The ARC1 E3 ubiquitin ligase was previously shown to be required for self-pollen rejection in Brassica, and this report shows that its function is conserved in other Brassicaceae species. ARC1 was found to be required for self-pollen rejection in Arabidopsis lyrata and was frequently deleted in genomes of Brassicaceae species that had lost this self-incompatibility trait.
Abstract
Self-pollen rejection is an important reproductive regulator in flowering plants, and several different intercellular signaling systems have evolved to elicit this response. In the Brassicaceae, the self-incompatibility system is mediated by the pollen S-locus Cys-Rich/S-locus Protein11 (SCR/SP11) ligand and the pistil S Receptor Kinase (SRK). While the SCR/SP11-SRK recognition system has been identified in several species across the Brassicaceae, less is known about the conservation of the SRK-activated cellular responses in the stigma, following self-pollen contact. The ARM Repeat Containing1 (ARC1) E3 ubiquitin ligase functions downstream of SRK for the self-incompatibility response in Brassica, but it has been suggested that ARC1 is not required in Arabidopsis species. Here, we surveyed the presence of ARC1 orthologs in several recently sequenced genomes from Brassicaceae species that had diversified ∼20 to 40 million years ago. Surprisingly, the ARC1 gene was deleted in several species that had lost the self-incompatibility trait, suggesting that ARC1 may lose functionality in the transition to self-mating. To test the requirement of ARC1 in a self-incompatible Arabidopsis species, transgenic ARC1 RNA interference Arabidopsis lyrata plants were generated, and they exhibited reduced self-incompatibility responses resulting in successful fertilization. Thus, this study demonstrates a conserved role for ARC1 in the self-pollen rejection response within the Brassicaceae.
INTRODUCTION
Many flowering plants have reproductive controls to avoid inbreeding when self-related male pollen appears on the female pistil. Self-incompatibility is one such system as it causes self-pollen rejection, and in so doing, allows pollination and fertilization by other genetically distinct plants. In the Brassicaceae (mustard family), the self-incompatibility response is regulated by the pistil-specific S Receptor Kinase (SRK) and the pollen S-locus Cys-Rich/S-locus Protein11 (SCR/SP11) ligand (Schopfer et al., 1999; Takasaki et al., 2000; Takayama et al., 2000; Silva et al., 2001). While polymorphic SRK and SCR/SP11 genes have been identified in several Brassicaceae species, including Arabidopsis lyrata (Kusaba et al., 2001; Schierup et al., 2001) and Capsella grandiflora (Paetsch et al., 2006; Boggs et al., 2009; Guo et al., 2009), their functions have been best defined in Brassica crop species (reviewed in Ivanov et al., 2010; Tantikanjana et al., 2010; Iwano and Takayama, 2012). Following self-pollination, the pollen grain contacts a stigmatic papilla at the top of the pistil, and the SCR/SP11 ligand on the pollen binds to the haplotype-specific SRK present in the papilla (Kachroo et al., 2001; Takayama et al., 2001; Shimosato et al., 2007). SRK then activates a pollen rejection pathway to rapidly block pollen hydration or pollen tube penetration into the stigmatic surface (reviewed in Chapman and Goring, 2010; Ivanov et al., 2010).
Downstream components required for self-pollen rejection in the SRK signaling pathway have also been identified in Brassica species, and two positive regulators that interact with SRK are the M-locus Protein Kinase (MLPK) and the ARM-Repeat Containing1 (ARC1) E3 ubiquitin ligase. Brassica rapa MLPK is required for the self-incompatibility response, and mlpk mutants are unable to reject self-pollen (Murase et al., 2004). MLPK belongs to the receptor-like cytoplasmic kinase subfamily and is located in the plasma membrane with SRK (Murase et al., 2004). MLPK has been shown to interact transiently with SRK at the plasma membrane, potentially with the inactive SRK complex before activation via SCR/SP11 (Kakita et al., 2007a, 2007b). B. napus ARC1 was originally identified through a yeast two-hybrid screen and binds to the phosphorylated SRK kinase through its C-terminal Armadillo (ARM) repeats (Gu et al., 1998). ARC1 is able to be phosphorylated in vitro by both SRK and MLPK, suggesting that perhaps ARC1 may be recruited to a SRK-MLPK complex at the plasma membrane (Samuel et al., 2008). ARC1 is a member of the Plant U-box (PUB) family of E3 ubiquitin ligases and is required for the self-incompatibility response (Stone et al., 1999, 2003; Azevedo et al., 2001). The U-box domain in ARC1 classifies it as an E3 ligase and has been shown to bind the E2 conjugating enzyme in the closely related Arabidopsis thaliana PUB14 protein (Stone et al., 2003; Andersen et al., 2004). In addition to the ARM repeat domain and the U-box domain, ARC1 contains a U-box N-terminal domain (UND) domain (Stone et al., 2003; Mudgil et al., 2004; Samuel et al., 2006). The UND domain is found in a subset of PUB proteins and may participate in protein–protein interactions (Mudgil et al., 2004; Samuel et al., 2006). More recently, the ARC1 UND domain was used in a yeast two-hybrid screen resulting in the identification of Exo70A1 as a target for ARC1 (Samuel et al., 2009). Exo70A1 is required in the stigma to accept compatible pollen and is proposed to function as part of the exocyst complex to deliver secretory vesicles to the stigmatic papillar plasma membrane under the pollen contact site, thereby providing resources to the compatible pollen for hydration and pollen tube penetration into the stigmatic surface. ARC1 is predicted to promote self-pollen rejection in the self-incompatibility response by negatively regulating Exo70A1 and blocking the delivery of secretory vesicles to the pollen contact site (Samuel et al., 2009).
Outside of the Brassica spp in the Brassicaceae, several population and functional studies have been performed using A. lyrata and A. thaliana. Surveys on self-incompatible A. lyrata populations across North America and Europe have shown a similar diversity in the polymorphic SCR/SP11 and SRK genes when compared with Brassica spp (Schierup et al., 2001, 2006; Mable et al., 2005; Prigoda et al., 2005; Mable and Adam, 2007). A. thaliana is a fully self-compatible species, and the loss of A. thaliana SCR/SP11 and SRK genes (and self-incompatibility) has been studied in various ecotypes through analyses on the genomic region encompassing these genes (Kusaba et al., 2001; Bechsgaard et al., 2006; Tang et al., 2007; Shimizu et al., 2008; Tsuchimatsu et al., 2010; Guo et al., 2011). Transgenic studies with SCR/SP11 and SRK genes have also been conducted to restore self-incompatibility in A. thaliana. The transformation of A. lyrata SRK and SCR/SP11 genes has been reported to confer self-incompatibility in some A. thaliana ecotypes, but not all ecotypes tested (Nasrallah et al., 2004; Boggs et al., 2009). Additionally, the transformation of a functional SCR/SP11a allele into the A. thaliana Wei-1 ecotype, which has an intact endogenous SRKa allele, resulted in a self-incompatibility phenotype (Tsuchimatsu et al., 2010). In these A. thaliana transgenic experiments, the signaling pathway activated by SRK is unknown as MLPK and ARC1 do not appear to function in the reconstituted self-incompatibility response in transgenic A. thaliana (Rea et al., 2010; Kitashiba et al., 2011). For example, while an ARC1 ortholog has been identified in the A. lyrata genome, ARC1 exists as a pseudogene in A. thaliana (Kitashiba et al., 2011). Thus, Kitashiba et al. (2011) proposed that an alternate SRK signaling pathway functions in Arabidopsis species, perhaps related to the fact that Brassica and Arabidopsis belong to different lineages that have been estimated to have diversified ∼20 to 40 million years ago (Franzke et al., 2011). However, it is difficult to know if the SRK signaling pathway(s) has become altered in A. thaliana with the transition to self-compatibility. In this study, we set out to investigate the presence of ARC1 orthologs in other members of the Brassicaceae and use RNA interference (RNAi) suppression to examine the role of ARC1 in the naturally occurring self-incompatible A. lyrata. The growing availability of sequenced genomes for members of the Brassicaceae allowed us to take an evolutionary genetics approach to conduct a more widespread examination of the ARC1 gene in regards to self-incompatibility. Our results indicate that ARC1 is frequently deleted in self-compatible Brassicaceae species and that ARC1 is required for the self-incompatibility response in A. lyrata.
RESULTS
Synteny of the ARC1 and PUB17 Genomic Regions in the Brassicaceae
Brassica ARC1 is a member of PUB family of E3 ubiquitin ligases and was previously found to be most closely related to A. thaliana PUB17 (At1g29340), a protein involved in plant defense responses (Azevedo et al., 2001; Yang et al., 2006). Brassica ARC1 was initially thought to be an ortholog of A. thaliana PUB17, but more recently, a potential Brassica ARC1 ortholog was identified in the A. lyrata genome, a self-incompatible species, while a corresponding pseudogene was found in the self-compatible A. thaliana Columbia-0 (Col-0) and C24 ecotypes (Rea et al., 2010; Kitashiba et al., 2011). A. lyrata is closely related to A. thaliana with speciation occurring ∼5 to 10 million years ago (Koch et al., 2001; Charlesworth and Vekemans, 2005; Bailey et al., 2006; Wang et al., 2011), so this raised the question of whether ARC1 orthologs have been lost in other Brassicaceae species and whether this loss was correlated with self-compatibility. We conducted a broader survey of potential ARC1 orthologs using genome sequences now available for several other Brassicaceae species: B. rapa (Wang et al., 2011); Thellungiella parvula (Dassanayake et al., 2011); Capsella rubella (Capsella rubella Genome Project 2011; www.phytozome.net/capsella); Thellungiella halophila (Thellungiella halophila Genome Project 2011; www.phytozome.net/thellungiella); and Sisymbrium irio, Leavenworthia alabamica race a4, and Aethionema arabicum (Value-Directed Evolutionary Genomics [VEGI] consortium; biology.mcgill.ca/vegi). Of the aforementioned species, B. rapa is another self-incompatible species, while the remaining sequenced genomes were from self-compatible plants. With PUB17 being the most closely related ARC1 paralog, its presence or absence was also surveyed.
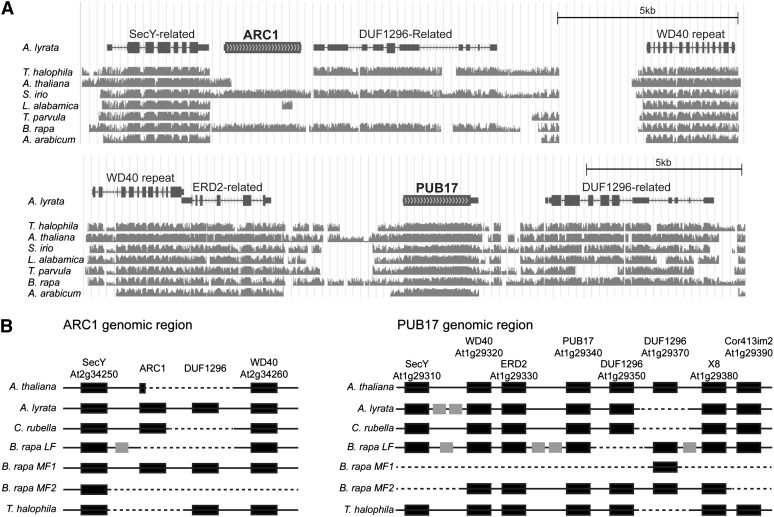
Predicted ARC1 and PUB17 genomic regions were first examined by synteny between A. lyrata, T. halophila, A. thaliana, S. irio, L. alabamica, T. parvula, B. rapa, and A. arabicum using the whole-genome orthologous alignment conducted by the VEGI consortium (http://grandiflora.eeb.utoronto.ca:8086) (Figure 1A). The most striking observation is that the predicted ARC1 gene is not found in five of the genomes (T. halophila, A. thaliana, L. alabamica, T. parvula, and A. arabicum), while the PUB17 gene is highly conserved in all eight genomes. This is unlikely to be an artifact of incomplete genome assemblies, as the flanking genes were identified in a syntenic block in all cases. Furthermore, all of the species that appear to be missing a predicted ARC1 gene are self-compatible, and S. irio is the only self-compatible species in this set that shows conservation to A. lyrata ARC1. Interestingly, a gene flanking ARC1, the predicted DUF1296-related gene, is also absent in four of the genomes. Figure 1B illustrates the predicted genes present in the ARC1 and PUB17 genomic regions assembled from annotated genomes in the public genome browsers (Tables 1 and 2). The ARC1 genomic regions can be identified by the flanking SecY-related (A. thaliana At2g34250) and WD40-repeat (A. thaliana At2g34260) genes, and these genes were found in a syntenic gene order with ARC1 missing. This analysis also includes another self-compatible species, C. rubella, and three regions from the triplicated B. rapa genome. In A. thaliana, the first 143 bp of the ARC1 coding region (with a missing start sequence) are present, but the rest of ARC1 and the DUF1296-related gene are both deleted. Interestingly, the C. rubella genome does contain a predicted ARC1 gene, despite being self-compatible, but the flanking DUF1296-related gene is deleted. In B. rapa, the MF1 subgenome has all four genes present, while the LF and MF2 subgenomes only have the flanking SecY-related/WD40-repeat genes and the SecY-related gene, respectively. Finally, as indicated in Figure 1A, the self-compatible T. halophila genome is missing ARC1 in this syntenic region, but still has the other three predicted genes. Thus, from these analyses, five of the seven self-compatible species appear to have lost the ARC1 ortholog, and S. irio is the only self-compatible species that does not appear to have undergone any large deletions in this region. These results seem to suggest a link between the loss of ARC1 and self-compatibility in these Brassicaceae species.
Figure 1.
Synteny of the ARC1 and PUB17 Genomic Regions in the Brassicaceae.
(A) Synteny in the predicted A. lyrata ARC1 and PUB17 genomic regions to T. halophila, A. thaliana, S. irio, L. alabamica, T. parvula, B. rapa, and A. arabicum. This image was generated in the University of California, Santa Cruz genome browser (Fujita et al., 2011) using a nine-species whole-genome orthologous alignment assembled by the VEGI consortium (http://grandiflora.eeb.utoronto.ca:8086). The A. lyrata annotated genes are shown at the top, and the degrees of similarity to the A. lyrata ARC1 or PUB17 genomic regions are shown in gray for each genome. Missing regions relative to the A. lyrata ARC1 or PUB17 genomic regions are shown as gaps.
(B) Schematic illustrating the gene conservation in the ARC1 and PUB17 genomic regions for A. thaliana, A. lyrata, C. rubella, B. rapa (LF, MF1, and MF2 subgenomes), and T. halophila. Annotated Brassicaceae genomes in the public genome databases were used to identify predicted genes present in the ARC1 and PUB17 genomic regions. The A. thaliana annotated genes are shown at the top, and the black boxes represent conserved orthologs annotated in the respective genomes. Gray boxes represent unconserved genes, and dashed lines represent deletions in the scaffolds.
Table 1. Gene Identifiers and Accession Numbers for the ARC1 and PUB17 Genomic Regions.
| Species | Gene Identifiers and Accession Numbers | |||
| A. thaliana | At2g34250; NP_180972 | ARC1 | DUF1296-related | At2g34260; NP_565782 |
| A. lyrata | 482367; XP_002879488 | 934022; XP_002879489 | 321144; XP_002881327 | 934024; XP_002879490 |
| C. rubella | 10023123 | 10025155 | – | 10023519 |
| B. rapa LF | Bra005441 | – | – | Bra005439 |
| B. rapa MF1 | Bra021898 | Bra021899 | Bra021900 | Bra021901 |
| T. halophila | 10016599 | – | 10016218 | 10016919 |
Dashes represent the absence of a corresponding orthologue in the ARC1 genomic region.
Table 2. Gene Identifiers and Accession Numbers for the PUB17 Genomic Region.
| Species | Gene Identifiers and Accession Numbers | |||||||
| A. thaliana | At1g29310; NP_174225 | At1g29320; NP_174226 | At1g29330; NP_564326 | At1g29340; NP_174228 | At1g29350; NP_174229 | At1g29370; NP_174230 | At1g29380; NP_174231 | At1g29390;NP_973936 |
| A. lyrata | 473141; XP_002890804 | 921968; XP_002890807 | 473143; XP_002893557 | 473144; XP_002890808 | 473146; XP_002893558 | – | 912742; XP_002890809 | 912743;XP_002893559 |
| C. rubella | 10009028 | 10009063 | 10010280 | 10008444 | 11008305 | – | 10011350 | 10010957 |
| B. rapa LF | Bra032303 | Bra032305 | Bra032306 | Bra032309 | Bra032310 | Bra032314 | Bra032315 | |
| B. rapa MF2 | – | Bra010836 | Bra010835 | Bra010834 | Bra010832 | Bra010833 | Bra010831 | – |
| T. halophila | 10007527 | 10007651 | 10008757 | 10006858 | 10006794 | – | 10008139 | 10008700 |
Dashes represent the absence of a corresponding orthologue in the PUB17 genomic region.
A similar analysis for the PUB17 region shows that an ortholog can be readily identified in all the genomes (Figure 1B, Table 2), including two copies in B. rapa (LF and MF1 subgenomes). PUB17 (At1g29340) is flanked on the right by a DUF1296-related gene (A. thaliana At1g20350), and in both A. thaliana (At1g29370) and the B. rapa LF subgenome, a second copy of a DUF1296-related gene is also present. Further to the right, two more conserved genes (A. thaliana At1g29380 and At1g29390) are present across all genomes surveyed. On the left side of PUB17, there are three conserved genes (A. thaliana At1g29310, At1g29320, and At1g29330) present across all genomes with the exception of the SecY-related gene absent from the B. rapa MF2 subgenome (and the MF1 subgenome that only contains a DUF1296-related gene). Thus, in contrast with the ARC1 genomic region, the PUB17 genomic region is highly conserved and present in all nine Brassicaceae species surveyed. Interestingly, the PUB17 genomic region also shares some synteny with the ARC1 genomic region. For example, DUF1296-related genes flank both ARC1 and PUB17 on the right side, and a SecY-related gene appears in the same orientation relative to ARC1 and PUB17. The WD40-repeat genes in the two genomic regions are paralogs, but in different positions relative to ARC1 and PUB17. The high degree of similarity of these two genomic regions likely originated from an ancient angiosperm whole-genome duplication (Jiao et al., 2011), followed by ARC1 and PUB17 acquiring divergent functions in the Brassicaceae (ARC1 in self-incompatibility; PUB17 in plant defense responses; Stone et al., 1999; Yang et al., 2006).
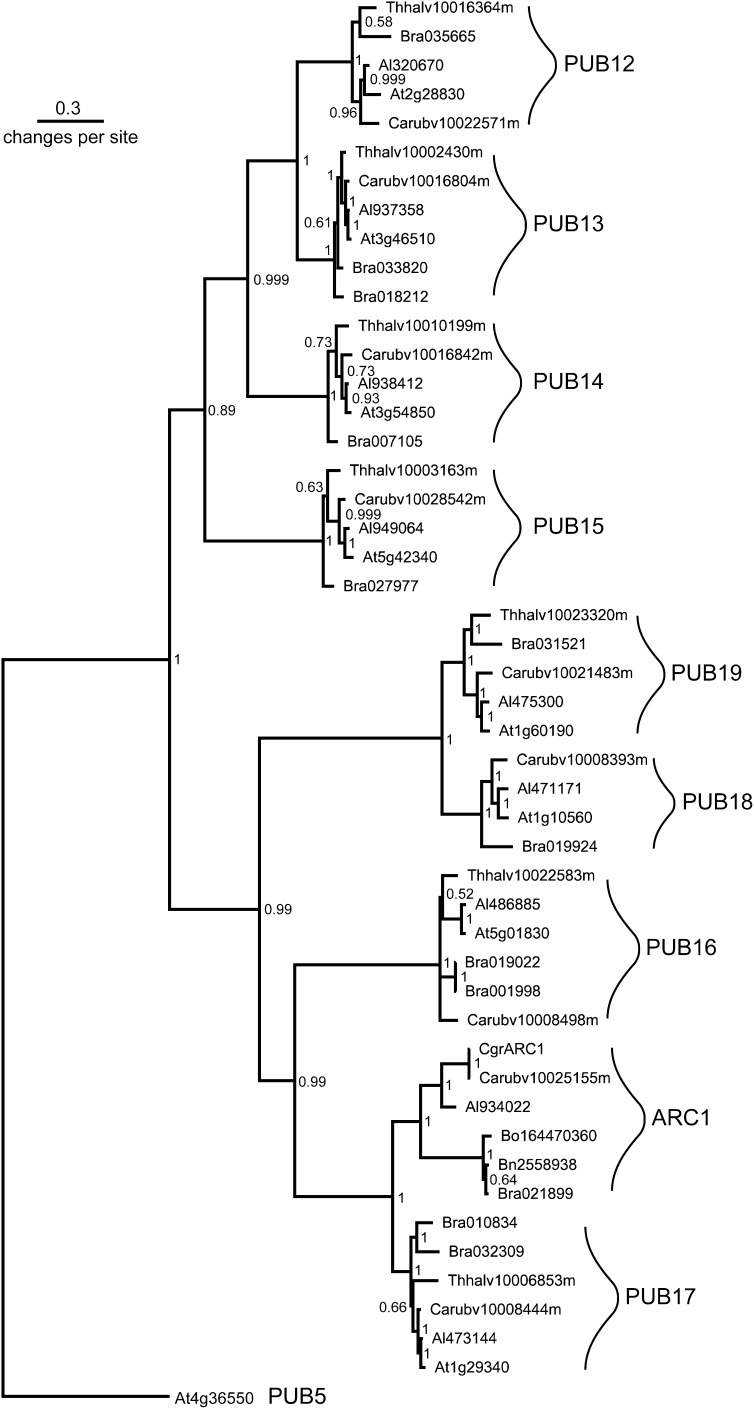
To confirm that the predicted ARC1 orthologs are in fact most closely related to the originally identified B. napus ARC1 (Gu et al., 1998), a phylogenetic analysis was conducted with a subset of eight most closely related U-box proteins, PUB12-19, that share the same domain organization as ARC1: a conserved UND domain followed by the U-box domain and the ARM repeat domain (Mudgil et al., 2004; Samuel et al., 2006). The full-length A. thaliana PUB12-PUB19 and B. napus ARC1 sequences were used to conduct BLAST analyses of sequenced genomes from A. lyrata (Hu et al., 2011), B. rapa (Wang et al., 2011), C. rubella (www.phytozome.net/capsella), and T. halophila (www.phytozome.net/thellungiella). All of the sequences that were identified in the BLAST search were full-length protein sequences that were then used for the phylogenetic analysis. Most species appeared to contain a single-copy gene for each of these PUB proteins (Figure 2, Table 3). The one exception was B. rapa, which appeared to have two copies of PUB11, PUB13, PUB16, and PUB17, likely resulting from the triplication of the genome (Wang et al., 2011). Predicted ARC1 proteins from two other self-incompatible plants, B. oleracea and C. grandiflora, were also included in this analysis. The phylogenetic tree generated by this analysis clearly resolves that the predicted PUB17 proteins are different from the predicted ARC1 proteins. The tree also shows that the ARC1 orthologs form a monophyletic clade, and the A. lyrata and Capsella ARC1 proteins cluster with the Brassica ARC1 proteins. As predicted from the synteny analysis in Figure 1, an ARC1 ortholog was not found in the self-compatible species, A. thaliana (Kitashiba et al., 2011) and T. halophila. Also, self-compatible C. rubella contains a copy of ARC1 that is very closely related to the ARC1 ortholog from the very closely related self-incompatible C. grandiflora.
Figure 2.
Bayesian Phylogeny Showing the Relationships between the ARC1 Proteins and the PUB5 and PUB12-19 Proteins from Different Brassicaceae Species.
Predicted amino acid sequences were identified from the A. lyrata (Al), C. rubella (Carub), T. halophila (Thhalv), and B. rapa (Bra) genomes by BLAST searches with the A. thaliana (At) PUB protein sequences. Predicted ARC1 sequences from B. oleracea (Bo), B. napus (Bn), and C. grandiflora (Cgr) were also included. ARC1 proteins are found in a clade sister to the most closely related PUB17 proteins, and PUB16 forms a single clade sister to the closely related ARC1 and PUB17 proteins. Node values = posterior probabilities.
Table 3. Gene Identifiers and Accession Numbers for the Predicted PUB Proteins Used in the Phylogenetic Analysis.
| Species | PUB Proteins | ||||
| PUB5 | PUB12 | PUB13 | PUB14 | PUB15 | |
| A. thaliana | At4g36550 | At2g28830 | At3g46510 | At3g54850 | At5g42340 |
| NP_195373 | NP_565676 | NP_190235 | NP_191045 | NP_199049 | |
| A. lyrata | 320670; XP_002881013 | 937358; XP_002877490 | 938412; XP_002876275 | 949064; XP_002863798 | |
| C. rubella | 1002257 | 10016804 | 10016842 | 10028542 | |
| B. rapa | Bra035665 | Bra033820; Bra018212 | Bra007105 | Bra027977 | |
| T. halophila | 10016364 | 10002430 | 10010199 | 10003163 | |
| PUB16 | PUB17 | PUB18 | PUB19 | ARC1 | |
| A. thaliana | At5g01830; NP_195803 | At1g29340; NP_174228 | At1g10560; NP_172526 | At1g60190; NP_176225 | – |
| A. lyrata | 486885; XP_002872992 | 473144; XP_002890808 | 471171; XP_002889817 | 475300; XP_002888147 | 934022; XP_002879489 |
| C. rubella | 10008498 | 10008444 | 10008393 | 10021483 | 10025155 |
| B. rapa | Bra019022; Bra001998 | Bra010834; Bra032309 | Bra019924 | Bra031521 | Bra021899 |
| T. halophila | 10022583 | 10008444 | 10008393 | 10023320 | – |
| B. napus | 2558938; AAB97738 | ||||
| B. oleracea | 164470360; ABY58019 | ||||
Dashes represent the absence of a corresponding ARC1 orthologue. For empty cells, the corresponding orthologue was not identified for the phylogenetic analysis.
Loss of the ARC1 Region in A. thaliana Is Conserved across All Ecotypes Surveyed
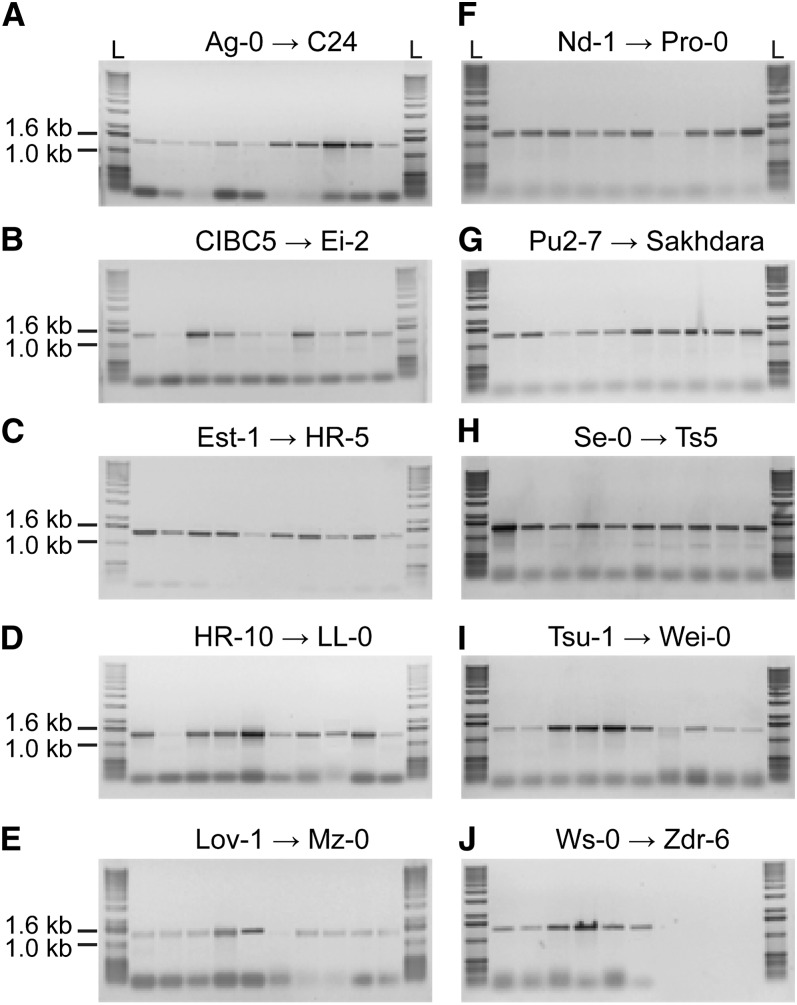
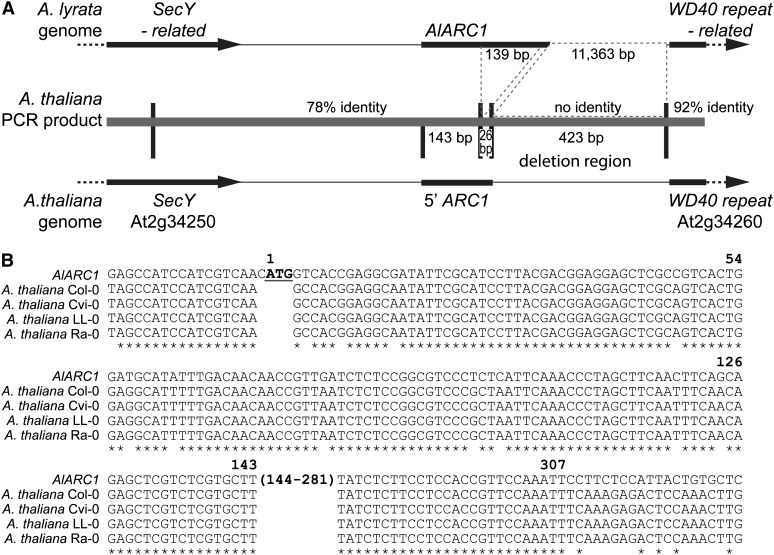
The source of self-compatibility in A. thaliana has been attributed to the loss of functional SRK and SCR genes (Kusaba et al., 2001; Tang et al., 2007; Tsuchimatsu et al., 2010). While A. lyrata SRK and SCR genes can be transformed into A. thaliana to confer self-incompatibility, the strength of the response can be quite variable, with some ecotypes showing a good self-incompatibility response to other ecotypes being completely self-compatible with a wild-type acceptance of pollen grains (Nasrallah et al., 2004; Boggs et al., 2009; Tsuchimatsu et al., 2010). Also, in an extensive survey of different A. thaliana ecotypes, several ecotypes were found to contain a functional copy of SRKa, and varying self-incompatibility phenotypes were observed when these A. thaliana pistils were pollinated with A. halleri SCRa pollen (Tsuchimatsu et al., 2010). With ARC1 being deleted from the A. thaliana Col-0 and C24 ecotypes, we were interested to determine if the ARC1 deletion is more widespread than previously reported (Kitashiba et al., 2011). A genomic PCR screen was conducted on two different natural accession sets: the Nordborg collection (96 ecotypes; Nordborg et al., 2005) and the Beck collection (261 ecotypes; Beck et al., 2008), which cover a wide geographic range of A. thaliana ecotypes and include ecotypes carrying a functional copy of SRKa (Tsuchimatsu et al., 2010). The PCR screen was designed such that if the ARC1 deletion region is present in an A. thaliana ecotype, a PCR product of 1.4 kb in size would be obtained (Figure 3). Thus, this screen served as a quick and straightforward method to examine the genetic diversity of the ARC1 deletion region across the A. thaliana ecotypes. The PCR screen revealed that all of the A. thaliana ecotypes surveyed contained the deletion region and therefore lacked a functional copy of ARC1 (Figure 3; see Supplemental Data Set 1 online). To verify that a similar deletion was present, PCR bands from 10 different A. thaliana ecotypes were selected for sequencing: C24, Kro-0, Ler-0, and Mrk-0 from Germany, Ra-0 from France, Br-0 from the Czech Republic, Wei-1 from Switzerland, LL-0 from Spain, Bur-0 from Ireland, Col-0 from the US, and Cvi-0 from Cape Verde Islands. The sequences from the PCR products indicated that the 10 ecotypes shared identical sequences for the ARC1 5′ coding region and the deletion breakpoint (Figure 4). A few single nucleotide polymorphisms were detected between the 10 different ecotypes in the region upstream of the A. thaliana ARC1 pseudogene. Thus, ARC1 has undergone a widespread deletion in A. thaliana, and we did not identify any ecotypes that carried an intact copy of ARC1. Furthermore, the deletion of the ARC1 region appears to have been a single event, given the conservation observed for the sequence and deletion breakpoint in the 10 diverse ecotypes.
Figure 3.
PCR Screen to Survey for the ARC1 Deletion Region in 96 A. thaliana Ecotypes.
The PCR results from the 96 Nordborg ecotype set are shown in alphabetical order with 10 samples per gel. The PCR product for the deleted ARC1 region is 1.4 kb, and all ecotypes were found to have the deletion, based on the presence of this PCR product. The different intensities in the PCR bands reflect only experimental variation. L, Gibco BRL 1-kb ladder.
(A) Ag-0, An-1, Bay-0, Bil-5, Bil-7, Bor-4, Br-0, Bur-0, and C24.
(B) CIBC-5, CIBC-17, Col-0, CS22491, Ct-1, Cvi-0, Eden-1, Eden-2, Edi-0, and Ei-2.
(C) Est-1, Fab-2, Fab-4, Fei-1, Ga-0, Got-7, Got-22, Gu-0, Gy-0, and HR-5.
(D) HR-10, Kas-2, Kin-0, Knox-10, Knox-18, Kondara, Kz-1, Kz-9, Ler-1, and LL-0.
(E) Lov-1, Lov-5, Lp2-2, Lp2-6, Lz-0, Mr-0, Mrk-0, Ms-0, Mt-0, and Mz-0.
(F) Nd-1, NFA-8, NFA-10, Nok-3, Omo2-1, Omo2-3, Oy-0, Pna-10, Pna-17, and Pro-0.
(G) Pu2-7, Pu2-23, Ra-0, Ren-1, Ren-11, Rmx-A02, Rmx-A180, RRS-7, RRS-10, and Sakhadara.
(H) Se-0, Sorbo, Spr1-2, Spr1-6, Sq-1, Sq-8, Tamm-2, Tamm-27, Ts-1, and Ts-5.
(I) Tsu-1, Ull2-3, Ull2-5, Uod-1, Uod-7, Van-0, Var2-1, Var2-6, Wa-1, and Wei-0.
(J) Ws-0, Ws-2, Wt-5, Yo-0, Zdr-1, and Zdr-6.
Figure 4.
ARC1 Deletion Region in the A. thaliana Genome.
(A) Schematic of the ARC1 region in A. lyrata and A. thaliana. The A. lyrata genome contains an intact ARC1 gene and when compared with A. thaliana shares sequences identity until the deletion break point in the A. thaliana ARC1 pseudogene. The At2g34250 3′ untranslated region and the ARC1 5′ untranslated region/start of the coding region are 78% identical at the DNA sequence level. This is followed by a 139-bp deletion in A. thaliana ARC1 and a small 26-bp sequence of ARC1 that is 100% identical between the two species genomes. The next 423 bp from A. thaliana has no sequence identity to the 11,363-bp region from A. lyrata. The sequence identity returns at 9 bp before the start of At2g34260, and the region is 92% similar up until the end of the sequencing read. The sequence identities were determined using the A. lyrata genome sequence and the sequenced PCR products from 10 different A. thaliana ecotypes.
(B) Sequence alignment of A. lyrata ARC1 to the corresponding region in A. thaliana. PCR products covering this region (shown in [A]) were sequenced for 10 ecotypes (Br-0, Bur-0, C24, Col-0, Cvi-0, Kro-0, Ler-0, LL-0, Mrk-0, Ra-0, and Wei-1). The 10 ecotypes shared identical sequences for the ARC1 5′ coding region and the deletion breakpoint, and sequences from four of these ecotypes are shown aligned to Al-ARC1. The start codon for Al-ARC1 is marked in bold and underlined and is absent in corresponding A. thaliana sequences.
Knockdown of ARC1 Expression in Transgenic A. lyrata Plants Leads to the Breakdown of Self-Incompatibility
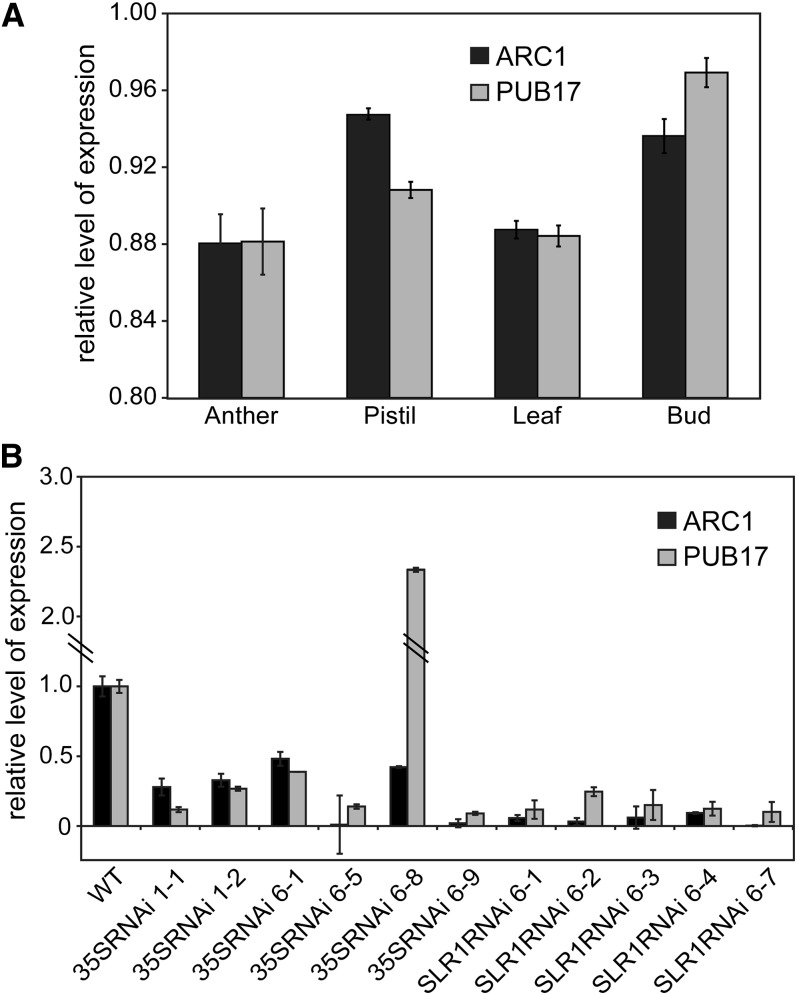
With our observation that the ARC1 genomic region has undergone distinct deletions in several self-compatible species, we were interested in testing ARC1’s role in a naturally occurring self-incompatible Arabidopsis species, A. lyrata. Previously, we found that when ARC1 expression was knocked down in transgenic B. napus plants, the self-incompatibility response was attenuated resulting in successful self-fertilization (Stone et al., 1999). Quantitative RT-PCR (qRT-PCR) was first used to examine the expression patterns of A. lyrata ARC1 (Al-ARC1), and it was found to be expressed at highest levels in the pistil and buds, and lower levels of expression could also be detected in the anthers and leaves (Figure 5A; see Supplemental Figure 1 online). The closely related Al-PUB17 was also tested, and it was expressed in all tissues with the highest level of expression detected in flower bud. Thus, Al-ARC1 was found to be expressed in the pistil where it could function in the self-incompatibility response, and the expression pattern is consistent with self-incompatibility genes often showing increased or specific expression in the pistil (e.g., SRK and B. napus ARC1; Goring et al., 1992; Gu et al., 1998).
Figure 5.
qRT-PCR Analysis of ARC1 and PUB17 Expression in Wild-Type and Transgenic ARC1 RNAi A. lyrata Lines.
(A) qRT-PCR analysis of ARC1 and PUB17 mRNA levels in anthers, pistil, leaves, and flower buds. ARC1 and PUB17 expression levels were normalized across tissue samples with TUB4 (Bassel et al., 2008), and data from two biological replicates are shown. Error bars indicate ± se.
(B) Quantitative PCR analysis of ARC1 and PUB17 expression levels in stigmas from the transgenic ARC1 RNAi A. lyrata lines. The expression levels of ARC1 and PUB17 are shown relative to a wild-type (WT) sample (set to a value of 1). ARC1 and PUB17 expression levels were normalized across the samples with Elf1α, and data from two biological replicates are shown. Error bars indicate ± se.
To test the requirement of Al-ARC1 for the self-incompatibility response, transgenic Al-ARC1 RNAi A. lyrata lines were generated to suppress Al-ARC1 expression. An Al-ARC1–specific hairpin RNAi driven by either the stigma-specific SLR1 promoter or the cauliflower mosaic virus 35S promoter (Franklin et al., 1996; Helliwell and Waterhouse, 2003) was transformed into self-incompatible perennial A. lyrata spp petraea (Kivimäki et al., 2007) using a modified Agrobacterium tumefaciens–mediated floral dip transformation method (Clough and Bent, 1998; Lu and Kang, 2008). As the A. lyrata plants were self-incompatible, flowers were manually pollinated with cross-compatible pollen following transformation. Eleven independent transgenic A. lyrata plants were generated, and stigma RNA from each line was analyzed by qRT-PCR to determine the degree of the ARC1 RNAi knockdown. The ARC1 mRNA levels were found to be reduced to varying degrees in all 11 independent transgenic lines (Figure 5B). While the 489-bp ARC1 fragment used in the hairpin RNAi vector was chosen for its specificity, this region does share 72% sequence identify to the closely related PUB17 gene and, as a result, could affect PUB17 expression as well. No other significant target genes were identified in BLAST searches with the 489-bp Al-ARC1 sequence. The qRT-PCR analysis indicated that PUB17 mRNA levels were also reduced in all lines, except the 35SRNAi 6-8 where PUB17 mRNA levels were increased (Figure 5B). These results were also verified for the transgenic A. lyrata 35SRNAi 6-8 line using a standard RT-PCR analysis (see Supplemental Figure 1 online).
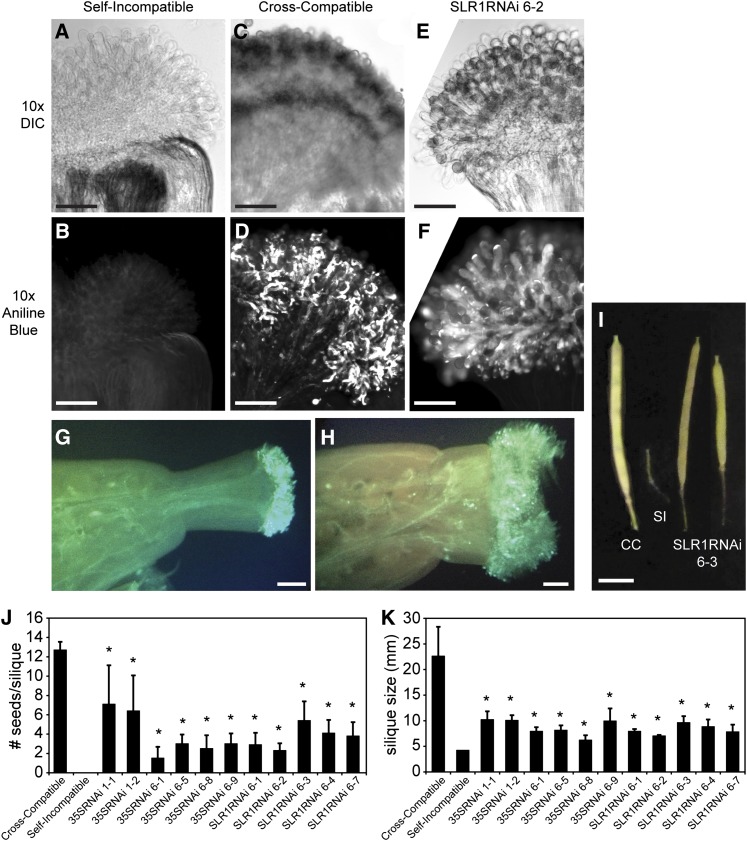
The 11 transgenic ARC1 RNAi A. lyrata lines with reduced ARC1 RNA levels were then examined for the robustness of their self-incompatibility response phenotypes. Similar to the Brassica species, self-pollen rejection in A. lyrata occurs very early as rejected pollen grains are unable to germinate and form pollen tubes. Therefore, the ability of the ARC1 RNAi plants to reject or accept self-pollen was measured by examining pollen grains and pollen tube penetration on the stigmatic surface. For the A. lyrata ARC1 RNAi lines, the pistils were stained with aniline blue to visualize the pollen tubes 24 h after pollination. Typically in the aniline blue staining process, only pollen grains that have been accepted remain on the surface. All rejected pollen grains would no longer remain on the stigmatic papillae as they would have been removed by the washes inherent in the aniline blue staining protocol. Therefore, in a successful self-incompatibility response, pollen grains are absent from the stigmatic surface (Figure 6A) and no pollen tubes can be observed (Figure 6B). By contrast, the cross-compatible pollinated stigma shows pollen grains on the stigmatic surface (Figure 6C) and numerous pollen tubes growing into the stigma (Figure 6D) and into the style and ovary (Figure 6G). For all 11 transgenic ARC1 RNAi A. lyrata lines, the self-pollen rejection response was impaired following self-pollination, and both pollen grains (Figure 6E; see Supplemental Figure 2 online) and pollen tubes could be observed growing into the stigmas (Figure 6F; see Supplemental Figure 2 online). Also, pollen tubes were observed growing into the style and ovary (Figure 6H). Thus, the reduction in Al-ARC1 mRNA levels was associated with a breakdown in the self-incompatibility response.
Figure 6.
Pollen Tube Growth and Seed Set in the Transgenic ARC1 RNAi A. lyrata Lines.
(A) to (F) Pollen grain attachment and pollen tube growth in A. lyrata stigmas from control self-incompatible cross ([A] and [B]), control cross-compatible cross ([C] and [D]), and SLR1RNAi 6-2 self-pollination ([E] and [F]). After pollination, pistils were stained with aniline blue to visualize the pollen tubes. DIC, differential interference contrast. Bars = 0.1 mm.
(G) and (H) Pollen tube growth in A. lyrata pistils from control cross-compatible cross (G) and SLR1RNAi 6-2 self-pollination (H). After pollination, pistils were stained with aniline blue to visualize the pollen tubes. Bars = 2 mm.
(I) Siliques from cross- and self-pollinated pistils. Wild-type cross-compatible (CC) pollinations result in a large silique while self-incompatible (SI) crosses result in a small undeveloped silique. Siliques from a self-pollinated SLR1RNAi 6-3 plant were much larger and developed. Bar = 5 mm
(J) Average number of seeds per silique following cross- and self-pollinations. All the transgenic ARC1 RNAi A. lyrata lines produced significant increases in the number of seeds/silique when compared with wild-type self-incompatible crosses (t test, P < 0.05). n = 10. Error bars indicate ± se.
(K) Average silique sizes following cross- and self-pollinations. All the transgenic ARC1 RNAi A. lyrata lines produced significant increases in silique lengths when compared with siliques from wild-type self-incompatible crosses (t test, P < 0.05). n = 10. Error bars indicate ± se.
To determine the extent of self-pollen acceptance in the Al-ARC1 RNAi lines, seed production, as a measure of successful pollen tube growth and fertilization, was examined. While cross-compatible pollinations led to well-developed siliques with ∼13 seeds/silique, the siliques from self-incompatible pollinations were very small and empty (Figures 6I to 6K). Following self-pollinations, transgenic ARC1 RNAi A. lyrata lines displayed a range of silique sizes, and some examples of larger siliques are shown for the SLR1RNAi 6-3 line in Figure 6I. The average silique sizes from the RNAi lines ranged from 6 mm in the 35SRNAi line 6-8 to10 mm in 35SRNAi line 1-1 and fell between the average 4-mm siliques from self-incompatible pollinations and 22.4-mm siliques from cross-compatible crosses (Figure 6K). Additionally, the number of seeds present in the siliques from the transgenic plants was variable and was usually less than the cross-compatible control (Figure 6J). Importantly though, all of the transgenic Al-ARC1 RNAi lines produced some seeds when self-pollinated while the wild-type self-incompatible pollinations failed to produce any seeds (Figure 6J). Thus, the transgenic ARC1 RNAi A. lyrata lines show clear evidence of breakdown in the self-incompatibility response following self-pollination. Also, the observed pseudo-self-compatibility phenotype was similar to that seen for B. napus ARC1 antisense lines (Stone et al., 1999).
DISCUSSION
ARC1 is an E3 ubiquitin ligase that functions in the pistil as part of the Brassica self-incompatibility pathway (Gu et al., 1998; Stone et al., 1999, 2003; Samuel et al., 2009), and this study demonstrates that ARC1 is also required for the A. lyrata self-incompatibility response. We found that the RNAi suppression of Al-ARC1 resulted in the acceptance of self-incompatible pollen, despite the presence of the upstream SCR/SP11 and SRK components. While PUB17, the most closely related paralog to ARC1, was also suppressed in many of these lines, PUB17 is unlikely to be responsible for this phenotype as one line, 35SRNAi 6-8, had increased PUB17 mRNA levels and still showed an attenuated self-incompatibility phenotype. Also, Rea et al. (2010) reported that when the A. lyrata SRK and SCR/SP11 genes were transformed into the A. thaliana pub17 mutant, the self-incompatible phenotype was unaffected, implying that PUB17 is not involved in the SRK signaling pathway. Finally, PUB17 has been previously reported to have a very different function where it is required for disease resistance and the hypersensitive response in tomato (Solanum lycopersicum) and A. thaliana (Yang et al., 2006).
With ARC1 being required for the self-incompatibility response in both B. napus and A. lyrata, ARC1 may play a more widespread signaling role in self-pollen rejection in the Brassicaceae than previously suggested by Kitashiba et al. (2011). Brassica and Arabidopsis belong to separate lineages in the Brassicaceae with Brassica in lineage II and Arabidopsis in lineage I (Franzke et al., 2011). As part of this study, we also investigated the occurrence of the ARC1 gene in other species in the Brassicaceae. This includes lineage I species (A. thaliana, A. lyrata, C. grandiflora, C. rubella, and L. alabamica), lineage II species (B. rapa, S. irio, T. halophila, and T. parvula), and a species outside of the core Brassicaceae (A. arabicum) (Franzke et al., 2011). Previously, ARC1 was reported to be present in the genome of self-incompatible A. lyrata but deleted from two self-compatible A. thaliana ecotypes, Col-0 and C24 (Kitashiba et al., 2011). A genome-wide survey of these sequenced genomes led to the intriguing observation that there were independent deletions in the ARC1 genomic region for most of the self-compatible species tested (A. thaliana, L. alabamica, T. halophila, T. parvula, and A. arabicum). In contrast with ARC1, the genomic region including PUB17 was highly conserved across all the genomes examined. The PUB17 gene was present in all the genomes and the structure and gene order in the PUB17 region was quite conserved.
While most of the self-compatible species did not carry a functional copy of the ARC1 gene, C. rubella was one self-compatible species that retained ARC1, and this may reflect the recent the transition to self-pollination that occurred for this species. The breakdown of self-incompatibility in C. rubella occurred ∼30,000 to 50,000 years ago (Foxe et al., 2009; Guo et al., 2009), while the breakdown of self-incompatibility of A. thaliana has been estimated to be much more distant, between ∼400,000 and 1 million years ago (Bechsgaard et al., 2006; Tang et al., 2007; Bomblies and Weigel, 2010). The speciation of C. rubella and the transition to a self-pollinating species was also a direct result of a population bottleneck, and the inactivation of the SCR/SP11-SRK genes was likely the initial event that allowed self-pollination to occur (Foxe et al., 2009; Guo et al., 2009).
Previously, it has been hypothesized that A. thaliana become a self-compatible species due to the pseudogenization of the SCR/SP11 and SRK genes (Kusaba et al., 2001). While the majority of the A. thaliana ecotypes surveyed to date carry pseudogenes for SCR/SP11 and SRK, some ecotypes were discovered to have intact copies of SRK or SCR/SP11 (Kusaba et al., 2001; Bechsgaard et al., 2006; Tang et al., 2007; Shimizu et al., 2008; Boggs et al., 2009; Tsuchimatsu et al., 2010; Guo et al., 2011). This indicated that the transition from self-incompatibility to self-compatibility in A. thaliana has occurred multiple times with independent SCR/SP11-SRK mutations. With the previous report that the ARC1 gene was deleted in the A. thaliana Col-0 and C24 ecotypes (Kitashiba et al., 2011), we also investigated how widespread this specific ARC1 deletion was in 357 A. thaliana ecotypes. The complete loss of the ARC1 gene in all 357 ecotypes tested, including those with a functional SCR/SP11 or SRK gene, suggests that ARC1 became a pseudogene before the inactivation of SCR/SP11 or SRK in A. thaliana. Tsuchimatsu et al. (2010) observed variations in the manifestation of the reconstituted self-incompatibility responses in different A. thaliana ecotypes, with the self-incompatibility response weakening in older flowers (pseudo-self-compatibility). They suggested that mutations conferring pseudo-self-compatibility could have occurred prior to the loss of the SCR/SP11 or SRK self-incompatibility genes and account for the pseudo-self-compatibility in A. thaliana. The widespread loss of A. thaliana ARC1 could be one such pseudo-self-compatibility mutation. In both B. napus and A. lyrata, we observed that the suppression of ARC1 expression is correlated with an incomplete breakdown of self-incompatibility. While this could be due to inefficient ARC1 suppression, it may also indicate that there are other elements functioning downstream of SRK (e.g., MLPK) and that loss of these elements, in addition to ARC1, would be required to produce a full breakdown of the self-incompatibility response. Thus, perhaps in A. thaliana, the weakening of the self-incompatibility response with the loss of ARC1 could have allowed for some self-pollination to occur, leading to conditions for further breakdown by the pseudogenization of the SCR/SP11 and SRK genes.
While we have described a role for ARC1 in a simple linear SRK signaling pathway (Samuel et al., 2009; Chapman, 2010), receptor kinase signaling pathways in animal systems are typically much more complex with multiple branches (Seet et al., 2006; Lemmon and Schlessinger, 2010). Thus, it would not be unexpected for SRK to signal through other unknown branches to cause self-pollen rejection, and the loss of one branch involving ARC1 may lead only to a pseudo-self-compatibility phenotype. This may also offer an explanation for the large ecotype-dependent range of self-incompatibility phenotypes observed in transgenic A. thaliana expressing the A. lyrata SCR/SP11 and SRK genes (Nasrallah et al., 2004; Boggs et al., 2009; Tsuchimatsu et al., 2010). The range of phenotypes from self-compatibility to stronger levels of self-incompatibility may reflect activity levels of other elements or pathways functioning downstream of SRK in these different ecotypes. In the future, it would be of interest to test whether the transformation of Al-ARC1 into transgenic A. thaliana displaying a pseudo-self-compatibility phenotype would strengthen the self-incompatibility response. In conclusion, we have shown that ARC1 has been independently deleted in the genomes of a number of self-compatible species, and its role in the self-incompatibility response is conserved between Arabidopsis and Brassica species.
METHODS
Synteny and Phylogenetic Analyses
For the nine-species whole-genome orthologous alignment assembled by the VEGI consortium (available on their genome browser at http://grandiflora.eeb.utoronto.ca:8086), the genomes of six species were obtained from the genome assemblers or from public databases (www.phytozome.org). Three additional species, Sisymbrium irio, Leavenworthia alabamica race a4 (Busch et al., 2011), and Aethionema arabicum were sequenced by the VEGI consortium using exclusively Illumina paired-end and mate-pair approaches and were assembled using a combination of Ray (Boisvert et al., 2010) for contiging and SOAPdenovo (Li et al., 2010) for scaffolding. A whole-genome multiple alignment of the nine genomes was produced using a progressive alignment pipeline similar to that previously published for the alignment of vertebrate genomes (Miller et al., 2007). Briefly, the procedure starts by computing pairwise alignments between each species and the Arabidopsis lyrata genome, which is used as a reference, using the lastZ program (Schwartz et al., 2003). A progressive multiple alignment procedure is then applied to assemble a multiple alignment, using the Mutliz program (Blanchette et al., 2004). In the case of the Brassica rapa and L. alabamica genomes, which have both undergone whole-genome triplications, only the best conserved, up to three remaining paralogous regions, was retained for the alignment.
The nine-species whole-genome orthologous alignment was accessed using the Synteny viewer in the University of California, Santa Cruz Genome Browser (Fujita et al., 2011) to construct the gene syntenies for the ARC1 and PUB17 genomic regions from the Arabidopsis thaliana, A. lyrata, S. irio, L. alabamica, Thellungiella halophila, Thellungiella parvula, B. rapa, and A. arabicum genomes. The synteny viewers in The Arabidopsis Information Resource (TAIR; Rhee et al., 2003) and the Brassica database (BRAD; Cheng et al., 2011) were also used in these analyses. Finally, A. lyrata, Capsella rubella, B. rapa, and T. halophila genomic sequences were extracted and protein BLAST searches were conducted using Phytozome v8.0 (Altschul et al., 1990; Goodstein et al., 2012). Gene identifiers are listed in Tables 1 and 2.
For the phylogenetic analysis, the A. thaliana PUB12-19 and A. lyrata ARC1 amino acid sequences were used in TBLASTN searches to identify orthologs from the A. lyrata, C. rubella, T. halophila, and B. rapa genomes. Default TBLASTN parameters were used in the Phytozome and BRAD databases (expect threshold −1, comparison matrix BLOSUM62, word length 3, allowed for gaps). The synteny viewers from TAIR (Rhee et al., 2003) and BRAD (Cheng et al., 2011) were also used to identify the genes encoding A. lyrata and B. rapa PUB10-19 and ARC1 proteins. Gene identifiers are listed in Table 3. The ARC1 sequences from B. napus (2,558,938) and B. oleracea (164,470,360) were obtained from GenBank, and the Capsella grandiflora ARC1 sequence was extracted from an RNASeq analysis (K. Hazzouri and S.I. Wright, unpublished data). Full-length PUB5, PUB12-19, and ARC1 amino acid sequences were aligned using MAFFT (Katoh et al., 2005). The alignment is available as Supplemental Data Set 2 online. The protein FASTA alignment file was used in MrBayes for a Bayesian phylogenetic analysis, with the mixed model option and 300,000 generations, and two runs used to check for convergence (Ronquist et al., 2012).
Plant Material
The A. lyrata plants used in this study were from the self-incompatible perennial A. lyrata spp petraea P6 and P7 populations collected by Kivimäki et al. (2007) in Northern Sweden. Plants from the P6 population were cross-compatible with the P7 population. The A. thaliana ecotypes screened for the ARC1 deletion were obtained from the ABRC and were from the Nordborg collection (96 ecotype set; Nordborg et al., 2005) and Beck collection (261 ecotype set; Beck et al., 2008). All plants were grown under long-day growth chamber conditions consisting of a 16-h-light/8-h-dark photoperiod at 22°C. Flowering was induced in the A. lyrata plants by vernalizing rosettes of ∼12 cm in size for 30 d on a short-day cycle of an 8-h-light/16-h-dark photoperiod at 5°C (Kuittinen et al., 2008).
Nucleic Acid Extractions and PCR Assays
For PCR genotyping of the transgenic A. lyrata plants and screening for the presence of the ARC1 deletion in the A. thaliana ecotypes, genomic DNA was extracted from young leaves using the Plant DNeasy kit (Qiagen). The A. thaliana ARC1 deletion region was PCR amplified using primers to the last exon of At2g34250 and the first exon of At2g34260. For the RT-PCR assays, RNA was extracted from A. lyrata anthers, leaves, buds, pistils, and stigmas tissues using of the Plant RNeasy kit (Qiagen). The RNA was quantified with a Nanodrop (GE Nanovue Plus) and then treated with DNase I (amplification grade; Invitrogen) to remove any contaminating genomic DNA. The DNase I was removed from the RNA samples by acid-phenol:chloroform (Ambion) extraction, followed by a phenol-chloroform extraction and nucleic acid precipitation (Sambrook and Russel, 2001). cDNA synthesis was performed on 50 ng/μL of RNA per tissue using the Super Script III reverse transcriptase (Invitrogen). The cDNA was tested by PCR reaction with A. lyrata Elf1α-specific primers. RT-PCR and qRT-PCR assays were then conducted on the cDNA samples. The qRT-PCR on the transgenic ARC1 RNAi A. lyrata lines was performed as previously described (Bassel et al., 2008; Indriolo et al., 2010) using stigma cDNA and 2× Power SYBR green (Applied Biosystems). All primers were optimized before use, and the ARC1 and PUB17 expression levels were normalized to Elf1α (for the different RNAi lines) or TUB4 (for the different tissues). The standard PCR conditions used for all reactions were a 2-min denaturation at 94°C, followed by a three-step cycle of 30 s 94°C denaturation, 30 s at the appropriate annealing temperature for each primer pair, and then extension at 72°C with a time of 1 kb/1 min for 25 to 35 cycles, followed by a final extension of 10 min at 72°C. Tsg polymerase (Biobasics) was used for all PCR reactions. See Supplemental Table 1 online for all primers used in this study.
A. lyrata ARC1 RNAi Constructs and Transformation
The Al-ARC1 hairpin RNAi construct was designed using an Al-ARC1 fragment covering nucleotides 1260 to 1748 of the coding sequence. The cauliflower mosaic virus 35S promoter:Al-ARC1 RNAi construct was made by amplifying the 489-bp Al-ARC1 fragment with Advantage 2 polymerase (Clontech) using a forward primer with BamHI and XhoI sites and a reverse primer with ClaI and KpnI sites. The PCR product was subcloned into pGEMT-easy (Promega) and was verified by sequencing. The Al-ARC1 hairpin was assembled in the pKannibal vector (XhoI and KpnI for the sense fragment, and BamHI and ClaI for the antisense fragment) and then transferred into the pART27 plant transformation vector using NotI (Helliwell and Waterhouse, 2003). To generate the SLR1 promoter:Al-ARC1 RNAi construct, the Al-ARC1 hairpin was amplified from pKannibal using a forward primer with an XmaI site and a reverse primer with an EcoRI site. The PCR product was then digested with XmaI and EcoRI and ligated into the pORE3 vector (Coutu et al., 2007) containing the SLR1 promoter (Chapman, 2010). For the transformation of the Al-ARC1 hairpin RNAi plant transformation vectors into A. lyrata, the Agrobacterium tumefaciens–mediated floral dip transformation method for A. thaliana (Clough and Bent, 1998) was modified with a Camilina sativa protocol that included placing the dipped plants under vacuum for 5 min (Lu and Kang, 2008). As the A. lyrata plants were self-incompatible, flowers were manually pollinated with cross-compatible pollen for 1 week after dipping.
Crosses and Pollination Assays
For the A. lyrata crosses, pistils from 1- to 2-d-old flowers were pollinated with pollen from 2- to 3-d-old flowers as this was determined to be the optimal timing for pistil receptivity and pollen viability using flower development in A. thaliana as a guide (Smyth et al., 1990). Aniline blue staining for pollen tubes was conducted on pistils that had been pollinated for 24 h as previously described (Samuel et al., 2009). Siliques from self-incompatible and cross-compatible crosses were characterized by measuring length and the number of seeds present in each silique for 10 randomly selected siliques from each control and transgenic ARC1 RNAi A. lyrata line.
Accession Numbers
Sequence data from this article can be found in the TAIR, BRAD, and Phytozome data libraries under the gene identifiers listed in Tables 1–3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RT-PCR Analysis of ARC1 Expression in Wild-Type and Transgenic ARC1 RNAi A. lyrata Lines.
Supplemental Figure 2. Pollen Grain Attachment and Pollen Tube Growth in Pistils from the Transgenic ARC1 RNAi A. lyrata Lines.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. List of A. thaliana Ecotypes Identified by PCR to Contain the ARC1 Deletion Region.
Supplemental Data Set 2. Text File of the Alignment Used for the Phylogenetic Analysis in Figure 2.
Acknowledgments
We thank Jon Agren for the A. lyrata seeds and the ABRC for the A. thaliana ecotype seeds. Genome sequence data for T. halophila and C. rubella were produced by the U.S. Department of Energy Joint Genome Initiative in collaboration with the user community. Genome sequence data for A. arabicum, L. alabamica, S. irio, and C. grandiflora were produced by the VEGI consortium and their collaborators. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada to D.R.G. and S.I.W., a Canada Research Chair to D.R.G., and an Ontario Government Early Researcher Award to S.I.W.
AUTHOR CONTRIBUTIONS
E.I. designed the research, performed research, analyzed data, and wrote the article. P.T. performed research and analyzed data. S.I.W. designed the research, contributed new computational tools, and analyzed data. D.R.G. designed the research, analyzed data, and wrote the article.
Glossary
- UND
U-box N-terminal domain
- RNAi
RNA interference
- Col-0
Columbia-0
- qRT-PCR
quantitative RT-PCR
- TAIR
The Arabidopsis Information Resource
- BRAD
Brassica database
- VEGI
Value-Directed Evolutionary Genomics
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Andersen P., Kragelund B.B., Olsen A.N., Larsen F.H., Chua N.H., Poulsen F.M., Skriver K. (2004). Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 279: 40053–40061 [DOI] [PubMed] [Google Scholar]
- Azevedo C., Santos-Rosa M.J., Shirasu K. (2001). The U-box protein family in plants. Trends Plant Sci. 6: 354–358 [DOI] [PubMed] [Google Scholar]
- Bailey C.D., Koch M.A., Mayer M., Mummenhoff K., O’Kane S.L., JrWarwick S.I., Windham M.D., Al-Shehbaz I.A. (2006). Toward a global phylogeny of the Brassicaceae. Mol. Biol. Evol. 23: 2142–2160 [DOI] [PubMed] [Google Scholar]
- Bassel G.W., Fung P., Chow T.F., Foong J.A., Provart N.J., Cutler S.R. (2008). Elucidating the germination transcriptional program using small molecules. Plant Physiol. 147: 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechsgaard J.S., Castric V., Charlesworth D., Vekemans X., Schierup M.H. (2006). The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 23: 1741–1750 [DOI] [PubMed] [Google Scholar]
- Beck J.B., Schmuths H., Schaal B.A. (2008). Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Mol. Ecol. 17: 902–915 [DOI] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Haussler D., Miller W. (2004). Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14: 708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs N.A., Dwyer K.G., Shah P., McCulloch A.A., Bechsgaard J., Schierup M.H., Nasrallah M.E., Nasrallah J.B. (2009). Expression of distinct self-incompatibility specificities in Arabidopsis thaliana. Genetics 182: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S., Laviolette F., Corbeil J. (2010). Ray: Simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J. Comput. Biol. 17: 1519–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K., Weigel D. (2010). Arabidopsis and relatives as models for the study of genetic and genomic incompatibilities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch J.W., Joly S., Schoen D.J. (2011). Demographic signatures accompanying the evolution of selfing in Leavenworthia alabamica. Mol. Biol. Evol. 28: 1717–1729 [DOI] [PubMed] [Google Scholar]
- Chapman L. (2010). The Role of Sec15b and Phosphatidylinositol-4-Phosphate in Early Pollen-Pistil Interactions. M.Sc. Thesis, University of Toronto.
- Chapman L.A., Goring D.R. (2010). Pollen-pistil interactions regulating successful fertilization in the Brassicaceae. J. Exp. Bot. 61: 1987–1999 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Vekemans X. (2005). How and when did Arabidopsis thaliana become highly self-fertilising. Bioessays 27: 472–476 [DOI] [PubMed] [Google Scholar]
- Cheng F., Liu S., Wu J., Fang L., Sun S., Liu B., Li P., Hua W., Wang X. (2011). BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 11: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coutu C., Brandle J., Brown D., Brown K., Miki B., Simmonds J., Hegedus D.D. (2007). pORE: A modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res. 16: 771–781 [DOI] [PubMed] [Google Scholar]
- Dassanayake M., Oh D.-H., Haas J.S., Hernandez A., Hong H., Ali S., Yun D.-J., Bressan R.A., Zhu J.-K., Bohnert H.J., Cheeseman J.M. (2011). The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxe J.P., Slotte T., Stahl E.A., Neuffer B., Hurka H., Wright S.I. (2009). Recent speciation associated with the evolution of selfing in Capsella. Proc. Natl. Acad. Sci. USA 106: 5241–5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin T.M., Oldknow J., Trick M. (1996). SLR1 function is dispensable for both self-incompatible rejection and self-compatible pollination processes in Brassica. Sex. Plant Reprod. 9: 203–208 [Google Scholar]
- Franzke A., Lysak M.A., Al-Shehbaz I.A., Koch M.A., Mummenhoff K. (2011). Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant Sci. 16: 108–116 [DOI] [PubMed] [Google Scholar]
- Fujita P.A., Rhead B., Zweig A.S., Hinrichs A.S., Karolchik D., Cline M.S., Goldman M., Barber G.P., Clawson H., Coelho A., Diekhans M., Dreszer T.R., Giardine B.M., Harte R.A., Hillman-Jackson J., Hsu F., Kirkup V., Kuhn R.M., Learned K., Li C.H., Meyer L.R., Pohl A., Raney B.J., Rosenbloom K.R., Smith K.E., Haussler D., Kent W.J. (2011). The UCSC genome browser database: Update 2011. Nucleic Acids Res. 39(Database issue): D876–D882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40(Database issue): D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goring D.R., Banks P., Fallis L., Baszczynski C.L., Beversdorf W.D., Rothstein S.J. (1992). Identification of an S-locus glycoprotein allele introgressed from B. napus ssp. rapifera to B. napus ssp. oleifera. Plant J. 2: 983–989 [PubMed] [Google Scholar]
- Gu T., Mazzurco M., Sulaman W., Matias D.D., Goring D.R. (1998). Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 95: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.L., Bechsgaard J.S., Slotte T., Neuffer B., Lascoux M., Weigel D., Schierup M.H. (2009). Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl. Acad. Sci. USA 106: 5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.L., Zhao X., Lanz C., Weigel D. (2011). Evolution of the S-locus region in Arabidopsis relatives. Plant Physiol. 157: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C., Waterhouse P. (2003). Constructs and methods for high-throughput gene silencing in plants. Methods 30: 289–295 [DOI] [PubMed] [Google Scholar]
- Hu T.T., et al. (2011). The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriolo E., Na G., Ellis D., Salt D.E., Banks J.A. (2010). A vacuolar arsenite transporter necessary for arsenic tolerance in the arsenic hyperaccumulating fern Pteris vittata is missing in flowering plants. Plant Cell 22: 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov R., Fobis-Loisy I., Gaude T. (2010). When no means no: Guide to Brassicaceae self-incompatibility. Trends Plant Sci. 15: 387–394 [DOI] [PubMed] [Google Scholar]
- Iwano M., Takayama S. (2012). Self/non-self discrimination in angiosperm self-incompatibility. Curr. Opin. Plant Biol. 15: 78–83 [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Kachroo A., Schopfer C.R., Nasrallah M.E., Nasrallah J.B. (2001). Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293: 1824–1826 [DOI] [PubMed] [Google Scholar]
- Kakita M., Murase K., Iwano M., Matsumoto T., Watanabe M., Shiba H., Isogai A., Takayama S. (2007a). Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 19: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakita M., Shimosato H., Murase K., Isogai A., Takayama S. (2007b). Direct interaction between the S-locus receptor kinase and M-locus protein kinase involved in Brassica self-incompatibility signaling. Plant Biotechnol J. 24: 185–190 [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. (2005). MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitashiba H., Liu P., Nishio T., Nasrallah J.B., Nasrallah M.E. (2011). Functional test of Brassica self-incompatibility modifiers in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 18173–18178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivimäki M., Kärkkäinen K., Gaudeul M., Løe G., Agren J. (2007). Gene, phenotype and function: GLABROUS1 and resistance to herbivory in natural populations of Arabidopsis lyrata. Mol. Ecol. 16: 453–462 [DOI] [PubMed] [Google Scholar]
- Koch M., Haubold B., Mitchell-Olds T. (2001). Molecular systematics of the Brassicaceae: Evidence from coding plastidic matK and nuclear Chs sequences. Am. J. Bot. 88: 534–544 [PubMed] [Google Scholar]
- Kuittinen H., Niittyvuopio A., Rinne P., Savolainen O. (2008). Natural variation in Arabidopsis lyrata vernalization requirement conferred by a FRIGIDA indel polymorphism. Mol. Biol. Evol. 25: 319–329 [DOI] [PubMed] [Google Scholar]
- Kusaba M., Dwyer K., Hendershot J., Vrebalov J., Nasrallah J.B., Nasrallah M.E. (2001). Self-incompatibility in the genus Arabidopsis: Characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13: 627–643 [PMC free article] [PubMed] [Google Scholar]
- Lemmon M.A., Schlessinger J. (2010). Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20: 265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Kang J. (2008). Generation of transgenic plants of a potential oilseed crop Camelina sativa by Agrobacterium-mediated transformation. Plant Cell Rep. 27: 273–278 [DOI] [PubMed] [Google Scholar]
- Mable B.K., Adam A. (2007). Patterns of genetic diversity in outcrossing and selfing populations of Arabidopsis lyrata. Mol. Ecol. 16: 3565–3580 [DOI] [PubMed] [Google Scholar]
- Mable B.K., Robertson A.V., Dart S., Di Berardo C., Witham L. (2005). Breakdown of self-incompatibility in the perennial Arabidopsis lyrata (Brassicaceae) and its genetic consequences. Evolution 59: 1437–1448 [PubMed] [Google Scholar]
- Miller W., et al. (2007). 28-Way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 17: 1797–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y., Shiu S.H., Stone S.L., Salt J.N., Goring D.R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Shiba H., Iwano M., Che F.S., Watanabe M., Isogai A., Takayama S. (2004). A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303: 1516–1519 [DOI] [PubMed] [Google Scholar]
- Nasrallah M.E., Liu P., Sherman-Broyles S., Boggs N.A., Nasrallah J.B. (2004). Natural variation in expression of self-incompatibility in Arabidopsis thaliana: Implications for the evolution of selfing. Proc. Natl. Acad. Sci. USA 101: 16070–16074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetsch M., Mayland-Quellhorst S., Neuffer B. (2006). Evolution of the self-incompatibility system in the Brassicaceae: Identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity (Edinb.) 97: 283–290 [DOI] [PubMed] [Google Scholar]
- Prigoda N.L., Nassuth A., Mable B.K. (2005). Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol. Biol. Evol. 22: 1609–1620 [DOI] [PubMed] [Google Scholar]
- Rea A.C., Liu P., Nasrallah J.B. (2010). A transgenic self-incompatible Arabidopsis thaliana model for evolutionary and mechanistic studies of crucifer self-incompatibility. J. Exp. Bot. 61: 1897–1906 [DOI] [PubMed] [Google Scholar]
- Rhee S.Y., et al. (2003). The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russel D. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press)
- Samuel M.A., Chong Y.T., Haasen K.E., Aldea-Brydges M.G., Stone S.L., Goring D.R. (2009). Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel M.A., Mudgil Y., Salt J.N., Delmas F., Ramachandran S., Chilelli A., Goring D.R. (2008). Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 147: 2084–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel M.A., Salt J.N., Shiu S.H., Goring D.R. (2006). Multifunctional arm repeat domains in plants. Int. Rev. Cytol. 253: 1–26 [DOI] [PubMed] [Google Scholar]
- Schierup M.H., Bechsgaard J.S., Nielsen L.H., Christiansen F.B. (2006). Selection at work in self-incompatible Arabidopsis lyrata: Mating patterns in a natural population. Genetics 172: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup M.H., Mable B.K., Awadalla P., Charlesworth D. (2001). Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158: 387–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer C.R., Nasrallah M.E., Nasrallah J.B. (1999). The male determinant of self-incompatibility in Brassica. Science 286: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. (2003). Human-mouse alignments with BLASTZ. Genome Res. 13: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet B.T., Dikic I., Zhou M.M., Pawson T. (2006). Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 7: 473–483 [DOI] [PubMed] [Google Scholar]
- Shimizu K.K., Shimizu-Inatsugi R., Tsuchimatsu T., Purugganan M.D. (2008). Independent origins of self-compatibility in Arabidopsis thaliana. Mol. Ecol. 17: 704–714 [DOI] [PubMed] [Google Scholar]
- Shimosato H., Yokota N., Shiba H., Iwano M., Entani T., Che F.S., Watanabe M., Isogai A., Takayama S. (2007). Characterization of the SP11/SCR high-affinity binding site involved in self/nonself recognition in Brassica self-incompatibility. Plant Cell 19: 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva N.F., Stone S.L., Christie L.N., Sulaman W., Nazarian K.A.P., Burnett L.A., Arnoldo M.A., Rothstein S.J., Goring D.R. (2001). Expression of the S receptor kinase in self-compatible Brassica napus cv. Westar leads to the allele-specific rejection of self-incompatible Brassica napus pollen. Mol. Genet. Genomics 265: 552–559 [DOI] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Anderson E.M., Mullen R.T., Goring D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Arnoldo M., Goring D.R. (1999). A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286: 1729–1731 [DOI] [PubMed] [Google Scholar]
- Takasaki T., Hatakeyama K., Suzuki G., Watanabe M., Isogai A., Hinata K. (2000). The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403: 913–916 [DOI] [PubMed] [Google Scholar]
- Takayama S., Shiba H., Iwano M., Shimosato H., Che F.S., Kai N., Watanabe M., Suzuki G., Hinata K., Isogai A. (2000). The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 97: 1920–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Shimosato H., Shiba H., Funato M., Che F.S., Watanabe M., Iwano M., Isogai A. (2001). Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413: 534–538 [DOI] [PubMed] [Google Scholar]
- Tang C., Toomajian C., Sherman-Broyles S., Plagnol V., Guo Y.L., Hu T.T., Clark R.M., Nasrallah J.B., Weigel D., Nordborg M. (2007). The evolution of selfing in Arabidopsis thaliana. Science 317: 1070–1072 [DOI] [PubMed] [Google Scholar]
- Tantikanjana T., Nasrallah M.E., Nasrallah J.B. (2010). Complex networks of self-incompatibility signaling in the Brassicaceae. Curr. Opin. Plant Biol. 13: 520–526 [DOI] [PubMed] [Google Scholar]
- Tsuchimatsu T., Suwabe K., Shimizu-Inatsugi R., Isokawa S., Pavlidis P., Städler T., Suzuki G., Takayama S., Watanabe M., Shimizu K.K. (2010). Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464: 1342–1346 [DOI] [PubMed] [Google Scholar]
- Wang X., et al. ; Brassica rapa Genome Sequencing Project Consortium (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Yang C.W., González-Lamothe R., Ewan R.A., Rowland O., Yoshioka H., Shenton M., Ye H., O’Donnell E., Jones J.D., Sadanandom A. (2006). The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]