomatal complexes. New Phytol. 161: 613–639 [DOI] [PubMed] [Google Scholar]
- Gallagher K., Smith L.G. (2000). Roles for polarity and nuclear determinants in specifying daughter cell fates after an asymmetric cell division in the maize leaf. Curr. Biol. 10: 1229–1232 [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Gönczy P. (2008). Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9: 355–366 [DOI] [PubMed] [Google Scholar]
- Grefen C., Lalonde S., Obrdlik P. (2007). Split-ubiquitin system for identifying protein-protein interactions in membrane and full-length proteins. Curr. Protoc. Neurosci. 5: 27 [DOI] [PubMed] [Google Scholar]
- Humphries J.A., Vejlupkova Z., Luo A., Meeley R.B., Sylvester A.W., Fowler J.E., Smith L.G. (2011). ROP GTPases act with the receptor-like protein PAN1 to polarize asymmetric cell division in maize. Plant Cell 23: 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C.T., Kirienko D.H., Sylvester A.W., Peter G.F., McCarty D.R., Koch K.E. (2012). Cellulose Synthase-Like D1 is integral to normal cell division, expansion, and leaf development in maize. Plant Physiol. 158: 708–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S., et al. (2010). A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front Physiol 1: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Liu S., Chen H.D., Makarevitch I., Shirmer R., Emrich S.J., Dietrich C.R., Barbazuk W.B., Springer N.M., Schnable P.S. (2010). High-throughput genetic mapping of mutants via quantitative single nucleotide polymorphism typing. Genetics 184: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llompart B., Castells E., Río A., Roca R., Ferrando A., Stiefel V., Puigdomenech P., Casacuberta J.M. (2003). The direct activation of MIK, a germinal center kinase (GCK)-like kinase, by MARK, a maize atypical receptor kinase, suggests a new mechanism for signaling through kinase-dead receptors. J. Biol. Chem. 278: 48105–48111 [DOI] [PubMed] [Google Scholar]
- Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. (2002). The protein kinase complement of the human genome. Science 298: 1912–1934 [DOI] [PubMed] [Google Scholar]
- Menke F.L., Scheres B. (2009). Plant asymmetric cell division, vive la différence! Cell 137: 1189–1192 [DOI] [PubMed] [Google Scholar]
- Min X., Lee B.H., Cobb M.H., Goldsmith E.J. (2004). Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure 12: 1303–1311 [DOI] [PubMed] [Google Scholar]
- Mohanty A., Yang Y., Luo A., Sylvester A.W., Jackson D. (2009). Methods for generation and analysis of fluorescent protein-tagged maize lines. Methods Mol. Biol. 526: 71–89 [DOI] [PubMed] [Google Scholar]
- Nakayama M., Kikuno R., Ohara O. (2002). Protein-protein interactions between large proteins: Two-hybrid screening using a functionally classified library composed of long cDNAs. Genome Res. 12: 1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer M.G. (1994). Mutagenesis. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 212–219. [Google Scholar]
- Nimchuk Z.L., Tarr P.T., Meyerowitz E.M. (2011). An evolutionarily conserved pseudokinase mediates stem cell production in plants. Plant Cell 23: 851–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Peterson K.M., Horst R.J., Torii K.U. (2011). Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell 23: 3260–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Torii K.U. (2012). Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63: 591–614 [DOI] [PubMed] [Google Scholar]
- Rajakulendran T., Sicheri F. (2010). Allosteric protein kinase regulation by pseudokinases: Insights from STRAD. Sci. Signal. 3: pe8. [DOI] [PubMed] [Google Scholar]
- Rasmussen C.G., Humphries J.A., Smith L.G. (2011). Determination of symmetric and asymmetric division planes in plant cells. Annu. Rev. Plant Biol. 62: 387–409 [DOI] [PubMed] [Google Scholar]
- Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.K., Hofhuis H., Lee M.M., Clark S.E. (2008). Key divisions in the early Arabidopsis embryo require POL and PLL1 phosphatases to establish the root stem cell organizer and vascular axis. Dev. Cell 15: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G.L., Shah S.S. (1960). Developmental studies of cell differentiation in the epidermis of monocotyledons. II. Cytological features of stomatal development in the Gramineae. Dev. Biol. 2: 477–500 [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Walker K.L., Müller S., Moss D., Ehrhardt D.W., Smith L.G. (2007). Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr. Biol. 17: 1827–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wülfing C., Tskvitaria-Fuller I., Burroughs N., Sjaastad M.D., Klem J., Schatzle J.D. (2002). Interface accumulation of receptor/ligand couples in lymphocyte activation: Methods, mechanisms, and significance. Immunol. Rev. 189: 64–83 [DOI] [PubMed] [Google Scholar]